Loss of spr-5 bypasses the requirement for the C.elegans presenilin sel-12 by derepressing hop-1 (original) (raw)
Abstract
Presenilins are part of a protease complex that is responsible for the intramembraneous cleavage of the amyloid precursor protein involved in Alzheimer’s disease and of Notch receptors. In Caenorhabditis elegans, mutations in the presenilin sel-12 result in a highly penetrant egg-laying defect. spr-5 was identified as an extragenic suppressor of the sel-12 mutant phenotype. The SPR-5 protein has similarity to the human polyamine oxidase-like protein encoded by KIAA0601 that is part of the HDAC–CoREST co-repressor complex. Suppression of sel-12 by spr-5 requires the activity of HOP-1, the second somatic presenilin in C.elegans. spr-5 mutants derepress hop-1 expression 20- to 30-fold in the early larval stages when hop-1 normally is almost undetectable. SPR-1, a C.elegans homologue of CoREST, physically interacts with SPR-5. Moreover, down-regulation of SPR-1 by mutation or RNA interference also bypasses the need for sel-12. These data strongly suggest that SPR-5 and SPR-1 are part of a CoREST-like co-repressor complex in C.elegans. This complex might be recruited to the hop-1 locus controlling its expression during development.
Keywords: Alzheimer’s disease/C.elegans/Notch/presenilins/suppressor genetics
Introduction
Mutations in the presenilins PS1 and PS2 account for the majority of early-onset familial Alzheimer’s disease (Selkoe, 2001). These mutations lead to the aberrant processing of the amyloid precursor protein (APP) and the generation of increasing amounts of the highly amyloidogenic form of Aβ, the amyloid β-peptide. Aβ is the predominant component of the plaques found in the brains of Alzheimer’s patients. Presenilins are polytopic transmembrane proteins that are part of the proteolytic γ-secretase complex that liberates Aβ. Apart from their role in APP processing, presenilins are also required for proteolytic processing of the Notch receptors in their transmembrane domain. Ligand-induced cleavage and release of the intracellular domain of the Notch receptor (NICD) are crucial for nuclear Notch signalling (Struhl and Adachi, 1998). In agreement with these results, in all organisms tested so far, the loss of presenilin activity leads to phenotypes that resemble that of Notch loss-of-function mutants (Fortini, 2001).
Similarly to humans, Caenorhabditis elegans has two somatically expressed presenilin genes, hop-1 and sel-12 (Levitan and Greenwald, 1995; Li and Greenwald, 1997). hop-1 and sel-12, like PS1 and PS2, show redundant activities since only double mutants display phenotypes associated with a complete loss of Notch signalling in C.elegans (Li and Greenwald, 1997; Westlund et al., 1999). A sel-12 null mutant can be rescued by transgenic expression of hop-1 (as well as either of the human PS1 or PS2). sel-12 is expressed rather uniformly at all developmental stages, while hop-1 expression is very weak and could not be detected by reporter gene fusions (Westlund et al., 1999). Probably as a consequence of these different levels of expression, hop-1 mutants are viable with no obvious phenotype, whereas sel-12 mutants display, along with other more subtle defects, an egg-laying (Egl) defect. The sel-12 Egl phenotype is caused by reduced LIN-12/Notch signalling, resulting in the failure to form a proper vulva–uterine connection (Cinar et al., 2001) and in sex-muscle patterning defects (Eimer et al., 2002).
One approach to identify new molecules involved in the regulation of presenilin activity or molecules that are able to bypass the need for presenilin activity is to perform genetic screens for suppressors of the Egl defect of sel-12 mutant animals. Two sel-12 suppressors have been described already. Mutations in the C.elegans gene sel-10 directly influence LIN-12/Notch signalling by enhancing the half-life of the intracellular signalling domain of LIN-12 (Wu et al., 1998). The F-box/WD40 repeat protein SEL-10 is part of a ubiquitin ligase complex that mediates the ubiquitylation and degradation of the activated nuclear NICD (Lai, 2002). In contrast, the suppressor mutant spr-2 does not influence LIN-12/Notch signalling directly but requires HOP-1 activity in a manner that has not been characterized (Wen et al., 2000) but obviously does not affect the level of its transcription. SPR-2 belongs to a family of nuclear assembly proteins (NAPs) which have been implicated in chromatin remodelling and cell cycle control. The different modes of action of sel-10 and spr-2 function suggest that there exist distinct mechanisms to suppress sel-12.
The functional conservation of presenilins suggests that a third possible mechanism to suppress the sel-12 mutant phenotype could involve transcriptional up-regulation of the expression of the second presenilin, in this case hop-1. Therefore, second site suppressors of sel-12 could include mutations in transcriptional activators that render them hyperactive, or mutations in repressors or repressor complexes that eliminate the down-regulation of hop-1.
It is becoming increasingly obvious that histone deacetylase (HDAC)-containing repressor complexes are required for both transient and persistent transcriptional silencing of selected genes. Notably, HDAC complexes play an important role in the Notch-mediated transcriptional silencing of downstream genes (reviewed in Mumm and Kopan, 2000). HDACs associate with several distinct complexes, including the Sin-associated protein (SAP) complex and the nucleosome remodelling and histone deacetylation (NURD) complex, which exhibits ATP-dependent chromatin-remodelling activity (Jepsen and Rosenfeld, 2002). Recently, yet another HDAC complex was identified containing the transcriptional co-repressor CoREST and protein KIAA0601, which is a putative flavin–adenine dinucleotide (FAD)-dependent enzyme of still unknown function.
We report here the identification of two new suppressors of the Egl defect of sel-12 mutant C.elegans animals, spr-5 and spr-1. The analysis of spr-5 revealed that it acts by de-repession of hop-1 expression in developmental stages where hop-1 normally is not expressed. Therefore, sel-12 presenilin activity is replaced by hop-1 activity. SPR-5 exhibits homology to the human polyamine oxidase (PAO)-like protein KIAA0601, which is an integral component of the CoREST co-repressor complex. Our screens also identified by133, an spr-1 allele that, like RNA interference (RNAi) inhibition of a C.elegans homologue of CoREST, phenocopies the spr-5 suppressor function. Furthermore, we show that D1014.8/SPR-1 forms a complex with SPR-5. We suggest that a CoREST-like co-repressor complex also exists in C.elegans and participates in the transcriptional repression of the presenilin hop-1 during development.
Results
spr-5 suppresses the sel-12 Egl phenotype in a non-allele-specific way
In a screen using the mutator strain sel-12(ar171) unc-1(e538); mut-7(pk204), we recovered a mutation on chromosome I, by101, that suppressed the Egl defect of sel-12(ar171) hermaphrodites. Genetic mapping of the mutant loci placed the suppressor in the interval between unc-101 and unc-59 close to unc-59 (Figure 1; for details see Materials and methods). As it complements the previously mapped but not yet cloned presenilin suppressor spr-4(ar208) on LGI (Wen et al., 2000), the mutant locus we found corresponds to a new gene, which we will refer to as spr-5. Subsequently, we identified five additional alleles of spr-5 in similar screens using chemical mutagens (details of the experimental pro cedures will be published elsewhere; B.Lakowski and R.Baumeister, unpublished data).
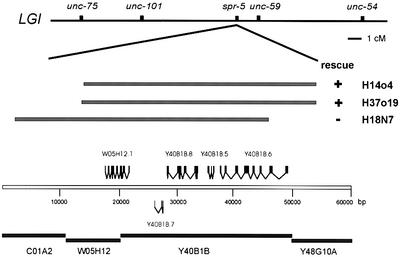
Fig. 1. Physical map of the spr-5 region on LGI. The location and the extension of the fosmids H14o4, H37o19 and H18N7 relative to the Y40B1B locus, as well as their rescuing activity, are indicated. The scale bar below represents base pairs.
All spr-5 mutants result in a very strong suppression of all aspects of the sel-12 Egl defect. Eighty-five to 95% of all spr-5; sel-12 animals are non-Pvl and non-Egl (Table I). Furthermore, the brood size of these animals is much larger than that of sel-12 and is in the range of that of the wild type (Table I). spr-5 mutations suppress all sel-12 alleles tested (Table I): ar171, a truncation after the fifth transmembrane domain (W225stop); ar131, a C60S missense mutation (Levitan and Greenwald, 1995); and lg1401, a deletion of sel-12 and part of the promoter that is a clear null allele (Eimer et al., 2002; Table I). This indicates that the mechanism of suppression is not allele specific and does not depend on the presence of SEL-12 protein.
Table I. spr-5 bypasses the need for sel-12 in a non-allele-specific way.
| Genotype | Pvl (%) | Egl (%) | Brood size | n |
|---|---|---|---|---|
| Wild-type N2 | 0 | 0 | 316 ± 8 | 20 |
| sel-12(ar171) | 80 | 100 | 62 ± 7 | 25 |
| sel-12(ar171); spr-5(by113) | 0 | 5 | 188 ± 15 | 20 |
| sel-12(ar171); spr-5(by128) | 10 | 15 | 230 ± 20 | 20 |
| sel-12(ar171); spr-5(by134) | 10 | 10 | 219 ± 12 | 20 |
| sel-12(ar171); spr-5(by101) | 5 | 10 | 208 ± 15 | 40 |
| sel-12(lg1401); spr-5(by101) | 0 | 5 | 214 ± 12 | 19 |
| sel-12(ar131); spr-5(by101) | 0 | 5 | 231 ± 17 | 20 |
| spr-5(by101) | 0 | 0 | 276 ± 12 | 27 |
In addition, as opposed to sel-12 animals, spr-5; sel-12 hermaphrodites also lay eggs in response to the drugs serotonin and imipramine, suggesting that they have a functional egg-laying system that can be stimulated pharmacologically (Trent et al., 1983). spr-5 mutations also restore male mating that is defective in sel-12 males (Eimer et al., 2002). Therefore, spr-5 rescues not only the structural defects of the egg-laying system of sel-12 animals, but also the functional defects.
When genetically separated from sel-12 alleles, spr-5 mutants alone display no obvious phenotype, suggesting that spr-5 is a specific suppressor of sel-12. Neither egg-laying, egg motility nor other behaviours were different from those of wild-type animals under the conditions tested (data not shown). In addition, the brood size of all spr-5 alleles is in the range of wild type (Table I).
spr-5 mutants do not enhance LIN-12/Notch signalling directly
In order to determine the mechanism of suppression, we first tested whether lin-12 is the prime target of spr-5 genetic interactions. Since sel-12 mutations reduce LIN-12 signalling, spr-5 mutations might act by increasing LIN-12 expression or activity. From other experiments not reported here, we had concluded that mRNA levels expressed from both C.elegans Notch genes, lin-12 and glp-1, did not differ between wild type and the spr mutants we had recovered in our screens (B.Lakowski and R.Baumeister, unpublished data). Therefore, it is unlikely that spr-5 mutants act by transcriptional up-regulation of Notch expression. The lin-12(n676n930) reduction of function allele displays a temperature-sensitive Egl defect (Sundaram and Greenwald, 1993), which is similar to the sel-12 Egl phenotype (Eimer et al., 2002). Therefore, we wondered whether spr-5 might suppress the Egl defect of lin-12(n676n930) at 25°C. However, spr-5 did not rescue any aspect of the lin-12(n676n930) Egl defect (Table II). We also tested whether spr-5 is able to enhance another well-characterized process that is defective in lin-12 mutants, the AC/VU decision (Greenwald et al., 1983). In the AC/VU decision, two initially equipotent cells of the somatic gonad adopt different cell fates through a LIN- 12-dependent process called lateral inhibition. The lin-12(n302) gain-of-function allele is vulva-less and therefore Egl, because it develops two VU cells at the expense of an AC (Greenwald et al., 1983). As the lin-12(n302) mutation is semi-dominant, 50% of the hetero zygote animals are Egl. spr-5 mutants show no increase in the 0 AC phenotype in heterozygotes, indicating that the LIN-12-dependent lateral inhibition is also not affected in spr-5 mutants (Table II). From these results, we conclude that spr-5 does not directly influence any of the characterized modes of LIN-12 signalling. Therefore, we consider it unlikely that spr-5 suppresses sel-12 by directly up-regulating LIN-12 signalling.
Table II. Genetic interactions of spr-5 with lin-12 and hop-1.
| sel-12 suppression by spr-5 is dependent on hop-1 | |
|---|---|
| Genotype | No. of sterile/total (%) |
| sel-12(ar171); _hop-1(lg1501)_a | 36/36 (100%) |
| sel-12(ar171); _hop-1(lg1501) spr-5(by101)_a | 43/43 (100%) |
| No. of Egl/total (%) | |
| sel-12(ar171); _hop-1(lg1501)_b | 50/50 (100%) |
| sel-12(ar171); _hop-1(lg1501) spr-5(by101)_b | 48/49 (98%)c |
| No effect on the 0 AC defect caused by elevating lin-12 activity | |
| Genotype | No. of Egl/total (%) |
| lin-12(n302)/ unc-32(e189) | 40/79 (51%) |
| lin-12(n302)/ unc-32(e189); spr-5(by101) | 77/141 (55%) |
| No enhancement of a lin-12 hypomorphic mutation | |
| Relevant genotype | No. of Egl/total (%) at 25°C |
| _lin-12(n676n930)_d | 50/50 (100%) |
| _lin-12(n676n930)_d; spr-5(by101) | 50/50 (100%) |
The suppressor activity of spr-5 requires functional HOP-1 presenilin
To determine whether spr-5 bypasses the need for functional presenilins in LIN-12 signalling or may act through modulating the activity of the second presenilin hop-1, we constructed hop-1 spr-5; sel-12 triple mutants. Both sel-12 and hop-1 have partial maternal effects, so the phenotype of hop-1; sel-12 double mutants depends on how they are constructed. Double mutant animals that have maternally supplied hop-1 are sterile, whereas, if sel-12 is provided maternally, hop-1; sel-12 mutants become Egl, with embryos that do not hatch (Westlund et al., 1999). hop-1 spr-5; sel-12 triple mutants, with maternally supplied sel-12, are also Egl (Table II) and only produce dead embryos. Furthermore, triple mutants with maternally supplied hop-1 are sterile like the similarly constructed sel-12; hop-1 double mutants (Table II). We therefore conclude that spr-5 mutants require _hop_-1 activity for sel-12 suppression.
Molecular cloning of spr-5
Initially, spr-5 was mapped near unc-59 on the right arm of chromosome I (Figure 1). A Tc3 transposon was found to co-segregate with the suppression phenotype in spr-5(by101) (for details see Materials and methods). Sequencing of the genomic DNA flanking the transposon revealed that the Tc3 had inserted into the seventh exon in codon 606 of the predicted reading frame Y40B1B.6 (Figure 1). When injected into sel-12; spr-5 animals, fosmids H14o4 (four out of four transgenic lines) and H37o19 (five out of six transgenic lines) that contain the entire coding region of Y40B1B.6 rescued the spr-5 mutant phenotype, and restored a sel-12 phenotype (Egl). In contrast, fosmid H18N7 that terminates after the sixth exon of Y40B1B.6 was not able to rescue the suppression by spr-5 (none out of three transgenic lines), suggesting that Y40B1B.6 indeed corresponds to spr-5 (Figure 1).
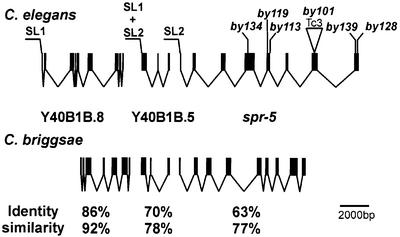
Further analysis of the genomic organization of the open reading frames (ORFs) on Y40B1B indicated that the genes Y40B1B.8, Y40B1B.5 and Y40B1B.6/spr-5 belong to an operon consisting of three genes (Figure 2). In operons, the downstream genes are _trans_-spliced to a 22 nucleotide splice leader SL2 (Spieth et al., 1993), whereas the most upstream gene is _trans_-spliced to an SL1 splice leader. We used RT–PCR with SL1 and SL2 primers to determine the 5′ ends of all three putative ORFs. We found that Y40B1B.8 is spliced exclusively to SL1, Y40B1B.5 is _trans_-spliced to a mixture of SL1 and SL2, and Y40B1B.6/spr-5 is spliced primarily to SL2 (Figure 2). The existence of a three-gene operon containing spr-5 is supported further by DNA array data (Blumenthal et al., 2002). Based both on their orientation and their encoded amino acid sequence, these three genes are highly conserved in the related nematode Caenorhabditis briggsae, consistent with being in an operon. However, the exon–intron structure of the C.elegans and C.briggsae genes differs to some extent (Figure 2). Although the genes are highly conserved on a protein level, Y40B1B.8 contains seven exons in C.elegans compared with nine in C.briggsae, and the fourth exon of Y40B1B.6/spr-5 is represented by two exons in C.briggsae (Figure 2).
Fig. 2. Exon–intron structure of the spr-5 operon on LGI. The specific splice leader and the locations of the mutations in the different spr-5 alleles are shown. In addition, the organization of the homologous operon in C.briggsae is included, along with the identity and similarity scores of the C.elegans and C.briggsae proteins.
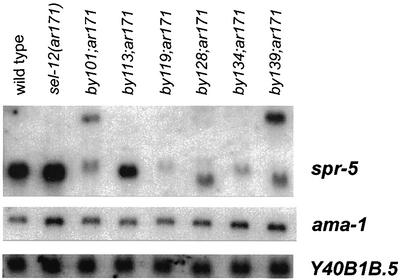
We examined in all spr-5 alleles the expression of the two downstream reading frames of the operon. On a mixed stage northern blot, most spr-5 alleles show clear alterations in either the size and/or the intensity of bands detected with an Y40B1B.6 probe, and no obvious differences in the transcript of Y40B1B.5 (Figure 3). To support this notion further, we performed RNAi experiments against each of the three genes in this operon by bacterial feeding of double-stranded RNA (dsRNA) (Timmons and Fire, 1998; Timmons et al., 2001). Although it has been reported that RNAi may also target the pre-mRNA (Bosher et al., 1999), only the dsRNAi against spr-5/Y40B1B.6 led to suppression of the Egl defect of sel-12 animals that is typical for the different spr-5 alleles (Table III). dsRNAi against any of the upstream genes was not able to suppress the Egl defect of sel-12 (Table III). Therefore, we conclude that spr-5 is Y40B1B.6. Subsequently, we confirmed the genomic structure predicted in Wormbase by sequencing. Y40B1B.6/spr-5 encodes a protein of 770 amino acids (Figure 4A). The positions of all mutations and alterations found in the various spr-5 alleles are shown in Figures 2 and 4A.
Fig. 3. Northern blot of mixed staged total RNA from the different alleles probed with spr-5, Y40B1B.5, and ama-1 as a loading control.
Table III. Summary of dsRNAi feeding experiments.
| RNAi construct | RNAi phenotype in strains | ||
|---|---|---|---|
| Wild type | sel-12(ar171) | sel-12(ar131) | |
| – | wt (15/15) | Egl (10/10) | Egl (10/10) |
| spr-5 | wt (10/10) | Non-Egl (39/40)a | Non-Egl (20/20) |
| Y40B1B.5 | wt (10/10) | Egl (20/20) | Egl (20/20) |
| Y40B1B.8 | wt (10/10)b | Egl (20/20)b | Egl (20/20)b |
| T08D10.2 | wt (11/11) | Egl (20/20) | Egl (20/20) |
| D1014.8 | wt (10/10) | Non-Egl (30/30) | ND |
| Y74C9A.4 | wt (10/10) | Egl (30/30) | ND |
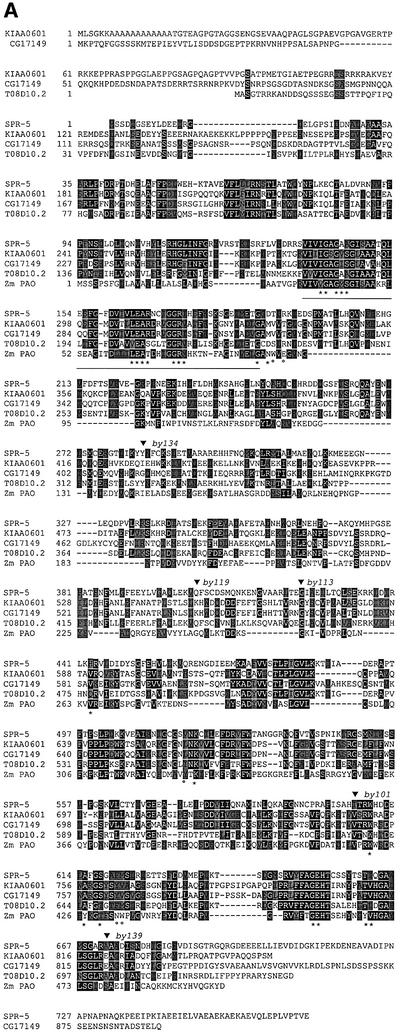
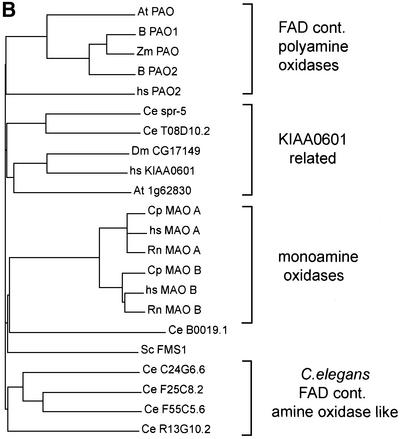
Fig. 4. (A) Alignment of SPR-5 with the human KIAA0601 (DDBJ/EMBL/GenBank accession No. BAA25527), Drosophila melanogaster CG17149 (accession No. AAF49051), C.elegans T08D10.2 and Zea mays PAO (accession No. CAA05249). Identical residues are shaded in black, whereas similar residues are shaded in grey. Asterisks indicate the residues contacting the FAD cofactor (Binda et al., 1999), and open circles the residues contacting the substrate (Binda et al., 2001) in the maize PAO. Underlined is the signature motif characteristic for the flavoprotein family (Dailey and Dailey, 1998). The positions of the spr-5 mutations are indicated by black triangles above the sequence. (B) ClustalW-aligned tree of the different protein families showing homologies to PAOs. SPR-5 defines a distinct subgroup including KIAA0601, DmCG17149 and T08D10.2. For details on nomenclature, see Supplementary data available at The EMBO Journal Online.
spr-5 is homologous to amine oxidases found in transcriptional repressor complexes
SPR-5 exhibits strong similarity to a large class of FAD-dependent amine oxidases. SPR-5 is most similar to the class of FAD-dependent PAOs (Figure 4). The PAOs are found in all organisms from bacteria to mammals and plants, and catalyse the oxidation of secondary amine groups of straight chain aminoalkanes (Sebela et al., 2001). Despite the differences in the site of action on the secondary amine group between plants and mammals, PAOs are monomeric soluble enzymes with non-covalently bound FAD cofactor (Seiler, 1995). All PAOs have a two-domain organization, with one domain binding the FAD cofactor while the other binds the substrate (Binda et al., 1999, 2001). Even though they only share 20–30% identity, the maize PAO has the same three-dimensional structure as the vertebrate monoamine oxidase, known to act on primary amines (Binda et al., 2002). SPR-5 and the maize PAO share the same FAD-binding signature motif (Dailey and Dailey, 1998), and the residues known to bind the FAD cofactor in maize PAO are conserved (Binda et al., 1999), while those residues that recognize the substrate are not conserved (Binda et al., 2001). Therefore, we conclude that it is likely that SPR-5 binds FAD as a cofactor while the substrate specificity may have diverged.
SPR-5 defines a subfamily of the PAOs along with a second C.elegans protein T08D10.2, the Drosophila melanogaster protein encoded by expressed sequence tag (EST) CG17149 and the human protein encoded by EST KIAA0601 (Nagase et al., 1998). Members of this subfamily of proteins are more similar to each other than to the maize PAO (Figure 4A and B). SPR-5 has 27% amino acid identity (45% similarity) to hKIAA0601 and Dm CG17149, whereas it has 44% identity (62% similarity) to the C.elegans paralogue. The protein that corresponds to KIAA0601 was co-purified as an integral component of the human CoREST–HDAC complex (Tong et al., 1998; Humphrey et al., 2001; You et al., 2001). The CoREST complex was shown to be a functional co-repressor that is required for REST-mediated repression of neuronal genes in non-neuronal cells (Andres et al., 1999; Ballas et al., 2001; Griffith et al., 2001). Additional components of the human CoREST complex are HDACs 1 and 2 and the SANT domain protein CoREST (Humphrey et al., 2001; You et al., 2001), which binds to the zinc finger factor REST (Andres et al., 1999; Ballas et al., 2001). REST recruits the CoREST complex to specific DNA sites, suggesting that the complex may regulate individual genes.
Loss of spr-5 leads to derepression of hop-1 expression
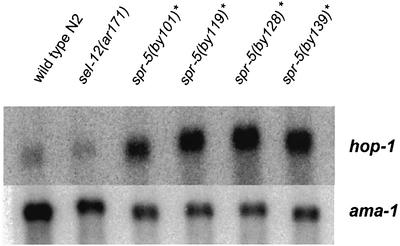
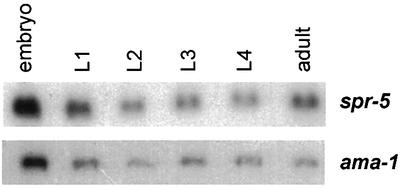
Due to the sequence similarity of SPR-5 to a member of a complex involved in transcriptional repression, it is possible that SPR-5 may have a similar function in C.elegans. Thus, we searched for candidate target genes in the Notch signalling pathway that may be regulated by spr-5. The suppression of sel-12 by spr-5 was shown not to up-regulate LIN-12/Notch signalling but was dependent on hop-1 activity. Since hop-1 and sel-12 are functionally redundant, hop-1 was an attractive candidate for _spr-5_-mediated transcriptional regulation. Therefore, we analysed the stage-specific transcriptional regulation of hop-1 by northern blot analysis. sel-12 is ubiquitously expressed at high levels throughout all developmental stages (Baumeister et al., 1997). In contrast, hop-1 expression dramatically changes throughout development. hop-1 is almost undetectable in the L1 and L2 larvae and its expression gradually increases throughout further development and reaches a maximum at the adult stage (B.Lakowski and R.Baumeister, unpublished data). As a consequence of the high expression levels in the adult stage, probably a high level of hop-1 mRNA is supplied to the embryo maternally. In sel-12 mutant animals, hop-1 expression levels are indistinguishable from wild-type levels, suggesting that sel-12 does not control hop-1 expression at the transcriptional level. Next, we investigated hop-1 expression in spr-5; sel-12 suppressor strains. hop-1 expression was detectable already in the L1 stage, while it is almost absent in wild-type L1 animals (Figure 6). The up-regulation of hop-1 expression was between 20- and 30-fold depending on the spr-5 allele tested and independent of the presence of sel-12 (data not shown). spr-5 is expressed in all stages at nearly the same level, indicating that it may act as a general co-repressor (Figure 5).
Fig. 6. Northern blot of L1-specific total RNA N2, sel-12(ar171) and different sel-12(a171); spr-5 double mutants probed with _hop-1_- and _ama-1_-specific probes. *Every spr-5 allele was in a sel-12(ar171) mutant background.
Fig. 5. Stage-specific northern blot of wild-type total RNA probed with spr-5, and ama-1 as a loading control.
These data suggest that spr-5 is required to repress hop-1 expression in the first larval stages and that loss of spr-5 leads to a derepression of hop-1 in these stages. Derepression of hop-1 expression is sufficient to replace the lack of sel-12 activity and, therefore, suppresses the Egl phenotype of the sel-12 mutants.
A CoREST-like complex might be involved in transcriptional repression of hop-1
If SPR-5 functions in a HDAC–CoREST complex of transcriptional regulation, then one might assume that manipulating the expression of other members of this complex should result in a phenotype similar to that of spr-5 mutants. We identified two predicted open reading frames, D1014.8 and Y74C9A.4, encoding proteins with similarity to CoREST. dsRNAi experiments were performed for both genes in a sel-12 mutant background (Table III). RNAi against Y74C9A.4 did not reveal any novel phenotype in a sel-12 background. Strikingly, however, dsRNAi against D1014.8 suppressed the sel-12 Egl defect as strongly as spr-5 (Table III). D1014.8 maps to a genomic region on LGV where another sel-12 suppressor, spr-1, was mapped previously (Wen et al., 2000), suggesting that D1014.8 corresponds to spr-1.
In our screens for sel-12 suppressors, we had identified a mutant with properties similar to spr-5 alleles. We had mapped this suppressor allele, by133, close to D1014.8, suggesting that it might encode an allele of spr-1. To determine if by133 is an allele of spr-1, we performed rescue experiments with the cosmid D1014 using an spr-1(by133); sel-12(ar171) strain. Seven of seven transgenic F2 lines clearly and profoundly rescued the _spr-1(by133)_-mediated suppression of sel-12. Subsequently, we sequenced the coding region of the by133 mRNA and found that it contains a T→A mutation at position +905, converting a TTA (leucine) to TAA (stop). This mutation truncates the protein at amino acid 301 after the first SANT domain, but before the second SANT domain. Thus, mutations in two members of the CoREST complex suppress the presenilin defect in C.elegans.
RNAi against the three C.elegans class I HDACs hda-1, hda-2 and hda-3, alone or in combination, resulted in a pleiotropic phenotype that was difficult to interpret. This result was not unexpected since it had been shown before that these HDACs in C.elegans are involved in a variety of different functions in different cell types (Dufourcq et al., 2002). Interestingly, RNAi against hda-1/gon-10 resulted in multiple defects affecting the egg-laying system, but also in sterility, which prevented the monitoring of sel-12 suppression.
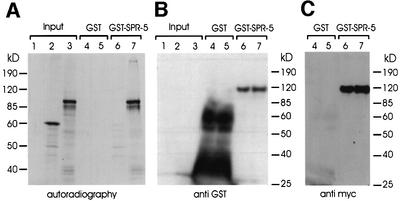
Complex formation is one prerequisite of the concerted activity of PAO and CoREST in human cells. Although our genetic data already strongly suggest a similar function for C.elegans SPR-1/CoREST and SPR-5/PAO, we tested whether both proteins are able to interact physically. A GST–MYC-SPR-5 protein fusion was used to probe protein interaction with SPR-1/CoREST by co-immunoprecipitation experiments. For this purpose, GST–SPR-5 expressed in Sf9 insect cells, immobilized on glutathione–Sepharose and incubated with 35S-labelled SPR-1 was in vitro translated in a reticulocyte lysate system. Protein binding was analysed by SDS–PAGE and western blots (Figure 7). Our results demonstrate that SPR-5/PAO binds specifically to SPR-1/CoREST, corroborating previous results (obtained with the human factors) that both proteins are components of the same protein complex. Interaction was subsequently confirmed by yeast two-hybrid experiments (S.Eimer, data not shown).
Fig. 7. SPR-5 and D1014.8/SPR-1 interact in vitro. (A) Autoradiography of 10% SDS–PAGE. Lanes 1–3 are input controls of 35S-labelled in vitro translation reactions: empty vector (lane 1), luciferase (lane 2; 61 kDa), D1014.8/SPR-1 (lane 3; 68 kDa). 35S-labelled luciferase and SPR-1 were incubated with GST (lanes 4 and 5), or with GST–SPR-5 (lanes 6 and 7), respectively and analysed for binding by SDS–PAGE and autoradiography as described in Materials and methods. Lanes 1–3 represent 12% of the input, and lanes 4 and 5, and lanes 6 and 7 represent 25%, each, of the binding reaction. (B) Detection of GST proteins on a western blot. The lanes are as described in (A). The blot was probed with an anti-GST antibody. Note that excess GST protein (100 µg; lanes 4 and 5) was used for the binding reaction compared with GST–SPR-5 (4 µg; lanes 6 and 7). (C) Detection of GST–SPR-5 fusion protein on a western blot. Since the GST–SPR-5 (115 kDa) fusion protein also contains a myc epitope tag, the blot was probed in addition with an anti-myc tag antibody. The lanes are as described in (A).
Discussion
From a screen for extragenic suppressors of the Egl defect of sel-12 mutant animals, we identified spr-5. The quality of suppression by spr-5 is remarkably high and is dependent on the activity of the second C.elegans presenilin gene, hop-1. However, spr-5 shows no genetic interaction with lin-12/Notch. It is therefore unlikely that spr-5 mutations act by up-regulating the lin-12/Notch pathway as was demonstrated for sel-10. Interestingly, the quality of suppression by spr-5 mutants is significantly higher than that of sel-10 mutants. It has been shown that the inability of sel-12 mutant animals to lay eggs is caused by two separate defects in the egg-laying apparatus. sel-12 mutants do not form a proper vulva–uterine connection (Cinar et al., 2001) and have impaired sex-muscles (Eimer et al., 2002). Therefore, the action of a suppressor is required in two unrelated tissues at different time points in development. We suggest that full suppression of the sel-12 presenilin defect would be rather difficult to accomplish by adjusting the different levels of LIN-12/Notch signalling in both tissues. However, the same effect can be accomplished if the defective presenilin is replaced with the (functionally equivalent) hop-1 by increasing the expression of the latter. Therefore, suppressor screens like we and others have performed are more likely to reveal _hop-1_-dependent suppressor mutants.
We demonstrate that spr-5 acts by derepressing hop-1 expression. hop-1 expression normally is repressed in the first two larval stages and then gradually is expressed more strongly until adulthood, where expression is strongest (B.Lakowski and R.Baumeister, unpublished data). In contrast, sel-12 is ubiquitously expressed at all stages. Most of the known LIN-12/Notch signalling events take place during the L2 to late L3 stage, including the AC/VU specification, the π-cell specification for a proper vulva– uterine connection, and sex-muscle morphogenesis (Li and Chalfie, 1990; Newman et al., 1995; Greenwald, 1998; Sharma-Kishore et al., 1999). Therefore, loss of sel-12 activity during L2 and L3 is most critical and is the cause of the Egl phenotype. Why do sel-12 mutants not show a lin-12 null phenotype, including a defective AC/VU decision? This is probably because different thresholds for the amount of LIN-12/Notch signalling are required for each developmental decision (Eimer et al., 2002). The AC/VU cell fate decision most probably needs less LIN-12/Notch activity than π-cell induction and sex-muscle morphogenesis and, therefore, the low hop-1 expression levels during larval stages, or the maternally supplied hop-1, are sufficient to maintain LIN-12 activity.
As shown here, loss of spr-5 function leads to a 20- to 30-fold increase in hop-1 expression in the first larval stages (Figure 6) most probably due to a removal of a repression of hop-1 expression in those stages. Therefore, hop-1 derepression occurs at precisely those stages at which lack of sel-12 presenilin activity would be most critical. We do not know why derepression is most significant in L1–L3, although spr-5 obviously is expressed at all developmental stages. Probably the timing of spr-5 dependent derepression is regulated by other, not yet identified factors.
The mechanism of the derepression is not clear, but might affect chromatin structure and remodelling. Another known suppressor of the sel-12 Egl defect, spr-2, encodes a protein with similarity to the Set/TAF-Iβ oncoprotein found in the INHAT (inhibitor of acetyltransferase) complex which is able to inhibit histone acetyltransferase (HAT) activities by p300/CBP and PCAF through histone masking (Seo et al., 2001). Genetically, spr-2 behaves similarly to spr-5 and was also shown to be dependent on hop-1 activity (Wen et al., 2000). However, in spr-2 mutants, hop-1 transcription does not increase, suggesting that spr-2 and spr-5 function through separate mechanisms. Interestingly, both proteins might be associated with chromatin complexes.
The proteins that are most similar to SPR-5, except for T08D10.2, a C.elegans paralogue, are the human KIAA0601/ p110b and a predicted protein from D.melanogaster encoded by CG17149. All those proteins share regions of similarity with FAD-dependent PAOs. The mutation in spr-5(by113) results in an exchange of a conserved glycine residue to arginine at position 423. This allele is phenotypically indistinguishable from the deletion alleles, suggesting that the G423R point mutation interferes with an essential function of the protein. Interestingly, in the maize PAO crystal structure, this position is located in the FAD-binding domain, close to the FAD-binding pocket (Binda et al., 1999). The human KIAA0601 protein was found to be an integral component of the CoREST co-repressor complex (Tong et al., 1998; Humphrey et al., 2001; You et al., 2001). This may indicate that the mutation interferes with an enzymatic function of SPR-5 required for the repressor activity of such a complex.
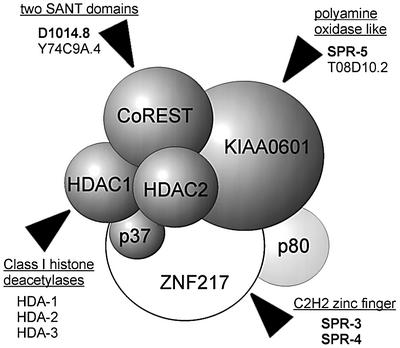
In addition to the PAO, other HDAC–CoREST complex components are HDAC1, HDAC2 and the SANT domain protein CoREST (Humphrey et al., 2001; You et al., 2001; Figure 8). CoREST, together with REST/NRSF (RE1 silencing transcription factor/neural-restrictive silencing factor), acts to repress neuronally expressed genes in non-neuronal cells (Andres et al., 1999; Ballas et al., 2001). However, REST can act through multiple deacetylase complexes, only one of them being CoREST (Griffith et al., 2001). The existence of a CoREST complex in C.elegans is corroborated further by the fact that our screens also identified a mutant of spr-1/CoREST. The similar phenotypes of spr-5 and spr-1 suggest a similar function of both encoded proteins in the repression of early hop-1 transcription. Our co-immunoprecipitation experiments also strongly indicate that SPR-1 and SPR-5 proteins interact in C.elegans, as was shown previously for their human homologues. Furthermore, in additional screens not reported here, we have identified two other sel-12 suppressors, spr-3 and spr-4, whose closest human homologue is REST (B.Lakowski and R.Baumeister, unpublished data). Therefore, mutations in at least four proteins similar to components of the CoREST–HDAC complex are able to suppress sel-12 by up-regulating the activity of the second presenilin, hop-1.
Fig. 8. Schematic representation of the human CoREST complexes as proposed (Humphrey et al., 2001). The components that are overlapping in both purifications are highlighted in grey. The molecular identities of the different proteins and the homologues found in C.elegans are indicated. The C.elegans proteins involved in sel-12 suppression are highlighted in bold. SPR-3 and SPR-4 will be described elsewhere.
The fact that there exist two reading frames encoding proteins with obvious homology to KIAA0601 (SPR-5 and T08D10.2) and CoREST protein (D1014.8 and Y74C9A.4), respectively, indicates that at least two independent CoREST complexes might exist in C.elegans that have different roles. Only one of them is involved in the regulation of hop-1.
What is the mechanism of hop-1 de-repression by spr-5?
The FAD-binding motif of SPR-5 is well conserved and places SPR-5 into the superfamily of FAD-dependent oxidases (Dailey and Dailey, 1998). It is probably inactive in the spr-5(by113) mutant (see above). The amino acid sequence identities between different members of this superfamily are normally quite low and range between 20 and 30% (Binda et al., 1999). In contrast to the FAD domain, the substrate recognition domain is not conserved among various members of this family and, therefore, different oxidative reactions are catalysed by individual amine oxidases. Although different substrates are bound, the overall three-dimensional structures of the substrate recognition domains of PAOs are strikingly similar to those of monoamine oxidases (Binda et al., 1999, 2002). It is possible, therefore, that despite its divergent substrate recognition domain, SPR-5 might have retained a comparable enzymatic activity. However, in the absence of functional data, one can only speculate about a function for this class of PAO.
A number of different proteins with enzymatic activities have been identified recently in HDAC complexes regulating transcriptional repression. For example, both HAT and HDAC complexes are involved in controlling transcriptional regulation mediated by the Notch intracellular domain (reviewed in Mumm and Kopan, 2000). However, our data clearly show that SPR-1 and SPR-5 do not regulate lin-12/Notch signalling directly. A recently discovered family of co-repressor proteins, the C-terminal binding proteins (CtBPs), exhibits similarity to dehydrogenase enzymes (Chinnadurai, 2002). CtBP adopts different conformations dependent on the cofactor bound (NAD+ or NADH), modulating its affinity for partner proteins and, thus, the level of repression (Zhang et al., 2002). It is possible that, upon FAD binding, a similar mechanism is induced in the SPR-5/PAO in the CoREST complexes. It has been suggested that KIAA0601 could, in principle, have an enzymatic activity that involves the oxidation of amines or amino groups, such as for example the methylation of lysine or arginine side chains on modified histone tails (Chinenov, 2002). The methylation of lysine residues in the histone tails has been shown to modulate the interaction of repressor complexes with specific regulatory sequences tails (Zegerman et al., 2002).
In summary, our data strongly indicate that spr-5 encodes a PAO-like factor that is part of a transcriptional repressor complex similar to the human CoREST complex. We describe for the first time a target gene that is controlled genetically by a CoREST-associated PAO. Caenorhabditis elegans SPR-5, most probably in a complex with CoREST/SPR-1, regulates the repression of hop-1 presenilin at early developmental stages. The presence of two homologous proteins of each component of the CoREST complex in C.elegans indicates that there may exist more co-repressor complexes of this type in the nematode, only one of them being involved in hop-1 regulation. Based on the dsRNAi experiment, it is possible that additional CoREST complexes might function in other regulatory processes not related to hop-1.
Materials and methods
Strains, plasmids and molecular techniques
All strains and mutants were maintained at 20°C according to standard procedures, if not stated otherwise. LGX, sel-12(ar171), sel-12(ar131), sel-12(lg1401); LGI, hop-1(lg1501), unc-73(e936), _glp-4(bn2_ts); LGII, unc-4(e120); LGIII, mut-7(pk204) (Ketting et al., 1999), lin-12(n676n930) unc-32(e189), unc-32(e189), lin-12(n302); LGIV, _fem-1(hc17_ts); LGVI, him-5(e1489).
Sequences of oligonucleotides and details on plasmid constructions are listed in the Supplementary data available at The EMBO Journal Online.
Screens for extragenic suppressors of the sel-12(ar171) Egl phenotype
A sel-12(ar171) unc-1(e538); mut-7(pk204) strain was constructed and was maintained at 15°C and for mutagenesis shifted to 20–23°C. The strain was tested for the appearance of an Egl and Him phenotype and sterility at 25°C, as described previously (Ketting et al., 1999). During the mutagenesis, single worms were transferred to 3.5 cm plates and kept at 20°C for two generations. The F2 progeny were transferred to 9 cm plates and screened for the appearance of eggs on the plate. The eggs or their non-Egl, Unc mothers were picked to new plates and tested for the inheritance of the non-Egl phenotype. Two independent mutations were discovered in a screen corresponding to ∼9600 haploid genomes. spr-3(by110) X will be described elsewhere (B.Lakowski and R.Baumeister, unpublished data); spr-5(by101)I was outcrossed with N2 and subsequently was analysed.
Transposon display and cloning of spr-5
The spontaneous mutants that showed a heritable suppression of the sel-12 Egl defect were outcrossed with N2 six times. The remaining transposons with their flanking sequences were displayed by PCR as described previously (Wicks et al., 2000). Genomic DNA from the sel-12(ar171) and sel-12(ar171); spr-5(by101) strains was prepared and digested with _Sau_3A. The digested DNA was ligated to an adaptor, and transposons were displayed following the Transposon Insertion Display protocol (http://genomics.niob.knaw.nl/) and identified Y40B1B.6 as the insertion locus of the transposon. To isolate a fosmid including Y40B1B.6, we hybridized a fosmid filter grid (kindly provided by Alan Coulson, Cambridge) with Y40B1B.6-specific radioactively labelled PCR products. Three positive fosmids were found that contain the identical genomic region except for differences around Y40B1B.6. H14o4 and H37o19 contain the whole coding region of Y40B1B.6 (DDBJ/EMBL/GenBank accession No. AY152852), whereas H18N7 terminates after the sixth exon of Y40B1B.6 (Figure 1).
Transgenic lines and rescue
As there was only one yeast artificial chromosome, Y40B1, available containing the genomic region of spr-5, we screened a C.elegans fosmid filter grid kindly provided by Alan Coulson for fosmids including spr-5. For that purpose, the filter grid was probed with two PCR products corresponding to the 5′ (RB927/RB928) and 3′ end (RB828/RB944) of the spr-5 cDNA. Two fosmids, H14o4 and H37o19, were found to hybridize to both probes, whereas H18N7 was only positive for the 5′-specific probe. To determine whether the _spr-5_-containing fosmids are able to revert the Egl suppression by sel-1(ar171); spr-5(by101) double mutants, they were injected individually at 10 ng/µl along with the rol-6(d) co-injection marker pRF4, at 100 ng/µl, using standard transformation techniques (Mello et al., 1991).
Identification of spr-1
Genetic mapping confirmed that by133 is an allele of spr-1 (Wen et al., 2000; S.Jarriault and I.Greenwald, personal communication). To rescue spr-1, we injected a spr-1(by133); sel-12(ar171) strain with the cosmid D1014 at 20 ng/µl with 100 ng/µl pRF4 as a co-transformation marker. We screened the transgenic progeny for appearance of Egl-defective animals indicating solid rescue of the _spr-1_-mediated Egl suppression.
Northern blotting
RNA was isolated from mixed stage plates or staged plates, and prepared with an RNAeasy kit (Qiagen) according to the manufacturer’s instructions. For most northern blots, 5 µg of total RNA per lane was denatured at 65°C for 5 min and then loaded onto a 0.8% agarose RNA gel. The gel was run overnight to separate fragments and blotted onto Hybond N+ membranes according to the protocol of Sambrook et al. (1989). For the L1 northern blots, 20 µg of total RNA was used per lane. Probes were labelled with [α-32P]dCTP using a Megaprime labelling kit according to the manufacturer’s instructions (Amersham, Freiburg, Germany). Blots were hybridized and washed according to the procedure of Church and Gilbert (1984) at 65°C. As a loading control, all northern blots were probed with an _ama-1_-specific probe using the PCR primers RB1186/RB1187 (Johnstone and Barry, 1996).
dsRNAi by feeding
Genes were transiently inactivated by RNAi through feeding of the Escherichia coli strain HT115(DE3) expressing dsRNA of the gene of interest (Timmons and Fire, 1998; Timmons et al., 2001). The dsRNA expression was induced as described previously (Kamath et al., 2000) and the worms were transferred as L4 larvae onto seeded plates containing 50 µg/ml ampicillin and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and kept at 20°C. The progeny were scored for the ability to lay eggs. A plate was scored as non-Egl if the majority of the progeny showed no protruding vulva (Pvl), solid egg-laying over 3 days and did not die with a bag of worm phenotype.
GST purification of the GST–SPR-5 fusion protein
Sf9 lysates from GST- and GST–SPR-5-expressing cells, respectively, were incubated with glutathione–Sepharose beads (Pharmacia, Freiburg) for 2 h at room temperature and washed with IP buffer (25 mM HEPES pH 7.6, 100 mM NaCl, 1 mM MgCl2, 0,1% NP-40, 10% glycerol, 1 mM dithiothreitol, 0.5 mM EDTA, 30 µM FAD) twice. Incubation with 35S -labelled SPR-1 produced in vitro using the TNT Coupled Reticulocyte Lysate System (Promega, Mannheim) was carried out for 2 h at room temperature. Subsequently, GST beads were collected by centrifugation and washed three times with 1 ml of IP buffer, resuspended in 40 µl of Laemmli sample buffer, and analysed by SDS–PAGE, western blotting and autoradiography.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank past and present members of the Baumeister lab for discussions and support. We are particularly grateful to Henri van Luenen and Ronald Plasterk for their help with the transposon display. We also thank Peter Becker and his lab for help and suggestions, Iva Greenwald and EleGene AG for strains, Tom Blumenthal and Iva Greenwald for providing unpublished results, Alan Coulson for the fosmid grid and fosmid clones, Yuji Kohara for providing EST clones, and Bob Barstead for the cDNA library. Some strains used in this work were provided by the Caenorhabditis Genetics Center. B.L. was supported by an EMBO fellowship, and R.B. was supported by grants from the EC (DIADEM), the DFG and the BMBF (NGFN network).
References
- Andres M.E., Burger,C., Peral-Rubio,M.J., Battaglioli,E., Anderson,M.E., Grimes,J., Dallman,J., Ballas,N. and Mandel,G. (1999) CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl Acad. Sci. USA, 96, 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N. et al. (2001) Regulation of neuronal traits by a novel transcriptional complex. Neuron, 31, 353–365. [DOI] [PubMed] [Google Scholar]
- Baumeister R., Leimer,U., Zweckbronner,I., Jakubek,C., Grunberg,J. and Haass,C. (1997) Human presenilin-1, but not familial Alzheimer’s disease (FAD) mutants, facilitate Caenorhabditis elegans Notch signalling independently of proteolytic processing. Genes Funct., 1, 149–159. [DOI] [PubMed] [Google Scholar]
- Binda C., Coda,A., Angelini,R., Federico,R., Ascenzi,P. and Mattevi,A. (1999) A 30-Ångstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Struct. Fold. Design, 7, 265–276. [DOI] [PubMed] [Google Scholar]
- Binda C., Angelini,R., Federico,R., Ascenzi,P. and Mattevi,A. (2001) Structural bases for inhibitor binding and catalysis in polyamine oxidase. Biochemistry, 40, 2766–2776. [DOI] [PubMed] [Google Scholar]
- Binda C., Newton-Vinson,P., Hubalek,F., Edmondson,D.E. and Mattevi,A. (2002) Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol., 9, 22–26.11753429 [Google Scholar]
- Blumenthal T. et al. (2002) A global analysis of Caenorhabditis elegans operons. Nature, 417, 851–854. [DOI] [PubMed] [Google Scholar]
- Bosher J.M., Dufourcq,P., Sookhareea,S. and Labouesse,M. (1999) RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics, 153, 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. (2002) A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem. Sci., 27, 115–117. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell, 9, 213–224. [DOI] [PubMed] [Google Scholar]
- Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar H.N., Sweet,K.L., Hosemann,K.E., Earley,K. and Newman,A.P. (2001) The SEL-12 presenilin mediates induction of the Caenorhabditis elegans uterine π cell fate. Dev. Biol., 237, 173–182. [DOI] [PubMed] [Google Scholar]
- Dailey T.A. and Dailey,H.A. (1998) Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J. Biol. Chem., 273, 13658–13662. [DOI] [PubMed] [Google Scholar]
- Dufourcq P., Victor,M., Gay,F., Calvo,D., Hodgkin,J. and Shi,Y. (2002) Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol. Cell. Biol., 22, 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer S., Donhauser,R. and Baumeister,R. (2002) The C.elegans presenilin sel-12 is required for mesodermal patterning and muscle function. Dev. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Fortini M.E. (2001) Notch and presenilin: a proteolytic mechanism emerges. Curr. Opin. Cell Biol., 13, 627–634. [DOI] [PubMed] [Google Scholar]
- Greenwald I. (1998) LIN-12/Notch signaling: lessons from worms and flies. Genes Dev., 12, 1751–1762. [DOI] [PubMed] [Google Scholar]
- Greenwald I.S., Sternberg,P.W. and Horvitz,H.R. (1983) The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell, 34, 435–444. [DOI] [PubMed] [Google Scholar]
- Griffith E.C., Cowan,C.W. and Greenberg,M.E. (2001) REST acts through multiple deacetylase complexes. Neuron, 31, 339–340. [DOI] [PubMed] [Google Scholar]
- Humphrey G.W., Wang,Y., Russanova,V.R., Hirai,T., Qin,J., Nakatani,Y. and Howard,B.H. (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem., 276, 6817–6824. [DOI] [PubMed] [Google Scholar]
- Jepsen K. and Rosenfeld,M.G. (2002) Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci., 115, 689–698. [DOI] [PubMed] [Google Scholar]
- Johnstone I.L. and Barry,J.D. (1996) Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J., 15, 3633–3639. [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos,M., Zipperlen,P., Fraser,A.G. and Ahringer,J. (2000) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol., 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C.elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Lai E.C. (2002) Protein degradation: four e3s for the notch pathway. Curr. Biol., 12, R74–R78. [DOI] [PubMed] [Google Scholar]
- Levitan D. and Greenwald,I. (1995) Facilitation of _lin-12_-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature, 377, 351–354. [DOI] [PubMed] [Google Scholar]
- Li C. and Chalfie,M. (1990) Organogenesis in C.elegans: positioning of neurons and muscles in the egg-laying system. Neuron, 4, 681–695. [DOI] [PubMed] [Google Scholar]
- Li X. and Greenwald,I. (1997) HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc. Natl Acad. Sci. USA, 94, 12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J.S. and Kopan,R. (2000) Notch signaling: from the outside in. Dev. Biol., 228, 151–165. [DOI] [PubMed] [Google Scholar]
- Nagase T., Ishikawa,K., Miyajima,N., Tanaka,A., Kotani,H., Nomura,N. and Ohara,O. (1998) Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res., 5, 31–39. [DOI] [PubMed] [Google Scholar]
- Newman A.P., White,J.G. and Sternberg,P.W. (1995) The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development, 121, 263–271. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sebela M., Radova,A., Angelini,R., Tavladoraki,P., Frebort,I.I. and Pec,P. (2001) FAD-containing polyamine oxidases: a timely challenge for researchers in biochemistry and physiology of plants. Plant Sci., 160, 197–207. [DOI] [PubMed] [Google Scholar]
- Seiler N. (1995) Polyamine oxidase, properties and functions. Prog. Brain Res., 106, 333–344. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J. (2001) Alzheimer’s disease: genes, proteins and therapy. Physiol. Rev., 81, 741–766. [DOI] [PubMed] [Google Scholar]
- Seo S.B., McNamara,P., Heo,S., Turner,A., Lane,W.S. and Chakravarti,D. (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell, 104, 119–130. [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R., White,J.G., Southgate,E. and Podbilewicz,B. (1999) Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development, 126, 691–699. [DOI] [PubMed] [Google Scholar]
- Spieth J., Brooke,G., Kuersten,S., Lea,K. and Blumenthal,T. (1993) Operons in C.elegans: polycistronic mRNA precursors are processed by _trans_-splicing of SL2 to downstream coding regions. Cell, 73, 521–532. [DOI] [PubMed] [Google Scholar]
- Struhl G. and Adachi,A. (1998) Nuclear access and action of notch in vivo. Cell, 93, 649–660. [DOI] [PubMed] [Google Scholar]
- Sundaram M. and Greenwald,I. (1993) Genetic and phenotypic studies of hypomorphic lin-12 mutants in Caenorhabditis elegans. Genetics, 135, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L. and Fire,A. (1998) Specific interference by ingested dsRNA. Nature, 395, 854. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court,D.L. and Fire,A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene, 263, 103–112. [DOI] [PubMed] [Google Scholar]
- Tong J.K., Hassig,C.A., Schnitzler,G.R., Kingston,R.E. and Schreiber,S.L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921. [DOI] [PubMed] [Google Scholar]
- Trent C., Tsung,N. and Horvitz,H.R. (1983) Egg-laying defective mutants of the nematode C.elegans. Genetics, 104, 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Levitan,D., Li,X. and Greenwald,I. (2000) spr-2, a suppressor of the egg-laying defect caused by loss of sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily. Proc. Natl Acad. Sci. USA, 97, 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlund B., Parry,D., Clover,R., Basson,M. and Johnson,C.D. (1999) Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signaling. Proc. Natl Acad. Sci. USA, 96, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S.R., de Vries,C.J., van Luenen,H.G. and Plasterk,R.H. (2000) CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol., 221, 295–307. [DOI] [PubMed] [Google Scholar]
- Wu G., Hubbard,E.J., Kitajewski,J.K. and Greenwald,I. (1998) Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein and SEL-12 presenilin. Proc. Natl Acad. Sci. USA, 95, 15787–15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A., Tong,J.K., Grozinger,C.M. and Schreiber,S.L. (2001) CoREST is an integral component of the CoREST–human histone deacetylase complex. Proc. Natl Acad. Sci. USA, 98, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Canas,B., Pappin,D. and Kouzarides,T. (2002) Histone H3 lysine 4 methylation disrupts the binding of the nucleosome remodelling and deacetylase (NuRD) repressor complex. J. Biol. Chem., 277, 11621–11624. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Piston,D.W. and Goodman,R.H. (2002) Regulation of corepressor function by nuclear NADH. Science, 295, 1895–1897. [DOI] [PubMed] [Google Scholar]