Identification of the Flavonoid Hydroxylases from Grapevine and Their Regulation during Fruit Development (original) (raw)
Abstract
Flavonoids are important secondary metabolites in many fruits, and their hydroxylation pattern determines their color, stability, and antioxidant capacity. Hydroxylation of the B-ring of flavonoids is catalyzed by flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H), and may also require cytochrome _b_5. We report the identification of genes encoding F3′H, F3′5′H, and a putative cytochrome _b_5 from grapevine (Vitis vinifera L. cv Shiraz) and their transcriptional regulation in fruit. Functionality of the genes VvF3_′_H and VvF3_′_5_′_H1 was demonstrated by ectopic expression in petunia (Petunia hybrida), which altered flower color and flavonoid composition as expected. VvF3_′_H was expressed in grapes before flowering, when 3′-hydroxylated flavonols are made, and all three genes were expressed after flowering, when proanthocyanidins (PAs) are synthesized. In berry skin, expression of all three genes was low at the onset of ripening (véraison) but increased after véraison concomitant with the accumulation of 3′- and 3′,5′-hydroxylated anthocyanins. VvF3_′_H and VvCytoB5 were expressed in seeds but not VvF3_′_5_′_H1, consistent with the accumulation of 3′-hydroxylated PAs in this tissue. VvCytoB5 expression was correlated with expression of both VvF3_′_H and VvF3_′_5_′_H1 in the different grape tissues. In contrast to red grapes, where VvF3_′_H, VvF3_′_5_′_H1, and VvCytoB5 were highly expressed during ripening, the expression of VvF3_′_5_′_H1 and VvCytoB5 in white grapes during ripening was extremely low, suggesting a difference in transcriptional regulation. Our results show that temporal and tissue-specific expression of VvF3_′_H, VvF3_′_5_′_H1, and VvCytoB5 in grapes is coordinated with the accumulation of the respective hydroxylated flavonols and PAs, as well as anthocyanins. Understanding the regulation of flavonoid hydroxylases could be used to modify flavonoid composition of fruits.
Plants synthesize a wide variety of flavonoid compounds with important functions in disease resistance, protection from UV radiation, and coloration of flowers and fruits (Harborne and Grayer, 1993; Mol et al., 1998; Bieza and Lois, 2001). Flavonoids are present in many fruits and plant products (wine, fruit juices and teas, etc.) and contribute to their color, taste, and nutritional value. In grapevine (Vitis vinifera), anthocyanins, proanthocyanidins (PAs, condensed tannins), and flavonols are the predominant flavonoids and play important roles in the color, quality, and health benefits of wine (Glories, 1988; Mateus et al., 2002). The hydroxylation pattern of the B-ring is one of the main structural features of flavonoids and is an important determinant of their coloration, stability, and antioxidant capacity (Forkmann, 1991; Croft, 1998). From a nutritional and food quality perspective, it is important to understand the mechanisms leading to the hydroxylation of plant flavonoids.
The biochemistry of the flavonoid pathway has been characterized in several plant species and many of the structural and regulatory genes have been cloned (Gerats and Martin, 1992; Holton and Cornish, 1995; Boss et al., 1996b; Winkel-Shirley, 2001). The microsomal monooxygenases flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) belong to the cytochrome P450 family and catalyze the hydroxylation of flavonoids, either at the 3′ or at both the 3′ and 5′ positions of the B-ring leading to the respective hydroxylated flavonols, anthocyanins, and PAs (Fig. 1; Forkmann, 1991; Winkel-Shirley, 2001). After the first isolation and functional characterization of the genes encoding F3′H and F3′5′H in petunia (Petunia hybrida; Holton et al., 1993; Brugliera et al., 1999), the genes were subsequently isolated from various plant species. F3_′_H was characterized in Arabidopsis (Arabidopsis thaliana; Schoenbohm et al., 2000), and F3_′_5_′_H cDNAs were isolated from a number of plants, including Solanum melongena (Toguri et al., 1993), Gentiana triflora (Tanaka et al., 1996), Eustoma grandiflorum (Nielsen and Podivinsky, 1997), Eustoma rusellianum (Shimada et al., 1999), Catharanthus roseus (Kaltenbach et al., 1999), Campanula medium (Okinaka et al., 2003), and Vinca major (Mori et al., 2004). Some plants, including Arabidopsis and important members of the Rosaceae such as apples (Malus spp.) and roses (Rosa spp.), do not have functional F3′5′H enzymes and produce only 3′-hydroxylated and 3′,4′-hydroxylated flavonoids (Forkmann, 1991). In petunia, an additional gene DifF was identified that encodes a cytochrome _b_5 that modulates and enhances F3′5′H activity but did not have an obvious effect on other cytochrome P450 enzymes (de Vetten et al., 1999). To our knowledge, there is no evidence yet for a similar interaction between cytochrome _b_5 and F3′5′H in other plant species.
Figure 1.
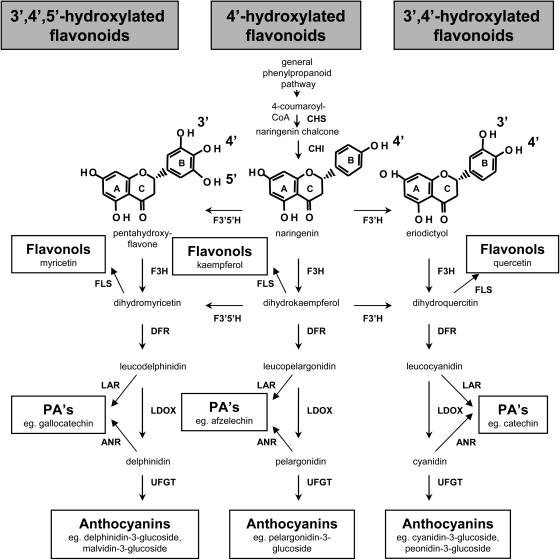
Schematic representation of the flavonoid biosynthetic pathway. Enzymes involved in the pathway shown are CHS, chalcone synthase; CHI, chalcone isomerase; F3′H; F3′5′H; F3H, flavanone-3_β_-hydroxylase; DFR; LDOX; FLS, flavonol synthase; LAR; ANR; and UFGT. Examples for the different hydroxylation patterns of the flavonoid B-ring are given for naringenin (4′-hydroxylated), eriodictyol (3′,4′-hydroxylated), and pentahydroxyflavone (3′,4′,5′-hydroxylated).
As the hydroxylation pattern of the anthocyanins in flowers strongly influences their coloration, there has been significant interest in studying the flavonoid hydroxylases in ornamentals. F3_′_5_′_H was intensively studied because some ornamental plants, such as roses and carnations (Dianthus caryophyllus), lack this activity and therefore are not able to develop blue or violet flowers (Mol et al., 1999). Recently, commercially available blue- and purple-flowering carnations and roses were developed by heterologous expression of F3_′_5_′_H (Florigene; http://www.florigene.com.au/news; Holton and Tanaka, 1994; Mol et al., 1999). Although anthocyanins are important in flowers, the hydroxylation pattern of flavonols and PAs is also determined by the activity of F3′H and F3′5′H, leading to flavonols and PAs with different biochemical and antioxidant properties (Fig. 1; Croft, 1998). For forage crops like legumes, the hydroxylation patterns of PAs have been shown to influence ruminant nutrition (Barahona et al., 2003; Min et al., 2003). Nevertheless, little is known about the role of F3′H and F3′5′H in determining flavonoid composition in fruits and leaves, where flavonols and PAs can be more important than anthocyanins.
As flavonoids are important quality components of grape berries and many other fruits, we isolated the genes encoding F3′H and F3′5′H from grapevine cv Shiraz and showed their functionality in petunia. To gain further insight into the role of F3′H, F3′5′H, and cytochrome _b_5 in fruit flavonoid metabolism, we analyzed the expression of genes encoding these enzymes in grapes during fruit development and related it to the accumulation of flavonols, anthocyanins, and PAs. Our results show that expression of the genes encoding these three enzymes is regulated in a temporal and tissue-specific manner that relates to the synthesis of anthocyanins, flavonols, and PAs in the different grape berry tissues during fruit development. The knowledge of their specific regulation could be important for future attempts to engineer flavonoid composition in fruit.
RESULTS
Cloning and Sequence Analysis of VvF3_′_H and VvF3_′_5_′_H1
Degenerate oligonucleotides based on conserved regions in F3′Hs and F3′5′Hs, respectively, were used for PCR amplification of a partial cDNA sequence from the corresponding grapevine genes (see “Materials and Methods”). The 5′ and 3′ ends of the cDNAs were identified by RACE. The VvF3_′_H cDNA (accession no. AJ880357) is 1,733 bp long and contains a putative open reading frame (ORF) of 1,527 bp encoding a 509-amino acid protein. The length of the VvF3_′_5_′_H1 cDNA (accession no. AJ880356) is 1,792 bp and contains a putative ORF of 1,524 bp coding for a protein of 508 amino acids. BLASTP searches with the deduced amino acid sequences of VvF3_′_H and VvF3_′_5_′_H1 in the GenBank database showed homologies to several flavonoid hydroxylases. The protein sequence VvF3′H (accession no. CAI54278) showed a close relationship with AtF3′H from Arabidopsis (66% identity, 80% similarity, accession no Q9SD85; Schoenbohm et al., 2000) and petunia Ht1 (76% identity, 85% similarity, accession no. AAD56282; Brugliera et al., 1999). VvF3′5′H1 (accession no. CAI54277) shares 75% identity and 86% similarity with the petunia F3′5′H sequences Hf1 and Hf2 (accession nos. CAA80265 and CAA80266; Holton et al., 1993). Comparative expressed sequence tag (EST) analysis of VvF3_′_5_′_H1 and VvF3_′_H within The Institute for Genomic Research (TIGR) grape index (Quackenbush et al., 2000; http://www.tigr.org/tdb/tgi/) identified the Tentative Consensus (TC) sequences TC45860 and TC42042, which contain several ESTs identical with the VvF3_′_5_′_H1 and VvF3_′_H sequences, respectively. Additionally, we identified within the TIGR grape index a single partial EST sequence (CF415436) 97% identical to VvF3_′_5_′_H1 within the predicted coding region but with a different 3′-untranslated region (UTR). As we were not able to detect significant amounts of this cDNA by real-time PCR or 3′-RACE, the gene corresponding to CF415436 may not be expressed in the tissues used in this study.
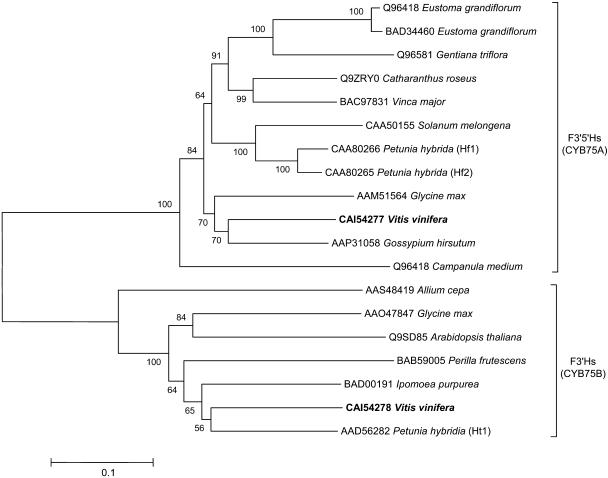
Phylogenetic analysis was performed using the deduced amino acid sequences VvF3′H and VvF3′5′H1, as well as a range of catalytically verified and putative flavonoid hydroxylases (Fig. 2). The phylogenetic tree places all analyzed F3′Hs and VvF3′H in the cytochrome P450 family CYP75B and all F3′5′Hs and VvF3′5′H1 in the family CYP75A. It also indicates that VvF3′H is most similar to F3′H from petunia (Ht1), whereas VvF3′5′H1 is most closely related to a putative F3′5′H from Gossypium hirsutum (accession no. AAP31058). The two P450 families are clearly separated with VvF3′H being more closely related to other F3′H sequences than to VvF3′5′H (Fig. 2).
Figure 2.
Phylogenetic tree showing selected flavonoid hydroxylases from GenBank or EMBL database grouping into the CYB75A and CYB75B cytochrome P450 family, respectively. The grapevine hydroxylases VvF3′H (CAI54278) and VvF3′5′H1 (CAI54277) are shown in bold. The ClustalW multiple sequence alignment was formed using the default parameters of the MEGA package (Kumar et al., 2004). The tree was constructed from the ClustalW alignment using the UPGMA method by the MEGA program. The scale bar represents 0.1 substitutions per site, and the numbers next to the nodes are bootstrap values from 100 replicates.
Functional Analysis of Grapevine Genes Encoding F3′H and F3′5′H
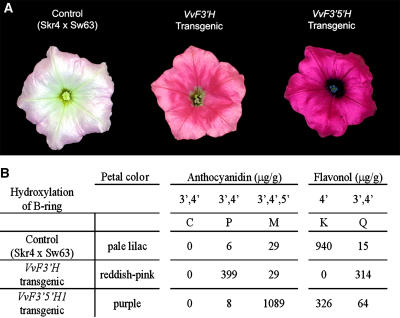
The petunia line Skr4 × Sw63 (Brugliera et al., 1999) is homozygous recessive for the loci Ht1 (encoding F3′H), Hf1, and Hf2 (encoding F3′5′H) and was therefore chosen to test functionality of the grapevine genes VvF3_′_H and VvF3_′_5_′_H1. For this purpose Skr4 × Sw63 was separately transformed with the cDNAs encoding VvF3′H and VvF3′5′H1 under the control of the 35S cauliflower mosaic virus promoter. Several transgenic lines were generated for each construct, and the petals, anthers, and pollen of the flowers showed visible color differences when compared with the nontransgenic control (Fig. 3A). Due to a leaky hf1 mutation, Skr4 × Sw63 produces pale lilac flowers with white anthers and pollen. Plants transformed with VvF3_′_H produced violet hued buds and flowers with magenta (reddish-pink) limbs, a tube with violet patterning, and violet anthers and pollen, whereas transgenic lines carrying VvF3_′_5_′_H1 had dark purple buds, flowers with bright purple limbs, a tube with dark purple patterning, and purple-blue anthers and purple pollen (Fig. 3A). HPLC analysis of acid-hydrolyzed extracts from the corolla limbs revealed that the nontransgenic control flowers produced mainly the 4′-hydroxylated flavonol, kaempferol, and small amounts of anthocyanins, whereas the F3_′_H transgenic flowers accumulated high levels of the 3′,4′-hydroxylated flavonoids peonidin (anthocyanin) and quercetin (flavonol; Fig. 3B). The lines transgenic for VvF3_′_5_′_H1 produced high levels of the 3′,4′,5′-hydroxylated anthocyanin, malvidin, and very little 3′,4′-hydroxylated anthocyanins. The transgenic lines containing VvF3_′5′_H1 also showed a shift from kaempferol to quercetin when compared to the nontransgenic control, although the transgenic lines with either of the gene constructs had lower total flavonols and greatly increased amounts of anthocyanins compared to the untransformed control (Fig. 3B). The color changes and the accumulation of the respective anthocyanins and flavonols in the transgenic VvF3_′_H and VvF3_′_5_′_H1 lines clearly demonstrate their function as F3′H and F3′5′H, respectively.
Figure 3.
Functional characterization of VvF3′H and VvF3′5′H1 by their ectopic expression in the petunia ht1 mutant line Skr4 × Sw63. A, Flowers of untransformed control Skr4 × Sw63 (pale lilac) and lines transgenic for VvF3_′_H (reddish-pink) and VvF3_′_5_′_H1 (purple). B, HPLC analysis of anthocyanidins and flavonols from flowers of the control line Skr4 × Sw63, the line H1.2 transgenic for VvF3_′_H, and O2.2 transgenic for VvF3_′_5_′_H1 showing the accumulation of the respective hydroxylated flavonoid. At least three additional independent Skr4 × Sw63 lines transgenic for VvF3_′_H or VvF3_′_5_′_H1 were analyzed and showed a similar phenotype and HPLC profile as the presented lines O2.2 or H1.2, respectively. The anthocyanidins delphinidin and pelargonidin as well as the flavonol myricetin were not detectable in any of the analyzed samples. C, Cyanidin; P, peonidin; M, malvidin; K, kaempferol; Q, quercetin.
The TC Sequence TC45693 Encodes a Putative Cytochrome _b_5 Protein from Grapevine
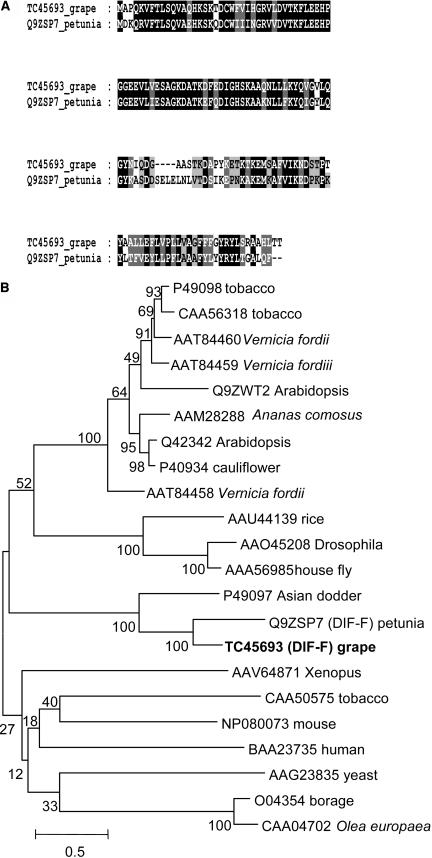
Searching the TIGR grape index (http://www.tigr.org/tdb/tgi/) by the BLAST algorithm with the sequence of the cytochrome _b_5 protein DIF-F (Q9ZSP7) from petunia (de Vetten et al., 1999) identified the TC sequence TC45693 as coding sequence of VvCytoB5, a putative cytochrome _b_5 homolog from grapevine. The length of TC45693 was 648 bp and contained a putative ORF of 441 bp coding for a protein of 147 amino acids. The protein sequence VvCytoB5 showed 60% identity and 72% similarity to the protein sequence of cytochrome _b_5 (Q9ZSP7) from petunia (Fig. 4A). Cytochrome _b_5 proteins are composed of a less conserved hydrophobic tail anchoring the protein to the membrane and an N-terminal heme-binding domain interacting with its redox partner cytochrome P450 (Vergeres and Waskell, 1995). This N-terminal domain (amino acids 1–82 of VvCytoB5) was 80% identical and 93% similar to petunia DIF-F and contained the heme-binding domain signature [FY]-[LIVMK]-{I}-{Q}-H-P-[GA]-G (PROSITE, PS00191) of the cytochrome _b_5 family. The COOH-terminal membrane anchor is less conserved, but the grape and petunia proteins shared a similar hydropathicity plot (data not shown). EMBL and GenBank database searches showed that VvCytoB5 is most closely related to DIF-F from petunia and had significant similarity to a range of cytochome _b_5 proteins from plants, animals, and yeast (Fig. 4B). Taken together, these data suggest that VvCytoB5 is a cytochrome _b_5 protein presumably interacting with similar redox partners as DIF-F, which was shown to affect the activity of the cytochrome P450 protein F3′5′H in petunia (de Vetten et al., 1999).
Figure 4.
Deduced VvCytoB5 (TC45693) amino acid sequence and comparison with the cytochrome _b_5 protein DIF-F (Q9ZSP7) from petunia and other related sequences. A, Sequence comparison of the deduced amino acid sequence of TC45693 from grapevine and the cytochrome _b_5 protein DIF-F (Q9ZSP7) from petunia. Identical amino acids are boxed in black, and similar amino acids are boxed in gray. B, Phylogenetic tree constructed with several cytochrome _b_5-related sequences recovered by using the deduced amino acid sequence of TC45693 in a BLAST algorithm on the GenBank and EMBL databases. VvCytoB5 (TC45693) is shown in bold, and the tree was constructed as described in Figure 2. The scale bar represents 0.5 substitutions per site, and the numbers next to the nodes are bootstrap values from 100 replicates.
Expression of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR during Grape Berry Development
The expression of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR was investigated throughout grape berry development (grapevine cv Shiraz) during the season 2000 to 2001 by real-time PCR (Fig. 5). VvUbiquitin1 (accession no. BN000705) was chosen for normalization of gene expression because its expression was found to be relatively constant throughout grape berry development (Downey et al., 2003b). Grape berries have two phases of growth, separated by a lag phase. The onset of ripening, termed véraison by viticulturalists, commences at the end of the lag phase, and anthocyanin accumulation in red grapes commences soon after véraison and continues through the ripening phase. Expression of the grapevine gene encoding dihydroflavonol 4-reductase (DFR), which is required for synthesis of both PAs and anthocyanins (Fig. 1), was determined to monitor flavonoid pathway activity. Real-time expression analysis of VvDFR resembles the expression patterns determined previously by northern-blot analysis (Boss et al., 1996b) and was similar to the expression patterns of the grapevine genes encoding leucoanthocyanidin dioxygenase (LDOX) and anthocyanidin reductase (ANR; Bogs et al., 2005).
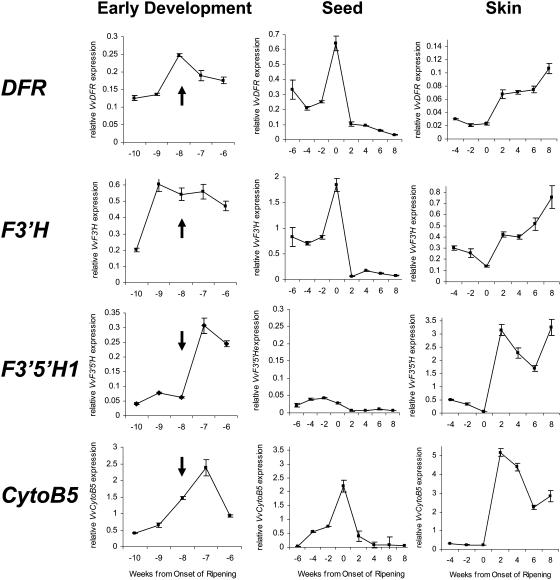
Figure 5.
Gene expression of VvDFR, VvF3_′_H, VvF3_′_5_′_H1, and VvCytoB5 (TC45693) in grapes during the early stages of berry development, in seeds, and in berry skin. Flowering is marked by an arrow and occurred 8 weeks before véraison, which is the onset of ripening in grapes. Transcript levels were determined by real-time PCR and are shown relative to expression of VvUbiquitin1 in each sample. All data are presented as mean of three replicates.
Expression of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR during flower and early berry development, when the skins and seeds could not be readily separated, is shown in Figure 5. Flowering, which occurred 8 weeks before véraison, is indicated by the arrows. The grapevine genes encoding F3′5′H, F3′H, CytoB5, and DFR were expressed throughout flower and early berry development. However, VvF3_′_H transcription was maximal 1 week before flowering, VvDFR expression at flowering, and VvF3_′_5_′_H1 and VvCytoB5 1 week after flowering.
From 6 weeks before véraison the grape berries were large enough to allow the seeds to be separated, and from 4 weeks before véraison it was possible to separate skin and seeds to obtain RNA from the respective tissues. In seeds, VvF3_′_H, VvCytoB5, and VvDFR were expressed before véraison with a maximum at véraison and declined to very low levels 2 weeks after véraison (Fig. 5). VvF3_′_5_′_H1 transcript levels in seeds also decreased after véraison but were generally very low compared to its 10- to 15-fold higher expression in skin (Fig. 5). In skin, expression of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR decreased from 4 weeks before véraison to low levels at véraison (Fig. 5). After véraison, concomitant with the beginning of anthocyanin synthesis during ripening (Boss et al., 1996b), transcription of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR was increased and transcripts were abundant during berry ripening until at least 8 weeks after véraison (Fig. 5).
White and Red Grape Varieties Exhibit Different Temporal Expression of VvF3_′_5_′_H1 and VvCytoB5
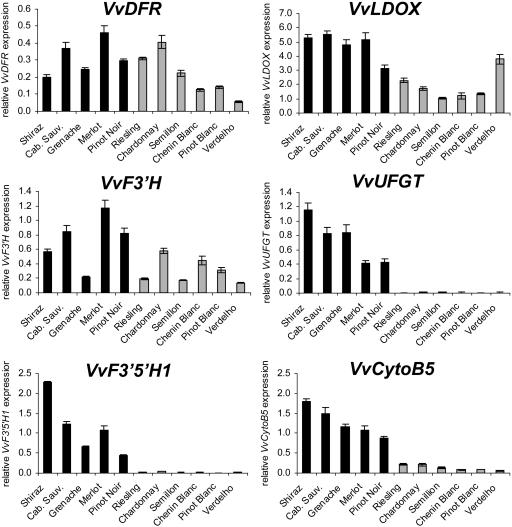
The difference between grapevine cultivars with red and white fruit is the absence of anthocyanins in the berry skin of white cultivars, which was shown to be associated with very low transcript levels of VvUFGT, encoding UDP-Glc 3-_O_-flavonoid:glucosyltransferase (UFGT), an enzyme that catalyzes one of the last steps of anthocyanin biosynthesis (Boss et al., 1996a). To further investigate gene expression differences in white and red grapevine cultivars, we compared transcript levels of VvF3_′_H, VvF3_′_5_′_H1, and VvCytoB5 during berry ripening. Additionally we analyzed expression of VvUFGT and the genes VvDFR and VvLDOX, which are required for synthesis of PAs and anthocyanidins (Fig. 1). Berries of five red grapevine cultivars (Shiraz, Cabernet Sauvignon, Grenache, Merlot, and Pinot Noir) and six white cultivars (Riesling, Chardonnay, Semillon, Chenin Blanc, Pinot Blanc, and Verdelho) were harvested 2 weeks after véraison, when anthocyanins are being synthesized in red grapes, and gene expression was analyzed (Fig. 6). The genes VvDFR, VvLDOX, and VvF3_′_H were transcribed in all red and white grapevine cultivars, although transcript levels varied from 1- to 4-fold when comparing all cultivars. In contrast, no significant expression of VvF3_′_5_′_H1 and VvUFGT was measured in any of the white grape varieties, whereas high transcript levels were detected in all red varieties we analyzed (Fig. 6), suggesting a related regulation of VvF3_′_5_′_H1 and VvUFGT during berry ripening. Expression of VvCytoB5 was detected in white grapes but the transcript levels were also strongly reduced (5- to 20-fold) in all white grape varieties when compared to expression in red varieties.
Figure 6.
Gene expression of VvDFR, VvLDOX, VvF3_′_H, VvUFGT, VvF3_′_5_′_H1, and VvCytoB5 (TC45693) was measured in red (dark bars) and white (light bars) grape cultivars by real-time PCR. Gene expression of the indicated genes was analyzed at 2 weeks after berry ripening (véraison) in different red and white grape varieties. Transcript levels were determined by real-time PCR and are shown relative to expression of VvUbiquitin1 in each sample. All data are presented as mean of three replicates.
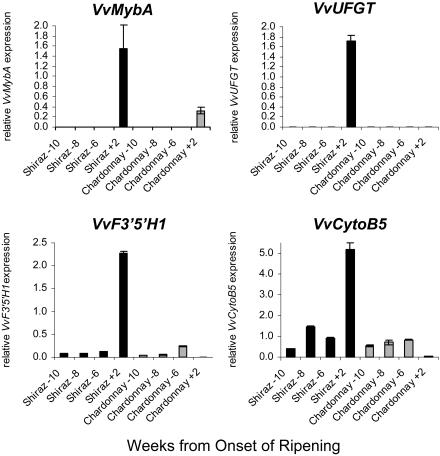
Although VvF3_′_5_′_H1 was not expressed after véraison in the white cultivars and expression of VvCytoB5 was very low, both genes were expressed in Chardonnay grapes prior to véraison (Fig. 7), whereas expression of VvUFGT was not detected in Chardonnay grapes at any time during ripening (Boss et al., 1996a). Additionally we analyzed transcript levels of VvMybA, gene products of which regulate VvUFGT expression. VvMybA is expressed only after véraison, and its transcript levels were relatively low in Chardonnay when compared to Shiraz grape. These results suggest a differential regulation of VvF3_′_5_′_H1 and VvCytoB5 compared to VvUFGT during early berry development, when flavonols and PAs are synthesized, and berry ripening when anthocyanins are accumulating.
Figure 7.
Transcript levels of VvMybA, VvUFGT, VvF3_′_5_′_H1, and VvCytoB5 in the red grape cultivar Shiraz (dark bars) and the white cultivar Chardonnay (light bars) at early berry development (10–6 weeks before véraison) and 2 weeks after véraison. Gene expressions were determined by real-time PCR and are shown relative to expression of VvUbiquitin1 in each sample. All data are presented as mean of three replicates.
DISCUSSION
The enzymes F3′H and F3′5′H and the genes that encode them have been intensively studied in flowers of various plant species (e.g. petunia, carnation, or rose) because of the importance of anthocyanin hydroxylation in determining flower coloration, However, little is known about regulation of these flavonoid hydroxylases in fruit, where 3′- and 3′,5′-hydroxylated PAs (condensed tannins) and flavonols are also synthesized, in addition to anthocyanins (Fig. 1). These flavonoids all contribute to the quality and health benefits of fruit. The aim of this work was to isolate F3_′_H and F3_′_5_′_H from grapes, to analyze their tissue- and temporal-specific regulation in fruit and to compare it with the accumulation of the respective hydroxylated anthocyanins, PAs, and flavonols.
The genes VvF3_′_H and VvF3_′_5_′_H1 were isolated by a PCR-based strategy from grapevine, and their deduced amino acid sequences showed high identity to F3′H (Ht1; Brugliera et al., 1999) and the F3′5′Hs (Hf1 and Hf2) from petunia, respectively (Holton et al., 1993). The various F3′Hs and F3′5′Hs clearly cluster into two separate groups, the CYB75B gene family encoding F3′Hs and the CYB75A family encoding F3′5′Hs (Fig. 2). The proposed function of the isolated clones VvF3_′_H and VvF3_′_5_′_H1 was verified by complementation of the petunia line Skr4 × Sw63 (Fig. 3), which is homozygous recessive for the loci Ht1, Hf1, and Hf2 (Brugliera et al., 1999). Similar to the complementation of Skr4 × Sw63 with the petunia genes encoding F3′H and F3′5′H (Holton et al., 1993; Brugliera et al., 1999), the expression of VvF3′H and VvF3′5′H1 in Skr4 × Sw63, which is pale lilac due to a leaky hf1 mutation, changed the flower color to reddish-pink and purple, respectively (Fig. 3A), and altered flavonoid composition as expected (Fig. 3B).
F3′H and F3′5′H are cytochrome P450 proteins whose activities are dependent on associated proteins, such as cytochrome P450 reductase and cytochrome _b_5, catalyzing the transfer of electrons to their prosthetic heme group (Vergeres and Waskell, 1995). In petunia the cytochrome _b_5 DIF-F was shown to modulate F3′5′H activity, and we identified a putative cytochrome _b_5 VvCytoB5 from grapevine by searching the TIGR grape index with DIF-F. Because of the close relationship of the amino acid sequence of VvCytoB5 and DIF-F, the presence of the heme-binding domain signature of the cytochrome _b_5 family in VvCytoB5 and the highly conserved N-terminal domain of VvCytoB5 and DIF-F (80% identical and 93% similar to DIF-F), which is interacting with the cytochrome P450 redox partner, VvCytoB5 was considered to be a putative homolog of DIF-F (Fig. 4). Therefore, we analyzed its expression in grapevine and found a close correlation with the expression of both VvF3_′_H and VvF3_′_5_′_H1 (Figs. 5–7, discussed below). Although VvCytoB5 is the putative cytochrome _b_5 modulating F3′5′H and F3′H activity in grapevine, its functionality has still to be shown. As complementation of the dif-f mutant from petunia is not practical due to its revertant phenotype (de Vetten et al., 1999), further studies expressing VvCytoB5 and F3′5′H in carnation will be required to determine its functionality.
The main products of the flavonoid pathway in grape berries are anthocyanins, flavonols, and PAs (Fig. 1), and regulation of this pathway must occur to coordinate synthesis of different hydroxylated flavonoids in different tissues of the grape berry during early development and ripening. Grape berry development occurs in two successive growth phases (Robinson and Davies, 2000; Coombe and McCarthy, 2000). During the first growth phase, from bloom until the onset of berry ripening (véraison), berry growth is initiated and berry expansion and seed formation occur in parallel. During this phase, flavonols are synthesized in the berry skin and PAs accumulate in the skin and in the developing seeds, but there is negligible accumulation of anthocyanins. The second berry growth phase starts at véraison and is characterized by accumulation of sugar, decreased acid levels, softening of the berry, and accumulation of anthocyanins and flavor compounds. PA accumulation in grapes starts before flowering and continues during the early stages of berry growth (Bogs et al., 2005). In the berry skin, PA synthesis is completed before véraison, whereas in grape seeds PA accumulation starts after flowering and continues until 1 to 2 weeks after véraison (Bogs et al., 2005). Grape seeds do not accumulate significant amounts of anthocyanins or flavonols, which are mostly found in the skin of the berry.
As hydroxylation of the B-ring of flavonoids is carried out by F3′H and F3′5′H, the composition of anthocyanins, flavonols, and PAs will be determined by the relative activities of these enzymes and their substrate specificity (Fig. 1). There are no reports of 4′-hydroxylated anthocyanins in grapes, which generally accumulate 3′,4′,5′-hydroxylated anthocyanins (based on malvidin, delphinidin, and petunidin) and lesser amounts of 3′,4′-hydroxylated pigments (based on cyanidin and peonidin; Table I). In contrast, flavonols in grapes are predominantly the 3′,4′-hydroxylated quercetin glycosides, with only minor amounts of 4′-hydroxylated (kaempferol) and 3′,4′,5′-hydroxylated (myricetin) flavonols observed (Table I). Grapes do not generally contain 4′-hydroxylated PA subunits (afzechelin), but the seeds and skins have different PA composition (Table I). All of the PA subunits in the seeds and the terminal subunits and free monomers of the skin PAs comprise only 3′,4′-hydroxylated units (catechin and epicatechin), whereas the extension subunits of PA in the berry skins also contain the 3′,4′,5′-hydroxylated epigallocatechin (Downey et al., 2003a).
Table I.
Accumulation of 4′-, 3′,4′-, and 3′,4′,5′-hydroxylated flavonols, anthocyanins, and PAs in grapevine (Mazza, 1995; Kennedy et al., 2001; Downey et al., 2003a, 2003b)
| Flavonoid | 4′-Hydroxylated | 3′,4′-Hydroxylated | 3′,4′,5′-Hydroxylated |
|---|---|---|---|
| Anthocyanins | − | + | + |
| Flavonols | − | + | − |
| Skin PAs | − | + | + |
| Seed PAs | − | + | − |
In grapes, most genes of the flavonoid pathway are expressed at flowering and during early berry development, when flavonols and PAs accumulate, then expression declines 4 to 6 weeks after flowering before increasing again at véraison, when anthocyanin synthesis commences (Boss et al., 1996b; Downey et al., 2003a, 2003b). In this study, expression of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR occurred early in berry development, when flavonols and PAs are synthesized, and again after véraison in the berry skin when genes required for anthocyanin synthesis are up-regulated (Boss et al., 1996a, 1996b). In grape seeds, where PAs and flavonols accumulate before véraison and no anthocyanins are being synthesized, VvF3_′_H, VvCytoB5, and VvDFR were also highly expressed up until véraison and were strongly down-regulated after véraison (Fig. 5). A similar pattern of expression of flavonoid pathway genes has been observed in bilberry (Jaakola et al., 2002), with two separate phases of gene expression coinciding with synthesis of flavonols and PAs early in fruit development and anthocyanins later.
The pattern of expression of VvF3_′_H, VvF3_′_5_′_H1, and VvCytoB5 during berry development and ripening was largely consistent with the timing of accumulation and the composition of the different flavonoids in each of the fruit tissues. During the first phase of berry development the concentrations of the predominant flavonol in grapes, quercetin glycoside (3′,4′-hydroxylated), was found to be highest just before flowering (9 weeks before véraison; Downey et al., 2003b), which correlates with a strong induction of VvF3_′_H 9 weeks before véraison (Fig. 5). In contrast, expression of VvF3_′_5_′_H1 was very low until flowering (Fig. 5), reflecting that no significant amounts of the 3′,4′,5′-hydroxylated flavonol myricetin were detected during flower and early berry development (Downey et al., 2003b). The expression of the gene encoding flavonol synthase and the amount of flavonols in grapes were both found to be highest 1 week before flowering and strongly decreased after fertilization (Downey et al., 2003b). However, grapevine flowers and young berries also contain significant levels of PAs and the genes encoding leucoanthocyanidin reductase (LAR) and ANR, which catalyze the synthesis of the 3′,4′- and 3′,4′,5′-hydroxylated flavan-3-ol monomers required for formation of PA polymers (Fig. 1), were shown to be induced just after flowering (Downey et al., 2003b; Bogs et al., 2005). Coincident with VvLAR1 and VvANR induction, VvF3_′_5_′_H1 expression was also strongly induced and VvF3_′_H was still highly expressed (Fig. 5), reflecting the accumulation of 3′,4′- and 3′,4′,5′-hydroxylated PAs after flowering. VvCytoB5 was expressed throughout early development with its highest transcript levels 7 weeks before véraison when VvF3_′_5_′_H1 also reached its maximum expression. The accumulation of PAs and the expression of ANR and LAR were detected in berry skin and seeds from fruit set until the onset of ripening (Downey et al., 2003b; Bogs et al., 2005). From 6 weeks before véraison, it is possible to analyze grape skins and seeds separately. Interestingly, the composition of PAs is quite different in berry skin and seed with respect to their hydroxylation pattern and polymer length. Whereas seed PAs are mainly derived from the 3′,4′-hydroxylated flavan-3-ols catechin and epicatechin, in berry skin almost 50% of the PA extension subunits are derived from the 3′,4′,5′-hydroxylated flavan-3-ol epigallocatechin (Downey et al., 2003b). The absence of epigallocatechin in seeds and its high concentration in skin correlates with the generally very low expression of VvF3_′_5_′_H1 in seeds and its relatively high expression in berry skin (Fig. 5). Coincident with the accumulation of 3′,4′-hydroxylated PAs in skin and seed until véraison, VvF3_′_H and VvCytoB5 were also highly expressed in these tissues. This is consistent with the expression of VvDFR (Fig. 5) and other flavonoid pathway genes at this stage (Boss et al., 1996b), which would be required for the synthesis of both PAs and flavonols (Fig. 1). Concomitant with the beginning of the second phase of berry development and the accumulation of anthocyanins, the expression of VvF3_′_H, VvF3_′_5_′_H1, VvCytoB5, and VvDFR in berry skin was induced (Fig. 5), similar to the expression of most of the other flavonoid pathway genes required for anthocyanin synthesis in grape and bilberry (Boss et al., 1996b; Jaakola et al., 2002). In flowers, expression of genes encoding F3′H and F3′5′H are often coordinated, as both are required for anthocyanin synthesis, whereas in grape berries the genes encoding these two hydroxylases show quite different patterns of expression, which correlate with the different types of flavonoids made in each of the fruit tissues. It is interesting to note that the expression of VvCytoB5 overlaps expression of both VvF3_′_H and VvF3_′_5_′_H1, indicating it may have a role in modulating the activity of both hydroxylases. This contrasts with the situation in petunia, where cytochrome b5 appears to affect only F3′5′H (de Vetten et al., 1999).
In various plant species the structural genes involved in the flavonoid pathway have been shown to be regulated by a complex of at least three transcription factors composed of a MYB, a basic helix-loop-helix, and a WD40 type protein (for review, see Koes et al., 2005). Mutational analysis in Arabidopsis has revealed that the genes involved in synthesis of anthocyanins, PAs, and flavonols are specifically regulated by the MYB-type transcription factors PAP1 or PAP2, TT2, and MYB12, respectively (Borevitz et al., 2000; Nesi et al., 2001; Mehrtens et al., 2005). The synthesis of anthocyanin in grapes is linked to the expression of the gene encoding UFGT, which catalyzes one of the last steps in the pathway. In grapes, the gene encoding UFGT is regulated independently of the other genes in the pathway. In white grapes, VvUFGT is not expressed, whereas most of the general flavonoid pathway genes are still expressed and are required for synthesis of other flavonoids, including flavonols and PAs (Boss et al., 1996a, 1996b). Expression of VvUFGT is controlled by two closely related MybA genes, which are not functional in white grape berries and therefore prevent the expression of VvUFGT but not other flavonoid pathway genes (Boss et al., 1996a; Kobayashi et al., 2002; A.R. Walker, E. Lee, J. Bogs, D. McDavid, M.R. Thomas, and S.P. Robinson, unpublished data). Interestingly, we found that VvF3_′_5_′_H1, like VvUFGT, was also expressed in red grapes after véraison but not in white grapes (Fig. 6). Other flavonoid pathway genes, including VvDFR, VvLDOX, VvCytoB5, and VvF3_′_H, were still expressed in white grapes after véraison although expression of VvCytoB5 was strongly reduced (5- to 20-fold) in all white grape varieties when compared to expression in red varieties. These results suggest there may be common regulators for VvF3_′_5_′_H1 and VvUFGT in grape berries. However, unlike VvUFGT, VvF3_′_5_′_H1 was also expressed before véraison in both white and red grapes (Fig. 7) when it would be required for PA synthesis (Fig. 1), so there must be separate regulators for VvF3_′_5_′_H1 and VvUFGT in the developing grape berry.
Significant amounts of anthocyanins, flavonols, and PAs are synthesized in the different tissues of developing grape berries, and the flavonoid pathway has complex temporal and tissue-specific regulation to achieve this. The expression patterns observed for VvF3_′_H and VvF3_′_5_′_H1 are consistent with their involvement in hydroxylation of not only anthocyanins, but also flavonols and PAs in grape berries. The data indicate that expression of the genes encoding these two enzymes is also regulated in developing grapes and this influences the composition of all three types of flavonoids, with potential impacts on fruit and wine quality.
MATERIALS AND METHODS
Plant Material
Grapevine tissues (Vitis vinifera L. cv Shiraz) were collected from a commercial vineyard during the 2000 to 2001 season. Samples were collected at weekly intervals throughout berry development from floral initiation until harvest, as described by Downey et al. (2003a). Whole flowers were sampled and analyzed until 6 weeks before véraison, when seeds could be readily separated from the remainder of the berry. In the following week, 5 weeks prevéraison, it was also possible to readily separate the skin from the berry. Samples were immediately frozen in liquid nitrogen upon dissection in the field and stored at −80°C until analyzed.
Isolation of VvF3_′_H and VvF3_′_5_′_H1
Degenerate primers were designed to conserved regions of F3′Hs and F3′5′Hs, which were identified by aligning the petunia (Petunia hybrida) F3′H (AAD56282) and F3′5′Hs (CAA80266 and CAA80265) with their respective homologs from the GenBank database (Fig. 2). The respective degenerate primers allowed the amplification and cloning of fragments of the grapevine F3_′_H and F3_′_5_′_H homologs. Standard RACE PCR reactions were used to identify the 5′ and 3′ ends of the coding sequence, and the full-length clones were subsequently cloned into pGem-T Easy (Promega) using standard protocols and primers designed to, and including, the initiation and termination codons of the cDNA. An 836-bp PCR fragment of VvF3_′_H was amplified from Shiraz cDNA with the degenerate primers F3H3, 5′-ATGGTIGTIGARATGATG-3′, and F3H6R, 5′-CCRTAIGCYTCYTCCATRTT-3′, and a 910-bp PCR fragment of VvF3_′_5_′_H1 was amplified with the degenerated primers F35H1, 5′-AAIATGGCIAARAARTAYGG-3′, and F35H5R, 5′-AAIGTYTCYTTRCADATIGC-3′. The sequence of these PCR fragments was used to design primers for the amplification of the 5′ and 3′ ends of VvF3_′_H and VvF3_′_5_′_H1 from Shiraz skin cDNA using RACE-PCR (GeneRacer, Invitrogen) according to the supplier's protocol. The final full-length sequences of VvF3_′_H and VvF3_′_5_′_H1 were then amplified from Shiraz skin cDNA using PfuTurbo polymerase (Stratagene) and gene-specific primers directed to the putative 5′ and 3′ UTR of the respective gene. All described PCR products were tailed using A-Addition kit (Qiagen), cloned into pGem-T Easy (Promega), and subjected to DNA sequencing.
Petunia Transformations
The F1 hybrid petunia Skr4 × Sw63 recessive for both F3_′_H and F3_′_5_′_H loci and producing pale lilac flowers was used for transformations. The F1 hybrid was obtained from Florigene and was produced from the original parents obtained from the Institut National de la Recherche Agronomique, Cedex, France. Surface-sterilized leaves from glasshouse-grown plants were transformed using the method of Brugliera et al. (1999). Putatively transformed plants were deflasked into soil and hardened off in a temperature-controlled glasshouse. Just fully expanded flowers were collected and dissected to separate tube, limb, and reproductive parts. Corolla limb tissue was then frozen in liquid nitrogen and stored at −80°C until analyzed by HPLC.
HPLC Analysis
To obtain flavonoids for HPLC analysis, floral tissue from corolla limbs was ground to a powder in liquid nitrogen. One sample consisted of combined tissue from five flowers. Ground tissue was extracted in acidulated methanol (Downey et al., 2004). This extract was then acid hydrolyzed by adding a half-volume of 3 n HCl and heating to 100°C for 2.5 h (Nakatsuka et al., 2005). Flavonoid content was determined by the method of Bogs et al. (2005) and use of commercial standards for delphinidin chloride, peonidin chloride, and malvidin chloride (Extrasynthese).
Preparation of cDNAs
Total RNA was isolated from the various plant tissues as described by Downey et al. (2003b). The quality of RNA was verified by demonstration of intact ribosomal bands following agarose gel electrophoresis in addition to the absorbance ratios (_A_260/280) of 1.8 to 2.0. For cDNA synthesis, 4 _μ_g of total RNA were reverse transcribed using oligo d(T)18 and SuperScript III reverse transcriptase (Invitrogen Life Technologies) following the protocol of the supplier.
Expression Analysis by Real-Time PCR
Gene expression analysis was measured by real-time PCR, using SYBR green method on a Rotor-Gene 2000 (version 4.2) real-time cycler (Corbett Research). Each PCR reaction (15 _μ_L) contained the following: 266 nm primer (each), cDNA (diluted 1:60), and 1× ABsolute QPCR SYBR Green ROX mix (ABgene House). The thermal cycling conditions were 95°C for 15 min followed by 95°C for 30 s, 58°C for 25 s, and 72°C for 25 s for 35 cycles, followed by a melt cycle from 50°C to 96°C. The primers F3HP1F (5′-CCAAGTTTTCGGGAAGTAAATG-3′) and F3HP2R (5′-TACCCCTTGAGAATCATCGTTT-3′) were designed to amplify a 171-bp PCR product from the 3′-UTR of the VvF3_′_H cDNA. The primers F35H1F (5′-GCATGGATGCAGTTAAGTAGAAAA-3′) and F35H1R (5′-ATATGGCTTGGTGGTAGAATGAAACGA-3′) were used for real-time PCR of VvF3_′_5_′_H1 amplifying a fragment of 113 bp from its 3′-UTR. Primers CytoB5f (5′-ACAAGGAAACAAAGACCAAAGA-3′) and CytoB5R (5′-GAACATACAATGCAGCAAGAAA-3′) were used for real-time PCR of VvCytoB5 (TIGR grape index accession no. TC45693) amplifying a 238-bp DNA fragment including its 3′-UTR. The primers LDOXF1 (5′-ACCTTCATCCTCCACAACAT-3′) and LDOXR2 (5′-AGTAGAGCCTCCTGGGTCTT-3′) were used for real-time PCR of VvLDOX (accession no. X75966) amplifying a 340-bp DNA fragment. The primers DFRO2F (5′-GGCTTTCTAGCGAGAGCGTA-3′) and DFRO2R (5′-ACTCTCATTTCCGGCACATT-3′) were used for real-time PCR of VvDFR (accession no. X75964) amplifying a 151-bp DNA fragment from its 3′-UTR. With all cDNAs used, the primer sets gave single PCR products, which were verified by determining the melt curves for the products at the end of each run, by analysis of the products using gel electrophoresis, and by comparing the DNA sequence of the PCR products with the gene sequence. For real-time PCR expression analysis of the three VvMybA isoforms (VvMybA1 [accession no. AB097923], VvMybA2 [accession no. AB097924], and VvMybA3 [accession no. AB097925]), the primers MybAF (5′-GAGGGTGATTTTCCATTTGAT-3′) and MybAR (5′-CAAGAACAACTTTTGAACTTAAACAT-3′) were designed to amplify 115-bp PCR products from their 3′-UTR. The efficiency of the primers was tested in preliminary experiments with dilutions of the purified PCR product and maintained an _r_2 value ≥ 0.98. As VvUbiquitin1 (accession no. BN000705) expression was found to be relatively constant throughout grape berry development (Downey et al., 2003b), it was chosen for normalization of gene expression. The VvUbiquitin1 transcripts were detected by amplifying a 182-bp product with the primers VvUbiquitin Forward (5′-GTGGTATTATTGAGCCATCCTT-3′) and VvUbiquitin Reverse (5′-AACCTCCAATCCAGTCATCTAC-3′). All samples were measured in triplicate, every run included the VvUbiquitin1 control for each sample, and experiments were repeated. The difference between the cycle threshold (Ct) of the target gene and the Ct of Ubiquitin, ΔCt = CtTarget − CtUbiquitin, was used to obtain the normalized expression of target genes, which corresponds to 2−ΔCt. The Rotor Gene 2000 software (Corbett Research) and the Q-Gene software (Muller et al., 2002) were used to calculate the mean normalized expression of the genes.
1
This work was supported by the Australian Government's Cooperative Research Centres Program and the Grape and Wine Research and Development Corporation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Simon P. Robinson (simon.robinson@csiro.au).
References
- Barahona R, Lascano CE, Narvaez N, Owen E, Morris P, Theodorou MK (2003) In vitro degradability of mature and immature leaves of tropical forage legumes differing in condensed tannin and non-starch polysaccharide content and composition. J Sci Food Agric 83**:** 1256–1266 [Google Scholar]
- Bieza K, Lois R (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126**:** 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Downey M, Harvey JS, Ashton AR, Tanner GJ, Robinson SP (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139**:** 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12**:** 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996. a) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32**:** 565–569 [DOI] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996. b) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111**:** 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugliera F, Barri-Rewell G, Holton TA, Mason JG (1999) Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 19**:** 441–451 [DOI] [PubMed] [Google Scholar]
- Coombe BG, McCarthy MG (2000) Dynamics of grape berry growth and physiology of ripening. Aust J Grape Wine Res 6**:** 131–135 [Google Scholar]
- Croft KD (1998) The chemistry and biological effects of flavonoids and phenolic acids: towards prolongation of the healthy life span. Ann NY Acad Sci 854**:** 435–442 [DOI] [PubMed] [Google Scholar]
- de Vetten N, ter Horst J, van Schaik HP, de Boer A, Mol J, Koes R (1999) A cytochrome b(5) is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA 96**:** 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003. a) Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust J Grape Wine Res 9**:** 15–27 [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003. b) Synthesis of flavonols and expression of flavonol synthase genes in developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). Aust J Grape Wine Res 9**:** 110–121 [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2004) The effect of bunch shading on berry development and flavonoid accumulation in Shiraz grapes. Aust J Grape Wine Res 10**:** 55–73 [Google Scholar]
- Forkmann G (1991) Flavonoids as flower pigments: the formation of the natural spectrum and its extension by genetic engineering. Plant Breed 106**:** 1–26 [Google Scholar]
- Gerats AGM, Martin C (1992) Flavonoid synthesis in Petunia hybrida: genetics and molecular biology of flower colour. In HA Stafford, RK Ibrahim, eds, Phenolic Metabolism in Plants. Plenum Press, New York, pp 165–199
- Glories Y (1988) Anthocyanins and tannins from wine: organoleptic properties. Prog Clin Biol Res 280**:** 123–134 [PubMed] [Google Scholar]
- Harborne JB, Grayer RJ (1993) Flavonoids and insects. In JB Harborne, ed, The Flavonoids: Advances in Research Since 1986. Chapman & Hall, London, pp 589–618
- Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu CY, Farcy E, Stevenson TW, Cornish EC (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366**:** 276–279 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7**:** 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton TA, Tanaka Y (1994) Blue roses: a pigment of our imagination. Trends Biotechnol 12**:** 40–42 [Google Scholar]
- Jaakola L, Maatta K, Pirttila AM, Torronen R, Karenlampi S, Hohtola A (2002) Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol 130**:** 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach M, Schroder G, Schmelzer E, Lutz V, Schroder J (1999) Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J 19**:** 183–193 [DOI] [PubMed] [Google Scholar]
- Kennedy JA, Hayasaka Y, Vidal S, Waters EJ, Jones GP (2001) Composition of grape skin proanthocyanidins at different stages of berry development. J Agric Food Chem 49**:** 5348–5355 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215**:** 924–933 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10**:** 236–242 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5**:** 150–163 [DOI] [PubMed] [Google Scholar]
- Mateus N, Silva AMS, Santos-Buelga C, Rivas-Gonzalo JC, de Freitas V (2002) Identification of anthocyanin-flavanol pigments in red wines by NMR and mass spectrometry. J Agric Food Chem 50**:** 2110–2116 [DOI] [PubMed] [Google Scholar]
- Mazza G (1995) Anthocyanins in grapes and grape products. Crit Rev Food Sci Nutr 35**:** 341–371 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138**:** 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min BR, Barry TN, Attwood GT, McNabb WC (2003) The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review. Anim Feed Sci Technol 106**:** 3–19 [Google Scholar]
- Mol J, Cornish E, Mason J, Koes R (1999) Novel coloured flowers. Curr Opin Biotechol 10**:** 198–201 [DOI] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3**:** 212–217 [Google Scholar]
- Mori S, Kobayashi H, Hoshi Y, Kondo M, Nakano M (2004) Heterologous expression of the flavonoid 3′,5′-hydroxylase gene of Vinca major alters flower color in transgenic Petunia hybrida. Plant Cell Rep 22**:** 415–421 [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32**:** 1372–1378 [PubMed] [Google Scholar]
- Nakatsuka T, Nishihara M, Mishiba K, Yamamura S (2005) Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants. Plant Sci 168**:** 1309–1318 [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13**:** 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KM, Podivinsky E (1997) cDNA cloning and endogenous expression of a flavonoid 3′,5′-hydroxylase from petals of lisianthus (Eustoma grandiflorum). Plant Sci 129**:** 167–174 [Google Scholar]
- Okinaka Y, Shimada Y, Nakano-Shimada R, Ohbayashi M, Kiyokawa S, Kikuchi Y (2003) Selective accumulation of delphinidin derivatives in tobacco using a putative flavonoid 3′,5′-hydroxylase cDNA from Campanula medium. Biosci Biotechnol Biochem 67**:** 161–165 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J (2000) The TIGR Gene Indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res 28**:** 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP, Davies C (2000) Molecular biology of grape berry ripening. Aust J Grape Wine Res 6**:** 175–188 [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381**:** 749–753 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Nakano-shimada R, Ohbayashi M, Okinaka Y, Kiyokawa S, Kikuchi Y (1999) Expression of chimeric P450 genes encoding flavonoid-3′,5′-hydroxylase in transgenic tobacco and petunia plants. FEBS Lett 461**:** 241–245 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Yonekura K, Fukuchi-Mizutani M, Fukui Y, Fujiwara H, Ashikari T, Kusumi T (1996) Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora. Plant Cell Physiol 37**:** 711–716 [DOI] [PubMed] [Google Scholar]
- Toguri T, Kobayashi O, Umemoto N (1993) The cloning of eggplant seedling cDNAs encoding proteins from a novel cytochrome-P-450 family (Cyp76). Biochim Biophys Acta 1216**:** 165–169 [DOI] [PubMed] [Google Scholar]
- Vergeres G, Waskell L (1995) Cytochrome B(5), its functions, structure and membrane topology. Biochimie 77**:** 604–620 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126**:** 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]