Chlorobium tepidum Mutant Lacking Bacteriochlorophyll c Made by Inactivation of the bchK Gene, Encoding Bacteriochlorophyll c Synthase (original) (raw)
Abstract
The gene encoding bacteriochlorophyll (BChl) c synthase was identified by insertional inactivation in the photosynthetic green sulfur bacterium Chlorobium tepidum and was named bchK. The bchK mutant of C. tepidum was rusty-orange in color and completely lacked BChl c. Because of the absence of the BChl c antenna, the mutant grew about seven times slower than the wild type at light intensities that were limiting to the wild type (<90 μmol m−2 s−1). Various pheophorbides, which probably represent precursors of BChl c which had lost magnesium, accumulated in the mutant cells. A small fraction of these pheophorbides were apparently esterified by the remaining chlorophyll (Chl) a and BChl a synthases in cells. The amounts of BChl a, Chl a, isoprenoid quinones, carotenoids, Fenna-Matthews-Olson protein, and chlorosome envelope protein CsmA were not significantly altered on a cellular basis in the mutant compared to in the wild type. This suggests that the BChl a antennae, photosynthetic reaction centers, and remaining chlorosome components were essentially unaffected in the mutant. Electron microscopy of thin sections revealed that the mutant lacked normal chlorosomes. However, a fraction containing vestigial chlorosomes, denoted “carotenosomes,” was partly purified by density centrifugation; these structures contained carotenoids, isoprenoid quinones, and a 798-nm-absorbing BChl a species that is probably protein associated. Because of the absence of the strong BChl c absorption found in the wild type, the bchK mutant should prove valuable for future analyses of the photosynthetic reaction center and of the roles of BChl a in photosynthesis in green bacteria. An evolutionary implication of our findings is that the photosynthetic ancestor of green sulfur bacteria could have evolved without chlorosomes and BChl c and instead used only BChl _a_-containing proteins as the major light-harvesting antennae.
All photoautotrophic organisms rely on chlorophyll (Chl)- or bacteriochlorophyll (BChl)-based photosynthesis. Chls and BChls are magnesium-chelating porphyrins, and several types are found naturally (35, 36, 40). These tetrapyrroles are found in a variety of light-harvesting antennae and always constitute the primary electron donors in photosynthetic reaction centers (16). The photosynthetic green bacteria, comprising the green sulfur bacteria (Chlorobiaceae) and the green filamentous bacteria (Chloroflexaceae), are characterized by having large light-harvesting antennae called chlorosomes (for reviews, see references 2, 14, 25, and 42). The primary pigment in chlorosomes is BChl c, or the related BChl d or e, and these BChl species are only found in chlorosomes. Chlorosomes confer a great competitive advantage to the green bacteria because they allow growth at very low light intensities. In fact, green sulfur bacteria can grow at light intensities that are too low to support growth of any other phototrophs. Recent estimates suggest that chlorosomes can contain up to ∼215,000 BChl c molecules (23), and thus the molecular ratio of BChl c to reaction center P840 could be in the range of 10,000 to 20,000:1. It is clear that chlorosomes and BChl c have evolved to form the largest and most efficient light-harvesting antennae known.
BChl c, d, and e differ from all other (B)Chls by having a hydroxyl group in the 31 position and by lacking a carboxymethyl group in the 132 position of the porphyrin ring (35). These structural features are directly related to the characteristic pigment-pigment interactions of these BChls in the chlorosome, which is an important structural feature of this antenna system (2, 25, 39, 41). Thus, it appears that the evolution of these specialized BChls and the evolution of chlorosomes are closely linked. Additionally, green sulfur bacteria methylate their BChl c, d, and e on the 8 and 12 substituents of the porphyrin ring, which is not observed for other (B)Chls in other organisms (35).
The biosynthetic pathways of all types of (B)Chls are thought to emanate from magnesium protoporphyrin IX and to vary only by the modifications to the porphyrin ring and by the esterifying alcohol attached (35, 40). The biosynthetic pathways of Chl a and BChl a are well known (40) but that of BChl c is not. A significant step forward in resolving this pathway was the complete sequencing of the genome of Chlorobium tepidum (7). These data suggest that several genes in the BChl a biosynthetic pathway have been duplicated in the BChl c biosynthetic pathway. Especially striking was the observation that the genome encodes three potential (B)Chl synthases (CT1270, CT1610, and CT1992), since this matches the presence of the three types of (B)Chl in C. tepidum (25): BChl c in the chlorosomes; BChl a in the chlorosomes, Fenna-Matthews-Olson (FMO) protein, and reaction center; and Chl _a_PD (Chl a esterified with Δ2,6-phytadienol) in the reaction center (19). [The abbreviations used in this paper for the various (B)Chl species follow the recommendations of Smith (36).] In the present study we show that insertional inactivation of the open reading frame (ORF) for one of these putative (B)Chl synthases (CT1992) resulted in the complete absence of BChl c in C. tepidum and that this ORF, named bchK, thus encodes BChl c synthase. Since BChl d and e closely resemble BChl c in structure, function, and occurrence, the synthases producing these BChls in other green sulfur bacteria are most likely very similar to BchK in C. tepidum.
MATERIALS AND METHODS
Organism and growth conditions.
The strain of C. tepidum used was the plating strain WT2321 (47) derived from strain ATCC 49652 (46). Growth media (CL and CP) and growth conditions were as previously described (11) except that transformation and transfer of cells on solid media were done at 42°C and all other growth experiments in liquid media were done at 47°C. After segregation, mutants were cultivated in liquid medium without antibiotics. Wild-type and mutant C. tepidum cells were grown in 0.5-liter bottles under incandescent illumination (160 μmol s−1 m−2) unless otherwise stated. For growth rate determinations, cultures were grown in 25-ml screw-cap tubes (diameter, 20 mm) mounted in a radial rotator (model RD4512; Glas-Col, Terre Haute, Ind.) in a thermostatically controlled incubator illuminated from the outside. The tubes were shaded with paper to obtain different light intensities. For growth rate determinations, optical densities at 600 nm for the tube cultures were measured directly with a Spectronic 20 spectrophotometer. (Optical densities recorded in the mid-exponential growth phase, for which the transient appearance of sulfur globules excreted by the cells contributes significantly to the measured optical density, were omitted from the growth rate calculations.)
Freeze-storage of cells.
Late-exponential-phase wild-type and mutant cultures of C. tepidum were preserved for long-term storage by adding 10% (vol/vol) glycerol from an autoclaved 50% (vol/vol) glycerol stock directly to the culture. Alternatively, 10% (vol/vol) glycerol was added to a cell suspension concentrated fivefold by centrifugation (6,000 × g, 4 min, approximately 30°C) and resuspended in fresh CL medium. In either case cell suspensions supplemented with glycerol were immediately transferred to storage at −80°C. Cultures were revived by inoculating a small aliquot of thawed stock into 10 to 20 volumes of fresh CL medium; the remaining glycerol stock cell suspension could be refrozen for later use.
Inactivation of the bchK gene.
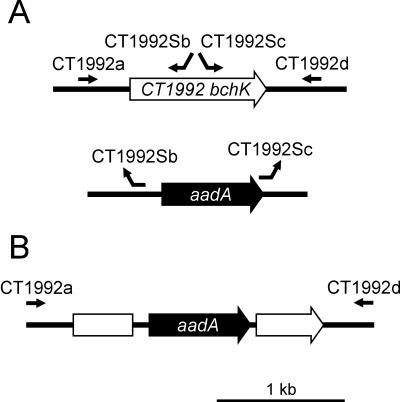
Initial attempts to clone the complete bchK coding sequence in Escherichia coli failed, probably because BchK is toxic to E. coli (S. Yang, N.-U. Frigaard, and D. A. Bryant, unpublished data). Therefore, an in vitro method denoted double megaprimer PCR (or long flanking homology region PCR) (45) was employed to synthesize the bchK inactivation construct using a high-fidelity polymerase (Platinum Pfx DNA polymerase; Invitrogen, Carlsbad, Calif.). A 0.83-kb upstream flanking region of the bchK gene was amplified from C. tepidum genomic DNA using primers CT1992a (5′-AAAAGGCGAGGCTTCAGG_-3′) and CT1992Sb (5′-GTTACCACCGCTGCGTT_C AGGAGCGAACCAACGAA_-3′), and a 0.93-kb downstream flanking region of bchK was amplified using primers CT1992Sc (5′-CAAGGTAGTCGGCAAATAATG_TCT _TCTTTTCCGCTCCAGC_-3′) and CT1992d (5′-_CCGTGATGTTCAGGTAGTGC_-3′) under the following PCR conditions: 94°C for 4 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min; and a final step of 68°C for 10 min. The CT1992Sb and CT1992Sc primers were constructed such that their 5′ ends were complementary to regions upstream and downstream from the aadA marker gene (see below; the italicized sequences of the primers are homologous to C. tepidum DNA, and the underlined sequences are homologous to regions flanking the aadA gene). The binding sites for the primers are depicted in Fig. 1. A double megaprimer PCR was then performed in a reaction mixture that contained the two gel-purified PCR products from preceding reactions as megaprimers, primers CT1992a and CT1992d, and _Ahd_I-digested plasmid pSRA2 under the following conditions: 94°C for 2 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 4 min; and a final step of 68°C for 10 min. (Plasmid pSRA2 is derived from plasmid pHP45Ω [30] and contains the aadA gene, which encodes streptomycin and spectinomycin resistance [N.-U. Frigaard and D. A. Bryant, unpublished data].) This PCR amplification produced a product in which the bchK gene was inactivated by insertion of a 0.98-kb DNA fragment encoding aadA (including promoter) flanked by a 0.83-kb and a 0.93-kb sequence of C. tepidum genomic DNA (Fig. 1B).
FIG. 1.
(A) Diagram showing how the primers used for synthesizing the bchK inactivation construct bind to the template DNA. The primers are shown as arrows pointing in the 5′ to 3′ direction. (B) The final product used for insertional inactivation of the bchK gene.
Transformation of C. tepidum and gene inactivation by homologous recombination was performed on solid CP medium at 42°C as previously described (11). The selective CP plates contained both streptomycin and spectinomycin (300 μg ml−1 and 150 μg ml−1, respectively).
Analytical PCR.
Genomic DNA was prepared from 1-ml aliquots of dense cell culture using the method of Bickley and Owen (1). Segregation was tested by PCR using Platinum Pfx DNA polymerase (Invitrogen) under the following conditions: 94°C for 4 min; 35 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 3 min; and a final step of 68°C for 10 min. The aadA marker gene was detected as a 400-bp PCR product using the primers _aadA_946 (5′-TACCAAGGCAACGCTATGTTC-3′) and _aadA_947 (5′-ATCAGAGGTAGTTGGCGTCAT-3′).
Preparation of vestigial chlorosomes (“carotenosomes”).
Harvested cells of the bchK mutant were resuspended in cold isolation buffer (50 mM Tris-HCl, 10 mM sodium ascorbate, 2 M NaSCN, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, pH 8) and passed through a cold French press three times at 19,000 lb/in2. The cell extract was clarified by a low-speed centrifugation step (13,000 × g, 20 min, 4°C) followed by a high-speed centrifugation step (215,000 × g, 2 h, 4°C). The supernatant was collected, supplemented with solid CsCl to a final concentration of 0.4 g ml−1, and centrifuged again under the same conditions (215,000 × g, 2 h, 4°C). The orange-red, carotenoid-rich fraction that floated on top of the solution was collected, diluted in cold isolation buffer containing 0.4 g of CsCl ml−1, and centrifuged again under the same conditions (215,000 × g, 2 h, 4°C). The orange-red top fraction was collected and dialyzed (12,000 to 14,000 molecular weight cutoff) against 50 mM Tris-HCl, pH 8.
Pigment analysis.
Pigments and quinones were extracted from pelleted cells by sonication with cold acetone-methanol (7:2 [vol/vol]). The pigment composition of the clarified extract was determined as previously described (12) by absorption spectroscopy in methanol using the following absorption coefficients: BChl c, 86 liters g−1 cm−1 at 669 nm (37); BChl a, 60 liters g−1 cm−1 at 770 nm (28); carotenoids, 265 liters g−1 cm−1 at 490 nm (18). The same values were used for high-pressure liquid chromatography (HPLC) analysis (except that BChl c was detected at 667 nm and carotenoids at 491 nm) in addition to the following: Chl a, 79.2 liters g−1 cm−1 at 664 nm (21); menaquinone-7, 26 liters g−1 cm−1 at 270 nm (12); chlorobiumquinones, 17 liters g−1 cm−1 at 270 nm (12). Chlorobiumquinone (1′-oxomenaquinone-7) and 1′-hydroxymenaquinone-7, collectively known as chlorobiumquinones (12), were not well separated in our HPLC system; the absorption coefficient used for the chlorobiumquinones was based on a 5:1 ratio of chlorobiumquinone to 1′-hydroxymenaquinone-7, which was close to the composition in the material analyzed in this study (data not shown). The absorption coefficient of bacteriopheophytin (BPhe) c in methanol was estimated to be 48 mM cm−1 at the Qy peak (668 nm) by pheophytinizing HPLC-purified BChl c in a methanol solution with citric acid and recording the change in absorption (13). This corresponds to about 83 liters g−1 cm−1 for the corresponding bacteriopheophorbide and to about 56 liters g−1 cm−1 for the corresponding bacteriopheophytin (assuming a molecular weight of about 850 g mol−1 for the bacteriopheophytin). The absorption of carotenoids at 490 nm in methanol extracts was corrected for the absorption of pheophorbides by assuming that the relative absorbance of pheophorbides at 490 nm is 10% of the absorption at the Qy band near 670 nm. The acetone-methanol pigment extracts were dried over anhydrous Na2SO4 and supplemented with 0.1 volume of 1 M ammonium acetate before injection into the HPLC column. The solvent system employed was that previously described (12) except that solvent B was held constant at 100% for 10 min rather than 6 min. The HPLC column was a 25-cm-by-4.6-mm 5-μm Discovery C18 (Supelco, Bellafonte, Pa.) fitted to a 1,024-element diode array detector (model G1315B, 1100 Series; Agilent Technologies, Palo Alto, Calif.) controlled with Agilent ChemStation software for HPLC. The pump (model 2350) and gradient programmer (model 2360) were from ISCO (Lincoln, Nebr.). Chl a esterified with phytol (Chl _a_P) for use as a standard in the HPLC measurements was extracted with acetone-methanol as described above from the cyanobacterium Synechococcus sp. strain PCC 7002. Bacteriopheophytin c esterified with farnesol (BPhe _c_F) for use as a standard in the HPLC measurements was prepared by acidifying a methanol extract of wild-type C. tepidum cells with citric acid as previously described (13).
Protein analysis.
Total cell protein was determined on the residual cell material remaining after organic extraction (see above) by a modified Lowry method as previously described (44) with bovine serum albumin as a standard. The protein composition of cellular and subcellular fractions of C. tepidum was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using the Tris-Tricine buffer system described by Schägger and von Jagow (33), with modifications as described by Vassilieva et al. (43, 44). Gels were stained with silver (3, 44) and immunoblotting was performed (43, 44) as previously described. Immunoreactive proteins were detected using goat anti-rabbit immunoglobulin G secondary antibodies (Sigma, St. Louis, Mo.) conjugated with horseradish peroxidase for the enhanced chemiluminescence assay (Amersham Pharmacia Biotech, Piscataway, N.J.). Digitized images of stained protein gels and developed immunoblots were manipulated into figures using Corel Photo-Paint, version 7.
Electron microscopy.
Cells were fixed by adding glutaraldehyde and paraformaldehyde directly to the culture medium (final concentrations, 2.5 and 1.5% [wt/vol], respectively) and incubating for 1 h at room temperature in the dark. The cells were postfixed with 2% (wt/vol) OsO4, stained en bloc with 2% uranyl acetate, dehydrated, and embedded in Spurr's resin by standard methods. Thin sections were poststained with 2% uranyl acetate followed by 0.45% lead citrate and images were photographed using a JEOL 1200 EXII transmission electron microscope (Peabody, Mass.). The photographs were digitized and manipulated into figures using Corel Photo-Paint, version 7.
Sequence analysis.
Phylogenetic analysis of protein sequences was performed with MacVector software (version 7.0; Genetics Computer Group, Madison, Wis.) using BLOSUM matrices in combination with manual editing for sequence alignments and a neighbor-joining method for generating trees.
RESULTS
Identification of the Chl and BChl synthases in C. tepidum.
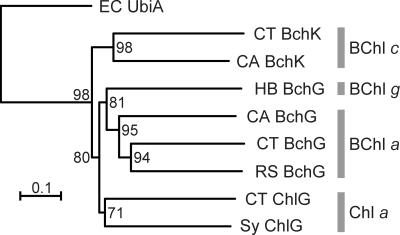
The complete genomic sequence of C. tepidum (7) contains three ORFs with strong sequence similarity to bchG and chlG genes encoding BChl a and Chl a synthases, respectively, of other organisms. These sequences were aligned to one another and to related protein sequences from other organisms. The UbiA sequence of E. coli, encoding a distantly related prenyltransferase involved in ubiquinone biosynthesis, was included as an outgroup. Figure 2 shows a phylogenetic tree showing the relationships among these sequences. The tree shows that the (B)Chl synthases clearly group according to the (B)Chl species synthesized. This is consistent with the importance of the tetrapyrrole substrate in determining the reaction characteristics of the esterifying enzyme (27, 34, 40).
FIG. 2.
A neighbor-joining distance tree of various (B)Chl synthases rooted with E. coli UbiA. The bootstrap values based on 1,000 replications are also shown. CA, Chloroflexus aurantiacus; CT, C. tepidum; EC, E. coli; HB, Heliobacillus mobilis; RS, Rhodobacter sphaeroides; Sy, Synechocystis sp. strain PCC 6803.
Further supporting the results shown in Fig. 2, the genes encoding BChl a synthase (bchG) in C. tepidum (identical to CT1610 [7]) and Chloroflexus aurantiacus have previously been identified by functional complementation of a bchG mutant of Rhodobacter capsulatus (48). Figure 2 shows that one of the C. tepidum ORFs, CT1270, exhibits a very high degree of sequence similarity to the Chl a synthase (ChlG) of the cyanobacterium Synechocystis sp. strain PCC 6803. It thus seemed most likely that ORF CT1270, denoted chlG, encodes the Chl _a_PD synthase. Consistent with this reasoning and with the important role of Chl _a_PD as the primary electron acceptor in reaction centers (19), initial attempts to inactivate CT1270 in C. tepidum have failed (A. Gomez Maqueo Chew, N.-U. Frigaard, and D. A. Bryant, unpublished data).
The third ORF with strong sequence similarity to BchG and ChlG proteins is encoded by CT1992. This ORF has strong sequence similarity to BchG2 of Chloroflexus aurantiacus (Fig. 2) (22), which like C. tepidum has chlorosomes and can synthesize BChl c. Based upon this simple analysis, it seemed most probable that ORF CT1992 encodes the BChl c synthase of C. tepidum, and thus, this gene, denoted bchK, was chosen for insertional inactivation.
Construction and verification of a bchK mutant of C. tepidum.
Transformation of C. tepidum with the bchK inactivation construct (see Materials and Methods and Fig. 1) yielded small orange-brown colonies that appeared after about 10 days on selective plates. (Colonies of the wild type normally appear as dark green colonies after 3 to 4 days on nonselective plates.) The observation that some of the colonies that appeared on the first selective plate after transformation were orange-brown suggested that segregation of the wild-type and mutant bchK alleles had occurred rapidly in these transformants. Selected colonies were restreaked a few more times on selective plates to allow the isolation of individual colonies. Several phenotypically homogeneous isolates were obtained, and one of these, denoted BGC2, was selected for more extensive characterization. Segregation of the bchK and bchK::aadA alleles in strain BGC2 was confirmed by PCR on genomic DNA with primers CT1992a and CT1992d, which only yielded the expected 2.7-kb product with strain BGC2 and a 1.8-kb product with the wild-type strain (data not shown; Fig. 1). PCR with primers _aadA_946 and _aadA_947 yielded a 0.40-kb product with strain BGC2 but no product with the wild-type strain (data not shown). These results show that strain BGC2 is homozygous for the bchK::aadA allele and that BchK is not required for viability of C. tepidum.
Appearance and growth of the bchK mutant.
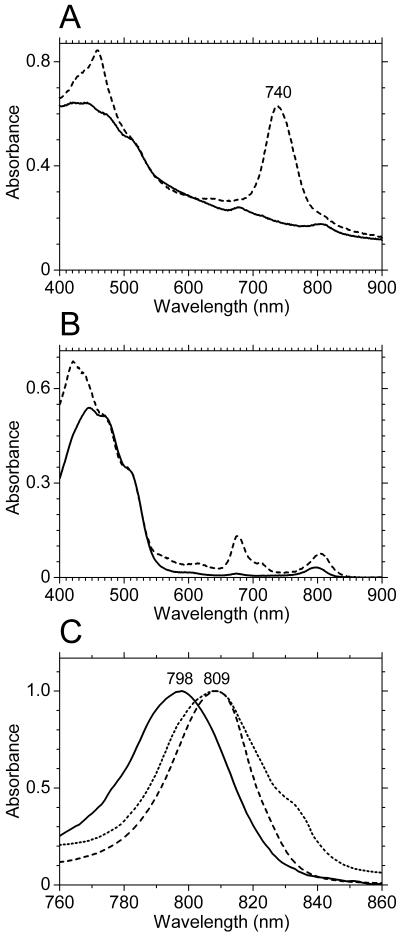
Figure 3A show absorption spectra of cell suspensions of the wild type and the bchK mutant strain BGC2. The BChl c Qy absorption peak at 740 nm was clearly missing in the bchK mutant, as was the associated BChl c Soret band at around 460 nm. The small absorption peak at about 675 nm in the bchK mutant resulted from pheophorbides that accumulated in the bchK mutant cells (see below) and which were not found in the wild type. In the absorption spectrum of the wild type, the BChl _a_-binding FMO protein was detectable as a shoulder at about 810 nm. However, in the bchK mutant strain, absorption due to the FMO protein produced a clearly resolved peak (Fig. 3A and B).
FIG. 3.
(A) Absorption spectra of washed, whole cells of the wild type (dashed line) and the bchK mutant (solid line) of C. tepidum normalized at 600 nm. (B) Absorption spectra of broken whole cells of the bchK mutant (dashed line) and isolated vestigial chlorosomes from the bchK mutant (carotenosomes; solid line) normalized at 500 nm. (C) Normalized absorption spectra showing the Qy absorption regions of the 798-nm-absorbing species found in the carotenosomes (solid line), the BChl _a_-binding FMO protein (dashed line), and a preparation of photosynthetic reaction centers (dotted line) (I. R. Vassiliev, J. H. Golbeck, and H. V. Scheller, unpublished data).
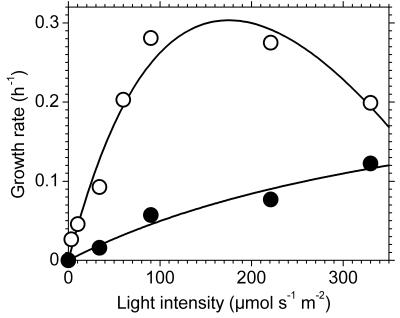
Figure 4 shows the effect of light intensity on the growth rates of the wild-type and bchK mutant strains. The growth rate of the wild type increased as the light intensity increased up to about 220 μmol m−2 s−1 and decreased significantly above that intensity. The minimal doubling time of 2.5 h, observed at 90 and 220 μmol m−2 s−1, was similar to the fastest growth rate reported, a doubling time of ∼2 h (46). In contrast, the growth rate of the bchK mutant increased as the light intensity increased throughout the entire range of values tested (30 to 330 μmol m−2 s−1). At the highest light intensity tested (330 μmol m−2 s−1), the bchK mutant had a doubling time of 5.7 h. This is only 2.3-fold longer than the fastest doubling time (2.5 h) for the wild type. At light intensities that were limiting for the wild type (<90 μmol m−2 s−1), the wild type grew about sevenfold faster than the bchK mutant (Fig. 4). This effect of complete removal of the BChl c antennae on growth is similar to the effect of completely removing the phycobilisome antennae from the cyanobacterium Synechococcus sp. strain PCC 7002 (49). In this cyanobacterium, mutants completely lacking the ability to assemble phycobilisomes have doubling times that are six- to sevenfold longer than the wild type. The bchK mutant probably relies mostly on the remaining BChl a antennae in the cell (i.e., chlorosomal BChl a, FMO protein, and the core antennae of the reaction centers) for its photosynthetic light harvesting. Further studies will be required to establish whether the large quantities of carotenoids (see below) associated with the vestigial chlorosomes and the photosynthetic membranes contribute substantially to light energy harvesting for photosynthesis in the bchK mutant.
FIG. 4.
Growth rate of wild-type (open circles) and bchK mutant (closed circles) strains of C. tepidum as a function of light intensity. The growth rate at each light intensity was independently determined at least twice, and the plotted points are the averages of those rate determinations.
Pigment analysis of the bchK mutant_._
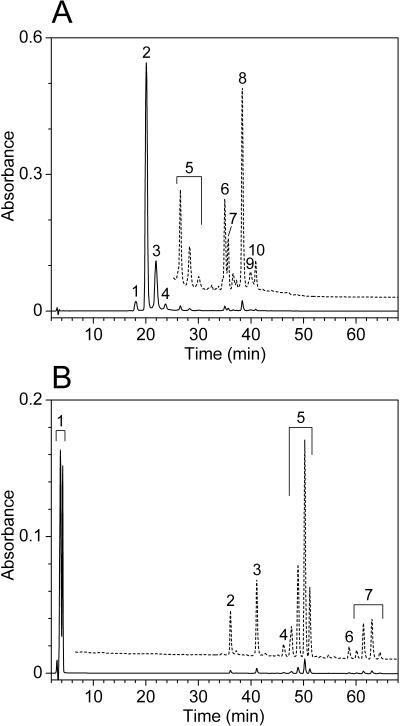
HPLC analyses confirmed that the bchK mutant was completely devoid of detectable BChl c and bacteriochlorophyllide c (Fig. 5). (The pigment contents reported in this and the next paragraph represent the averages of at least two separate measurements in which the standard deviation did not exceed 15% of the average.) The wild type contained about 160 μg of BChl c per mg of cell protein. About 95% of this BChl c was esterified with farnesol (peaks 1, 2, 3, and 4 in Fig. 5A are [E,M]BChl _c_F, [E,E]BChl _c_F, [Pr,E]BChl _c_F, and [I,E]BChl _c_F, respectively [38]), whereas the remaining BChl c was esterified with more hydrophobic alcohols (peaks 5 through 10 in Fig. 5A). No components with an absorption spectrum corresponding to BChl c were found in the bchK mutant. Instead, the mutant accumulated hydrophilic pheophorbides (roughly 30 μg per mg of cell protein; peak 1 in Fig. 5B) and small amounts of several hydrophobic pheophytins (roughly 4 μg per mg of cell protein; peaks 4 through 7 in Fig. 5B). HPLC analyses with better resolution of the hydrophilic components showed that the pheophorbides (peak 1 in Fig. 5B) consisted of at least 15 different species. All of these had a Qy peak near 670 nm, smaller absorption peaks near 620, 550, and 520 nm, and a large Soret band near 410 nm (data not shown). These probably represent precursors of BChl c or precursors of BChl c which had lost magnesium. A similar mixture of pheophorbides was also excreted by the cells into the culture medium (data not shown). Semisynthetically prepared BPhe c homologs esterified with farnesol eluted between 27 and 33 min (data not shown), which means that the hydrophobic pheophytins eluting between 46 and 65 min (peaks 4 through 7 in Fig. 5B) were esterified with alcohols more hydrophobic than farnesol, such as phytol or geranylgeraniol. These hydrophobic pheophytins were probably formed by esterification of the hydrophilic pheophorbides with such long-chain alcohols by the two remaining (B)Chl synthases in the cell (BChl a synthase and Chl _a_PD synthase). Such nonspecific esterification may have occurred because the pheophorbide concentration in the mutant was at least 3 orders of magnitude higher than in the wild type. The multitude of pheophorbide and pheophytin species, of which some subsets had identical absorption spectra but slightly different elution times (e.g., subsets 5 and 7 in Fig. 5B), indicates that these species were methylated to varying extents in the C-8, C-12, and C-20 positions on the porphyrin ring. For example, the four peaks marked as subset 5 in Fig. 5B all had an absorption spectrum that was identical to that of BPhe c.
FIG. 5.
HPLC analysis of a whole-cell pigment extract from wild-type (A) and bchK mutant (B) strains of C. tepidum. The chromatograms were recorded at 666 nm; enlarged chromatograms are shown with dashed lines (enlarged 20 times in panel A and 15 times in panel B). See text for details.
The content of other pigments varied little between the wild type and the bchK mutant. The carotenoid content of the wild type was 12 μg per mg of cell protein while the carotenoid content of strain BGC2 was 17 μg per mg of cell protein. The increased carotenoid content in the bchK mutant could be due to the slower growth rate of the mutant (see above), since similar carotenoid changes have been observed in wild-type C. tepidum grown at different light intensities (5). The distribution of BChl a homologs was similar in the wild type and the bchK mutant, with about 95% being BChl _a_P (BChl a esterified with phytol), which eluted between peaks 6 and 7 in Fig. 5A and as peak 2 in Fig. 5B. Only one Chl a homolog was identified (Chl _a_PD; peak 10 in Fig. 5A and peak 3 in Fig. 5B); Chl _a_PD eluted 2.5 min earlier than the Chl _a_P standard extracted from Synechococcus sp. strain PCC 7002. The BChl a and Chl _a_PD contents were similar in the wild type (4.3 and 0.43 μg per mg of cell protein, respectively) and the bchK mutant (4.6 and 0.38 μg per mg of cell protein). These results suggest that the amounts of FMO protein, other BChl a antennae, and photosynthetic reaction centers were similar in the wild type and the bchK mutant.
The content of isoprenoid quinones was about 6 μg of menaquinone-7 and 5 μg of chlorobiumquinones (1′-oxomenaquinone-7 and 1′-hydroxymenaquinone-7) per mg of protein in the wild type and about 3 μg of menaquinone-7 and 6 μg of chlorobiumquinones per mg of protein in the mutant. The difference in menaquinone-7 content between the wild type and the bchK mutant is probably related to the normal variation in content and distribution of isoprenoid quinones in green sulfur bacteria observed as a function of growth rate and growth state (N.-U. Frigaard, unpublished data).
Ultrastructural analysis of the bchK mutant.
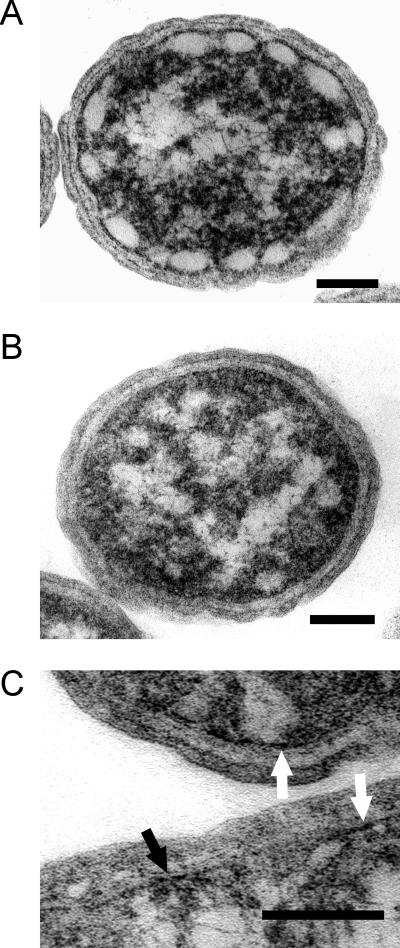
The cellular morphology of the wild type and the bchK mutant appeared indistinguishable by light microscopy (data not shown). Additionally, the overall cell shape and dimensions of the bchK mutant appeared identical to the wild type in electron microscopy of thin sections. However, typical chlorosomes were clearly absent from the bchK mutant (Fig. 6B). Chlorosomes in the wild type appeared as very lightly stained structures surrounding the periphery of the cytoplasm, with dimensions of roughly 60 by 30 nm (Fig. 6A). (These relatively small dimensions suggest that the chlorosomes were predominantly sectioned perpendicular to the long axis of the chlorosome.) The chlorosomes were clearly separated from the cytoplasmic membrane by a more heavily stained region with a thickness of roughly 5 nm. Although no obvious vestigial chlorosome structures were consistently visible along the inner surface of the cytoplasmic membrane of the bchK mutant, localized areas containing more heavily stained regions could occasionally be identified in some sections of the mutant cells (Fig. 6C). If the chlorosome envelope collapses in the absence of BChl c, such regions could represent empty chlorosome envelopes and their associated FMO protein layer.
FIG. 6.
Transmission electron micrographs of thin sections of the wild type (A) and the bchK mutant (B) of C. tepidum. The sections shown were sectioned approximately perpendicular to the long axis of the cell. (C) Enlargement showing the heavily stained regions appressed to the cytoplasmic membrane that may represent FMO protein and vestigial chlorosomes. For additional details, see text. Bars represent 100 nm.
Chlorosomes and FMO protein content of the bchK mutant.
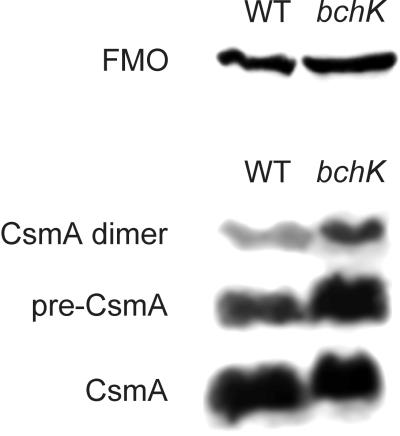
Although BChl c was eliminated from the bchK mutant, the FMO protein and the remaining components of the chlorosome seemed likely to be retained. An immunoblotting analysis of the FMO protein and the major protein of the chlorosome envelope, CsmA, in whole-cell extracts showed that the contents of these proteins were indeed similar in the wild-type and bchK mutant strains (Fig. 7). Based upon this result and the observation that the total BChl a content was also similar in the wild type and the mutant, it can be concluded that the contents of the BChl _a_-binding FMO protein and reaction centers were similar in the wild-type and bchK mutant strains.
FIG. 7.
Immunoblot analysis of the BChl _a_-binding FMO protein and the chlorosome envelope protein CsmA in whole-cell extracts of the wild type (WT) and the bchK mutant. Equal amounts of total cell protein (180 μg) were loaded in each lane. Immunoreactive proteins were detected by enhanced chemiluminescence.
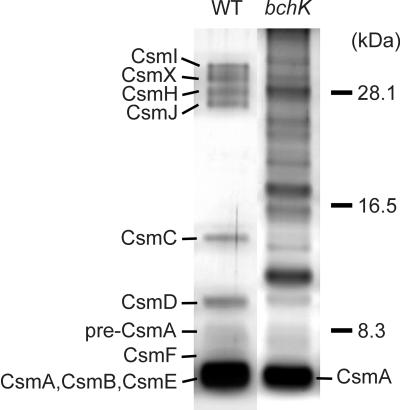
A method to isolate a fraction enriched in vestigial chlorosome structures from the bchK mutant was developed (see Materials and Methods for details). This procedure yielded an orange fraction that floated on top of the solution after centrifugation. The contents of this fraction have tentatively been named carotenosomes because of the orange-red coloration and high content of carotenoids. The absorption spectrum of the carotenosome fraction is shown in Fig. 3B and C. In addition to the large carotenoid absorption band between 400 and 500 nm, this fraction contained a minor absorption band at 798 nm. This absorption band is clearly distinct from that of the FMO protein (absorption maximum at 809 nm) and the reaction centers of the cytoplasmic membrane (absorption maximum around 810 nm with a shoulder around 835 nm; Fig. 3C). This 798-nm absorption must arise from interactions between more than one BChl a molecule or between BChl a and protein or both, since the spectrum of this BChl a is significantly shifted toward red relative to the spectrum of monomeric BChl a in organic solvents, which has an absorption maximum around 770 to 780 nm (28). The 798-nm BChl a species in our carotenosomes is probably similar to the 794-nm species derived from Chlorobium limicola chlorosomes that has been described by Gerola and Olson (15). SDS-PAGE analysis of the carotenosome fraction clearly indicated that this fraction contained CsmA in addition to several other proteins (Fig. 8). HPLC analysis and absorption spectroscopy of methanol extracts showed that the carotenosome fraction also contained 0.13 μg of BChl a, 0.58 μg of chlorobiumquinones, 0.18 μg of menaquinone-7, and about 0.05 μg of BPhe c per μg of carotenoid. A complete biochemical and spectroscopic analysis of the carotenosome fraction will be the subject of a future paper.
FIG. 8.
SDS-PAGE analysis of the protein composition of chlorosomes isolated from the wild-type (WT) strain and partially purified carotenosomes isolated from the bchK mutant strain. Wild-type chlorosomes corresponding to 0.5 μg of carotenoid and bchK carotenosomes corresponding to 1.5 μg of carotenoid were loaded in each lane. Proteins were detected by silver staining.
DISCUSSION
The results presented in this paper establish that ORF CT1992 encodes the BChl c synthase of C. tepidum. To avoid future confusion associated with the ambiguous and inappropriate locus assignments previously suggested, bchG2 and bchGc (22, 48), which would imply that CT1992 is allelic with the bchG gene encoding BChl a synthase when in fact their products catalyze different reactions, we suggest that the locus designation bchK be used to specify genes encoding BChl c synthases.
The BChl c synthases from C. tepidum and Chloroflexus aurantiacus show 34% identity and 55% similarity whereas the BChl a synthases show 39% identity and 57% similarity. In contrast, BchK and BchG from C. tepidum show 24% identity and 42% similarity while BchK and BchG from Chloroflexus aurantiacus show 30% identity and 51% similarity. The Chl a synthases (ChlG) from C. tepidum and Synechocystis sp. strain PCC 6803 show 35% identity and 55% similarity. Both BChl synthases of Chloroflexus aurantiacus have been overproduced in E. coli and enzymatically characterized (34). The results from these studies support the conclusion that BchG2 is a BChl c synthase while BchG is a BChl a synthase. Since our studies clearly establish that BchK is a BChl c synthase and since BchG2 is most closely related in sequence and function to BchK, it seems appropriate that the bchG2 gene of Chloroflexus aurantiacus should also be redesignated bchK as discussed above. Final confirmation of the identity of the bchG2 gene product in Chloroflexus aurantiacus could be obtained by using it in complementation studies of the bchK mutant of C. tepidum described here. Such complementation would also give interesting information on the specificity for the esterifying alcohol, since BChl c in Chloroflexus aurantiacus is predominantly esterified with stearol (2, 31) while BChl c in C. tepidum is predominantly esterified with farnesol (38).
Although it cannot be ruled out that minor modifications such as the methylations at the C-8, C-12, and C-20 positions of the tetrapyrrole ring occur after the esterification, the esterification catalyzed by BchK is probably the last step in the BChl c biosynthetic pathway by analogy with the Chl a and BChl a biosynthetic pathways (40). Since the pheophorbides in the bchK mutant seemed to be methylated to a variable extent, the C-8 and C-12 methyltransferases probably act on nonesterified precursors of BChl c. BChl c is found to be esterified with a variety of long-chain alcohols in green sulfur bacteria, although species esterified with farnesol are typically most abundant (6, 38). However, the substrate specificity of the BChl c synthase of C. tepidum for long-chain alcohols appears not to be very high, since other alcohols such as phytol or geranylgeraniol are easily incorporated into BChl c if these alcohols are supplied to the cells in the culture medium (38). The predominance of farnesol as the esterifying alcohol of BChl c under standard laboratory growth conditions may thus simply reflect that this alcohol is abundant in cells under such growth conditions.
The observations that BChl c is nonessential and that various pheophorbide intermediates accumulate which are not toxic to C. tepidum allow the possibility that all other genes unique for BChl c biosynthesis may also be identified by gene inactivation. These genes most likely include a BchE cyclase specific for BChl c, a C-20 methyltransferase (for BChl d to BChl c conversion), a BchF hydratase for the introduction of the hydroxyl group at the C-31 position, and C-8 and C-12 methyltransferases which probably also belong to the BchE family (7).
BChl c biosynthesis is very strongly inhibited by anaesthetic gases, such as ethylene, acetylene, and nitrous oxide (10, 26). When Chlorobium sp. cultures are treated with such inhibitors, cellular levels of BChl c can drop to as little as 10% of the levels found in control cells. A recent study has shown that the synthesis and accumulation of chlorosome envelope proteins are essentially unchanged in cells treated with acetylene (43). These results, as well as studies by Oelze and coworkers in Chloroflexus aurantiacus (8, 9), are consistent with the idea that chlorosome envelopes and their component proteins are constitutively synthesized and then filled with BChls as required by the cells. The results reported here indicate that chlorosome envelopes containing CsmA and perhaps other chlorosome proteins are synthesized in the bchK mutant and that carotenoids, isoprenoid quinones, and BChl a are imported into these carotenosomes even in the complete absence of BChl c.
The bchK mutant has some immediate advantages to offer in the study of green bacteria. The high BChl c absorption normally found in the wild type obscures the absorption from the various minor BChl a species. The bchK mutant is therefore suitable for optical studies of these species. For example, electron and excitation transfer kinetics in the reaction center and peripheral antennae may be more easily investigated in whole cells and subcellular preparations of the bchK mutant. The ability to perform measurements on whole cells is important, since breakage of the cells is likely to disrupt at least some of the normal electron and excitation transfer pathways (this has already been shown to be a problem with the FMO antenna protein and reaction centers [20, 24]). In addition, the possibility of isolating chlorosomes without BChl c will allow new approaches to investigate the roles and localization of carotenoids and BChl a in chlorosomes and to determine whether the 798-nm-absorbing BChl a species of chlorosomes is protein bound as suggested by Sakuragi et al. (32).
All green sulfur bacteria isolated so far from natural sources possess chlorosomes and BChl c, d, or e. However, our finding that BChl c is not necessary for photosynthetic growth of C. tepidum suggests that there may naturally exist photosynthetic “orange sulfur bacteria.” These bacteria would be closely related to green sulfur bacteria, would lack chlorosomes and BChl c, and would thus mainly rely on BChl a proteins as light-harvesting antennae. Such bacteria could thrive in sulfide-rich environments similar to those preferred by green sulfur bacteria except that they would possibly require a much higher light intensity to sustain reasonable growth rates. Interestingly, such bacteria that are phylogenetically related to the green filamentous bacteria have already been identified and isolated (4, 17, 29). Heliothrix oregonensis (29) and Roseiflexus castenholzii (17) are photoheterotrophs isolated from hot springs in Oregon and Japan, respectively. These organisms share many properties with Chloroflexus aurantiacus but lack chlorosomes and BChl c, synthesize only carotenoids and BChl a as photosynthetic pigments, and are distantly related to Chloroflexus aurantiacus on the basis of molecular taxonomic markers. Other red, anoxygenic photoheterotrophs have more recently been described in alkaline hot springs of Yellowstone National Park (4).
It seems likely that chlorosomes may have been exchanged by horizontal gene transfer between the otherwise distantly related green sulfur bacteria and green filamentous bacteria. However, it is not clear whether it was an ancestor of Chlorobium or of Chloroflexus which first evolved chlorosomes. The low sequence similarity and early branching in the phylogenetic tree of BchK between C. tepidum and Chloroflexus aurantiacus (Fig. 2) suggest that the chlorosome was exchanged early in evolution between the ancestors of these two bacteria. This idea is also supported by the low sequence similarity of the predominant chlorosome protein CsmA in these bacteria (only 27% identical) and by the fact that the composition of the remaining chlorosome proteins is significantly different (14). Since ancestors of Chloroflexus spp. could probably grow chemoorganotrophically as well as photoheterotrophically and photoautotrophically, we favor the hypothesis that chlorosomes were evolved by an ancestor of Chlorobium sp. that was phenotypically similar to the bchK mutant of C. tepidum. Since a _Chlorobium_-like, obligate photoautotroph with properties similar to those of the bchK mutant of C. tepidum would have been much more constrained metabolically than a _Chloroflexus_-like ancestor, such an organism would have been subject to very powerful selection pressure to evolve a more efficient light-harvesting antenna as oxygen began to appear on Earth.
Acknowledgments
This work was supported by U.S. Department of Energy grant DE-FG02-94ER20137 to D.A.B. N.-U.F. was supported by The Danish Natural Science Research Council.
We thank The Institute for Genomic Research for prepublication access to the genome sequence of C. tepidum. We also thank Rosemary A. Walsh and Missy L. Hazen from the Electron Microscopy Facility for the Life Sciences, The Pennsylvania State University, for assistance with the transmission electron microscopy. Finally, we thank Robert E. Blankenship (Arizona State University) for the antiserum to the FMO protein of C. tepidum, I. R. Vassiliev, J. H. Golbeck (The Pennsylvania State University), and H. V. Scheller (The Royal Veterinary and Agricultural University, Copenhagen, Denmark) for the absorption spectra of isolated photosynthetic reaction centers (Fig. 3C), and John G. Ormerod (University of Oslo, Oslo, Norway) for comments on the manuscript.
REFERENCES
- 1.Bickley, J., and R. J. Owen. 1995. Preparation of bacterial genomic DNA. Methods Mol. Biol. 46**:**141-147. [DOI] [PubMed] [Google Scholar]
- 2.Blankenship, R. E., J. M. Olson, and M. Miller. 1995. Antenna complexes from green photosynthetic bacteria, p. 399-435. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8**:**93-99. [Google Scholar]
- 4.Boomer, S. M., D. P. Lodge, B. E. Dutton, and B. K. Pierson. 2002. Molecular characterization of novel red green nonsulfur bacteria from five distinct hot spring communities in Yellowstone National Park. Appl. Environ. Microbiol. 68**:**346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrego, C. M., P. D. Gerola, M. Miller, and R. P. Cox. 1999. Light intensity effect on pigment composition and organization in the green sulfur bacterium Chlorobium tepidum. Photosynth. Res. 49**:**159-166. [Google Scholar]
- 6.Caple, M. B., H.-C. Chow, and C. E. Strouse. 1978. Photosynthetic pigments of green sulfur bacteria. The esterifying alcohols of bacteriochlorophyll c from Chlorobium limicola. J. Biol. Chem. 253**:**6730-6737. [PubMed] [Google Scholar]
- 7.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, J. C. Venter, T. M. Gruber, K. A. Ketchum, H. Tettelin, D. A. Bryant, and C. M. Fraser. The complete genome sequence of the green sulfur bacterium Chlorobium tepidum. Proc. Natl. Acad. Sci. USA, in press.
- 8.Foidl, M., J. R. Golecki, and J. Oelze. 1994. Bacteriochlorophyll c formation and chlorosome development in Chloroflexus aurantiacus. Photosynth. Res. 41**:**145-150. [DOI] [PubMed] [Google Scholar]
- 9.Foidl, M., J. R. Golecki, and J. Oelze. 1998. Chlorosome development in Chloroflexus aurantiacus. Photosynth. Res. 55**:**109-114. [DOI] [PubMed] [Google Scholar]
- 10.Frigaard, N.-U., and J. G. Ormerod. 1995. Hydrophobic modification of antenna chlorophyll in Chlorobium during growth with acetylene, p. 163-166. In P. Mathis (ed.), Photosynthesis: from light to biosphere. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Frigaard, N.-U., and D. A. Bryant. 2001. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl. Environ. Microbiol. 67**:**2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frigaard, N.-U., S. Takaichi, M. Hirota, K. Shimada, and K. Matsuura. 1997. Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch. Microbiol. 167**:**343-349. [Google Scholar]
- 13.Frigaard, N.-U., K. Matsuura, M. Hirota, M. Miller, and R. P. Cox. 1998. Studies of the location and function of isoprenoid quinones in chlorosomes from green sulfur bacteria. Photosynth. Res. 58**:**81-90. [Google Scholar]
- 14.Frigaard, N.-U., E. V. Vassilieva, H. Li, K. J. Milks, J. Zhao, and D. A. Bryant. 2001. The remarkable chlorosome, article S1-003. In PS2001 Proceedings: 12th International Congress on Photosynthesis. CSIRO Publishing, Melbourne, Australia.
- 15.Gerola, P. D., and J. M. Olson. 1986. A new bacteriochlorophyll _a_-protein complex associated with chlorosomes of green sulfur bacteria. Biochim. Biophys. Acta 848**:**69-76. [DOI] [PubMed] [Google Scholar]
- 16.Golbeck, J. H. 1993. Shared thematic elements in photochemical reaction centers. Proc. Natl. Acad. Sci. USA 90**:**1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. 2002. Roseiflexus castenholzii gen. and sp. nov., a thermophilic filamentous photosynthetic bacterium which lacks chlorosomes. Int. J. Syst. E vol. Microbiol. 52**:**187-193. [DOI] [PubMed] [Google Scholar]
- 18.Holo, H., M. Broch-Due, and J. G. Ormerod. 1985. Glycolipids and the structure of chlorosomes in green bacteria. Arch. Microbiol. 143**:**94-99. [Google Scholar]
- 19.Kobayashi, M., H. Oh-oka, S. Akutsu, M. Akiyama, K. Tominaga, H. Kise, F. Nishida, T. Watanabe, J. Amesz, M. Koizumi, N. Ishida, and H. Kano. 2000. The primary electron acceptor of green sulfur bacteria, bacteriochlorophyll 663, is chlorophyll a esterified with delta 2,6-phytadienol. Photosynth. Res. 63**:**269-280. [DOI] [PubMed] [Google Scholar]
- 20.Kramer, H., T. J. Aartsma, and J. Amesz. 1996. Excited states and charge separation in membranes of the green sulfur bacterium Prosthecochloris aestuarii. Photochem. Photobiol. 64**:**26-31. [Google Scholar]
- 21.Lichtenthaler, H. K. 1987. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148**:**350-382. [Google Scholar]
- 22.Lopez, J. C., S. Ryan, and R. E. Blankenship. 1996. Sequence of the bchG gene from Chloroflexus aurantiacus: relationship between chlorophyll synthase and other polyprenyltransferases. J. Bacteriol. 178**:**3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montaño, G. A., B. P. Bowen, J. T. LaBelle, N. W. Woodbury, V. B. Pizziconi, and R. E. Blankenship. 2001. Determination of the number of bacteriochlorophyll molecules per chlorosome light-harvesting complex in Chlorobium tepidum, article S1-020. In PS2001 Proceedings: 12th International Congress on Photosynthesis. CSIRO Publishing, Melbourne, Australia.
- 24.Neerken, S., H. P. Permentier, C. Francke, T. J. Aartsma, and J. Amesz. 1998. Excited states and trapping in reaction center complexes of the green sulfur bacterium Prosthecochloris aestuarii. Biochemistry 37**:**10792-10797. [DOI] [PubMed] [Google Scholar]
- 25.Olson, J. M. 1998. Chlorophyll organization and function in green photosynthetic bacteria. Photochem. Photobiol. 67**:**61-75. [Google Scholar]
- 26.Ormerod, J. G., T. Nesbakken, and S. I. Beale. 1990. Specific inhibition of antenna bacteriochlorophyll synthesis in Chlorobium vibrioforme by anaesthetic gases. J. Bacteriol. 172**:**1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oster, U., C. E. Bauer, and W. Rüdiger. 1998. Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J. Biol. Chem. 272**:**9671-9676. [DOI] [PubMed] [Google Scholar]
- 28.Permentier, H. P., K. A. Schmidt, M. Kobayashi, M. Akiyama, C. Hager-Braun, S. Neerken, M. Miller, and J. Amesz. 2000. Composition and optical properties of reaction centre core complexes from the green sulfur bacteria Prosthecochloris aestuarii and Chlorobium tepidum. Photosynth. Res. 64**:**27-39. [DOI] [PubMed] [Google Scholar]
- 29.Pierson, B. K., S. J. Giovannoni, D. A. Stahl, and R. W. Castenholz. 1985. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch. Microbiol. 142**:**164-167. [DOI] [PubMed] [Google Scholar]
- 30.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29**:**303-313. [DOI] [PubMed] [Google Scholar]
- 31.Risch, N., H. Brockmann, and A. Gloe. 1979. Structuraufklarung von neuartigen bacteriochlorphyllen aus Chloroflexus aurantiacus. Liebigs Ann. Chem., p. 408-418.
- 32.Sakuragi, Y., N.-U. Frigaard, K. Shimada, and K. Matsuura. 1999. Association of bacteriochlorophyll a with the CsmA protein in chlorosomes of the photosynthetic green filamentous bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 1413**:**172-180. [DOI] [PubMed] [Google Scholar]
- 33.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis of the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166**:**368-379. [DOI] [PubMed] [Google Scholar]
- 34.Schoch, S., U. Oster, K. Mayer, R. Feick, and W. Rüdiger. 1999. Substrate specificity of overexpressed bacteriochlorophyll synthase from Chloroflexus aurantiacus, p. 213-216. In J. H. Argyroudi-Akoyunoglou and H. Senger (ed.), The chloroplast: from molecular biology to biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 35.Senge, M. O., and K. M. Smith. 1995. Biosynthesis and structures of the bacteriochlorophylls, p. 137-151. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 36.Smith, K. M. 1994. Nomenclature of the bacteriochlorophylls c, d, and e. Photosynth. Res. 41**:**23-26. [DOI] [PubMed] [Google Scholar]
- 37.Stanier, R. Y., and J. H. C. Smith. 1960. The chlorophylls of green bacteria. Biochim. Biophys. Acta 41**:**478-484. [DOI] [PubMed] [Google Scholar]
- 38.Steensgaard, D. B., R. P. Cox, and M. Miller. 1996. Manipulation of the bacteriochlorophyll c homolog distribution in the green sulfur bacterium Chlorobium tepidum. Photosynth. Res. 48**:**385-393. [DOI] [PubMed] [Google Scholar]
- 39.Steensgaard, D. B., H. Wackerbarth, P. Hildebrandt, and A. R. Holzwarth. 2000. Diastereoselective control of bacteriochlorophyll e aggregation. 31-_S_-Bchl e is essential for the formation of chlorosome-like aggregates. J. Phys. Chem. B 104**:**10379-10386. [Google Scholar]
- 40.Suzuki, J. Y., D. W. Bollivar, and C. E. Bauer. 1997. Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet. 31**:**61-89. [DOI] [PubMed] [Google Scholar]
- 41.Van Rossum, B.-J., D. B. Steensgaard, F. M. Mulder, G. J. Boender, K. Schaffner, A. R. Holzwarth, and H. J. M. de Groot. 2001. A refined model of the chlorosomal antennae of the green bacterium Chlorobium tepidum from proton chemical shift constraints obtained with high-field 2-D and 3-D MAS NMR dipolar correlation spectroscopy. Biochemistry 40**:**1587-1595. [DOI] [PubMed] [Google Scholar]
- 42.Vassilieva, E. V., N.-U. Frigaard, and D. A. Bryant. 2000. Chlorosomes: the light-harvesting complexes of the green bacteria. Spectrum 13**:**7-13. [Google Scholar]
- 43.Vassilieva, E. V., J. G. Ormerod, and D. A. Bryant. 2002. Biosynthesis of chlorosome proteins is not inhibited in acetylene-treated cultures of Chlorobium vibrioforme. Photosynth. Res. 71**:**69-81. [DOI] [PubMed]
- 44.Vassilieva, E. V., V. L. Stirewalt, C. U. Jakobs, N.-U. Frigaard, K. Inoue-Sakamoto, M. A. Baker, A. Sotak, and D. A. Bryant. 2002. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH and CsmX. Biochemistry 41**:**4358-4370. [DOI] [PubMed] [Google Scholar]
- 45.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12**:**259-265. [DOI] [PubMed]
- 46.Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs. Chlorobium tepidum sp. nov. Arch. Microbiol. 156**:**81-90. [Google Scholar]
- 47.Wahlund, T. M., and M. T. Madigan. 1995. Genetic transfer by conjugation in the thermophilic green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 177**:**2583-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong, J., W. M. Fischer, K. Inoue, M. Nakahara, and C. E. Bauer. 2000. Molecular evidence for the early evolution of photosynthesis. Science 289**:**1724-1730. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, J., J. Zhou, and D. A. Bryant. 1992. Energy transfer processes in phycobilisomes as deduced from analyses of mutants of Synechococcus sp. PCC 7002, p. 25-32. In N. Murata (ed.), Research in photosynthesis, vol. I. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]