XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands (original) (raw)
Abstract
The secretory function of cells relies on the capacity of the endoplasmic reticulum (ER) to fold and modify nascent polypeptides and to synthesize phospholipids for the subsequent trafficking of secretory proteins through the ER–Golgi network. We have previously demonstrated that the transcription factor XBP-1 activates the expression of certain ER chaperone genes and initiates ER biogenesis. Here, we have rescued the embryonic lethality of XBP-1 deficient fetuses by targeting an XBP-1 transgene selectively to hepatocytes (XBP-1−/−;LivXBP1). XBP-1−/−;LivXBP1 mice displayed abnormalities exclusively in secretory organs such as exocrine pancreas and salivary gland that led to early postnatal lethality from impaired production of pancreatic digestive enzymes. The ER was poorly developed in pancreatic and salivary gland acinar cells, accompanied by decreased expression of ER chaperone genes. Marked apoptosis of pancreatic acinar cells was observed during embryogenesis. Thus, the absence of XBP-1 results in an imbalance between the cargo load on the ER and its capacity to handle it, leading to the activation of ER stress-mediated proapoptotic pathways. These data lead us to propose that XBP-1 is both necessary and sufficient for the full biogenesis of the secretory machinery in exocrine cells.
Keywords: apoptosis, endoplasmic reticulum, exocrine pancreas, XBP-1, unfolded protein response
Introduction
The endoplasmic reticulum (ER) is the intracellular organelle, where folding and post-translational modifications of transmembrane and secretory proteins (as well as proteins destined for transport to other organelles) occur. Properly folded proteins exit the ER for targeting to destined compartments, while misfolded proteins are degraded by an ER-associated protein degradation (ERAD) mechanism (reviewed in Ellgaard and Helenius, 2003). Highly secretory cells such as pancreatic acinar cells and plasma B cells display an elaborate ER structure. However, the mechanism by which ER biogenesis and expansion of the secretory machinery are induced in these cells is poorly understood.
Eukaryotic cells have developed a specialized pathway, the unfolded protein response (UPR) to cope with the stress of unfolded proteins in the ER. The transcription factor XBP-1 is activated by IRE1 in response to the accumulation of unfolded/misfolded proteins in the ER (Shen et al, 2001; Yoshida et al, 2001; Calfon et al, 2002; Lee et al, 2002). IRE1, a type I ER transmembrane protein cleaves XBP-1 mRNA to remove 26 nucleotides by using an endoribonuclease activity in its cytoplasmic domain. The processed XBP-1 mRNA encodes a potent transcriptional transactivator, XBP-1s. Ectopic XBP-1s expression in vitro induced multiple secretory pathway genes, increased cell size, expanded the ER and elevated total protein synthesis (Lee et al, 2003; Shaffer et al, 2004; Sriburi et al, 2004). Thus, XBP-1 enforces changes in cellular structure and function consistent with the requirements of professional secretory cells.
XBP-1 mRNA is abundantly expressed in osteoblasts in the skeletal system and in acinar cells in exocrine glands in the mouse embryo, a finding of particular interest given the highly secretory nature of these cells (Clauss et al, 1993). A fundamental question in developmental biology is the identity of those organs most reliant on the UPR for proper development. However, the function of XBP-1 in secretory organ systems has been largely unknown because of the embryonic lethality of XBP-1−/− fetuses from liver apoptosis (Reimold et al, 2000). To circumvent the lethal liver phenotype of XBP-1−/− mice, we targeted an XBP-1 transgene back to liver using a liver-specific promoter. XBP-1−/−;LivXBP1 mice lacking XBP-1 in all organs except the liver died shortly after birth from a severe impairment in the production of pancreatic digestive enzymes leading to hypoglycemia and death. Expansion of the ER and the expression of certain ER chaperone genes were severely impaired in pancreatic exocrine secretory cells and, to a lesser degree, in salivary gland acinar cells lacking XBP-1. Taken together with the requirement for XBP-1 in plasma cell differentiation (Reimold et al, 2001), our findings suggest that XBP-1 is essential for the development of highly secretory exocrine cells.
Results
Rescue of embryonic lethality by liver specific expression of an XBP-1 transgene
Transgenic mice that expressed XBP-1 in the liver were generated by using a cDNA encoding the unprocessed XBP-1 mRNA, which can be spliced by the endogenous IRE1α, driven by an apolipoprotein E promoter (Figure 1A) (Simonet et al, 1993; Miyake et al, 2001). Northern blot analysis of offspring from a transgene positive founder, LivXBP1, revealed that XBP-1 mRNA expression was increased by about two-fold in transgenic liver (Figure 1B). WT mice express an ∼2.0 kb XBP-1 mRNA. An additional mRNA species of ∼2.5 kb arising from the targeted allele is also present in XBP-1+/− mice but does not encode a functional protein (Lee et al, 2003). LivXBP1 mice on both a WT and heterozygous XBP-1+/− background expressed an XBP-1 transgene mRNA at the expected size of ∼1.7 kb, as well as another species of ∼2.2 kb, which likely contains an intron from the vector. XBP-1+/+;LivXBP1 and XBP-1+/−;LivXBP1 mice had no discernible abnormalities in any organs including the liver (Figure 1L and M, Supplementary Figure 1 and data not shown).
Figure 1.
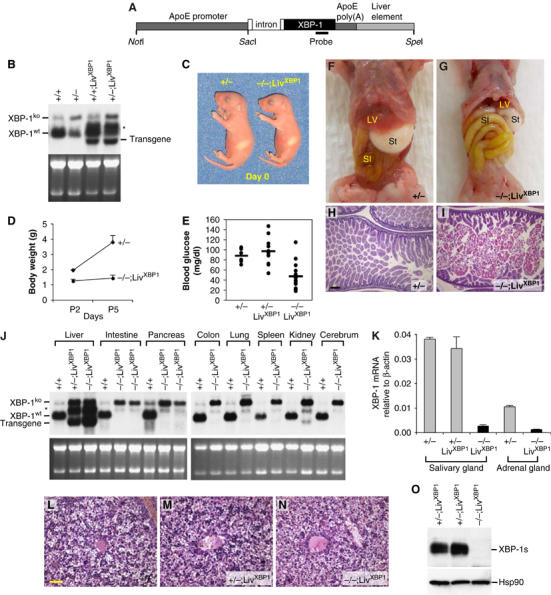
Rescue of embryonic lethality by liver specific expression of an XBP-1 transgene. (A) Transgenic construct to drive liver-specific XBP-1 expression. A mouse XBP-1 cDNA was inserted into the pLiv.7 vector using the promoter and the polyadenylation element of the apolipoprotein E gene (Miyake et al, 2001; Simonet et al, 1993). (B) Total RNA was isolated from the liver of 2-day-old mice of different genotypes for Northern blot analysis. XBP-1 mRNA species produced from the WT and the XBP-1 null allele are indicated. LivXBP1 mice expressed transgenic XBP-1 mRNA at the expected size of ∼1.7 kb as well as another species of ∼2.2 kb, which could be generated by alternative mRNA splicing. (C) A pair of newborn littermates, XBP-1+/− and XBP-1−/−;LivXBP1 mice. The milk spot on their left sides indicates that both have fed since birth. (D) Body weights of XBP-1+/− and XBP-1−/−;LivXBP1 mice were measured at day 2 and 5 after birth (_n_⩾4). (E) Blood glucose levels of XBP-1+/− (_n_=6), XBP-1+/−;LivXBP1 (_n_=11), and XBP-1−/−;LivXBP1 (_n_=14) mice measured with FreeStyle™ glucometer.(F, G) Abdominal contents of 3-day-old control and XBP-1−/−;LivXBP1 mice. Liver (Lv), stomach (St), small intestine (SI) are indicated. In XBP-1−/−;LivXBP1 mice, the small intestine is markedly distended (resulting in the rostral displacement of the liver) and pale in color, reflecting large amounts of undigested milk. (H, I) Hematoxylin and eosin (H&E) staining of the duodenum of heterozygous XBP-1+/− and XBP-1−/−;LivXBP1 mice. Note the eosinophilic staining of undigested milk in the XBP-1−/−;LivXBP1 mice (I). (J) Total RNAs isolated from various organs of WT and the XBP-1−/−;LivXBP1 mice were subjected to Northern blot analysis to measure the expression level of XBP-1. Bands corresponding to the WT, null and the transgenic XBP-1 mRNA are indicated. Ethidium bromide staining of the gel is shown at the bottom as a loading control. (K) Total RNA was isolated from the submandibular salivary gland and the adrenal gland. XBP-1 mRNA levels were measure by real time PCR analysis. The primers used do not recognize the mutant mRNA produced from the XBP-1 null allele. (L–N) H&E staining of E 18.5 liver of XBP-1+/− (L), XBP-1+/−;LivXBP1 (M) and XBP-1−/−;LivXBP1 rescued mice (N) reveals normal liver histology. (O) Pancreas lysates were tested for the expression of XBP-1s protein by Western blot analysis using anti XBP-1 antibody. Scale (H, I) 150 μm, (L–N) 40 μm.
To generate mice that express functional XBP-1 mRNA only in the liver, XBP-1+/− mice were crossed with LivXBP1 transgenic mice to produce XBP-1+/−;LivXBP1 mice, which were then intercrossed. The breeding scheme thus generated several genotypes all of which were examined in at least some of the experiments. The genotypes examined included three controls, XBP-1+/+, XBP-1+/−, and XBP-1+/−;LivXBP1 and we compared these three controls to the experimental genotype, XBP-1−/−;LivXBP1. Thus, we were careful to establish that expression of the XBP-1 transgene by itself in liver did not result in a phenotype that differed from the two other controls, XBP-1+/+ and XBP-1+/−. XBP-1+/−;LivXBP1 intercrosses resulted in close to expected Mendelian frequencies of XBP-1−/−;LivXBP1 offspring, with 39 null out of 236 mice (17%). Although XBP-1−/−;LivXBP1 mutant mice were born at a relatively normal frequency, the majority died within a few days after birth. Newborn mutant XBP-1−/−;LivXBP1 mice were slightly smaller than their littermates (body weight ∼80% of WT controls at day 0), but otherwise showed no gross abnormalities (Figure 1C). However, mutant mice exhibited severe growth retardation postnatally (Figure 1D). XBP-1−/−;LivXBP1 mice displayed marked hypoglycemia postnatally (Figure 1E) and depleted fat reserves (data not shown), symptoms of poor nutritional status that likely accounted for their growth retardation and perinatal lethality, despite being able to feed (Figure 1C and F). Mutant pups also exhibited distended loops of bowel filled with undigested milk (Figure 1F–I). Since XBP-1+/−, XBP-1+/+;LivXBP1 and XBP-1+/−;LivXBP1 mice were indistinguishable from WT mice in every parameter tested, including gross morphology, embryonic and postnatal growth, and histological features of various organs (data not shown), they were used as littermate controls for XBP-1−/−;LivXBP1 mice in several experiments.
Liver specific expression of XBP-1 in XBP-1−/−;LivXBP1 mice was confirmed by Northern blot and quantitative real time PCR analysis of RNA from various organs (Figure 1J and K). XBP-1−/−;LivXBP1 mice expressed three species of XBP-1 mRNA in the liver, one from the mutant allele (∼2.5 kb) and the other two from the transgene (∼1.7 and ∼2.2 kb). XBP-1 transgene mRNA levels in XBP-1−/−;LivXBP1 livers were similar to endogenous XBP-1 mRNA levels in WT liver, thus no overexpression of XBP-1 mRNA occurred in the rescued mice. Histological analysis revealed no apparent abnormality in XBP-1−/−;LivXBP1 livers, suggesting that the XBP-1 transgene efficiently rescued the liver abnormality (Figure 1L–N). In contrast, XBP-1 transgene mRNA was minimally expressed in all other organs tested, except for kidney, which expressed low levels of the transgene. The absence of XBP-1 protein in the pancreas of XBP-1−/−;LivXBP1 mice was also confirmed by Western blot (Figure 1O).
Impaired development of the exocrine pancreas with reduced production of pancreatic digestive enzymes in XBP-1−/−;LivXBP1 mice
The high level XBP-1 expression, we noted earlier in embryonic pancreas (Clauss et al, 1993) together with the presence of large quantities of undigested milk in the intestines of XBP-1−/−;LivXBP1 mice suggested disruption of exocrine pancreatic function. Gross morphologic examination revealed a small poorly developed pancreas in XBP-1−/−;LivXBP1 mice, whose size was approximately 10% of littermate controls based on total RNA and protein yields (Figure 2A). Histological analysis revealed that the exocrine compartment of the XBP-1−/−;LivXBP1 pancreas consisted of sparsely distributed acini in a loose mesenchymal background (Figure 2B and C). XBP-1−/−;LivXBP1 acinar cells lacked intense eosinophilic staining of the cytoplasm, suggesting fewer zymogen granules (Figure 2D and E). In contrast, islets of Langerhans were readily identified in XBP-1−/−;LivXBP1 mice, and the morphology of the islet cells was not distinguishable from WT (Figure 2D and E). A comparison of the histology of +/+, +/+;LivXBP1, +/−;LivXBP1 and +/−;LivXBP1 control pancreata is shown in Supplementary Figure 1, revealing normal and indistinguishable histology of the pancreas in all control genotypes.
Figure 2.
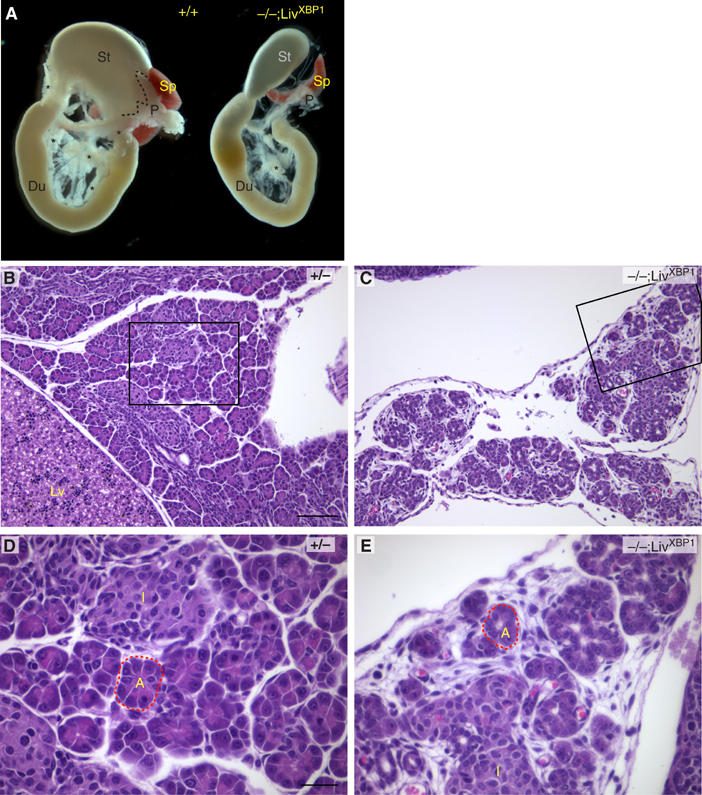
Development of the exocrine pancreas is impaired in the absence of XBP-1. (A) The stomach (St), duodenum (D), spleen (Sp) and pancreas (P) were removed en bloc from P2 mice for photography. Dashed lines outline the superior border of the control pancreas. Asterisks indicate additional areas of pancreatic parenchyma. (B–E) Histologic sections of control heterozygous XBP-1+/− (B, D) and XBP-1−/−;LivXBP1 (C, E) pancreas. Boxed regions in B and C are shown at higher magnifications in D and E, respectively. (L) Liver; (I) Islet. Dashed line outlines a representative acinar unit (A) in control and mutant. Scale (B,C) 100 μm, (D,E) 30 μm.
The fine structure of acinar cells was examined by transmission electron microscopy (TEM). WT acini consisted of cells organized to form a small central lumen into which apically located granules are secreted for transport of digestive enzymes to the duodenum (Grossman, 1984; Motta et al, 1997; Wasle and Edwardson, 2002) (Figure 3A). XBP-1−/−;LivXBP1 acinar cells were similarly situated around the lumen (Figure 3B). However, mutant acinar cells contained markedly fewer membranous granules around the lumen, and these were much smaller than those found in WT cells (Figure 3B). Interestingly, mutant acinar cells, but not WT cells, also contained small granules inside the lumen of the ER, which are likely to represent immature precursors (Figure 3C and D). Another striking difference between WT and mutant acinar cells was the development of the ER. WT acinar cells contained highly elaborate rough ER with multiple layers of closely spaced cisternae (Figure 3E). In stark contrast, the ER in the mutant acinar cells was poorly developed and had few, disorganized cisternae (Figure 3F).
Figure 3.
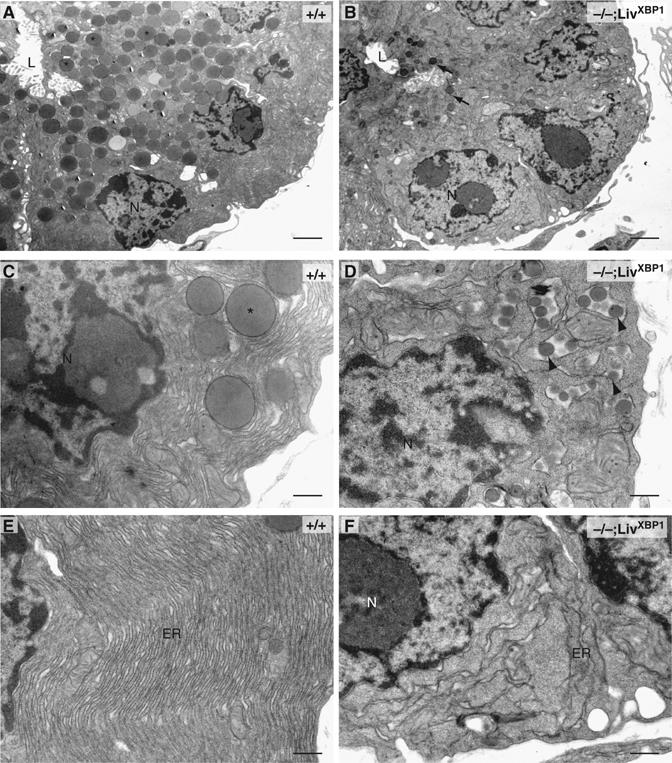
Ultrastructure of pancreatic acini reveals defects in zymogen granules and endoplasmic reticulum. Electron micrographs of sections of acini from WT and XBP-1−/−;LivXBP1 mice. (A–D) While pancreatic cells from WT mice (A, C) have abundant apically located membrane-bound zymogen granules (asterisks), pancreatic acinar cells of the XBP-1−/−;LivXBP1 mice have only a few, small apical granules (B, arrows), as well as immature granule precursors located inside of the endoplasmic reticulum (ER) lumen (D, arrowheads). N, nucleus. L, lumen. (E, F) Electron micrographs of the basolateral portion of acinar cells from WT and mutant mice. The ER contains elaborately organized long, thin, densely packed cisternae. In mutants, the ER is poorly organized and sparse. Scale, 2 μm in A and B; 500 nm in C–F.
The zymogen granules of pancreatic acinar cells contain digestive enzymes, including proteases, lipases, and enzymes, which digest carbohydrates. The presence of undigested milk in the duodenum and the striking decrease in number and size of mature granules suggested impaired pancreatic production of zymogens in XBP-1−/−;LivXBP1 mice. Indeed, amylase and trypsin, highly abundant protein species produced by the pancreas, were markedly decreased in XBP-1−/−;LivXBP1 pancreas (Figure 4A). Similarly, mRNAs encoding zymogens such as amylase, elastase I and II were also severely decreased in the mutant pancreas (Figure 4B). These findings suggest that XBP-1 is essential for the terminal differentiation of pancreatic acinar cells.
Figure 4.
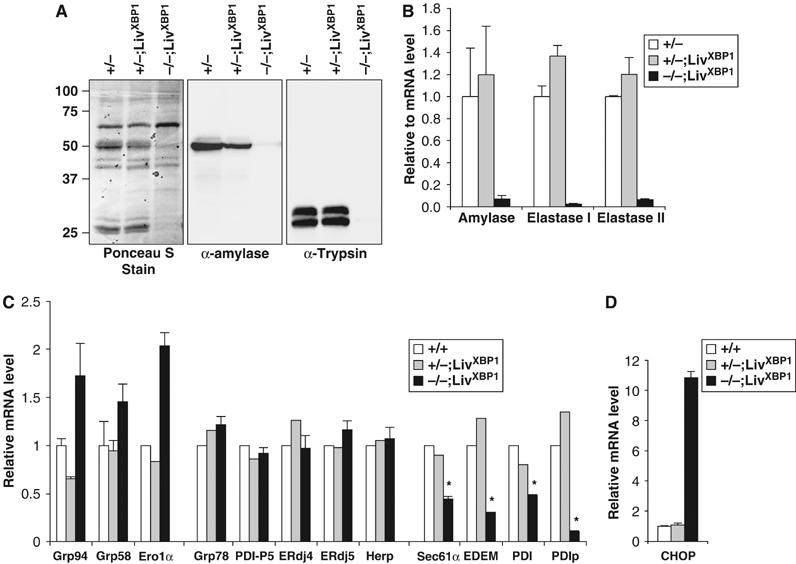
Reduced production of pancreatic digestive enzymes in XBP-1−/−;LivXBP1 mice. (A) Western blot analysis of whole-pancreas lysates to detect α-amylase and trypsin. The blot was stained with Ponceau S after transfer. (B) Measurement of zymogen mRNAs. Total RNAs were isolated from WT, XBP-1+/−;LivXBP1, and XBP-1−/−;LivXBP1 pancreas. Expression of the indicated zymogen mRNAs was measured by real time PCR analysis. Values were normalized to β-actin. (C) Expression of select XBP-1 target genes and pancreas specific chaperone genes, as well as (D) CHOP were measured by real time PCR analysis in WT and XBP-1−/−;LivXBP1 pancreas. The expression levels of each gene are shown as fold induction over WT. Asterisks indicate genes whose expression was significantly impaired in the absence of XBP-1. _N_=2–4 mice per group.
XBP-1 is essential for the expression of a subset of UPR target genes in the pancreas
Extensive gene array analysis in B cells and fibroblasts demonstrated that XBP-1 regulates expression of genes involved in protein folding in the ER and the subsequent secretion, as well as degradation, of misfolded proteins from the ER (Lee et al, 2003; Yoshida et al, 2003; Shaffer et al, 2004). To identify genes regulated by XBP-1 in the pancreas, we performed quantitative real time PCR analysis for select XBP-1 target genes identified in other systems as well as for several other ER chaperone proteins highly expressed in the pancreas. Expression of Sec61α, EDEM, PDI, and PDIp was significantly decreased in XBP-1−/−;LivXBP1 pancreas, suggesting that these genes might be partly responsible for the abnormalities in the exocrine pancreas observed (Figure 4C). Sec61α and EDEM are involved in the translocation of newly synthesized polypeptides across the ER membrane (Rapoport et al, 1996) and the degradation of misfolded ER proteins (Hosokawa et al, 2001), respectively. PDI and PDIp are ER-localizing enzymes that catalyze disulfide bond formation and isomerization of newly synthesized proteins (Wilkinson and Gilbert, 2004). PDIp is selectively expressed in the exocrine compartment of the pancreas and binds to zymogen-derived peptides, suggesting its importance for the folding of zymogens in acinar cells (Desilva et al, 1996; Volkmer et al, 1997). The very marked decrease in PDIp expression in XBP-1−/−;LivXBP1 pancreas suggests that it may be a critical XBP-1 target gene involved in the secretory function of pancreatic acinar cells. In contrast, expression of ER chaperones, Grp78 and Grp94 and ERdj4, and protein disulfide isomerases, ERdj5, PDI-P5 and Grp58 was not compromised in the absence of XBP-1. Given that the expression of ERdj4 and PDI-P5 are dependent on XBP-1 in both plasma cells and tunicamycin-treated fibroblast cells (Lee et al, 2003), our data suggest that the array of XBP-1 downstream target genes may be cell-type specific. It is also likely that absence of XBP-1 may itself result in ER stress that activates other branches of the UPR signaling pathway (Harding et al, 1999) and hence induces these genes. Indeed, CHOP, which is a downstream target gene of the PERK UPR signaling pathway, was markedly induced in XBP-1−/−;LivXBP1cells (Figure 4D).
XBP-1-deficient pancreatic acinar cells undergo marked apoptosis during embryonic development
In the absence of XBP-1, the terminal maturation program of pancreatic acinar cells might not occur at all during development. Alternatively, cells might die during maturation because they cannot handle the stress of abundant zymogens in the ER without an intact IRE1α/XBP-1 signaling pathway. The levels of pancreatic exocrine enzymes increase dramatically during the late phase of embryonic development. In rat, mRNA and protein concentrations of exocrine enzymes increase by approximately 1000 fold between E15 and 21, which corresponds to approximately E14 to 19 in the mouse (Han et al, 1986; Githens, 1993). We asked whether the sparsity of mature acinar cells was due to lack of specification or due to apoptosis of maturing acinar cells by examining the XBP-1−/−;LivXBP1 pancreas at different embryonic stages. At E15.5, WT pancreas contained cells organized to form acini, but clearly immature as judged by the minimal amounts of zymogen revealed by cytoplasmic staining (Figure 5A). XBP-1−/−;LivXBP1 pancreas displayed a similar organization of acinar cells at E15.5, suggesting that XBP-1 is not essential for the development of the pancreas to this stage (Figure 5B). At E18.5, WT pancreatic acinar cells displayed intense eosinophilic cytoplasmic staining (Figure 5C), concomitant with a dramatic increase in zymogen production. In contrast, the majority of acinar cells in the XBP-1−/−;LivXBP1 pancreas displayed a low or intermediate content of cytoplasmic protein (Figure 5D). Occasional mutant acinar cells had fragmented nuclei, indicating ongoing apoptosis. To visualize cell death more directly, we employed TUNEL staining. At E15.5, little apoptosis was present and no difference between WT and XBP-1−/−;LivXBP1 pancreas was noted (data not shown). In contrast, at E18.5, we observed significant numbers of apoptotic cells in XBP-1−/−;LivXBP1 but not in WT pancreas (Figure 5E and F). We conclude that pancreatic acinar cells undergo apoptosis in the absence of XBP-1 due to ER stress as they increase zymogen production during development leading postnatally to a marked decrease in pancreatic parenchyma with zymogen-depleted acini.
Figure 5.
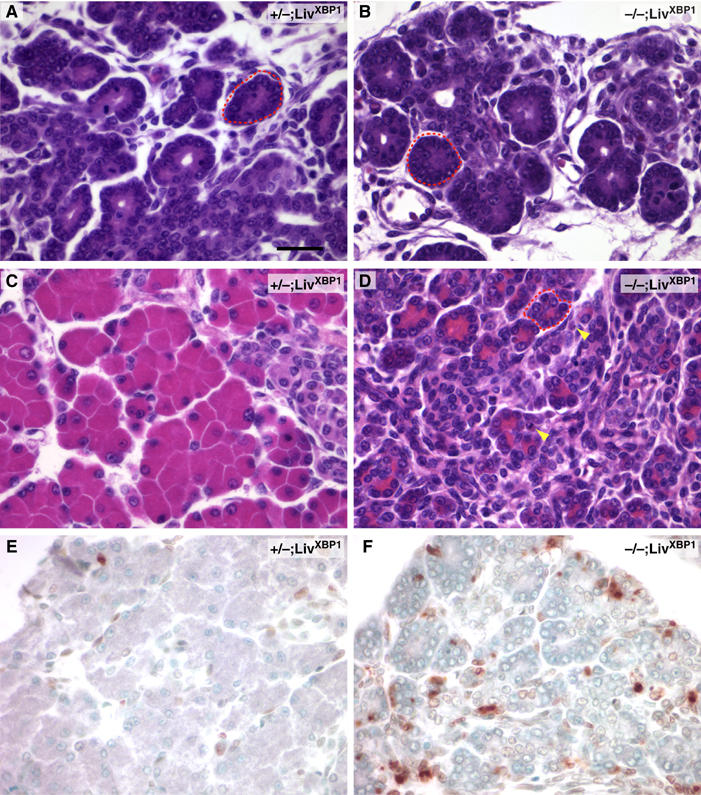
XBP-1-deficient pancreatic acinar cells undergo marked apoptosis during embryonic development. (A, B) Hematoxylin and eosin staining of XBP-1+/−;LivXBP1 control and XBP-1−/−;LivXBP1 E15.5 pancreas. At this stage, acini (red dashed circles) have begun to bud from ducts but granules have only begun to accumulate. (C, D) At E18.5, bright eosinophilic granules show extensive accumulation in the XBP-1+/−; LivXBP1 pancreas but are drastically reduced in mutant acini. Apoptotic cells can be seen in the mutant. (E, F) TUNEL staining of XBP-1+/−; LivXBP1 and XBP-1−/−;LivXBP1 E18.5 pancreas highlights numerous apoptotic acinar cells in the mutant but few in wild type. Scale bar (A–F), 30 μm.
Development of the endocrine pancreas is unaffected in the absence of XBP-1
The endocrine pancreas is also a secretory organ, comprised of islet cells producing insulin, glucagon and other peptide hormones. These endocrine cells also contain membranous granules, but are not so clearly polarized as acinar cells (Dean, 1973). Islets were readily identified in the XBP-1−/−;LivXBP1 pancreas by histological analysis and appeared similar to WT (Figure 2B–E). Consistently, cells producing insulin or glucagon were readily visualized in islets of both WT and XBP-1−/−;LivXBP1 mice (Figure 6A and B). Levels of insulin mRNA were also similar between WT and XBP-1−/−;LivXBP1 mice, as shown by real-time PCR analysis on total pancreatic RNA (Figure 6C). In contrast, glucagon mRNA levels were two- to three-fold higher in the mutant XBP-1−/−;LivXBP1 pancreas. These findings likely represent the result of poor nutritional status, limited food digestion and consequent hypoglycemia, as well as normalization adjustments needed to account for islet cell representation in the total pancreatic mRNA pool studied.
Figure 6.
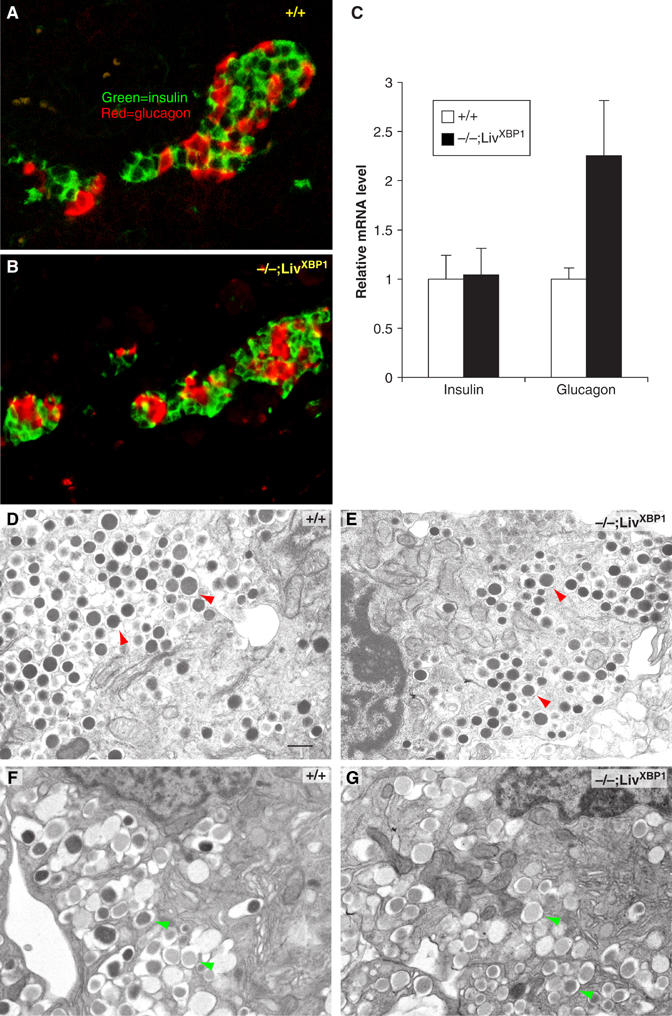
Normal development of the endocrine pancreas in the absence of XBP-1. (A, B) Immunostaining of WT and the XBP-1−/−;LivXBP1 islets with insulin (green) and glucagon (red) antibodies. (C) Expression level of insulin and glucagon mRNA. Total RNA was isolated from whole pancreas of mice with the indicated genotypes and transcript levels quantified by real time PCR analysis. Values were normalized to β-actin. (D, E) α-Cells containing numerous homogeneous electron-dense glucagon granules (red arrowheads) were identified by electron microscopy in WT and XBP-1−/−;LivXBP1 mice. (F, G) β-Cells containing slightly heterogeneous and angular insulin-containing granules (green arrowheads) could be found in both WT and mutant XBP-1−/−;LivXBP1 mice. Scale (D–G, 500 nm).
The fine structure of islet cells was also examined by TEM. Both α- and β-cells from WT mice contained large numbers of membranous granules in the cytoplasm composed of glucagon and insulin, respectively (Figure 6D–G). Notably, compared to acinar cell granules, islet cell granules are much smaller, and the ER in these cells is less well developed. However, we did not find any significant difference in the size or number of the granules in the α-cells of WT versus XBP-1−/−;LivXBP1 pancreas (Figure 6D and E). Similarly, XBP-1−/−;LivXBP1 β-cells were largely normal, although there were somewhat fewer electron dense granules than in the WT (Figure 6F and G). These data suggest that at the early postnatal stage, XBP-1 is not essential for the development and secretory function of the endocrine pancreas.
Impaired development of salivary glands in XBP-1−/−;LivXBP1 mice
The overall structure of the salivary gland is similar to that of the exocrine pancreas, such that secretory cells cluster as acini, composed of two different secretory cell types, serous and mucous acinar cells that produce granules containing amylase and mucin, respectively (Beaudoin and Grondin, 1991; Castle and Castle, 1993). Previous studies indicated that XBP-1 was highly expressed in salivary glands (Clauss et al, 1993). Histological examination of XBP-1−/−;LivXBP1 mice also revealed hypoplastic development of the serous salivary glands (Figure 7A and B). Serous acinar cells of the submandibular glands of the XBP-1−/−;LivXBP1 mice contained less cytoplasm than WT acinar cells, and greater intercellular space between secretory lobules. By TEM, WT salivary gland had well developed ER and apically located granules (Figure 7C). Interestingly, XBP-1−/−;LivXBP1 salivary gland acinar cells had significantly less ER than WT, although they still had abundant cytoplasmic granules (Figure 7D). To measure amylase production more quantitatively, we performed Western blots with whole lysates prepared from submandibular salivary glands. Analysis of two different animals showed decreased production of amylase in XBP-1−/−;LivXBP1 salivary glands (Figure 7E). We also examined the expression of UPR target genes in WT and XBP-1−/−;LivXBP1 salivary glands (Figure 7F). Interestingly, although the expression profiles of UPR target genes were similar between salivary gland and pancreas, the PERK-dependent gene CHOP was significantly less induced in the former, consistent with the much less profound effect of XBP-1 on its development.
Figure 7.
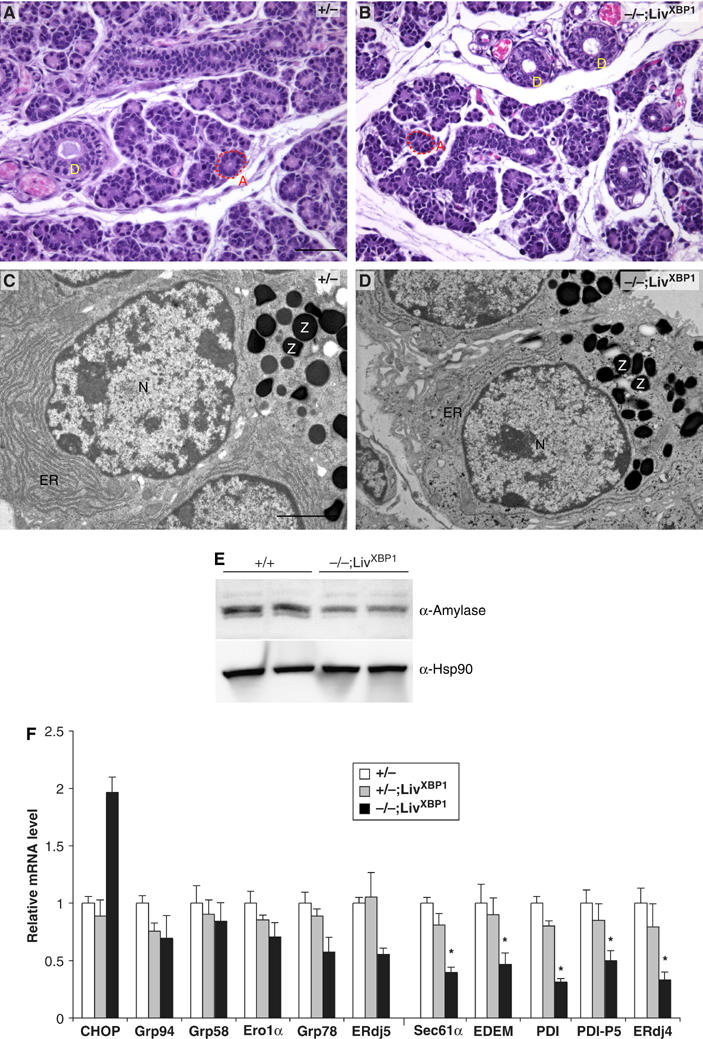
Impaired development of salivary glands in XBP-1−/−;LivXBP1 mice. (A, B) Hematoxylin and eosin staining of the submandibular salivary glands in the control XBP-1+/−;LivXBP1 and mutant XBP-1−/−;LivXBP1 mice. The lobules comprised of secretory acini (A, red dashed lines) appear hypoplastic in the mutant compared to controls. (D) Secretory ducts. (C, D) Electron micrographs of serous salivary glands of control and mutant XBP-1−/−;LivXBP1 mice. Similar to pancreatic acini, these salivary gland acini have well-developed endoplasmic reticulum and apically located zymogen granules (Z). In the mutant, the ER is less well developed but some zymogen granules are produced. (E) Whole-cell lysates of the submandibular salivary glands were subjected to Western blot analysis to measure amylase expression. Scale A and B, 50 μm; C and D, 2 μm. (F) Expression of select XBP-1 target genes and CHOP were measured by real time PCR analysis in WT and XBP-1−/−;LivXBP1 salivary glands. The expression levels of each gene are shown as fold induction over WT. Asterisks indicate genes whose expression was significantly impaired in the absence of XBP-1.
Forced expression of XBP-1 in progenitor B cells results in elevated immunoglobulin production in vivo
We next asked whether enforced expression of XBP-1 could increase the production of secretory proteins in vivo. Immunodeficient _Rag2_-deficient mice were reconstituted with bone marrow (BM) cells that had been transduced with the spliced active form of XBP-1 (RVGFP-XBP-1s) or with control retrovirus (RVGFP) (Figure 8). The peripheral B cell compartments of Rag2 mice that had received RVGFP-transduced BM contained approximately 50% GFP+ peripheral B cells while mice that received RVGFP-XBP-1s transduced BM exhibited GFP+ B cell populations in a range from 10 to 50% (data not shown). Despite the somewhat lower numbers of the transduced B cells, however, mice reconstituted with XBP-1s transduced BM cells secreted larger amounts of Ig of all isotypes tested, suggesting that higher XBP-1s level confers greater secretory capacity on primary B cells in vivo.
Figure 8.
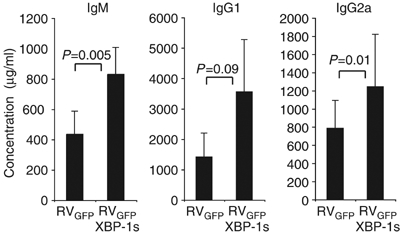
Forced expression of XBP-1 in progenitor B cells results in elevated immunoglobulin production in vivo. Lethally irradiated 8-week old _Rag2_−/− hosts were reconstituted with WT hematopoietic stem cells (HSCs) infected with RVGFP-XBP-1s retrovirus or empty RVGFP-control retrovirus, _N_=4–6 mice per group. Reconstituted animals were analyzed after 9 weeks by harvesting sera to measure levels of IgM, IgG1 and IgG2a by ELISA. The _P_-values are shown.
Discussion
The XBP-1 transcription factor is required for cellular responses to ER stress. Upon sensing ER stress, the spliced form of XBP-1 activates a variety of genes involved in protein maturation in the ER, ER associated degradation and ER expansion that ready the cell for efficient production and secretion of proteins (Lee et al, 2003; Yoshida et al, 2003; Shaffer et al, 2004; Sriburi et al, 2004). To further investigate the requirement for XBP-1 in secretory cells, we rescued XBP-1 homozygous mutant embryos from lethality by selectively targeting an XBP-1 transgene to liver. XBP-1−/−;LivXBP1 mice displayed severe abnormalities in the development and terminal differentiation of the pancreatic exocrine compartment, with an extreme impairment in production of digestive enzymes resulting in death shortly after birth from malnutrition. XBP-1−/−;LivXBP1 pancreatic acinar cells lacked a highly developed ER structure and underwent marked apoptosis during development accompanied by induction of the proaptotic gene, CHOP. This phenotype is likely due to failure to handle the ER stress caused by increasing zymogen production during development. XBP-1−/−;LivXBP1 mice also displayed qualitatively similar although more modest abnormalities in the salivary glands. Other organs were histologically normal. These data along with our previous studies in the plasma cell imply a critical role for the UPR in secretory pathways and provide strong evidence for an essential role of XBP-1 in particular in the development and function of professional secretory cells.
The functional homologue of XBP-1 in yeast is the transcription factor Hac1p, which is similarly activated through IRE1p-mediated mRNA splicing (Cox and Walter, 1996; Sidrauski and Walter, 1997). Hac1p not only activates the transcription of genes encoding ER-localizing chaperones, but also promotes membrane lipid synthesis (Cox et al, 1997; Travers et al, 2000). It appears that Hac1p antagonizes transcriptional repressor Opi1p, allowing the activation of genes encoding phospholipid biosynthesis enzymes. Our findings suggest that mammalian cells also depend on the UPR for ER expansion upon increases in client proteins, and that XBP-1 plays a central role in this process. However, it remains unclear how XBP-1 regulate membrane biogenesis. Augmentation of enzymatic activities in the phospholipid biosynthesis pathway could be one mechanism (Sriburi et al, 2004), although the targets of XBP-1 responsible for increased ER biogenesis are unknown.
Secretory cells are also present in various endocrine and exocrine systems in the body, such as pancreatic islets and the skeletal system. We noted a slight delay in bone formation in XBP-1−/−;LivXBP1 mice with decreased mineralization, although endochondral and intramembranous ossification did occur (data not shown). However, it is unclear whether this difference is due to an intrinsic osteoblast defect or secondary to the general growth retardation of XBP-1−/−;LivXBP1 mice. We also failed to find any significant abnormality in the pancreatic islet cells of XBP-1−/−;LivXBP1 mice, suggesting that XBP-1 is not essential for the embryonic development of the islet. It appears that dependence on XBP-1 correlates with the magnitude of the secretory load in a given cell. It will be of interest to use mice conditionally deficient in XBP-1 in each organ system to establish whether the sustained production of secretory proteins from these organs after birth causes ER stress and hence confers XBP-1 dependency.
We have previously used gene profiling to identify XBP-1 target genes in mouse embryonic fibroblasts (MEF) and in B cells (Lee et al, 2003; Shaffer et al, 2004), and here have examined the expression of select XBP-1 target genes in XBP-1−/−;LivXBP1 pancreas and salivary gland. Interestingly, we found significant differences across different cell types in the requirement for XBP-1 expression. For example, the uncompromised expression of ERdj4 and PDI-P5 mRNA in XBP-1−/−;LivXBP1 pancreas was particularly surprising, given that induction of these genes by ER stress was highly dependent on XBP-1 in both MEFs and B cells. The expression of Grp78, Grp94, Herp, Grp58 and ERO1α, which were upregulated by enforced expression of XBP-1 s in B cells was also unaltered in XBP-1−/−;LivXBP1 pancreas. Indeed, several of these UPR target genes were slightly induced, suggesting that even the small amount of secretory proteins present in XBP-1−/−;LivXBP1 acinar cells induced ER stress leading to activation of the alternate PERK and ATF6 UPR pathways. In contrast, expression of Sec61α, EDEM and PDI mRNA was significantly decreased in XBP-1−/− pancreas. Interestingly, PDIp, an ER-localizing exocrine pancreas-restricted enzyme that catalyzes disulfide bond formation and isomerization of newly synthesized proteins and binds to zymogen-derived peptides (Desilva et al, 1996; Volkmer et al, 1997; Wilkinson and Gilbert, 2004), was dramatically decreased in the absence of XBP-1. These data provide evidence that the IRE1α/XBP-1 UPR signaling pathway may control different programs of gene expression in different cell types.
The phenotypes caused by defects in the PERK/eIF2α and IRE1/XBP-1 pathways are not identical, suggesting both unique and overlapping functions (Delepine et al, 2000; Reimold et al, 2000, 2001; Harding et al, 2001; Zhang et al, 2002, 2005). Interestingly, UPR-mediated translational control through eIF2α phosphorylation is not required for B lymphocyte maturation and/or plasma cell differentiation as assessed by reconstitution of _Rag2_−/− mice with fetal liver cells from homozygous eIF2α knockin mutant (Ser51/Ala) embryos (Zhang et al, 2005). Further, although PERK deficient mice display abnormalities in the exocrine pancreas with somewhat decreased secretion of digestive enzymes, distended ER and increased apoptosis of acinar cells, the phenotype is rather modest especially in young mice as compared to XBP-1−/−;LivXBP1 mice (Harding et al, 2001; Zhang et al, 2002). Instead, PERK or eIF2α deficiency causes progressive loss of pancreatic islet β-cells and impaired bone formation after birth both in human and mouse, indicating a function for the PERK UPR pathway in β-cells and osteoblasts rather than other secretory cells (Delepine et al, 2000; Harding et al, 2001; Zhang et al, 2002; Scheuner et al, 2005). The differences in the requirement for PERK/eIF2α and IRE1/XBP-1 pathways could be ascribed to differences in the mode of activation and/or the downstream target genes. Although PERK and IRE1 share functionally similar luminal domains and both are activated in cells treated in vitro with ER stress inducers (Harding et al, 1999; Bertolotti et al, 2000; Liu et al, 2000), they clearly are selectively activated in vivo (Gass et al, 2002; Zhang et al, 2005). The PERK/eIF2α pathway activates a broad range of target genes, which is not surprising given that various cellular stresses signal through eIF2α phosphorylation (Clemens, 2001). In contrast, XBP-1 target genes largely increase the capacity of the ER and improve the quality control system (Lee et al, 2003; Yoshida et al, 2003; Shaffer et al, 2004).
Impaired or overactive ER function has been observed in many genetic diseases, where mature proteins are not produced due to mutations causing inefficient protein folding or to states leading to the accumulation of misfolded protein in the ER due to inefficient ERAD pathways (Aridor and Balch, 1999). The generation and analysis of the XBP-1−/−;LivXBP1 mice allowed us uncover a novel and essential function for XBP-1 in the function and development of secretory cells of exocrine organs. Given that XBP-1 is important not only for protein folding in the ER but also for degradation of misfolded proteins generated from the ER, compounds that modulate its activity are potential therapeutic agents for the treatment of such diseases.
Materials and methods
Generation of mice lacking XBP-1 in all organs except for liver (XBP-1−/−;LivXBP1)
To generate transgenic mice that express XBP-1 in the liver, we used the pLiv.7 vector (gift of Dr Roger Davis, San Diego State University) that contains the human ApoE gene promoter and hepatic control region of the apoE/C-I gene locus (Simonet et al, 1993; Miyake et al, 2001). The 1.2 kb mouse XBP-1 cDNA was inserted into the _Hpa_I site of pLiv.7 plasmid to generate pLiv-XBP1. The 7.1 kb fragment of the transgenic construct was obtained by digestion of pLiv-XBP1 plasmid with _Not_I and _Spe_I restriction enzymes, followed by gel purification by using the Qiaquick gel extraction kit (Qiagen). The transgenic construct was microinjected into C57BL/6J fertilized embryos and implanted into pseudopregnant females. Transgenic founder mice were identified by a PCR based genotyping of tail DNA with the following primers: 407S, 5′-ACACGCTTGGGAATGGACAC-3′; 577A, 5′-CCATGGGAAGATGTTCTGGG-3′. The LivXBP1 transgenic mice were distinguished from WT mice by the production of a 171 bp PCR product. XBP-1−/−;LivXBP1 mice that express the transgenic XBP-1 mRNA in the liver were generated by crossing the transgenic LivXBP1 strain onto heterozygous XBP-1+/− mice, which yielded XBP-1+/−;LivXBP1 mice, which were then intercrossed.
RNA and protein analysis
Total RNA was isolated from tissues using Trizol reagent (Invitrogen). For Northern blot analysis, 5 μg total RNA was electrophoresed on 1.2% agarose, 6% formaldehyde gels, transferred onto Genescreen Plus membrane (NEN), and then hybridized with 32P-radiolabeled probes by using the Ultrahyb reagent (Ambion) as described previously (Iwakoshi et al, 2003). cDNA was synthesized from RNA samples using the iScript cDNA synthesis kit containing oligo (dT) and random hexamer primers (Bio-Rad). Quantitative real-time PCR reactions employing SYBR green fluorescent reagent were run in an ABI PRISM 7700 system (Applied Biosystems). The relative amounts of mRNAs were calculated from the comparative threshold cycle (Cτ) values by using β-actin as control. Primer sequences designed by Primer Express software (Applied Biosystems) are shown in Supplementary Table 1. Whole-cell lysates of the pancreas and the submandibular salivary glands were prepared in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS). Western blot analysis was performed as described previously (Iwakoshi et al, 2003), with anti-XBP-1, anti-Amylase (Calbiochem) and anti-trypsin (Chemicon) antibodies, except that the blot was stained with Ponceau S reagent (Pierce) after the transfer.
Histology
Mouse pups and embryos were fixed in formalin or Bouin's fixative and sectioned transversely at 5 μm thickness. Sections were stained by conventional hematoxylin and eosin staining.
Immunohistochemistry and TUNEL staining
Pancreata isolated from 2-day old mice was fixed in 4% paraformaldehyde, dehydrated and embedded in paraffin. For immunostaining, 5 μm tissue sections were first treated with mouse monoclonal anti-insulin (clone E2E3, Signet Laboratories) and rabbit anti-glucagon (Signet Laboratories) antibodies. Glucagon signals were amplified by using the TSA biotin system (NEN) after treating sections with HRP-conjugated goat anti-rabbit antibodies. Goat anti-mouse IgG-Alexa 488 and streptavidin-Alexa 594 (Invitrogen) were used to visualize insulin and glucagon producing cells, respectively. TUNEL staining was performed on paraffin sections of E18.5 mice fixed in 10% buffered formalin solution by using the In situ Cell Death Detection Kit (Roche), according to the manufacturer's instructions.
TEM
Tissues were fixed with 1.25% formaldehyde, 2.5% glutaraldehyde, 0.03% picric acid in 100 mM sodium cacodylate buffer. After washing with 100 mM sodium cacodylate buffer, tissues were treated for 1 h with 1% osmium tetroxide and 1.5% potassium ferrocyanide, and then 30 min with 0.5% uranyl acetate in 50 mM maleate buffer, pH 5.15. After dehydration in ethanol, tissues were treated for 1 h in propylenoxide and then embedded in Epon/Araldite resin. Ultrathin sections were collected on EM grids and observed by using a JEOL 1200EX transmission electron microscope at an operating voltage of 60 kV.
Generation of bone marrow reconstituted chimeric mice overexpressing XBP-1s
For normal BM transfer experiments, recipient mice were irradiated with 600 rads 3–5 h before the procedure. BM cells were harvested aseptically from the tibia and femurs of donor mice, and 2 × 106 cells were injected intravenously in the recipients. For retroviral infections, BM cells were harvested from donor mice 5 days after they received an intraperitoneal injection of 5 mg 5-Fluorouracil (Sigma) in Dulbecco's PBS (Gibco/BRL). The cells were cultured for 4 days at a density of 2 × 106 cells/ml with 20 ng/ml rmIL-3, 50 ng/ml rmIL-6 and 50 ng/ml rmSCF (Peprotech, Rocky Hill, NJ) in DMEM containing 10% FCS. After 48 and 72 h, the cells were spin infected with control and XBP-1s-expressing retroviruses generated as described (Iwakoshi et al, 2003). After infections, the supernatant was removed and replaced with fresh media containing cytokines. Irradiated recipient mice were then injected with 1 × 106 infected BM cells. In all cases, irradiated mice were maintained on trimethaprim-sulfamethoxazole treated water in sterile cages for 6–12 weeks before analysis. Assay for immunoglobulin levels was carried out as described (Reimold et al, 2001).
Supplementary Material
Supplementary Figure 1
Supplementary Table 1
Acknowledgments
This study was supported by the National Institute of Health, Grant Numbers AI32412 and P01 AI56296 (LHG); and an Ellison Medical Foundation Grant (LHG). A-HL is a recipient of National Institute of Health Career Development Award, Grant Number 1P50CA100707; and NNI is supported by an Irvington Institute Postdoctoral Fellowship Award. We thank Dr Roderick Bronson, Harvard Medical School, for advice on pathological analyses, and Li Zhang of the Rodent Histopathology Core, Dana Farber/Harvard Cancer Center for expert technical assistance.
References
- Aridor M, Balch WE (1999) Integration of endoplasmic reticulum signaling in health and disease. Nat Med 5: 745–751 [DOI] [PubMed] [Google Scholar]
- Beaudoin AR, Grondin G (1991) Secretory pathways in animal cells: with emphasis on pancreatic acinar cells. J Electron Microsc Tech 17: 51–69 [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332 [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Castle JD, Castle AM (1993) Sorting and secretion of salivary proteins. Crit Rev Oral Biol Med 4: 393–398 [DOI] [PubMed] [Google Scholar]
- Clauss IM, Gravallese EM, Darling JM, Shapiro F, Glimcher MJ, Glimcher LH (1993) In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev Dyn 197: 146–156 [DOI] [PubMed] [Google Scholar]
- Clemens MJ (2001) Initiation factor eIF2 alpha phosphorylation in stress responses and apoptosis. Prog Mol Subcell Biol 27: 57–89 [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404 [DOI] [PubMed] [Google Scholar]
- Dean PM (1973) Ultrastructural morphometry of the pancreatic-cell. Diabetologia 9: 115–119 [DOI] [PubMed] [Google Scholar]
- Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C (2000) EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott–Rallison syndrome. Nat Genet 25: 406–409 [DOI] [PubMed] [Google Scholar]
- Desilva MG, Lu J, Donadel G, Modi WS, Xie H, Notkins AL, Lan MS (1996) Characterization and chromosomal localization of a new protein disulfide isomerase, PDIp, highly expressed in human pancreas. DNA Cell Biol 15: 9–16 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Gass JN, Gifford NM, Brewer JW (2002) Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem 277: 49047–49054 [DOI] [PubMed] [Google Scholar]
- Githens S (1993) Differentiation and Development of the Pancreas in Animals. New York: Raven Press Ltd [Google Scholar]
- Grossman A (1984) An overview of pancreatic exocrine secretion. Comp Biochem Physiol B 78: 1–13 [DOI] [PubMed] [Google Scholar]
- Han JH, Rall L, Rutter WJ (1986) Selective expression of rat pancreatic genes during embryonic development. Proc Natl Acad Sci USA 83: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D (2001) Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell 7: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K (2001) A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep 2: 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH (2003) Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol 4: 321–329 [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16: 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ (2000) Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem 275: 24881–24885 [DOI] [PubMed] [Google Scholar]
- Miyake JH, Doung XD, Strauss W, Moore GL, Castellani LW, Curtiss LK, Taylor JM, Davis RA (2001) Increased production of apolipoprotein B-containing lipoproteins in the absence of hyperlipidemia in transgenic mice expressing cholesterol 7alpha-hydroxylase. J Biol Chem 276: 23304–23311 [DOI] [PubMed] [Google Scholar]
- Motta PM, Macchiarelli G, Nottola SA, Correr S (1997) Histology of the exocrine pancreas. Microsc Res Tech 37: 384–398 [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem 65: 271–303 [DOI] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH (2000) An essential role in liver development for transcription factor XBP-1. Genes Dev 14: 152–157 [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (2001) Plasma cell differentiation requires transcription factor XBP-1. Nature 412: 300–307 [DOI] [PubMed] [Google Scholar]
- Scheuner D, Mierde DV, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ (2005) Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med 11: 757–764 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21: 81–93 [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107: 893–903 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Simonet WS, Bucay N, Lauer SJ, Taylor JM (1993) A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J Biol Chem 268: 8221–8229 [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW (2004) XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167: 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Volkmer J, Guth S, Nastainczyk W, Knippel P, Klappa P, Gnau V, Zimmermann R (1997) Pancreas specific protein disulfide isomerase, PDIp, is in transient contact with secretory proteins during late stages of translocation. FEBS Lett 406: 291–295 [DOI] [PubMed] [Google Scholar]
- Wasle B, Edwardson JM (2002) The regulation of exocytosis in the pancreatic acinar cell. Cell Signal 14: 191–197 [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Gilbert HF (2004) Protein disulfide isomerase. Biochim Biophys Acta 1699: 35–44 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K (2003) A time-dependent phase shift in the Mammalian unfolded protein response. Dev Cell 4: 265–271 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ (2005) The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest 115: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR (2002) The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol 22: 3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Table 1