Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling (original) (raw)
Abstract
In eukaryotic cells Rab/Ypt GTPases represent a family of key membrane traffic controllers that associate with their targeted membranes via C-terminally conjugated geranylgeranyl groups. GDP dissociation inhibitor (GDI) is a general and essential regulator of Rab recycling that extracts prenylated Rab proteins from membranes at the end of their cycle of activity and facilitates their delivery to the donor membranes. Here, we present the structure of a complex between GDI and a doubly prenylated Rab protein. We show that one geranylgeranyl residue is deeply buried in a hydrophobic pocket formed by domain II of GDI, whereas the other lipid is more exposed to solvent and is skewed across several atoms of the first moiety. Based on structural information and biophysical measurements, we propose mechanistic and thermodynamic models for GDI and Rab escort protein-mediated interaction of RabGTPase with intracellular membranes.
Keywords: membrane extraction, protein prenylation, RabGDP dissociation inhibitor, RabGTPases, vesicular transport
Introduction
Rab/Ypt proteins comprise the largest subgroup of the Ras GTPase superfamily, with more than 60 members identified in the human genome (Pfeffer, 2001; Stenmark and Olkkonen, 2001). These proteins function as molecular switches that mediate various events, including tethering, docking, fusion and motility of intracellular membranes, with many of these functions being conserved from yeast to humans (Segev, 2001; Zerial and McBride, 2001). The process of switching between the active GTP-bound conformation and the inactive GDP-bound conformation is controlled by a multitude of interacting and regulatory proteins. The active form of Rab proteins interacts with Rab effectors and GTPase-activating proteins, while the inactive conformation is recognised by guanine nucleotide exchange factors and by molecular chaperones such as Rab escort proteins (REPs) and Rab GDP dissociation inhibitors (GDIs). Unlike other Rab regulators, REPs and GDIs, which are related both structurally and functionally, show only limited preference for individual Rab proteins and are strictly essential. REP (Mrs6 in yeast nomenclature) is a component of the Rab prenylation machinery, and forms a stable complex with all newly synthesised RabGTPases, mediating their prenylation by presenting them to Rab geranylgeranyl transferase (RabGGTase) and subsequently delivering them to their specific membranes (Andres et al, 1993). Although GDI is structurally closely related to REP, it is unable to support Rab prenylation, but instead controls the distribution of the active GTP- and inactive GDP-bound forms between membranes and cytosol, respectively (Alory and Balch, 2001). GDI stably associates only with Rab/Ypt proteins that are both prenylated and GDP bound, ensuring retrieval of inactivated GTPases from the membrane at the end of their functional cycle and subsequent delivery to donor membranes for the beginning of another functional cycle (Sasaki et al, 1990). REP/GDI-mediated delivery of prenylated RabGTPases to membranes is believed to require a specialised factor referred to as GDI-displacement factor (GDF). This promotes dissociation of the stable GTPase:REP/GDI complex and mediates membrane insertion of the RabGTPase (Ayad et al, 1997; Dirac-Svejstrup et al, 1997). Recent work demonstrated that mammalian Yip3 protein is a catalytically acting GDF that promotes dissociation of the Rab9:GDI complex on endosomes (Sivars et al, 2003).
Despite a considerable interest in RabGTPases and the role of REP/GDI molecules in their activity cycle, the functional mechanism of the latter is poorly understood. Most importantly, the ability of GDI to extract RabGTPases from the membranes has not been explained either from a mechanistic or from a thermodynamic point of view. Neither is it clear why the Rab functional cycle cannot be governed by a single molecule combining the functions of GDI and REP molecules.
In an effort to elucidate the structural details of Rab:GDI interactions, a high-resolution structure of the latter was solved and revealed a tightly packed molecule composed of two domains tilted with respect to each other (Schalk et al, 1996). Structure-based mutational analysis defined regions in domain I that are involved in the association with Rab proteins, a putative membrane receptor, termed the Rab-binding platform and the mobile effector loop (MEL), but did not shed light on the location of the lipid binding site (Alory and Balch, 2003). Due to the critical importance of these data for understanding the mechanism of GDI action and the dynamics of membrane protein interplay in intracellular vesicular transport, the structure of the Rab:GDI complex was sought in subsequent years by several groups. Initially, a structure of mammalian α-GDI in complex with geranylgeranyl cysteine was solved and led to the suggestion that the lipid binding site is located on domain I above the MEL (An et al, 2003). According to this finding, the lipid binding site is near the L92P mutation in the α-GDI gene this mutation leads to X-linked nonsyndromic mental retardation in humans (Garrett et al, 1994; D'Adamo et al, 1998). The authors proposed that this mutation is located at the edge of the putative lipid binding site of RabGDI, and thus impairs its ability to extract RabGTPases from the membrane, leading to the observed defects.
Recently, we used a combination of organic synthesis and expressed protein ligation (EPL) to generate and crystallise the semisynthetic monoprenylated Ypt1:GDI complex (Rak et al, 2003). The 1.48 Å resolution structure of the complex revealed that in contrast with previous proposals, the conjugated geranylgeranyl group is harboured by a cavity formed in the hydrophobic core of domain II of the GDI. The cavity is formed as the result of an outward shift by one of the outer helices of this domain. The cavity is not present in apo-GDI. The apparently contradictory reports on the location of the lipid binding site led to speculation that they could operate sequentially (Pfeffer and Aivazian, 2004) or that the solved structure of monoprenylated Ypt1:GDI complex might be misleading due to the fact that Ypt1 is diprenylated in vivo.
In the present study, we describe the in vitro semisynthesis, purification, crystallisation and structure solution of the doubly prenylated Ypt1:GDI complex. Combined with the biophysical data on the interaction of RabGTPases with REP/GDI proteins, these structural data allow us to formulate mechanistic and thermodynamic models for the interaction of prenylated RabGTPases with RabGDI and membranes and to explain the basis of the functional segregation between RabGDI and REP.
Results and discussion
Construction of the doubly prenylated Ypt1:GDI complex using EPL
Attempts to determine the structure of prenylated Rab proteins in complex with GDI or other proteins have been hampered by difficulties associated with the production of preparative amounts of pure and homogeneously prenylated GTPase. In order to circumvent these problems, we developed an approach relying on the combination of recombinant protein production methods, chemical synthesis of lipidated peptides with precisely designed and readily alterable structures and a technique for peptide-to-protein ligation (Alexandrov et al, 2002; Durek et al, 2004). In this approach, a recombinant truncated version of the Rab protein is ligated to the synthetic prenylated peptide mimicking the missing C-terminal residues. Use of EPL ensures the formation of a native polypeptide bond upon joining the fragments (reviewed in Muir, 2003; Supplementary Figure S1). To obtain a complex of doubly prenylated Ypt1 GTPase with GDI, we ligated the Ypt1 protein truncated by three amino acids to a synthetic tripeptide, H2N-Cys-Cys(GG)-Cys(GG)-OH. The protein was complexed to recombinant yeast RabGDI and purified by gel filtration. Using mass spectrometric analysis, we confirmed that Ypt1 in complex with GDI was homogeneously diprenylated (Supplementary Figure S2B). The protein was subjected to crystallisation trials that yielded diffracting crystals under the conditions described in Materials and methods. Diffraction data were collected to 1.48 Å resolution. Initial X-ray phases were obtained by molecular replacement using the coordinates of the monoprenylated Ypt1:GDI complex (PDB code 1UKV). Cyclic rounds of model building and refinement resulted in a 1.48 Å resolution model of the Ypt1:GDI complex that has good values for stereochemistry and crystallographic validation parameters (Table I).
Table 1.
X-ray data collection and refinement statistics
| Data collection statistics | |
|---|---|
| X-ray source | ESRF ID14-2 |
| Wavelength (Å) | 0.934 |
| Resolution (Å) | 19.6–1.48 |
| Space group | P 21 |
| Unit cell a, b, c (Å) α, β, γ (deg) | 47.66, 120.23, 60.55 90.00 90.67 90.00 |
| Observations total/unique | 417 970/109 178 |
| Completenessa (%) (last shell) | 96.5 (75.8) |
| _R_syma,b (last shell) | 7.6 (30.4) |
| 〈_I_〉/σ(I)a (last shell) | 11.32 (2.88) |
| Refinement statistics | |
| Resolution (Å) | 19.6–1.48 |
| _R_workc/_R_freec | 0.163/0.192 |
| Rmsd bonds/angles (Å/deg) | 0.013/1.45 |
| Mean B (Å2) | 22.57 |
| aCompleteness, R_sym and 〈_I/σ(I)〉 are given for all data and for the highest-resolution shell: 1.49–1.48 Å. | |
| b_R_sym=∑j_∣_Ij_−〈_Ij_〉∣/∑_jIj, where 〈_Ij_〉 is the average intensity of reflection j for its symmetry equivalents; values in parentheses are for the highest-resolution shell. | |
| c_R_work=∑∣_F_obs∣−_k_∣_F_calc∣/∑∣_F_obs∣. In all, 5% of randomly chosen reflections were used for the calculation of _R_free. |
Overall structure of the doubly prenylated YPT1:GDI complex
The doubly prenylated Ypt1:GDI complex has a roughly cylindrical shape, with the proteins establishing an extensive interface of ca. 1880 Å2 area. The interface is formed by the Rab-binding platform and the C-terminal-binding region (CBR) of GDI and the Switch I/II regions and the C-terminus of Ypt1, respectively. A comparison between the recently determined structure of monoprenylated Ypt1:GDI complex and the structure of the diprenylated complex shows only minor conformational changes. The overall structure of the di- and monoprenylated complexes have an r.m.s.d. on Cα of 0.38 Å. Residues 197–206 of the Ypt1 C-terminus are not traceable in the electron density maps, indicating that this region is flexible and does not have a defined position (Figure 1A). However, both geranylgeranyl moieties could be identified in the electron density (Figure 1A) (see Materials and methods). Additional electron density, probably representing the connecting peptide, is detectable between the first carbons of the geranylgeranyl groups, but the quality of the map in this region is not sufficient to model it reliably (not shown).
Figure 1.
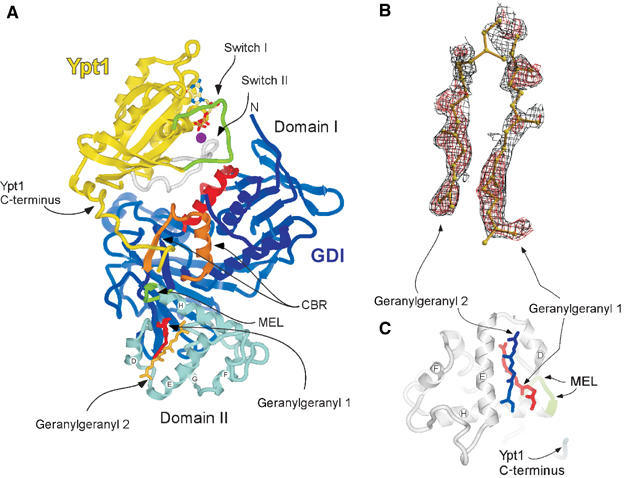
Structure of the doubly prenylated Ypt1:GDI complex. (A) Ribbon representation of GDI (blue) bound to Ypt1 (yellow). Domain I (dark blue), domain II (light blue), the Rab-binding platform (red), the C-terminus-binding region (CBR gold) and the mobile effector loop (MEL green) of GDI are highlighted. The Switch I and II regions of Ypt1 are highlighted in green and grey, respectively. The helices composing domain II of GDI are marked by letters. The isoprenoid moieties 1 (red) and 2 (dark yellow) are displayed in ball-and-stick representation. The GDP (atomic colours) and Mg2+ (magenta) ion are shown in the nucleotide-binding pocket in ball-and-stick and space filling representations, respectively. Unless otherwise indicated, this and other figures were prepared using ICM (Molsoft LLC). (B) Plot of a sigma A-weighted _F_o–_F_c map contoured at 2σ (red) or at 0.6σ (black) in the region of the geranylgeranyl. The maps were generated after several refinement rounds omitting the GG groups. The picture was generated with BobScript and RASTER3D (Merritt and Murphy, 1994; Esnouf, 1997). (C) Domain II of GDI from Ypt1GG:GDI complex displayed in ribbon representation (grey); the secondary structure elements are denoted as in (A). The last visible residues of the Ypt1 C-terminus and of the MEL of GDI are coloured blue and green, respectively. The geranylgeranyl moieties 1 (red) and 2 (blue) filling the lipid binding site are displayed in ball-and-stick representation.
Structure of the lipid binding site
The presented structure reveals that both geranylgeranyl moieties are harboured by the GDI lipid-binding pocket formed by α helices D, E and H of the domain II (Figure 1A and C). The average B-factor for the geranylgeranyl groups (51 Å2) is higher than that of the rest of the protein molecules (21 Å2) reflecting the greater flexibility of the isoprenoid moieties. The observed densities are likely to represent the two geranylgeranyl moieties rather than two alternative conformations of one lipid. Firstly, as mentioned above, the density present between the lipids is most likely to represent the connecting peptide. Secondly, no clashes are observed between closely positioned isoprenoids, suggesting that they represent two simultaneously coexisting entities. Finally, since geranylgeranyl moieties typically yield poor signals in X-ray diffraction and the observed signals for both isoprenoids are at the upper end of quality for typical isoprenoid density, it is unlikely that they represent alternative conformation of a single lipid, since the already weak signal would then be diluted further by distribution over these two conformations. The lipids are positioned in the lipid-binding cavity on top of each other with a skew of ca. 20° (Figure 1C). In this arrangement, the first and slightly bent lipid (GG1) protrudes into the core of domain II, anchoring to the hydrophobic binding site via the GDI residues V127, P128, A129, A134, L139, M140, M148, L152, F192, M197, C221, V224 and A225 (Figure 2A). The second geranylgeranyl moiety (GG2) is located on the surface aligned between helices D and E and forms a lid shielding a large part of the buried GG1 from the solvent (Figure 2B). The environment of GG2 is more hydrophilic than of the buried lipid and its binding site contains only seven hydrophobic amino acids: M148, L152, I155, I193, W200, Y205 and L218. The relative paucity of hydrophobic residues in the binding site is compensated by the hydrophobicity of GG1 that forms the bottom of the binding site for GG2 (Figure 2C).
Figure 2.
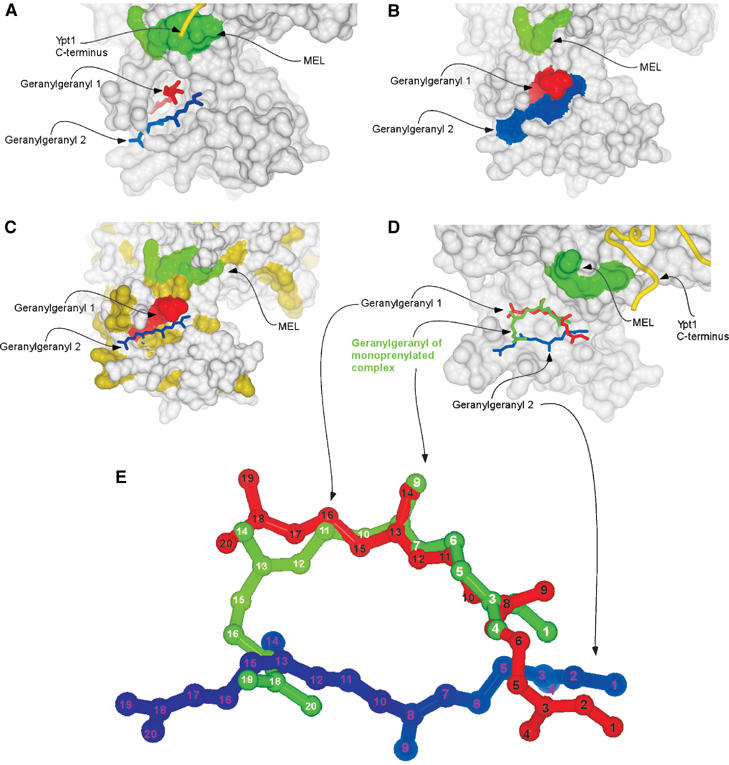
Organisation of the lipid binding site of the doubly prenylated Ypt1:GDI complex. (A) Surface representation of domain II of GDI. The MEL is coloured in green. The C-terminus of Ypt1 is displayed as a yellow worm. The geranylgeranyl moieties are displayed as in Figure 1C. (B) Surface representation of the doubly prenylated Ypt1:GDI complex. The MEL is coloured in green, while the geranylgeranyl moieties 1 and 2 are coloured red and blue, respectively. (C) Same as in (B) but solvent-exposed hydrophobic residues are coloured in yellow and geranylgeranyl 2 is displayed in ball-and-stick representation coloured in blue. (D) Superimposition of mono- and diprenylated Ypt1:GDI complexes. GDI from the diprenylated complex is displayed as in (A), but helix D forming the left wall of the lipid binding side is removed. Geranylgeranyl moieties of the doubly prenylated complex are displayed as in (A) and the geranylgeranyl of the monoprenylated Ypt1:GDI complex is displayed in ball-and-stick representation and coloured in green. (E) Superposition of the geranylgeranyl moieties of the mono- and diprenylated Ypt1:GDI complexes. The colouring is the same as in (D) but the carbon atoms are numbered.
Comparison of lipid binding site structures of mono- and diprenylated Ypt1:GDI complexes
The structure of the lipid binding site in the doubly prenylated Ypt1:GDI complex has undergone only minor transformations when compared with the monoprenylated Ypt1:GDI complex. The shape of the cavity changes slightly due to minor outward movements of helix D (∼0.35 Å) and helix E with the adjacent loop (∼0.55 Å). This movement is consistent with the flexible position of these helices that serve as gates to the lipid-binding cavity (Luan et al, 2000; Rak et al, 2003). In contrast to the protein part of the complex, the position of the conjugated isoprenoids differs significantly between the two structures. As can be seen in Figure 2D, in the monoprenylated complex the isoprenoid moiety is inserted in a bent conformation that exposes the end of the isoprenoid to the solvent while burying the middle of the molecule in the hydrophobic core of the protein. Superpositioning of this structure with the structure of the doubly prenylated complex shows that the geranylgeranyl group of the monoprenylated structure occupies a position overlapping with that of both GG1 and GG2. Its distal part, composed of carbons 16–20, is exposed to the surface and occupies a position similar to carbons 12–15 of GG2, while its cysteine-conjugated part and the middle of the molecule up to position 11 occupy a position very similar to that of carbons 7–20 of GG1 from the doubly prenylated complex (Figure 2D and E). Based on these observations, we conclude that the conformational transition of the GDI lipid binding site operates in an essentially binary modus switching between ‘open' or ‘closed' states. The extent of its dilation appears to be independent of the number of lipids conjugated to the C-terminus, possibly due to the fact that in monoprenylated complexes both lipid binding sites are occupied by a bent lipid that may provide the necessary spreading force to keep the binding site fully dilated. Alternatively, it is possible that dilation of the lipid binding site is induced by a long-range effect following the formation of GDI:Ypt protein:protein interface. In the case of the monoprenylated complex, the lipid moiety remains partially exposed to the solvent, which, presumably, enables its contact with the acceptor lipid or protein while still providing the sufficiently strong anchoring necessary for the formation of the stable GDI:Ypt1 complex. The monoprenylated GDI:Ypt structure probably represents a physiologically relevant complex for a subset of naturally monoprenylated Rab proteins such as Rab8, Rab13, Rab18, Rab23 and Rab28.
Mechanistic model of GDI-mediated membrane extraction of Rab proteins
The observed arrangement of the isoprenoid binding site can be rationalised from the perspective of the functional role of GDI. The GG2 positioned on the surface of the GDI molecule is exposed to the solvent and forms fewer hydrophobic contacts than GG1, which is buried in the core of domain II. It is expected to flip easily out of its shallow binding site to be transferred to an acceptor protein or lipid bilayer. This should promote relocation of GG1 into the binding site of GG2 and subsequently to the membrane (Figure 3A). GDI-mediated RabGTPase extraction from membranes is likely to be a reversal of the same sequence (Figure 3B). Initially, the GTPase domain of RabGTPases is recognised by GDI and a low-affinity complex is formed. The interaction of the CBR of GDI with the hydrophobic residues of the Rab C-terminus leads to the orientation of domain II on the membrane above the bilayer-buried geranylgeranyl moieties. It is likely that the first extracted lipid initially binds to the superficial lipid binding site resulting in the formation of a transient high-affinity complex still anchored in the membrane. This complex is converted into the soluble Rab:GDI complex by coordinated transfer of the GDI-bound lipid into the buried binding site and facilitated flipping of the second lipid from the membrane to the surface binding site (Figure 3B).
Figure 3.
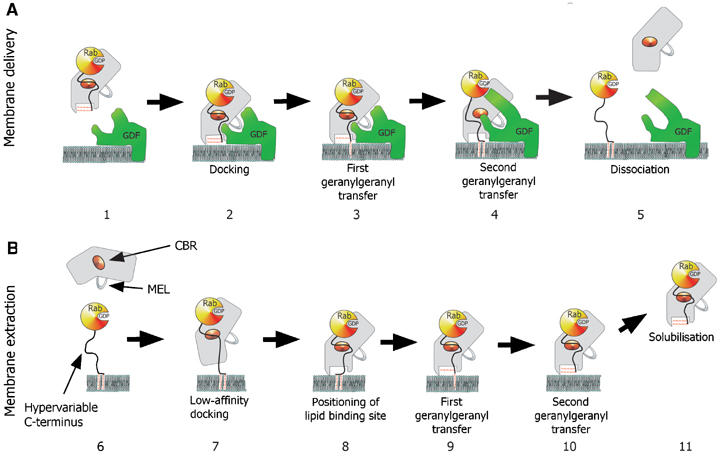
Model for the GDI-mediated Rab/Ypt interaction with the putative Rab receptors and membranes. (A) GDI-mediated delivery of prenylated RabGTPases to the membrane is proposed to involve docking of the Rab:GDI complex with a putative membrane Rab recruitment/GDF via a protein:protein interaction (2). The docked complex undergoes a conformational change, which leads to the transfer of the first and then the second geranylgeranyl moiety into the membrane and subsequently to the release of the Rab C-terminus from the CBR (3 and 4). Finally, GDI is released into the cytosol and the Rab protein enters its functional cycle. (B) GDI-mediated extraction of prenylated RabGTPases from the membrane. Initial recognition is mediated by the low-affinity interaction of the Rab-binding platform with the CBR of GDI (7). This leads to the positioning of GDI domain II on the lipid bilayer over the buried geranylgeranyl moieties of the Rab protein (8). The geranylgeranyl lipids are transferred from the membrane to the lipid binding sites on GDI in two consecutive steps (9 and 10), leading to dissociation of the complex from membrane (11).
Affinity of GDI for Ypt1 and Ypt7
The proposed model raises the question of the driving force behind GDI's ability to remove RabGTPases from the membrane and functional segregation between GDI and REP/Mrs6. To answer this question, one needs to know the affinities between RabGTPases and REP/GDI family members. Since the initial steps of the extraction process are likely to involve the interaction of GDI with only the GTPase domain of prenylated GTPases, we set out to characterise the interaction of unprenylated Ypt1 with yeast GDI using biophysical methods. Since we expected that the interaction might be too weak to be detected easily by spectroscopic methods, we chose to investigate the thermodynamic properties of GDI:Ypt1 interactions by means of isothermal titration calorimetry (ITC; Wiseman et al, 1989). In the experiment performed, 20 μM solutions of Ypt1 or Ypt7 were titrated with a 200 μM solution of GDI (Figure 4A and B). The data could be fitted using the approach outlined in Materials and methods to yield _K_a values of ca. 36 and 6 μM for Ypt1 and Ypt7, respectively, indicating that, as in the case of RabGTPase interaction with REP molecules, the affinities of individual interactions can vary over a significant range (Rak et al, 2004). Taking into account that exact numbers are very difficult to obtain due to the water insolubility of prenylated Rabs, the best available estimation indicates that prenylated Rab proteins bind GDI with a _K_d value equal to or lower than 23 nM (Shapiro and Pfeffer, 1995). Taking this value as an upper limit, it appears that the affinity of GDI for prenylated and unprenylated Rab protein differs by at least 3 orders of magnitude. The obtained affinities fit well with the established model of a dramatic preference of GDI for the prenylated forms of Rab proteins (Araki et al, 1990).
Figure 4.
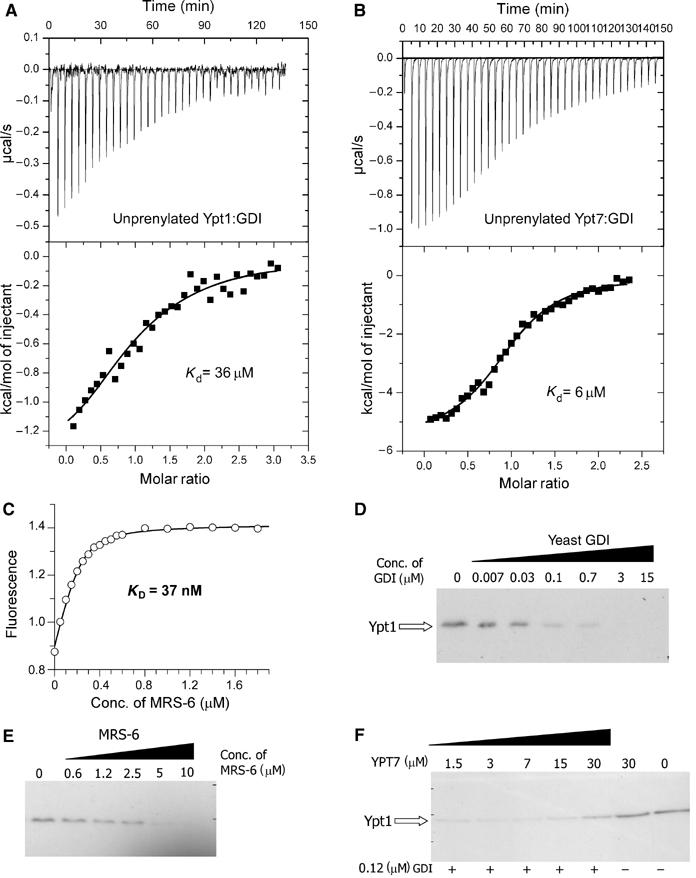
Interaction of GDI and Mrs6 with unprenylated and membrane-bound Ypt proteins. (A) ITC titration of Ypt1 with increasing concentrations of GDI. Fitting of the data led to a _K_d value of 36 μM. (B) Same as in (A) but with Ypt7 (_K_d=6 μM). (C) A representative fluorescence titration of dans_Ypt1 with Mrs6. The concentration of dans_YPT1 was 240 nM. The fluorescence of the dansyl group was excited at 280 nm via FRET from tryptophanes and the emission was collected at 490 nm. The data were corrected for unspecific fluorescence increase and analysed as described under ‘Materials and methods', leading to a _K_d value of 37 nM. (D) Extraction of Ypt1 from S. cerevisiae membranes with yeast GDI. The isolated membrane fraction was incubated with various concentrations of recombinant GDI and the fraction of membrane-associated Ypt1 was determined by Western blotting as described in ‘Materials and methods'. (E) Same as in (D) but the membranes were incubated with recombinant Mrs6. Complete extraction of Ypt1 at high concentrations is not in contradiction to the model calculations, since these were made for specific values of the effective membrane concentration and the affinity of geranylgeranyl residues for membranes, which are not likely to apply here. (F) Unprenylated Ypt7 is able to interfere with the ability of GDI to extract membrane-bound Ypt1. S. cerevisiae membranes were incubated with 120 nM GDI and increasing concentrations of Ypt7. The sample in lane 6 was incubated with 30 μM of Ypt7 and no GDI, while the sample in lane 7 was incubated with buffer alone.
Interaction of Mrs6 with Sec4/Ypt proteins
In order to obtain the necessary parameters for the development of a mechanistic and thermodynamic model of REP/Mrs6/GDI action in the Rab cycle, we analysed the interaction of Ypt proteins with Mrs6 using established fluorescent assays (Alexandrov et al, 2001). In these assays, the changes in the fluorescence intensity of fluorogenic labels attached either to GTPase-bound nucleotides or to the C-terminal cysteines were used to monitor Rab protein interaction with REP/Mrs6 molecules. We used this approach in order to determine the equilibrium binding and rate constants for the interaction of Mrs6 with Sec4, Ypt1, Ypt7, Ypt51 and Rab7 (Figure 4C) (Supplementary data). The results summarised in Table II demonstrate that, like their mammalian counterparts, yeast RabGTPases display a range of affinities for Mrs6 with _K_d values ranging from 4 nM in the case of Sec4 to >500 nM in the case of Ypt7. Thus, the pronounced heterogeneity of affinities of Rab:REP interactions observed in the mammalian system appears to be present but not protein specifically conserved throughout evolution. For instance, Rab7 is the tightest identified binder of mammalian REPs, while its closest homologue, Ypt7, appears to be the weakest interaction partner of Mrs6. Interestingly, Mrs6 displays an affinity for mammalian Rab7 with a _K_d value of 33 nM, suggesting that the Rab-binding interface on the REP/Mrs6 molecules is more conserved than the REP/Mrs6-binding interface of RabGTPases. Heterogeneity of the affinities in the Rab:REP interaction can be rationalised from the perspective that parallel coevolution of RabGTPases with their effectors and REP/GDI molecules exerts different pressure on the shared binding sites on the GTPases that may give rise to different affinities towards either of the components.
Table 2.
Dissociation constants determined for the interaction of Mrs6 and GDI with Rab/Ypt proteins
| Protein | Ypt1 | Ypt51 | Ypt7 | Sec4 | Rab7 |
|---|---|---|---|---|---|
| Mrs6 | 37±4 nM | 320±18 nM | 516±33 nM | 4.2±0.5 nM | 33±7 nM |
| GDI | 36±5 μM | 6±1 μM | |||
| The error margins represent the least-squares uncertainty of the fit. |
Thermodynamic basis of GDI-mediated solubilisation of membrane-associated Rab proteins
We previously showed that the enlargement of the Rab-binding interface on domain I and the emergence of the RabGGTase-binding interface on domain II of REP determines the structural and functional segregation between the REP and GDI families (Pylypenko et al, 2003; Rak et al, 2003, 2004). These structural data, in combination with quantitative characterisation of the interaction between Rabs and members of the REP/GDI superfamily, allow us to formulate a thermodynamic and functional model of Rab recycling. Before considering the details of the mechanism, we discuss here the thermodynamic basis of Rab membrane extraction by GDI and REP and the difference in their efficiency in this process. Beginning with the latter point, REP appears to be less efficient than GDI in Rab extraction, and this is in keeping with their respective biological roles, since REP is probably only involved in delivery of Rabs to membranes, whereas GDI, in addition to this property, must also be able to extract Rabs. These different properties can be explained qualitatively by considering the affinities of REP/GDI for unmodified and prenylated forms of Rab, respectively. Thus, whereas REP binds with high affinity to both unprenylated and prenylated Rabs, enabling it to present the unprenylated form to RabGGTase, GDI only binds prenylated Rabs with high affinity (Araki et al, 1990). The large increase in affinity of GDI to Rab on docking of the C-terminus and the prenyl groups appears to be the driving force for the extraction process. This can be shown in a more quantitative manner by examination of the formal extraction scheme shown in Figure 5B. The first step is the docking of REP/GDI onto the GTPase domain of the Rab molecule, occurring with an association constant designated as _K_gdi. Although it is clear that the process is not likely to happen as depicted, we then allow dissociation of the prenylated C-terminus from the membrane in a second step (dissociation constant _K_mem), followed by docking of the prenylated C-terminus onto its binding site on REP/GDI, defining the equilibrium constant for this step as _K_dock. Obviously, this is not meant to reflect the actual mechanism, since it involves the highly unfavourable step of spontaneous release of the geranylgeranyl groups from the membrane, but the equilibrium constant of the overall process must be the same regardless of the detailed mechanism, which will presumably involve some sort of merging of steps 2 and 3, and possibly other protein factors (Figure 5B). On the basis of this scheme, the following equation can be derived (see Supplementary data), which defines the fraction of the Rab molecules extracted from the membrane as a function of the REP/GDI concentration.
Figure 5.
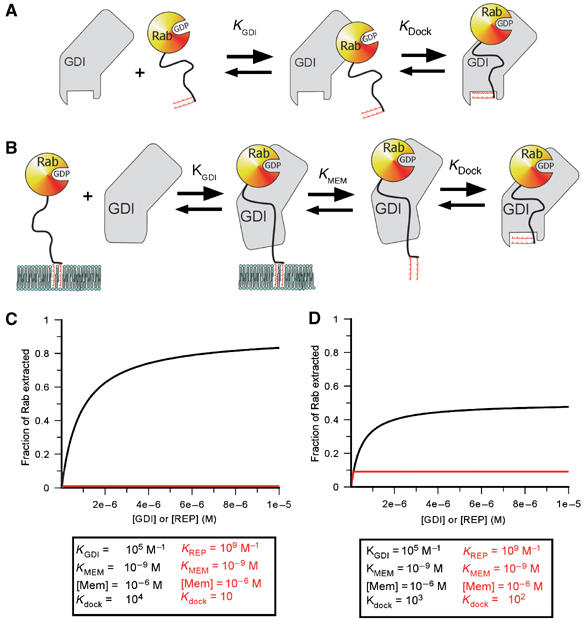
Quantitative simulation of the extraction of prenylated Rabs by GDI/REP based on a simple model. (A) Two-step binding of Rab to GDI/REP. The affinity between the two proteins would be given by _K_GDI(1+_K_dock). (B) Extension of the model to extraction from a membrane. This includes a step in which the dissociation of the C-terminal prenyl groups from the lipid bilayer occurs in a discrete step, which is mechanistically unrealistic but thermodynamically sound given coupling to the next step (docking of the lipid onto its GDI/REP binding site). (C) Simulation of the dependence of the degree of extraction of Rab as a function of the GDI/REP concentration using the constants given. For simulation, a value of 1 μM has been chosen for the effective membrane concentration, together with a value of 1 nM for the affinity of the prenylated C-terminus of Rab for the membrane. While these might not be an accurate reflection of the situation in reality, different values will have identical effects on both GDI and REP extraction efficiencies, but will not affect their relative efficiencies. (D) Same as in (C) but with a different set of constants differing less dramatically for GDI and REP.
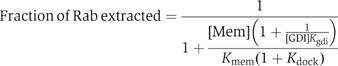
At a constant effective membrane concentration ([Mem]), this would lead to a hyperbolic dependence of the fraction of Rab removed from the membrane on the REP/GDI concentration. The concentration dependence of the degree of extraction is determined by _K_gdi, which reflects the affinity of REP/GDI if they only interact with the GTPase domain. The extent of extraction at saturating REP/GDI concentrations is not, in the general case, 100%, but is given by the expression:
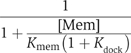
Thus, at given values for [Mem] and _K_mem, the fraction extracted at high REP/GDI concentrations will be dependent on _K_dock, the equilibrium constant for docking of the Rab C-terminus and lipid moieties onto domain II of REP/GDI when the RabGTPase domain is already bound to domain I. Thus, increasing the value of _K_dock will increase the degree of extraction, and Figure 5C and D shows simulations of the effect of different values of _K_dock, which approximately agree with those for GDI and REP, respectively. With a given value for the overall affinity of Rab for REP/GDI, expressed as _K_gdi(1+_K_dock), or _K_gdi_K_dock if _K_dock is ≫1, a high value of _K_dock can be achieved by choosing a small value of _K_gdi. This appears to be the situation for GDI, which has a low value of _K_gdi. Calculation based on the affinity values described above leads to a value of _K_dock of 104 if the overall affinity of GDI to prenylated Rabs is 109 M−1, or 103 for the more realistic value of 108 M−1 (Shapiro et al, 1993). Reduction of the equilibrium constant for the first step would lead to a higher value of _K_dock, but would also mean that higher concentrations of GDI would be needed to achieve maximal extraction affinity. Thus, there is a trade-off between these constants. In the case of REP, for which the increase in overall affinity due to interaction of the prenylated C-terminus with Rab appears to be small, the high affinity of the initial interaction means that maximal extraction would be reached at low concentration, but that this would be small compared with the maximal degree of extraction with GDI due to the low value of _K_dock.
The main point emerging from the calculations above can be summarised as follows: GDI is efficient at extracting Rabs from the membrane since there is a large difference in binding energy between the situations in which only the GTPase domain interacts with GDI and the situation in which the C-terminus and lipid moiety are also docked. The difference in binding energies provides the thermodynamic driving force for extraction from the membrane. In contrast, most of the binding energy in the case of REP comes from the interaction with the GTPase domain, with only very little driving force for the extraction provided by the interaction with the C-terminus. This provides a long sought explanation for the need for two related Rab chaperone molecules with nonoverlapping functions. Confirmation for this model comes from efforts to generate a variant of REP or GDI that could perform both functions. In accordance with the model, no molecule that could efficiently perform both functions could be designed or generated (Alory and Balch, 2003).
In order to test the proposed model directly in a system that simulates the in vivo situation, we tested the ability of GDI and Mrs6 to extract endogenous Ypt1 from isolated yeast membranes. In this assay, a fixed amount of Ypt1-containing yeast membranes were treated with increasing concentrations of recombinant GDI or Mrs6; the extracted membranes were separated from the soluble protein by floating on sucrose layer and analysed by Western blotting with Ypt1-specific antibodies. As expected, both GDI and Mrs6 were able to extract Ypt1 from the membranes (Figure 4D and E). However, in accordance with the proposed model, GDI was ca. 50 times more potent than Mrs6 in this process. The observed difference falls short of the affinity differences with unprenylated proteins, but this factor would only be expected if the difference in these affinities were exactly compensated for by the difference in the contributions of the docking process in order to produce the same overall affinity of prenylated Rab to GDI and Mrs6, but there is no evidence to suggest that the latter point actually obtains. Taken together with the fact that GDI is at least 10 times more abundant than REP, the presented data provides an explanation for the segregation of REP and GDI function in the GTPase cycle (Alory and Balch, 2001).
Since the model predicts that the initial recognition of membrane-bound Rab protein occurs only via GTPase domain of the GTPase, an unprenylated GTPase should function as a competitive inhibitor of GDI-mediated extraction of prenylated GTPases. To test this experimentally, we performed the extraction assay using a concentration of GDI that can extract 50% of Ypt1 from the membrane under the conditions used. On inclusion of increasing concentrations of competing Ypt7 protein, the extraction was progressively inhibited. As can be seen in Figure 4F, the presence of 30 μM Ypt7 inhibited the extraction of Ypt1 by ca. 50%, approximately in accordance with the measured affinity between GDI and Ypt7.
Mechanism of GDI-mediated membrane delivery of RabGTPases
The model outlined for GDI-mediated extraction of RabGTPases allows us to make several predictions with respect to the potential mechanism of GDI-mediated membrane delivery of Rab proteins. While this must be, at some level, the reverse of the extraction process, it seems likely on theoretical grounds and from experimental evidence that a facilitating factor is involved. Membrane-associated GDI-displacing activities, also termed ‘Rab receptors' or GDFs, were identified in several systems, a number of them belonging to the group of Yip/PRL proteins (Sivars et al, 2003; Pfeffer and Aivazian, 2004). These factors are expected to affect two regions of GDI: the lipid binding site and the Rab-binding platform that interacts with the Switch I/Switch II regions of GDI. A recent study suggests that the Yip3 protein can dislodge GDI, possibly by transferring conjugated isoprenoids to the membrane (detergent micelle in this case) or, less probably, transiently binding it (Sivars et al, 2003). In the case of GDI, this alone should be sufficient for dissociation of the Rab:GDI complex. However, in the case of REP, its affinity for the GTPase domain of Rab protein should remain very high even in the absence of the protein:lipid interaction. Therefore, it is likely that the membrane-associated recognition factor induces additional conformational changes in the Rab:GDI/REP complex that promotes its dissociation (Figure 3A). These considerations provide initial guidance for biochemical and biophysical experiments that should lead to the identification of high-affinity intermediates of this reaction, which could be subjected to biochemical and structural investigation.
Materials and methods
Synthesis of the digeranylgeranylated tripeptide
The geranylgeranylated tripeptide H2N-Cys(S_t_Bu)-Cys(GG)-Cys(GG)-OH was synthesised using solid-phase peptide chemistry essentially as described before (Watzke et al, 2005). Fmoc-Cys(S_t_Bu)-OH and Fmoc-Cys(GG)-OH were coupled to the chlorotrityl chloride resin under standard conditions employing _N_-hydroxybenzotriazole and diisopropylcarbodiimide as coupling reagents. The tripeptide was cleaved from the resin with 1% TFA/CH2Cl2 to yield Fmoc-Cys(S_t_Bu)-Cys(GG)-Cys(GG)-OH with a yield of 95%. Final Fmoc deprotection was performed by treatment with diethylamine/methylenechloride (1:4) to obtain the final dipeptide H2N-Cys(S_t_Bu)-Cys(GG)-Cys(GG)-OH in 85% yield. The N-terminal cysteine side chain remained protected until the ligation reaction where in situ deprotection occurs due to the excess of thiol reagent.
Protein expression and purification
Ypt1 truncated by three-amino-acid residues was C-terminally fused to an intein/chitin-binding domain assembly as implemented in the pTWIN-1 vector (New England Biolabs). Protein expression in Escherichia coli and purification of thioestertagged protein was performed as described (Durek et al, 2004). The resulting Ypt1Δ3-MESNA thioester protein was desalted on a PD-10 column (Amersham) equilibrated with 10 mM Na-phosphate, pH 7.5, 0.1 mM MgCl2, 2 μM GDP and concentrated to 20 mg/ml. Recombinant yeast RabGDI and Mrs6 were expressed in E. coli and purified as described before (Dursina et al, 2002; Rak et al, 2003). Recombinant Ypt1, Ypt7 and Rab7 proteins were expressed in E. coli and purified as described (Alexandrov et al, 1999; Vollmer et al, 1999). The open-reading frames of Sec4 and Ypt51 were amplified by PCR with specific primers using yeast genomic DNA as a template and PCR products were subcloned into the pET19-TEV vector. The expression in E. coli and purification was performed as described before (Alexandrov et al, 1999).
Preparation of the diprenylated Ypt1:GDI complex, protein ligation and complex formation
These were performed as described previously (Rak et al, 2003; Durek et al, 2004) and in Supplementary data.
Crystallisation and structure solution
Crystallisation was performed at 20°C using the vapour diffusion method in hanging drops. A measure of 1 μl of a 11 mg/ml protein solution was mixed with 1 μl of reservoir solution consisting of 14% MME-PEG 2000 and 100 mM Tris-acetate, pH 7.9. Crystals appeared overnight and reached their maximal size (200 × 100 × 1000 μm) in 2 days. The crystal drops were further equilibrated against 30% MME-PEG 2000 and 100 mM Tris-acetate, pH 7.9, after which the crystals were picked and flash frozen in liquid nitrogen without additional treatment with cryoprotecting agent. Beam line ID-14-2 at the ESRF (Grenoble) was used to collect a complete data set. Data to 1.48 Å resolution were processed using the XDS program suite (Kabsch, 1993).
The crystals are isomorphous to the previously reported complex of monoprenylated Ypt1:GDI (Rak et al, 2003). The structure was determined by refinement using Ypt1:GDI (PDB code 1UKV) as the initial model with water molecules and the geranylgeranyl lipid removed. The resulting σA-weighted difference maps were calculated using bulk solvent correction procedures revealing peaks in the lipid-binding pocket that clearly showed the position of the two geranylgeranyl moieties. The lipid moieties were fitted into the electron density, the complex model was rebuilt and the water molecules added. The complete model of the complex was iteratively rebuilt and refined. Refinement was carried out using Refmac 5 (Murshudov et al, 1997) and the model built using software package O (Jones et al, 1991). Data collection and refinement statistics are shown in Table I. The structure was deposited to PDB under accession code 2BCG.
In vitro Ypt1 membrane extraction assay
For GDI assays, Saccharomyces cerevisiae membranes were prepared as follows. Cells were disrupted using a Multi-Lab bead-mill (Bachofen, Switzerland) at 4°C in buffer (RB) (containing 20 mM HEPES, pH 7.5, 200 mM sorbitol, 50 mM KC2H3O2, 1 mM DTT, 1 mM EDTA, 5 mM MgCl2 and protease inhibitors) at a concentration corresponding to 20 (_A_600/ml). Cell extracts were cleared by centrifugation at 300 g for 5 min. The extract was then loaded onto a 1 ml cushion of sucrose (60% in RB), and centrifuged at 100 000 g for 1 h in a Sorvall S80 AT3 rotor. The buffer–sucrose interface was collected in a minimal volume, and the protein concentration was determined.
Standard assays contained 80 μg of membrane protein in the final assay volume of 150 μl, and standard assay conditions were 30°C for 15 min. After the assay, 1 ml of RB was added to each reaction and mixed. Diluted assay mixtures were centrifuged for 1 h at 100 000 g in a Sorvall S45A rotor. SDS–PAGE sample buffer was then added to each pellet and samples were resolved on 12% SDS–PAGE gel. Ypt1 was visualised by immunoblotting with anti-Ypt1 antibody and quantitated by densitometry.
Biophysical analysis of GDI/Mrs6/Ypt interaction
GDI/Ypt interactions were monitored using an isothermal titration calorimeter (MCS-ITC, MicroCal Inc.) as described elsewhere (Rudolph et al, 2001). The data were analysed using the manufacturer's software yielding the stoichiometry (N), the binary equilibrium association constant (_K_a) and the enthalpy of binding (Δ_H_0). Δ_H_0 was assumed to be independent of primary ligand concentration. From the relationship Δ_G_0=−RT ln _K_a and the Gibbs–Helmholtz equation, the free energy (Δ_G_0) and entropy of association (Δ_S_0) were calculated.
Fluorescence measurements were performed as described in Supplementary data.
Supplementary Material
Supplementary materials
Acknowledgments
G Holtermann is acknowledged for invaluable technical assistance. We thank the staff of beam-line 14-1, ESRF for help during data collection. We thank I Schlichting and W Blankenfeldt for help with data collection. D Gallwitz is acknowledged for a generous gift of anti-Ypt1 antibody. We are grateful to C Delon for comments on the manuscript. AR and OP were supported by DFG/EUIRI Grant RA 1364/1-1. KA was supported by Heisenberg Award of the Deutsche Forschungsgemeinschaft. ATC was supported by a Boehringer Ingelheim fellowship. This work was supported in part by Grant DFG AL 484/7-2 to KA and Grant I/77 977 of Volkswagen foundation to KA, RSG and HW.
Conflicts of interest The authors declare that they do not have any competing commercial interests in relation to the submitted work.
References
- Alexandrov K, Heinemann I, Durek T, Sidorovitch V, Goody RS, Waldmann H (2002) Intein-mediated synthesis of geranylgeranylated Rab7 protein in vitro. J Am Chem Soc 124: 5648–5649 [DOI] [PubMed] [Google Scholar]
- Alexandrov K, Scheidig AJ, Goody RS (2001) Fluorescence methods for monitoring interactions of Rab proteins with nucleotides, Rab escort protein, and geranylgeranyltransferase. Methods Enzymol 329: 14–31 [DOI] [PubMed] [Google Scholar]
- Alexandrov K, Simon I, Yurchenko V, Iakovenko A, Rostkova E, Scheidig AJ, Goody RS (1999) Characterization of the ternary complex between Rab7, REP-1 and Rab geranylgeranyl transferase. Eur J Biochem 265: 160–170 [DOI] [PubMed] [Google Scholar]
- Alory C, Balch WE (2001) Organization of the rab-gdi/chm superfamily: the functional basis for choroideremia disease. Traffic 2: 532–543 [DOI] [PubMed] [Google Scholar]
- Alory C, Balch WE (2003) Molecular evolution of the Rab-escort-protein/guanine-nucleotide-dissociation-inhibitor superfamily. Mol Biol Cell 9: 3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y, Shao Y, Alory C, Matteson J, Sakisaka T, Chen W, Gibbs RA, Wilson IA, Balch WE (2003) GDI-Rab GTPase recycling. Structure 11: 347–357 [DOI] [PubMed] [Google Scholar]
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73: 1091–1099 [DOI] [PubMed] [Google Scholar]
- Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y (1990) Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J Biol Chem 265: 13007–13015 [PubMed] [Google Scholar]
- Ayad N, Hull M, Mellman I (1997) Mitotic phosphorylation of rab4 prevents binding to a specific receptor on endosome membranes. EMBO J 16: 4497–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Adamo P, Menegon A, Lo NC, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu SK, Tandon A, Valtorta F, Balch WE, Chelly J, Toniolo D (1998) Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet 19: 134–139 [DOI] [PubMed] [Google Scholar]
- Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR (1997) Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J 16: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek T, Alexandrov K, Goody RS, Hildebrand A, Heinemann I, Waldmann H (2004) Synthesis of fluorescently labeled mono- and diprenylated Rab7 GTPase. J Am Chem Soc 126: 16368–16378 [DOI] [PubMed] [Google Scholar]
- Dursina B, Thoma NH, Sidorovitch V, Niculae A, Iakovenko A, Rak A, Albert S, Ceacareanu AC, Kolling R, Herrmann C, Goody RS, Alexandrov K (2002) Interaction of yeast rab geranylgeranyl transferase with its protein and lipid substrates. Biochemistry 41: 6805–6816 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15: 132–133 [DOI] [PubMed] [Google Scholar]
- Garrett MD, Zahner JE, Cheney CM, Novick PJ (1994) GDI1 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO J 13: 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Luan P, Heine A, Zeng K, Moyer B, Greasely SE, Kuhn P, Balch WE, Wilson IA (2000) A new functional domain of guanine nucleotide dissociation inhibitor (alpha-GDI) involved in Rab recycling. Traffic 1: 270–281 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy ME (1994) RASTER3D version 2 0 A program for photorealistic molecular graphics. Acta Crystallogr D 50: 869–873 [DOI] [PubMed] [Google Scholar]
- Muir TW (2003) Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem 72: 249–289 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Aivazian D (2004) Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 5: 886–896 [DOI] [PubMed] [Google Scholar]
- Pfeffer SR (2001) Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol 11: 487–491 [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Rak A, Reents R, Niculae A, Sidorovitch V, Cioaca MD, Bessolitsyna E, Thoma NH, Waldmann H, Schlichting I, Goody RS, Alexandrov K (2003) Structure of rab escort protein-1 in complex with rab geranylgeranyltransferase. Mol Cell 11: 483–494 [DOI] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K (2003) Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science 302: 646–650 [DOI] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K (2004) Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell 117: 749–760 [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Linnemann T, Grunewald P, Wittinghofer A, Vetter IR, Herrmann C (2001) Thermodynamics of Ras/effector and Cdc42/effector interactions probed by isothermal titration calorimetry. J Biol Chem 276: 23914–23921 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Kikuchi A, Araki S, Hata Y, Isomura M, Kuroda S, Takai Y (1990) Purification and characterization from bovine brain cytosol of a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to smg p25A a ras p21-like GTP-binding protein. J Biol Chem 265: 2333–2337 [PubMed] [Google Scholar]
- Schalk I, Zeng K, Wu SK, Stura EA, Matteson J, Huang M, Tandon A, Wilson IA, Balch WE (1996) Structure and mutational analysis of Rab GDP-dissociation inhibitor. Nature 381: 42–48 [DOI] [PubMed] [Google Scholar]
- Segev N (2001) Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol 13: 500–511 [DOI] [PubMed] [Google Scholar]
- Shapiro AD, Pfeffer SR (1995) Quantitative analysis of the interactions between prenyl Rab9 GDP dissociation inhibitor-alpha and guanine nucleotides. J Biol Chem 270: 11085–11090 [DOI] [PubMed] [Google Scholar]
- Shapiro AD, Riederer MA, Pfeffer SR (1993) Biochemical analysis of rab9 a ras-like GTPase involved in protein transport from late endosomes to the trans Golgi network. J Biol Chem 268: 6925–6931 [PubMed] [Google Scholar]
- Sivars U, Aivazian D, Pfeffer SR (2003) Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425: 856–859 [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM (2001) The Rab GTPase family. Genome Biol 2: 3007.1–3007.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer P, Will E, Scheglmann D, Strom M, Gallwitz D (1999) Primary structure and biochemical characterization of yeast GTPase-activating proteins with substrate preference for the transport GTPase Ypt7p. Eur J Biochem 260: 284–290 [DOI] [PubMed] [Google Scholar]
- Watzke A, Brunsveld L, Durek T, Alexandrov K, Rak A, Goody RS, Waldmann H (2005) Chemical biology of protein lipidation: semi-synthesis and structure elucidation of prenylated RabGTPases. Org Biomol Chem 7: 1157–1164 [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 179: 131–137 [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials