Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice (original) (raw)
Abstract
The abundance of repetitive DNA varies greatly across centromeres within an individual or between different organisms. To shed light on the molecular mechanisms of centromere repeat proliferation, we performed structural analysis of LTR-retrotransposons, mostly centromere retrotransposons of rice (CRRs), and phylogenetic analysis of CentO satellite repeats harbored in the core region of the rice chromosome 4 centromere (CEN4). The data obtained demonstrate that the CRRs in the centromeric region we investigated have been enriched more significantly by recent rounds of segmental duplication than by original integration of active elements, suggesting that segmental duplication is an important process for CRR accumulation in the centromeric region. Our results also indicate that segmental duplication of large arrays of satellite repeats is primarily responsible for the amplification of satellite repeats, contributing to rapid reshuffling of CentO satellites. Intercentromere satellite homogenization was revealed by genome-wide comparison of CentO satellite monomers. However, a 10-bp duplication present in nearly half of the CEN4 monomers was found to be completely absent in rice centromere 8 (CEN8), suggesting that CEN4 and CEN8 may represent two different stages in the evolution of rice centromeres. These observations, obtained from the only complex eukaryotic centromeres to have been completely sequenced thus far, depict the evolutionary dynamics of rice centromeres with respect to the nature, timing, and process of centromeric repeat amplification.
Despite the lack of conserved DNA sequence (Malik and Henikoff 2002; Jiang et al. 2003; Lamb et al. 2004; Henikoff and Dalal 2005), the centromeres from most multicellular eukaryotes, such as Arabidopsis (Copenhaver et al. 1999; Heslop-Harrison et al. 1999; Kumekawa et al. 2001), rice (Cheng et al. 2002; Nagaki et al. 2004; Wu et al. 2004; Zhang et al. 2004), maize (Ananiev et al. 1998; Jin et al. 2004), Drosophila (Sun et al. 1997, 2003), and human (Schueler et al. 2001; Rudd et al. 2003) share very similar structural features. It is well documented that, in addition to abundant transposable elements, large arrays of satellite DNA, for instance the pAL1 satellite repeat in Arabidopsis (Martinez-Zapater et al. 1986), the CentO satellite repeats in rice (Cheng et al. 2002), the CentC satellite repeat in maize (Ananiev et al. 1998), and the α satellite repeat in human (Willard and Waye 1987), are the most marked components of the centromeric regions. These satellite repeats are typically arranged in a tandem head-to-tail fashion, and are usually surrounded and interspersed by various repeats, primarily composed of long terminal repeat (LTR)-retrotransposons in plant centromeres (Cheng et al. 2002; Nagaki et al. 2004, 2005; Wu et al. 2004; Zhang et al. 2004). These centromere retrotransposons (CRs) were generally considered to have preferentially accumulated in centromeric regions by insertion of active elements mediated by RNA reverse transcription (Kumar and Bennetzen 1999).
The sizes of satellite monomers are relatively conserved across species and nearly uniform within a genome (Martinez-Zapater et al. 1986; Willard and Waye 1987; Ananiev et al. 1998; Cheng et al. 2002; Nagaki et al. 2004; Wu et al. 2004; Zhang et al. 2004). However, the copy numbers of satellite monomers vary dramatically across species, within an organism, or in a same chromosome from different subspecies or varieties (Cheng et al. 2002; Guy et al. 2003; Eichler et al. 2004), suggesting that the amount of centromere satellite DNA in a single centromere can be increased or reduced dramatically in a very short time frame.
Evolutionary mechanisms regarding the copy number oscillation of centromere satellite repeats remain largely unknown. Because satellite repeats are present in tens of thousands of copies in an organism, genome-wide homogenization of the satellite repeats by gene conversion and unequal crossover was postulated (Smith 1976; Dover 1982; Stephan 1986; Charlesworth et al. 1994). Based on this molecular driving model, it is expected that numerous DNA rearrangements must have occurred gradually in the process of satellite DNA homogenization. Hence, it is extremely challenging to define and identify individual events of satellite DNA rearrangements, especially when sequence “gaps” remain in centromere regions.
Due to large arrays of highly homogenized satellite repeats, assemblies of centromeric regions in higher eukaryotic organisms, including Arabidopsis, human, and rice, have proven to be extremely difficult. To date, only two centromeres, CEN4 and CEN8 from rice chromosomes 4 and 8, respectively, which contain the least CentO satellite DNA among the 12 rice centromeres (Cheng et al. 2002), have been completely sequenced and successfully assembled (Wu et al. 2004; Zhang et al. 2004). To understand the evolutionary dynamics of centromeric repetitive DNA, we have undertaken in-depth analyses of the core regions of the rice chromosome 4 centromere, including structural analysis of CRRs, and homology and phylogenetic analysis of CRRs and CentO repeats. We also compared CentO repeats from different centromeres of rice. We present here the nature and mechanisms of recent centromere repeat amplification that facilitated the structural variation and size expansion of rice centromeres.
Results
Classification and distribution of CRRs in the core region of CEN4
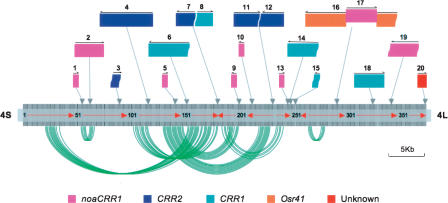
In the core region of chromosome centromere 4 (Zhang et al. 2004), we identified 20 LTR-retrotransposon elements (REs) (Fig. 1; Table 1), including four intact elements (REs2, 4, 17, and 18) and six solo LTRs (REs1, 5, 9, 10, 13, and 20). These 10 elements are flanked by five base-pair target site duplications (TSDs). The other 10 retroelements are truncated at one or both ends. In Zhang et al. (2004), two truncated elements, REs11 and 12, both sharing the same 5′ LTRs, were misannotated as one element, “CR4–11,” whereas RE16, a single element that was interrupted by RE17, was misannotated as two elements, “CR4–15” and “CR4–17.” In addition, RE20, a solo LTR, was not previously found. We categorized these 20 elements into five families/subfamilies by sequence homology comparison with previously described LTR-retrotransposon families/subfamilies in rice (McCarthy et al. 2002; Nagaki et al. 2005), including three centromere retrotransposon families/subfamilies (CRR1, noaCRR1, and CRR2) that account for 18 CRR elements, family Osr41, and an unknown family that has not been defined due to the lack of intact elements of this family in the complete rice genome (data not shown). These retroelements, which comprise 63 kb of DNA, are interspersed into ∼60 kb CentO satellites and separate CentO into 18 satellite blocks (Fig. 1).
Figure 1.
Arrangement and rearrangement of centromeric repeats in the core region of rice centromere 4. The intact LTR-retrotransposons and solo LTRs are represented by color-shadowed boxes, whereas the truncated retrotransposons are represented by color-shadowed boxes with one or both curved ends. The five families/subfamilies of retrotransposons are marked by five different colors. (Arrows and numbers above the shadowed boxes) Orientations and order of LTR-retrotransposons in the region analyzed, (dark vertical lines) CentO satellite monomers, (vertical arrows connected to the boxes) positions of LTR-retrotransposons, (red arrows) orientation of CentO blocks separated by LTR-retrotransposons. The most related pairs of monomers distributed in corresponding duplicated segments in conserved order (in the same or opposite orientations) are connected by curved green lines.
Table 1.
LTR-retrotransposons harbored in CentO satellites of rice chromosome 4
| Retroelement (REa) | Family or subfamily | Size (bp) | 5′ flanking sequence | 3′ flanking sequence | Structural feature |
|---|---|---|---|---|---|
| 1 | noaCRR1 | 794 | GTATT | GTATT | Solo LTR |
| 2 | noaCRR1 | 4291 | GTATT | GTATT | Intact element |
| 5 | noaCRR1 | 790 | CGCGC | CGCGC | Solo LTR |
| 9 | noaCRR1 | 792 | CGCGC | CGCGC | Solo LTR |
| 10 | noaCRR1 | 795 | CGCGC | CGCGC | Solo LTR |
| 13 | noaCRR1 | 793 | CGCGC | CGCGC | Solo LTR |
| 17 | noaCRR1 | 4368 | AAGGC | AAGGC | Intact element |
| 19 | noaCRR1 | 4292 | NA | NA | Truncated element |
| 3 | CRR2 | 1676 | NA | NA | Truncated element |
| 4 | CRR2 | 7740 | CGCAC | CGCAC | Intact element |
| 7 | CRR2 | 2723 | NA | CGCAC | Truncated element |
| 11 | CRR2 | 3268 | CGCAC | NA | Truncated element |
| 12 | CRR2 | 3906 | CGCAC | NA | Truncated element |
| 6 | CRR1 | 6252 | NA | TCCTC | Truncated element |
| 8 | CRR1 | 1472 | NA | TCCTC | Truncated element |
| 14 | CRR1 | 5772 | NA | TCCTC | Truncated element |
| 15 | CRR1 | 741 | NA | NA | Truncated element |
| 18 | CRR1 | 4998 | CGCGG | CGCGG | Truncated elementb |
| 16 | Osr41 | 6586 | NA | GAGTG | Truncated element |
| 20 | Unknown | 1338 | GATAT | GA-AT | Solo LTR |
“Preferential” insertions and insertion sites of CRRs in the core region of CEN4
Although CRRs were detected in all 12 rice centromeres (Cheng et al. 2002; Nagaki et al. 2005), such a high degree of CRR enrichment in the core region of centromere 4 is a truly exceptional observation. More interestingly, preferential insertion of four noaCRR1 retroelements, “CR4–5,”“CR4–9,”“CR4–10,” and “CR4–12” (i.e., RTs5, 9, 10, and 13) in four different but adjacent sites was previously described by Zhang and coworkers (2004). However, we found that these four noaCRR1 elements were situated in exactly the same position in four different CentO monomers and were flanked by identical 5-bp (CGCGC) TSDs (Table 1). All four noaCRR1 elements sharing the same TSDs are solo LTRs. In addition, two other noaCRR1 elements—a solo LTR, RE1, and an intact element, RE2—were found to share the same target sites (GTATT) (Table 1). We further analyzed the CRR1 and CRR2 families that contain mostly truncated elements. All three single-end truncated CRR1 elements (REs6, 8, and 14) were found to be flanked by “TCCTC” at their intact ends. All three single-end truncated CRR2 elements (REs7, 11, and 12) were flanked by the “CGCAC,” which was identical to the TSD flanking RE4, the only intact CRR2 element identified in the region studied. All the 5-bp flanking sequences (summarized in Table 1) are unique sites in the CentO monomers. The boundaries of the truncated ends of other CRRs were difficult to precisely define and thus were not investigated further.
The observations obtained above were unexpected and have not been previously reported in centromeres investigated so far. Theoretically, the frequency that two LTR-retrotransposons share an identical TSD in a genome is <0.1% (1/45). Thus, it is not surprising that no two of 1000 randomly chosen LTR-retrotransposons previously investigated in the rice genome were found to be flanked by an identical TSD (Ma et al. 2004). Even though CRRs were preferentially inserted into CentO satellite monomers in rice, the possibility that two CRRs inserted into an identical site of different monomers would be extremely low (theoretically, ∼1/155 or 1/165), unless a specific retrotransposon family or subfamily has preferential insertion sites in CentO monomers. However, the observation that the seven noaCRR1 elements have three different insertion sites in CentO monomers invalidates the possibility of a single preferential insertion site.
Segmental duplication of CRRs in the core region of CEN4
We hypothesized that the CRR elements that share the same TSDs have been duplicated by some process other than transposition mediated by RNA reverse transcription. If this is the case, then CentO monomers harboring the duplicated CRRs may have also been involved in the same duplication, and the duplicated monomers would share the highest sequence similarities as compared with other monomers. To test this hypothesis, we extracted all monomers harboring CRRs and BLAST-searched these monomers against all CEN4 monomer sequences. As expected, the best matched pairs or groups of monomers harbored the same family or subfamily CRR elements flanked by the same target insertion sites (Table 2). These data suggest that at least nine apparent segmental duplications of CRRs occurred in the region following six original insertions of CRRs.
Table 2.
Monomers harboring _CRR_s and their best matches
| REa | Family or subfamily | Monomer harboring CRR | Best matched monomerb |
|---|---|---|---|
| 1 | noaCRR1 | 55 | 62 |
| 2 | noaCRR1 | 62 | 55 |
| 4 | CRR2 | 115 | 179 |
| 5 | noaCRR1 | 140 | 199 |
| 7 | CRR2 | 179 | 115 |
| 8 | CRR1 | 180 | 245 |
| 9 | noaCRR1 | 199 | 140 |
| 10 | noaCRR1 | 211 | 140 |
| 11 | CRR2 | 230 | 179 |
| 12 | CRR2 | 231 | 230 |
| 14 | CRR1 | 245 | 180 |
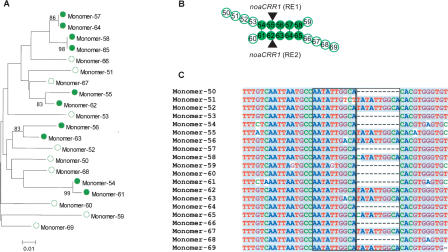
To further verify and detail the scenario regarding CRR amplification by segmental duplication, we initially performed phylogenetic analysis of 20 contiguous monomers surrounding two noaCRR1 elements, REs 1 and 2, that share the same TSDs. The 20 monomers were extracted and aligned using ClustalX (Thompson et al. 1997). Subsequently, a Neighbor-Joining phylogenetic tree was generated using MEGA3 (Kumar et al. 2004). Five pairs of the most related monomers supported by >80% bootstrap values are illustrated in Figure 2A. The sequence similarities between paired monomers vary from 98%–100%. Interestingly, the most related pairs of monomers are arranged in two monomer clusters in the same order and orientation (Fig. 2B). We also found that the most related pairs of monomers either both contain the 10-bp duplication or neither do (Fig. 2C), even though the parameter of “complete deletion” (of indels) was employed by MEGA3 for the phylogenetic analysis of the monomers. Moreover, the monomers that harbored the noaCRR1 elements, REs1 and 2, were found to be most related to each other (Fig. 2B). These data demonstrate that a segmental duplication of five contiguous CentO monomers and a noaCRR1 element, RE1 or RE2, occurred, and subsequently a solo LTR (RE1) was generated by intraelement homologous recombination between its two LTRs (Devos et al. 2002; Ma et al. 2004).
Figure 2.
Phylogenetic analysis of a cluster of CentO monomers surrounding the first two CRRs in the core region of CEN4. (A) Neighbor-Joining tree obtained with the monomer sequence data. (Filled circles) Closely related monomers with bootstrap values >80% based on 500 replicates, (open circles) other monomers without clear relationship. (B) A segmental duplication event revealed by the Neighbor-Joining tree. The closely related pairs of monomers are connected, respectively, by short vertical lines. The integration sites of the noaCRR1 elements are marked by dark arrows. (C) A subregion of the alignment of the monomers analyzed above showing a 10-bp duplication site.
Dates of CRR amplification by segmental duplications
LTR divergence has been used to date insertions of LTR-retrotransposons (SanMiguel et al. 1998). This method is based on the fact that the two LTR sequences of a single LTR-retrotransposon are usually identical upon integration. However, because segmental duplications seem to be a major process for LTR-retrotransposon amplification in this study, a retrotransposon, if duplicated, may have accumulated mutations between its two LTRs before the duplication event occurred. Hence, the insertion date estimated by this method would not likely be the amplification date of an LTR-retrotransposon.
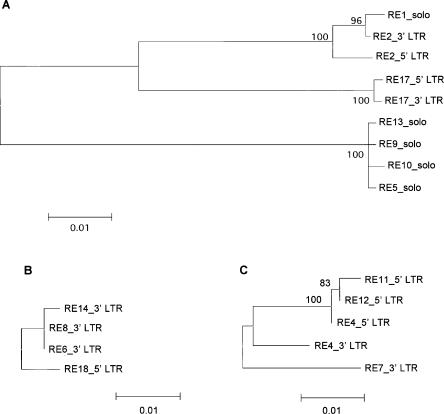
In order to outline the duplication processes and to date the duplication events, we first performed phylogenetic analysis of the CRR elements belonging to the three different families or subfamilies, respectively. Because most CRRs are truncated elements and solo LTRs, only LTRs from these CRR elements were extracted and used to generate Neighbor-Joining trees. The seven noaCRR1 elements clustered into three relatively distant groups, containing two, one, and four elements, respectively (Fig. 3A), whereas the four CRR1 elements and the four CRR2 elements were clustered into one respective group (Fig. 3B,C). The elements within each group were closely related to each other and were flanked by the same TSDs (Table 1). Hence, at least nine CRRs were amplified by segmental duplication if the most diverged element (showing greatest distance from others) within each group was considered to be the most ancestral copy.
Figure 3.
Amplification of CRRs revealed by phylogenetic analysis. (A,B,C) Neighbor-Joining trees obtained with LTR sequence data from noaCRR1, CRR1, and CRR2 elements, respectively. Numbers adjacent to nodes indicate bootstrap values >80% from the test of 500 replicates.
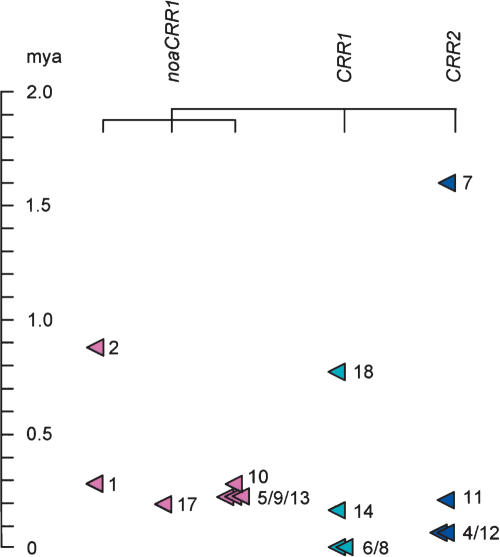
Under the assumption that the most diverged CRR element within a group is the originator of the other copies, the average distance between the most diverged CRR and each of the other additional copies was used to estimate the amplification time of the most diverged CRR. Similarly, other CRRs were dated by using the average distance between the more diverged copy and each of the other copies within a subgroup. RE17, an insertion into RE16, was dated by the divergence of its two LTRs. By using these strategies, the amplification (insertion or duplication) dates of 15 CRRs were calculated. Assuming the most diverged CRRs represent the originator of the other corresponding CRRs, all subsequent accumulation of CRRs by segmental duplication occurred <0.3 Mya, ∼0.2 Mya on average (Fig. 4).
Figure 4.
The times of CRR amplification estimated based on the phylogenies and pair-wise distances between LTRs. (Mya, million years ago).
Amplification of CentO satellites by segmental duplication
To further understand the dynamics of CentO satellite amplification, we performed phylogenetic analysis of all monomers that were identified in CEN4. By analyzing the most related monomers revealed by the Neighbor-Joining tree obtained (data not shown), we identified 50 additional pairs of monomers that are arranged in four duplicated clusters of monomers in either the same or inverted orientations (Fig. 1). Each pair of monomers between the duplicated segments has a very high degree of sequence similarity (ranging from 98% to 100%), indicating very recent events. This observation is consistent with our estimation of CRR amplification dates (Fig. 4). The largest duplicated segment detected in this study contains 23 monomers and a 7.5-kb intact CRR2 element (RE4) (Fig. 1; Table 1). Given that most duplicated segments were interrupted by a few to 130 CentO monomers and intact or truncated CRR elements, and that they were arranged in overlapping or nested patterns, the precise sizes of the segmental duplication could not be determined. These observations also suggest the rapid and dramatic rearrangement and reshuffling of the CentO satellites, which largely limited our capacity to track the boundaries of duplicated segments precisely. Except for the segmental duplication described above, we did not detect any more ordered or “higher-order repeat units,” which were observed in centromeric α satellite repeats of human and primate chromosomes (Warburton and Willard 1996). The identities between different monomers ranged from 90% to 98% across the CentO blocks, although the divergence of the monomers in both terminal CentO blocks was more apparent than others (Zhang et al. 2004). In addition, the CentO monomers within the duplicated arrays (see Fig. 2) do not show a precipitous or gradual change in sequence divergence.
Comparison of CentO satellite repeats across rice chromosomes
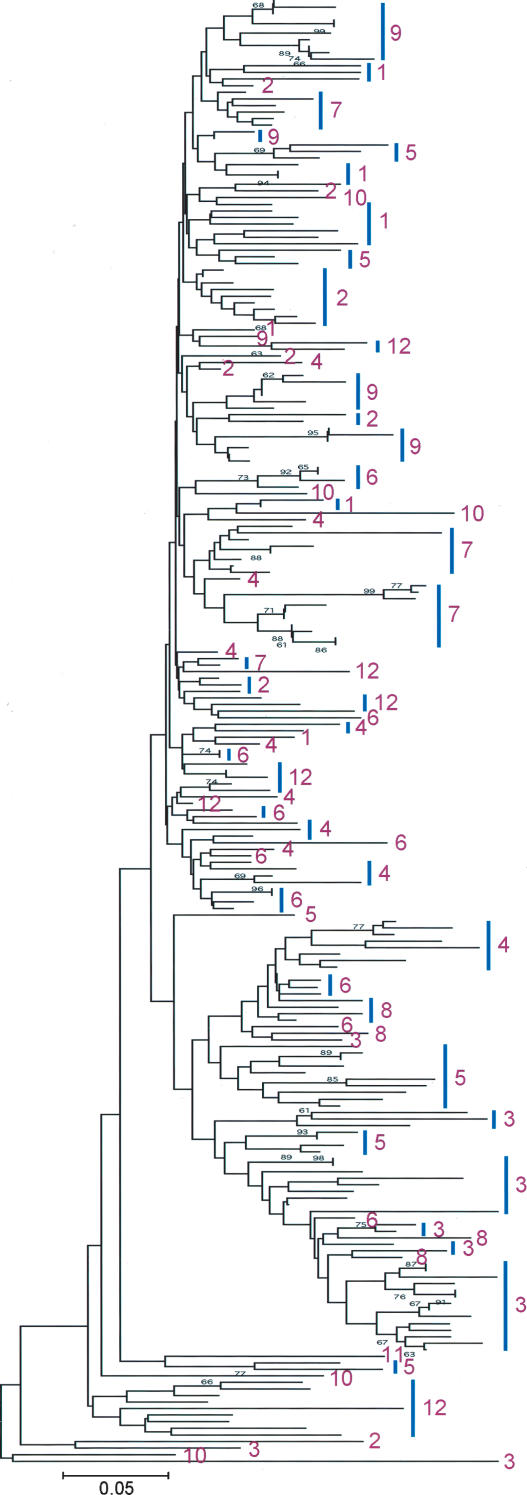
To understand the dynamic variation of CentO satellite repeats, we compared 226 CentO monomers from the 12 centromeres of rice (∼20 monomers per centromere, except CEN10 and CEN11, in which only a few monomers were sequenced), which were randomly chosen from 4464 monomers that we extracted from the latest genomic sequence of rice (Build 3.0 pseudomolecules) generated by IRGSP (International Rice Genome Sequencing Project 2005). The Neighbor-Joining tree obtained (Fig. 5) exhibited very low bootstrap values between most groups or branches, indicating the overall high degree of sequence similarity. Although the most closely related monomers were always found in the same centromere, some monomers, either within a single centromere or between different centromeres, showed very similar distances. This observation suggests that CentO satellites have undergone interchromosomal exchange and genome-wide homogenization. Similar to the results obtained from analyses of centromeric α satellite repeats in human and primate chromosomes (Warburton and Willard 1996), the average rate of interchromosomal monomer divergence is lower than that of intrachromosomal monomer divergence in rice (Fig. 5).
Figure 5.
Phylogenetic analysis of CentO satellite monomers across the 12 rice centromeres. The monomers from each centromeric region of rice were randomly chosen. The vertical bars mark groups of monomers from the same chromosomes. The numbers adjacent to individual branches or vertical bars indicate the chromosomes from which the monomers were chosen. The numbers adjacent to nodes indicate bootstrap values >60% from the test of 500 replicates.
An extremely intriguing observation is that the completely sequenced centromere 8 was devoid of 166-bp monomers that contain a 10-bp duplication, as shown in Figure 2C, whereas this 10-bp duplication was found in 161 out of the 372 monomers within CEN4. Monomers containing the 10-bp duplication were also found in all 10 other rice centromeres (Table 3), although their copy numbers cannot be determined due to the incompleteness of the centromere sequences. Moreover, the 166-bp monomers were found to be intermingled with 155-bp monomers across the CEN8 region (Fig. 3; Zhang et al. 2004). These findings raise a possibility that the 10-bp duplication may have occurred a long time ago and gradually spread into the different chromosomes. Thus, the rice CEN8 may represent an intermediate-stage centromere that originally formed in a non-centromeric region by recruiting and subsequently amplifying the 155-bp monomers, and, as a result, the CEN8 region has not been invaded by the 166-bp monomers to date. This inference is supported by the previous discovery of active genes in the CEN8 region (Nagaki et al. 2004).
Table 3.
Distribution of CentO satellite repeats containing 10-bp duplication
| Centromerea | # of monomers with 10-bp duplicationb | # of monomers without 10-bp duplicationb | # of monomers without target sitesc | # of monomers in rice previously estimatedd |
|---|---|---|---|---|
| CEN1 | 545 | 490 | 21 | 8900 |
| CEN2 | 59 | 275 | 8 | 4500 |
| CEN3 | 9 | 150 | 8 | 1100 |
| CEN4 | 161 | 196 | 15 | 700 |
| CEN5 | 74 | 251 | 3 | 1000 |
| CEN6 | 15 | 21 | 2 | 5100 |
| CEN7 | 104 | 476 | 3 | 2000 |
| CEN8 | 0 | 454 | 6 | 400 |
| CEN9 | 441 | 306 | 4 | 3900 |
| CEN10 | 1 | 5 | 3 | 2900 |
| CEN11 | 1 | 0 | 1 | 11,900 |
| CEN12 | 124 | 216 | 18 | 1000 |
Discussion
The preferential amplification of centromere-specific transposable elements, mostly retrotransposons, has been observed in most, if not all, centromeric regions of complex genomes investigated thus far (Copenhaver et al. 1999; Schueler et al. 2001; Cheng et al. 2002; Sun et al. 2003; Wu et al. 2004; Zhang et al. 2004; Nagaki et al. 2005). Hence, it is not surprising that 18 out of 20 LTR-retrotransposons identified in the core region of CEN4 were found to be CRRs in our study. The extensive segmental duplication mediated by _Alu–Alu_-mediated recombination (duplicative transposition) events has been observed in the pericentromeric regions of humans and across the human genome (Horvath et al. 2003; She et al. 2004; Locke et al. 2005). However, the preferential amplification of centromere retrotransposons by rounds of segmental duplication was an unexpected finding. Because the segmental duplication of CentO arrays that do not harbor CRR elements was observed in the CEN4 region (Fig. 1), and because no CRR or other LTR-retrotransposon elements were observed within the duplicated CentO arrays in the CEN8 region (J. Ma and J.L. Bennetzen, unpubl.), it is likely that the duplication of CRRs in the CEN4 region was mediated by the duplication of clusters of CentO monomers that harbor these CRRs.
Unequal homologous chromosome or sister chromatid exchange between tandem arrays (Smith 1976) could account for the amplification of the CentO satellite repeats and the CRRs involved. However, unequal crossover could lead to contraction of satellite arrays and remove both satellite repeats and the CRRs. Given the extensive accumulation of CRRs in the CEN4 region, there must be evolutionary force(s) counteracting potential loss of satellite repeats and CRRs due to unequal crossover and facilitating the accumulation of these centromere repeats. Recently, progressive expansion of the X chromosome centromere was observed in primate species (Schueler et al. 2005), although the major events of satellite DNA amplification in the X chromosome centromere were relatively older than those detected in the CEN4 region.
Because of the complete or nearly complete suppression of homologous chromosome recombination in all centromeric regions that have been investigated (Copenhaver et al. 1999; Wu et al. 2003), gene conversion has been suggested to be another potential mechanism for satellite DNA variation. Conversion events could partially explain the rapid homogenization of satellite repeats (Smith 1976; Dover 1982; Charlesworth et al. 1994). However, gene conversion may not have played a major role in amplification of CentO satellites and CRRs in rice. The largest duplicated segment found in this study is >11 kb, whereas very few conversion events that generated fragments >2 kb have been reported in all plant genomes investigated so far (Dooner and Martinez-Ferez 1997; Ma et al. 2005; Yandeau-Nelson et al. 2005). In addition, the CRRs and flanking CentO monomers generated by gene conversion events were mostly fragmented (Ma et al. 2005; Yandeau-Nelson et al. 2005), but the majority of segmental duplication boundaries detected in the CentO arrays end with, and are flanked by, intact monomers. Moreover, if unequal crossover between homologous chromosomes or between sister chromatids is suppressed in a centromere, it follows that gene conversion in the same centromere would be suppressed, too.
Theoretically, recombination by unequal crossover could also take place within different homologous regions of a single chromatid. This kind of recombination would eliminate DNA between the homologous regions, and thus it would not account for the amplification of centromere repeats. However, it is likely to be one of the processes responsible for the formation of solo LTRs, especially in centromeric regions. In this study, we identified six solo LTRs in the core region of CEN4, although four (RTs 5, 9, 10, and 13) out of the six solo LTRs appear to have been formed before the duplication events. Dozens of solo LTRs and a high ratio (2:1) of solo LTRs to intact retroelements were found in the core region of CEN8 (Nagaki et al. 2004; Wu et al. 2004; J. Ma and J.L. Bennetzen, unpubl.), indicating frequent intraelement unequal recombination in the centromeric regions of rice. Solo LTRs could also be generated by unequal crossover between homologous chromosomes and between sister chromatids, but the products containing three LTRs would be expected to be the counterparts of solo LTRs. However, based on two recent studies of LTR-retrotransposons in Arabidopsis (Devos et al. 2002) and rice (Ma et al. 2004), the two plant species that have been completely sequenced, the proposed three-LTR elements were not found in either species. This observation favors the intrachromatid unequal recombination model for formation of solo LTRs. Alternatively, gametophytes carrying the three LTR retrotransposons, if generated by unequal crossover, may have been efficiently eliminated under natural selection. Regardless of which proposed mechanisms are involved in the amplification of CentO satellites and CRRs, intracentromeric DNA recombination does not seem to be severely suppressed over evolutionary time.
The mechanisms proposed could result in direct segmental duplication but may not, however, explain other phenomena observed in CEN4, such as inverted segmental duplication of satellite arrays and the truncation of CRRs. Hence, multiple mechanisms, including unequal crossover, gene conversion, duplicative transposition, and satellite transposition (Alexandrov et al. 1988; Alkan et al. 2002; Bailey et al. 2003; Horvath et al. 2005), illegitimate recombination (Devos et al. 2002; Ma et al. 2004), and some other unknown ones, may be involved in centromeric DNA evolution.
In this study, at least nine CRRs were estimated to be duplicated in the core region of CEN4 ∼0.3 Mya, after the proposed divergence time (0.44 Mya) of the two subspecies of rice, japonica and indica (Ma and Bennetzen 2004). The amplification dates of a few intact retroelements were apparently overestimated previously by comparison of two LTRs from a single element (Zhang et al. 2004). However, we should point out that because the average divergence among LTRs in a group or subgroup, instead of a single duplication event, was used to date the segmental duplication of CRRs, the timing of the duplication of CRRs could also be slightly overestimated in this study. Therefore, significant differences regarding distribution and organization of CRRs and their flanking CentO satellites between indica and japonica would be expected. Recently, the CentO sequence was found to be absent in another Oryza species, O. brachyantha (Lee et al. 2005). Together, these observations suggest that not only the copy number and organization but also the sequence of centromere satellite repeats in rice are extremely variable.
Based on the finding that centromeres from an individual genome usually share the same types of satellite repeats and centromere retrotransposons, it is arguable that the centromere-specific repeats may be associated with centromere function. A number of studies have provided evidence in favor of this view (Tyler-Smith et al. 1993; Harrington et al. 1997; Nagaki et al. 2004). On the other hand, it has also been postulated that the centromere-specific repeats are not indispensable for centromere formation, based on the recent discoveries of active neocentromeres of human and Drosophila, which exhibit a complete absence of centromere satellite repeats (du Sart et al. 1997; Choo 2001; Maggert and Karpen 2001), and based on the recent finding that barley centromeres can move to new positions and that satellite DNA is not necessary for efficient centromere formation (Nasuda et al. 2005). Regardless of the potential roles of centromere repeats, it at least has been demonstrated that both CentO satellites and CRRs in centromeric regions of rice bind CENH3 and assemble functional kinetochores (Cheng et al. 2002), although not all the CentO satellites or CRRs in the centromeric regions could be associated with centromere function (Nagaki et al. 2004). Given the rapid amplification and reshuffling of centromeric DNA detected in CEN4, and previous observations in other centromeric regions of rice (Cheng et al. 2002; Nagaki et al. 2004; Wu et al. 2004), it is reasonable to believe that the specific arrangement of centromere repeats is not an important factor in formation of functional centromeres. However, as observed in the core region of CEN4, centromeric DNA can be dramatically accumulated and rearranged over a short time frame, resulting in a dynamic structure (e.g., enrichment of CRRs and CentO satellites) that could facilitate the binding of CENH3 and the formation of nucleosomes that enable the kinetochore assembly.
Methods
Characterization and classification of LTR-retrotransposons
A combined structural analysis and sequence homology comparison were employed to identify LTR-retrotransposons in the complete BAC sequence (GenBank accession number BX890594) that contain the centromeric region of rice chromosome 4. The intact elements were identified by the method previously described (Ma et al. 2004). Solo LTRs and truncated elements were identified by sequence homology searches against a rice LTR-retrotransposon database that was developed by collecting known LTR-retrotransposons (McCarthy et al. 2002; Ma et al. 2004; Nagaki et al. 2005) and by scanning the 371-Mb rice genome sequence generated by the International Rice Genome Sequencing Project (2005) (Build 3.0 pseudomolecules, accession numbers, AP008207–AP008209 and AP008211–AP008218) using a LTR-retrotransposon finding program, LTR_STRUC (McCarthy and McDonald 2003). The structures of all LTR-retrotransposon elements identified were confirmed by manual inspection. The LTR-retrotransposon elements were classified by sequence homology comparison, and individual families and subfamilies were defined by the criteria previously described (Ma et al. 2004; Nagaki et al. 2004).
Identification and isolation of CentO satellite monomers
The consensus sequence of CentO satellite monomers previously reported (Wu et al. 2004) was used to search against the BAC sequence BX890594 containing centromere 4 (CEN4) of rice, and the genomic sequences of rice (AP008207–AP008209 and AP008211–AP008218) containing other 11 centromeric regions by BLAST and CROSS_MATCH (http://www.phrap.org/phrap.docs/general.html). The boundaries of all monomers in the centromeric regions were manually inspected, and the monomer sequences were extracted and converted in uniform orientation by a perl program, feature_parse, that was provided by Canadian Bioinformatics Help Desk. The monomers that were interrupted by LTR-retrotransposons were inspected and rejoined manually.
Sequence alignments and Neighbor-Joining tree construction
The LTR sequences extracted from retrotransposons of individual families/subfamilies and the CentO monomers were aligned, respectively, using ClustalX (Thompson et al. 1997). The alignments were edited manually if necessary. MEGA3 (Kumar et al. 2004) was used to calculate pairwise transition and transversion mutations. The Neighbor-Joining trees were built using the Kimura two-parameter method (Kimura 1980).
Dating of insertion and duplication events
The LTR sequences were used for dating of insertion and/or duplication events. The amplification (insertion or duplication) time of a CRR was estimated by using average distances between the element and each of the other elements younger than it indicated by phylogenetic analysis. A mutation rate of 1.3 × 10-8 substitutions per base per year proposed for intergenic sequences of rice (Ma and Bennetzen 2004) was employed to convert into dates of the amplification events.
Acknowledgments
We thank Phillip SanMiguel for critical reading of this manuscript, Paul Stothard, Canadian Bioinformatics Help Desk, for his assistance in programming, Brian Abernathy for computational support, and three anonymous reviewers for their valuable comments. This work was supported by the National Science Foundation (Grant no. DBI-0227414 to S.A.J).
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4583106.
References
- Alexandrov, I.A., Mitkevich, S.P., and Yurov, Y.B. 1988. The phylogeny of human chromosome specific α satellites. Chromosoma 96 443-453. [DOI] [PubMed] [Google Scholar]
- Alkan, C., Bailey, J.A., Eichler, E.E., Sahinalp, S.C., and Tuzun, E. 2002. An algorithmic analysis of the role of unequal crossover in α-satellite DNA evolution. Genome Inform. Ser. Workshop Genome Inform. 13 93-102. [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. 1998. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. 95 13073-13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Liu, G., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplication. Am. J. Hum. Genet. 73 823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., Sniegowski, P., and Stephan, W. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371 215-220. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., Dong, F., Langdon, T., Ouyang, S., Buell, C.R., Gu, M., Blattner, F.R., and Jiang, J. 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14 1691-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1 165-177. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Nickel, K., Kuromori, T., Benito, M.I., Kaul, S., Lin, X., Bevan, M., Murphy, G., Harris, B., Parnell, L.D., et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468-2474. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K., and Bennetzen, J.L. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K. and Martinez-Ferez, I.M. 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9 1633-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover, G. 1982. Molecular drive: A cohesive mode of species evolution. Nature 299 111-117. [DOI] [PubMed] [Google Scholar]
- du Sart, D., Cancilla, M.R., Earle, E., Mao, J.I., Saffery, R., Tainton, K.M., Kalitsis, P., Martyn, J., Barry, A.E., and Choo, K.H. 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat. Genet. 16 144-153. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Clark, R.A., and She, X. 2004. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat. Rev. Genet. 5 345-354. [DOI] [PubMed] [Google Scholar]
- Guy, J., Hearn, T., Crosier, M., Mudge, J., Viggiano, L., Koczan, D., Thiesen, H.J., Bailey, J.A., Horvath, J.E., Eichler, E.E., et al. 2003. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10p. Genome Res. 13 159-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, J.J., Van Bokkelen, G., Mays, R.W., Gustashaw, K., and Willard, H.F. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15 345-355. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. and Dalal, Y. 2005. Centromeric chromatin: What makes it unique? Curr. Opin. Genet. Dev. 15 177-184. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S., Murata, M., Ogura, Y., Schwarzacher, T., and Motoyoshi, F. 1999. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11 31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, J.E., Gulden, C.L., Bailey, J.A., Yohn, C., McPherson, J.D., Prescott, A., Roe, B.A., de Jong, P.J., Ventura, M., Misceo, D., et al. 2003. Using a pericentromeric interspersed repeat to recapitulate the phylogeny and expansion of human centromeric segmental duplications. Mol. Biol. Evol. 20 1463-1479. [DOI] [PubMed] [Google Scholar]
- Horvath, J.E., Gulden, C.L., Vallente, R.U., Eichler, M.Y., Ventura, M., McPherson, J.D., Graves, T.A., Wilson, R.K., Schwartz, S., Rocchi, M., et al. 2005. Punctuated duplication seeding events during the evolution of human chromosome 2p11. Genome Res. 15 914-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. 2005. The map-based sequence of the rice genome. Nature 436 793-800. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Birchler, J.A., Parrott, W.A., and Dawe, R.K. 2003. A molecular view of plant centromeres. Trends Plant Sci. 8 570-575. [DOI] [PubMed] [Google Scholar]
- Jin, W., Melo, J.R., Nagaki, K., Talbert, P.B., Henikoff, S., Dawe, R.K., and Jiang, J. 2004. Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16 571-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111-120. [DOI] [PubMed] [Google Scholar]
- Kumar, A. and Bennetzen, J.B. 1999. Plant retrotransposons. Annu. Rev. Genet. 33 479-532. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 5 150-163. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. 2001. The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8 285-290. [DOI] [PubMed] [Google Scholar]
- Lamb, J.C., Theuri, J., and Birchler, J.A. 2004. What's in a centromere? Genome Biol. 5 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.-R., Zhang, W., Langdon, T., Jin, W., Yan, H., Cheng, Z., and Jiang, J. 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. 102 11793-11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Jiang, Z., Pertz, L.M., Misceo, D., Archidiacono, N., and Eichler, E.E. 2005. Molecular evolution of the human chromosome 15 pericentromeric region. Cytogenet. Genome Res. 108 73-82. [DOI] [PubMed] [Google Scholar]
- Ma, J. and Bennetzen, J.L. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. 101 12404-12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Devos, K.M., and Bennetzen, J.L. 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., SanMiguel, P., Lai, J., Messing, J., and Bennetzen, J.L. 2005. DNA rearrangement in orthologous Orp regions of the maize, rice and sorghum genomes. Genetics 170 1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert, K.A. and Karpen, G.H. 2001. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158 1615-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S. and Henikoff, S. 2002. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 12 711-718. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Estelle, M.A., and Somerville, C.R. 1986. A high repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204 417-423. [Google Scholar]
- McCarthy, E.M. and McDonald, J.F. 2003. LTR_STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics 19 362-367. [DOI] [PubMed] [Google Scholar]
- McCarthy, E.M., Liu, J., Gao, L.Z., and McDonald, J.F. 2002. Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 3 research0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138-145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Neumann, P., Zhang, D., Ouyang, S., Buell, C.R., Cheng, Z., and Jiang, J. 2005. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 22 845-855. [DOI] [PubMed] [Google Scholar]
- Nasuda, S., Hudakova, S., Schubert, I., Houben, A., and Endo, T.R. 2005. Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. 102 9842-9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, M.K., Schueler, M.G., and Willard, H.F. 2003. Sequence organization and functional annotation of human centromeres. Cold Spring Harb. Symp. Quant. Biol. 68 141-149. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20 43-45. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. 2001. Genomic and genetic definition of a functional human centromere. Science 294 109-115. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Dunn, J.M., Bird, C.P., Ross, M.T., Viggiano, L., Rocchi, M., Willard, H.F., and Green, E.D. 2005. Progressive proximal expansion of the primate X chromosome centromere. Proc. Natl. Acad. Sci. 102 10563-10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, X., Horvath, J.E., Jiang, Z., Liu, G., Furey, T.S., Christ, L., Clark, R., Graves, T., Gulden, C.L., Alkan, C., et al. 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430 857-864. [DOI] [PubMed] [Google Scholar]
- Smith, G.P. 1976. Manipulating the genetic code: Jurisprudential conundrums. Georgetown Law J. 64 697-733. [PubMed] [Google Scholar]
- Stephan, W. 1986. Recombination and the evolution of satellite DNA. Genet. Res. 47 167-174. [DOI] [PubMed] [Google Scholar]
- Sun, X., Wahlstrom, J., and Karpen, G. 1997. Molecular structure of a functional Drosophila centromere. Cell 91 1007-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Le, H.D., Wahlstrom, J.M., and Karpen, G.H. 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13 182-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith, C., Oakey, R.J., Larin, Z., Fisher, R.B., Crocker, M., Affara, N.A., Ferguson-Smith, M.A., Muenke, M., Zuffardi, O., and Jobling, M.A. 1993. Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nat. Genet. 5 368-375. [DOI] [PubMed] [Google Scholar]
- Warburton, P. and Willard, H. 1996. Evolution of centromeric α satellite DNA: Molecular organization within and between human and primate chromosomes. In Human genome evolution (eds. M. Jackson, T. Strachan, and G. Dover), pp. 121-145. BIOS Scientific, Guildford, UK.
- Willard, H.F. and Waye, J.S. 1987. Chromosome-specific subsets of human α satellite DNA: Analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 3 207-214. [DOI] [PubMed] [Google Scholar]
- Wu, J., Mizuno, H., Hayashi-Tsugane, M., Ito, Y., Chiden, Y., Fujisawa, M., Katagiri, S., Saji, S., Yoshiki, S., Karasawa, W., et al. 2003. Physical maps and recombination frequency of six rice chromosomes. Plant J. 36 720-730. [DOI] [PubMed] [Google Scholar]
- Wu, J., Yamagata, H., Hayashi-Tsugane, M., Hijishita, S., Fujisawa, M., Shibata, M., Ito, Y., Nakamura, M., Sakaguchi, M., Yoshihara, R., et al. 2004. Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16 967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandeau-Nelson, M.D., Zhou, Q., Yao, H., Xu, X., Nikolau, B.J., and Schnable, P.S. 2005. MuDR transposase increases the frequency of meiotic crossovers in the vicinity of a Mu insertion in the maize a1 gene. Genetics. 169 917-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Huang, Y., Zhang, L., Li, Y., Lu, T., Lu, Y., Feng, Q., Zhao, Q., Cheng, Z., Xue, Y., et al. 2004. Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32 2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]