The Mechanism of Nuclear Export of Smad3 Involves Exportin 4 and Ran (original) (raw)
Abstract
Transforming growth factor beta (TGF-β) receptors phosphorylate Smad3 and induce its nuclear import so it can regulate gene transcription. Smad3 can return to the cytoplasm to propagate further cycles of signal transduction or to be degraded. We demonstrate that Smad3 is exported by a constitutive mechanism that is insensitive to leptomycin B. The Mad homology 2 (MH2) domain is responsible for Smad3 export, which requires the GTPase Ran. Inactive, GDP-locked RanT24N or nuclear microinjection of Ran GTPase activating protein 1 blocked Smad3 export. Inactivation of the Ran guanine nucleotide exchange factor RCC1 inhibited Smad3 export and led to nuclear accumulation of phosphorylated Smad3. A screen for importin/exportin family members that associate with Smad3 identified exportin 4, which binds a conserved peptide sequence in the MH2 domain of Smad3 in a Ran-dependent manner. Exportin 4 is sufficient for carrying the in vitro nuclear export of Smad3 in cooperation with Ran. Knockdown of endogenous exportin 4 completely abrogates the export of endogenous Smad3. A short peptide representing the minimal interaction domain in Smad3 effectively competes with Smad3 association to exportin 4 and blocks nuclear export of Smad3 in vivo. We thus delineate a novel nuclear export pathway for Smad3.
Transforming growth factor beta (TGF-β) is a multipotent polypeptide which regulates cell proliferation, differentiation, and apoptosis (29). Upon exposure to TGF-β, its type II receptor phosphorylates and activates the type I receptor, which subsequently phosphorylates the receptor-activated Smads (R-Smads), Smad2 and Smad3, at their diserine C-terminal SXS motif. The activated R-Smads form a complex with Smad4 as they translocate to the nucleus, where they regulate expression of diverse groups of genes (5). We and others have shown that Smad3 contains a lysine-rich nuclear localization signal (NLS) in its N-terminal Mad homology 1 (MH1) domain that is specifically recognized by importin-β1 only when Smad3 is phosphorylated at its C-terminal serine residues by type I receptor kinase and then imported to the nucleus in a Ran-dependent manner (14, 36). On the other hand, Smad2 was shown to be imported to the nucleus independent of cytosolic factors (40). The ability of the C-terminal MH2 domain of Smad2 to directly recognize specific nucleoporins has been proposed as a major mechanism for the directed import of Smad2 to the nucleus (41). One of the reasons why these similar R-Smad proteins use different import mechanisms is a 30-amino-acid-long insert in the Smad2 MH1 domain, encoded by Smad2's unique exon 3, which hampers the interaction between Smad2 and importin-β1 (14).
After prolonged TGF-β treatment, ubiquitination and degradation of Smads and the TGF-β receptors have been observed, and several ubiquitin ligases and cofactors that mediate these processes have been identified (11). The activated TGF-β signaling pathway is thereby quenched. On the other hand, nuclear export of endogenous Smad proteins was observed after TGF-β stimulation in the presence of cycloheximide without significant downregulation or ubiquitination, suggesting the existence of alternative mechanisms that restore the ground state of the Smad pathway (24). Such nuclear export was recently proposed to occur after dephosphorylation of nuclear R-Smads and disruption of the R-Smad/Smad4 complex (10, 41).
In the absence of TGF-β signaling, Smad4 was shown to continuously shuttle between the nucleus and the cytoplasm (24, 32). Smad4 is exported by the specific exportin CRM1 (chromosome region maintenance 1)/exportin 1, which is directly inhibited by leptomycin B (LMB). Nucleocytoplasmic shuttling of Smad1, a bone morphogenetic protein (BMP)-specific R-Smad, was also found to be regulated by CRM1 (37, 38). In addition, Smad2 has been recently shown to shuttle in and out of the nucleus with distinct kinetics from Smad4, both in the ground state and after TGF-β signaling (21). The mechanism of nuclear export of Smad3 has not been studied in detail, but recent evidence suggested that this mechanism could not be blocked by LMB (10, 24, 37). In this study, we used microinjection and reverse import assays to analyze the mechanism of Smad3 nuclear export in vivo and in vitro. We identified a novel export mechanism of nuclear Smad3 by exportin 4 and the Ran GTPase. Exportin 4 is a novel member of the importin-β family that was previously shown to transport the eukaryotic translation initiation factor 5A (eIF-5A) to the cytoplasm via recognition of a complex nuclear export signal (NES) (18). Thus, our findings suggest that exportin 4 is not only a critical factor controlling protein synthesis but can also regulate Smad signaling.
MATERIALS AND METHODS
Cloning and recombinant protein expression.
Molecular cloning and expression of glutathione _S_-transferase (GST)-green fluorescent protein (GFP)- Smad3, -Smad3D, -Smad3A, -Smad3MH1, -Smad3Linker, and -Smad3MH2 as well as GST-Smad3, -Smad3MH1+Linker, -Smad3Linker+MH2, and GST-Smad4 has been described previously (14, 22). pGEX6P2-GFP-Smad3MH1-R74K/K81R was generated by PCR amplification of pcDNA3-FLAG-Smad3-R74K/K81R (20) and ligation of the PCR products into pGEX6P2-GFP (provided by S. Kuroda, Institute of Scientific and Industrial Research, Osaka, Japan). Restriction enzymes and DNA-modifying enzymes used were from New England BioLabs (Ipswich, MA) or Fermentas GmbH (St. Leon-Rot, Germany). The recombinant proteins were purified on glutathione beads using the glutathione _S_-transferase (GST) purification kit from GE Healthcare/Amersham Biosciences (Uppsala, Sweden) and either eluted with glutathione or cleaved by thrombin (Sigma-Aldrich, Stockholm, Sweden) or precision protease (GE Healthcare/Amersham Biosciences, Uppsala, Sweden). Proteins were further purified to homogeneity by high-performance liquid chromatography. His-tagged CRM1 and His-tagged exportin-t were provided by I. W. Mattaj (Heidelberg, Germany) and prepared as described previously (2, 6). Expression and purification of importin-α2 (PTAC 58) and mouse importin-β (PTAC 97) were performed as described previously (9, 12). Recombinant human p10/NTF2 protein was expressed and purified as described previously (31). Recombinant wild-type Ran, RanT24N, and RanQ69L carrying a pentahistidine (His) tag at their N termini were expressed, purified, and charged with GDP (Ran and RanT24N) or GTP (RanQ69L), as described previously (8). To construct pcDNA3-Flag-hExp4, pQE30-hExp4, and pZZ70-hExp4, cDNA clone KIAA1721, obtained from the Kazusa DNA institute (Chiba, Japan), was amplified by PCR and subcloned into (i) the HindIII and XhoI sites of pcDNA3-Flag vector, (ii) the BamHI and NotI sites of pQE30 vector (QIAGEN Nordic, West Sussex, United Kingdom), or (iii) the BamHI and XhoI sites of pZZ70 vector (16), respectively. His-tagged Exp4 and zz-tagged Exp4 were prepared as described previously (18). The following 15 transportin cDNA clones in pDNA3-Flag were obtained from A. Shimamoto (GeneCare Research Institute, Kanagawa, Japan): CRM1, exportin 5 (exp5), exportin-t (expt), importin-β (impβ), importin 4 (imp4), importin 13 (imp13), Ran binding protein 5 (RanBP5), Ran binding protein 7 (RanBP7), Ran binding protein 8 (RanBP8), Ran binding protein 11 (RanBP11), Ran binding protein 16 (RanBP16), Ran binding protein 17 (RanBP17), transportin 1 (Trsp1), transportin 2 (Trsp2), and transportin SR (TrspSR). To construct pGEX5X2-NLS-NES-GFP, oligonucleotides encoding the NLS of simian virus 40 large T antigen were inserted into the BamHI site of pGEX5X2-NES-GFP. The GST-NES-GFP, GST-NLS-NES-GFP, and GST-GFP fusion proteins were expressed and purified as described previously (31). GST-RanGAP1 was prepared as described previously (13). To prepare the injection marker for microinjection experiments, bovine serum albumin (BSA) was labeled with Cy3 and purified by gel filtration chromatography following the manufacturer's instructions (Invitrogen AB/Molecular Probes, Stockholm, Sweden). Aliquots of each recombinant protein and the labeled proteins were frozen in liquid nitrogen and stored at −70°C. To construct expression vectors for GFP-Smad3 deletion fragments, the Smad3 fragments were amplified by PCR and subcloned into pEGFPc2 or pEGFPc3 vector (Invitrogen AB/Clontech, Stockholm, Sweden).
Cell culture.
HeLa and tsBN2 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS), l-glutamine, and penicillin/streptomycin at 37°C and 33.5°C, respectively. NIH 3T3 cells were cultured in DMEM supplemented with 10% calf serum, l-glutamine, and penicillin/streptomycin at 37°C. Human embryonic kidney 293T cells and human immortalized HaCaT keratinocytes were cultured in DMEM supplemented with 10% FBS, l-glutamine, and penicillin/streptomycin at 37°C. All reagents for cell culture were from Sigma-Aldrich (Stockholm, Sweden). Recombinant mature TGF-β1 was from PeproTech EC Ltd. (London, United Kingdom).
Cell fractionation and immunoblotting.
tsBN2 monolayers were harvested after stimulation with TGF-β1 as indicated in the figures and two fractions were prepared, one enriched in cytosolic and the other in nuclear proteins, using the NE-PER kit from Pierce Biotechnology Inc. (Rockford, IL) and according to the protocol of the manufacturer. Nucleus-enriched extracts that were normalized for protein content were used for immunoblotting analysis after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described previously (15). In control experiments for estimation of degree of enrichment, both nucleus- and cytosol-enriched extracts that were normalized for protein content were analyzed, and enrichment of 90% or higher was always obtained (see Fig. 3E). Affinity-purified rabbit anti-phospho-Smad3 antibody was a gift from M. Reiss (New Brunswick, NJ); mouse monoclonal anti-Smad1/2/3 (H2), mouse monoclonal anti-PCNA (proliferating cell nuclear antigen; PC-10), rabbit polyclonal anti-Grb2 (C-23) antibodies, and the enhanced chemiluminescence detection system were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Secondary anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG coupled to horseradish peroxidase were from GE Healthcare/Amersham Biosciences (Uppsala, Sweden).
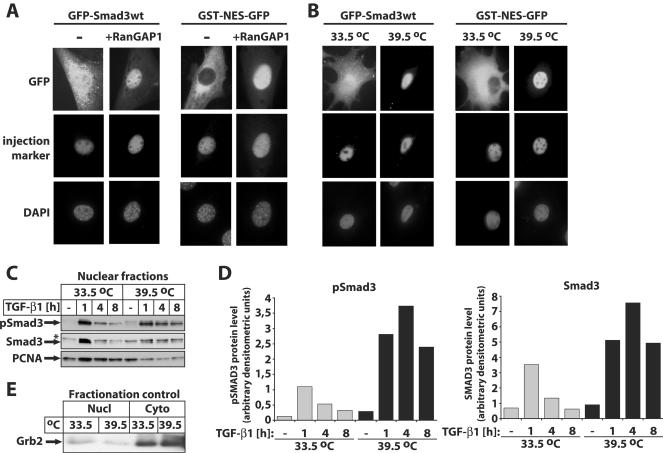
FIG. 3.
The GTP form of Ran is crucial for nuclear export of Smad3 in vivo. (A) GFP-Smad3 wild type (wt) (1 mg/ml) or GST-NES-GFP (1 mg/ml) was coinjected with buffer (−) or GST-RanGAP1 (5 mg/ml) and the injection marker Cy3-labeled BSA into nuclei of NIH 3T3 cells. After 1 h of incubation at 37°C, localization of the proteins was monitored by GFP autofluorescence, and DAPI staining of nuclei served as a control. (B) GFP-Smad3 wild type (wt) (1 mg/ml) or GST-NES-GFP (1 mg/ml) was microinjected together with the injection control Cy3-labeled BSA into the nucleus of tsBN2 cells. The cells were cultured either at the permissive (33.5°C) or nonpermissive (39.5°C) temperature for 24 h prior to injection. After 1 h of incubation at 37°C, localization of the proteins was monitored by GFP autofluorescence, and DAPI staining of nuclei served as a control. (C) Inactivation of the Ran GEF RCC1 leads to nuclear accumulation of Smad3. Immunoblot analysis of endogenous nuclear levels of phospho-Smad3 (pSmad3) and Smad3 in tsBN2 cells cultured at the permissive (33.5°C) and nonpermissive (39.5°C) temperatures for 24 h prior to stimulation with 2 ng/ml TGF-β1 for the indicated periods of time and prior to cell lysis and fractionation. The specific protein bands are shown with arrows, and a star indicates a background band. PCNA represents a nuclear protein marker. (D) Graphic representation of densitometric quantification of the protein bands in panel C, with phospho-Smad3 levels (graph on the left) or total Smad3 levels (graph on the right) normalized to the corresponding PCNA levels. Gray bars represent the permissive (33.5°C) temperature, and black bars represent the nonpermissive (39.5°C) temperature. (E) Immunoblot analysis of aliquots of the nuclear (Nucl) and corresponding cytoplasmic (Cyto) extracts used in the experiments of panel C for the cytoplasmic protein marker Grb2 (shown by an arrow).
Densitometric analysis of specific protein bands from the immunoblots was performed using AIDA software, version 3.10.039 (Fujifilm Sverige AB, Stockholm, Sweden).
In vitro reverse import assays in digitonin-permeabilized cells.
Transport assays were performed essentially as described previously (1, 9). Briefly, 1 × 105 HeLa cells/ml were plated on an 8-well multitest slide (MP Biochemicals, Irvine, CA) 24 to 48 h before use. Cells were rinsed twice with transport buffer and permeabilized for 5 min in ice-cold transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate) containing 2 mM dithiothreitol (DTT), protease inhibitors, and 40 to 60 μg/ml digitonin (EMD Biosciences, Inc./Calbiochem, San Diego, CA). After washing twice, the slides were immersed in transport buffer containing 2 mM DTT and protease inhibitors at room temperature for 6 min. Excess buffer was removed and cells were incubated with 10 μl/well of reaction mixture for 12 min at 25°C. After washing twice, the slides were immersed in transport buffer containing 2 mM DTT and protease inhibitors on ice for 5 min and further incubated with 10 μl/well of reaction mixture for 20 min (see Fig. 2) or 6 min (see Fig. 6) at 25°C. All reactions contained an ATP regeneration system (1 mM ATP, 5 mM creatine phosphate, and 20 U/ml creatine phosphokinase; Sigma-Aldrich, Stockholm, Sweden), 2% bovine serum albumin, 2 mM DTT, and protease inhibitors in transport buffer. For import assays in the presence of cytosol, Ehrlich ascites tumor cell total cytosol was prepared as described previously (9). For import of Smad3 proteins, 0.4 μM importin-β1, 1 μM p10, and 3 μM Ran-GDP were used with 0.5 μM cargo proteins. To deplete ATP, the reaction mixture was supplemented with 10 mM sodium azide. After incubation, the cells were rinsed with transport buffer and fixed in 3.7% formaldehyde in transport buffer for 15 min at 25°C. GFP fusion proteins were detected by autofluorescence microscopy. Photomicrographs were obtained either with a Zeiss Axioplan 2 microscope equipped with a Hammamatsu C4742-95 digital camera using the Zeiss Plan-neofluar 40×/0.75 objective lens and photographing at ambient temperature in the absence of immersion oil or with a confocal microscope (Leica TCS NT).
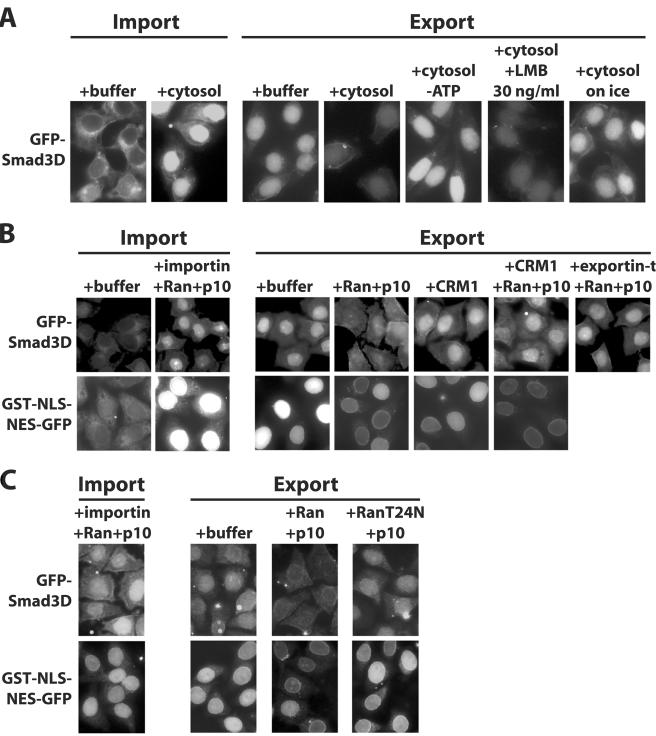
FIG. 2.
Smad3 requires cytosolic cofactors and Ran for nuclear export in vitro. (A) GFP-Smad3D was first imported into the nuclei of permeabilized HeLa cells in the presence of cytosol and an ATP-generating system. Twelve minutes later, cells were further incubated for 20 min under the conditions shown and the distribution of the protein was monitored by GFP autofluorescence. (B and C) GFP-Smad3D or GST-NLS-NES-GFP was first imported into the nuclei of permeabilized cells in the presence of an ATP-generating system, importin-β1, Ran-GDP, and p10 (for GST-NLS-NES-GFP, both importin-α2 and importin-β1 were used). After 12 min of incubation, cells were further incubated another 20 min (GFP-Smad3D) or 5 min (GST-NLS-NES-GFP) under the conditions shown, and the distribution of the protein was monitored by GFP autofluorescence. (B and C) Results from two different representative experiments; panel C compares the effects of Ran-GDP to those of its mutant derivative RanT24N-GDP. The concentrations of recombinant proteins added in the export reactions were 3 μM Ran-GDP (or RanT24N-GDP) plus 1 μM p10, 0.2 μM CRM1, and 0.1 μM exportin-t.
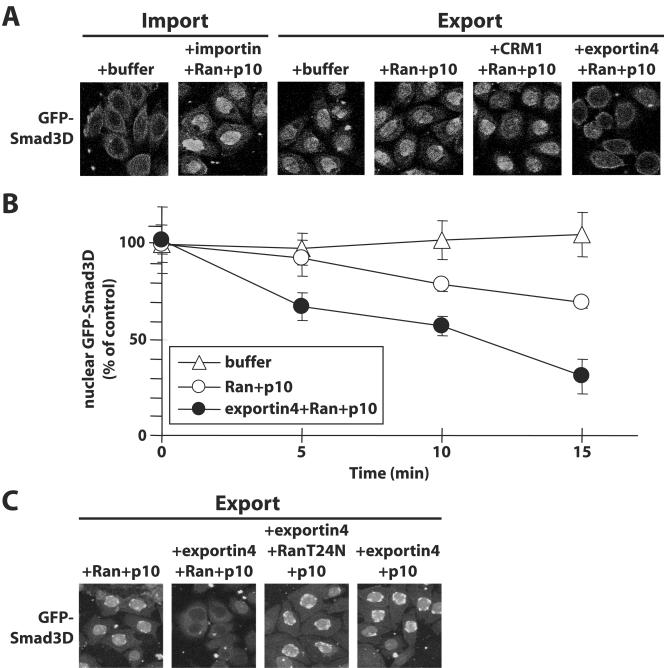
FIG. 6.
Exportin 4 is sufficient to carry nuclear export of Smad3 in vitro. (A) GFP-Smad3D was first imported into nuclei of permeabilized HeLa cells in the presence of an ATP-generating system, importin-β1, Ran-GDP, and p10. After 12 min of incubation, cells were further incubated another 6 min under the conditions shown, and the distribution of the protein was monitored by GFP autofluorescence. The concentrations of recombinant proteins added in the export reactions were 3 μM Ran-GDP plus 1 μM p10, 0.2 μM CRM1, and 0.1 μM exportin 4. (B) Graph showing kinetic data from in vitro reverse import assays identical to those described in panel A and performed by means of a time course between 0 and 15 min. Datum points represent the average number of nuclei that scored positive for GFP-Smad3D at each time point of transport reactions carried in the presence of transport buffer, Ran plus p10, or exportin 4 plus Ran plus p10, as indicated in the inset. Error bars indicate standard errors of the means derived from populations of 350 nuclei counted per condition. (C) Experimental repeat of panel A with the inclusion of two novel conditions that serve as specificity controls: constitutive mutant RanT24N-GDP in the place of wild-type Ran and the complete absence of exogenous Ran.
In vivo import assays in microinjected cells.
For microinjection of recombinant proteins, 1 × 105 NIH 3T3 cells were plated on coverslips in normal medium containing 50 mM HEPES (pH 7.3). On the next day, the recombinant proteins mixed with injection markers (Cy3-labeled bovine serum albumin [BSA], tetramethylrhodamine isothiocyanate [TRITC]-labeled IgG, or GST-GFP) and were injected through a glass capillary into the nucleus by using an Eppendorf (model 5246) automatic microinjector, and the cells were cultured for 1 h in the absence or presence of LMB as indicated in the figures. LMB was a kind gift from M. Yoshida (Tokyo, Japan). The same protocol was used for tsBN2 cells, except that cells were incubated for 24 h at the permissive (33.5°C) or nonpermissive (39.5°C) temperature prior to microinjection. To prepare homokaryon cells, NIH 3T3 cells were fused with polyethylene glycol as described previously (44). Homokaryons were incubated for 3 h at 37°C prior to microinjection. After incubation, the cells were rinsed with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde in PBS for 15 min at 25°C. GFP fusion proteins were detected by autofluorescence microscopy. To detect GST-tagged or Flag-tagged Smad proteins, fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min at 25°C, blocked with 5% FBS for 30 min, and then subjected to indirect immunofluorescence using a monoclonal anti-GST antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or a monoclonal anti-Flag antibody (M2; Sigma-Aldrich, Stockholm, Sweden), respectively. Anti-mouse Cy3- and anti-mouse TRITC-conjugated secondary antibodies were from Invitrogen AB/Molecular Probes (Stockholm, Sweden) and DakoCytomation (Glostrup, Denmark), respectively. To visualize nuclei, after antibody incubation permeabilized cells were incubated for 5 min at 25°C with 1 ng/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Stockholm, Sweden). All photomicrographs were obtained as described in the previous section.
Immunofluorescence microscopy of endogenous Smads and small interfering RNA (siRNA) transfection.
HaCaT cells were seeded on glass coverslips in 6-well plates and cultured in the absence of serum for 24 to 48 h prior to stimulation with 2 ng/ml TGF-β1 or vehicle (4 mM HCl-0.1% BSA) for 1 h. Stimulated cells were then washed extensively with plain DMEM, and fresh serum-free medium was added in the presence of dimethyl sulfoxide (DMSO) (control) or 2 μM LY580276 (a selective TGF-β type I receptor inhibitor [23]) for another 2 h. In some experiments, 8 ng/ml LMB was added together with LY580276 during the 2-h incubation. Stimulated cell monolayers were processed for immunofluorescence and confocal microscopy as described above for in vitro reverse import assays. The primary antibodies used were anti-Smad3 (rabbit polyclonal affinity-purified antibody 51-1500; Invitrogen AB/Zymed Laboratories, Stockholm, Sweden), anti-phospho-Smad3(Ser433/435) (rabbit polyclonal affinity-purified antibody 9514; Cell Signaling Technology, Inc., Beverly, MA), and anti-Smad4 (rabbit polyclonal antiserum H-552; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The secondary antibody used was Alexa Fluor 546 goat anti-rabbit IgG (Invitrogen AB/Molecular Probes, Stockholm, Sweden).
HaCaT cells cultured as described above were transiently transfected with Lipofectamine 2000 (Invitrogen AB, Stockholm, Sweden) using 25 to 100 nM of each RNA oligonucleotide pool according to the manufacturer's protocol. The human exportin 4 (Xpo4)-specific siRNA (accession no. NM_022459) was a pool of four distinct RNA oligonucleotides, termed SMARTpool (reagent number M-027196-00); siRNA against the luciferase reporter vector pGL2 (accession no. X65324) served as control; all siRNAs were from Dharmacon Research, Inc. (Boulder, CO). Six hours posttransfection the cells were cultured in complete medium, and after an overnight incubation the transfection was repeated a second time. Alternatively, HaCaT cells were transfected using Lipofectamine 2000 using GFP or GFP-Smad3(271-324) fusion vectors by a single transfection protocol according to the manufacturer's recommendation. Cell treatments with TGF-β1, DMSO, or LY80276 were as described above and were applied 48 h after the first and 24 h after the second siRNA transfection or 24 h after the GFP vector transfection, followed by immunofluorescence confocal microscopy as described above. Doubly transfected cell monolayers from the plastic dish surrounding the glass coverslips used for immunofluorescence microscopy were solubilized and used for immunoblotting with anti-exportin 4 (V-18) affinity-purified goat polyclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Transient transfection and in vitro binding assays.
293T cells were transiently transfected with full-length pcDNA3-6myc-Smad3 or its derivative deletion mutants as shown in the figures or with the pcDNA3-Flag-Smad3 wild type or alanine point mutant at the two C-terminal serines (2A) together or not with constitutively active TGF-β type I receptor (caALK5) that mimics ligand stimulation. Transfected cells were lysed in binding buffer (50 mM Tris-HCl, pH 7.5, 110 mM potassium acetate, 5 mM magnesium acetate, and protease inhibitors) by sonication and centrifuged for 5 min at 4°C. The supernatant was incubated with zz-Exp4-IgG Sepharose beads in the presence of 2 μM His-tagged RanQ69L-GTP for 2 h at 4°C. Beads were washed three times with the binding buffer, and bound proteins were eluted with 1.5 M MgCl2, 50 mM Tris-HCl, pH 7.5, precipitated with isopropanol and dissolved in sodium dodecyl sulfate sample buffer prior to electrophoresis and immunoblotting. For immunodetection of bound proteins, mouse monoclonal anti-myc (9E10) antibody was prepared in house from hybridoma cells, anti-Flag (clone M2 or M5) monoclonal antibodies were from Sigma-Aldrich (Stockholm, Sweden), affinity-purified rabbit anti-phospho-Smad3 antibody was a gift from M. Reiss (New Brunswick, NJ), and anti-histidine mouse monoclonal antibody (34660) was from QIAGEN Nordic (West Sussex, United Kingdom). Bacterially semipurified proteins were visualized by staining of immunoblots with Ponceau S.
RESULTS
Nuclear export of Smad3 is mediated by its MH2 domain.
In order to analyze the mechanisms of nuclear export of Smad3, bacterially expressed recombinant green fluorescent protein (GFP)-Smad3 fusion protein was microinjected into nuclei in the absence or presence of LMB. GFP-Smad3 was readily exported to the cytoplasm in the absence of LMB, much like the GST-Smad4 fusion protein that served as a positive control (Fig. 1A). Although the nuclear export of GST-Smad4 was completely inhibited by LMB as reported previously (32), LMB had no detectable effect on the export of GFP-Smad3 (Fig. 1A). We tested LMB concentrations up to 30 ng/ml in this experiment without any obvious effects. Nuclear export of Smad3 was also detected when the protein was tagged with GST, GST plus GFP, or Flag (Fig. 1B to D). These experiments confirmed previous reports, which suggested that Smad3 is exported from the nucleus in an LMB-independent manner.
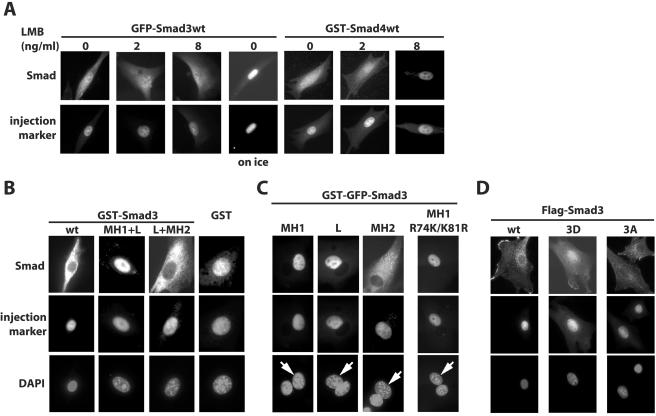
FIG. 1.
Nuclear export of Smad3 is constitutive and requires the C-terminal MH2 domain. (A) Recombinant GFP-Smad3 (1 mg/ml) or GST-Smad4 (1 mg/ml), both in their wild-type (wt) forms, were injected into the nuclei of NIH 3T3 cells together with the injection control protein TRITC-IgG or GST-GFP, respectively. After incubation for 1 h at 37°C in the absence or presence of the indicated concentrations of LMB, cells were fixed and the localization of GFP-Smad3 was examined by autofluorescence microscopy. GST-Smad4 was detected by indirect immunofluorescence with anti-GST antibody. (B) Recombinant Smad3 deletion mutants fused to GST were microinjected into the nuclei of NIH 3T3 cells together with the injection control Cy3-labeled BSA and incubated for 1 h at 37°C, and protein localization was assayed by indirect immunofluorescence with anti-GST antibody. (C) Recombinant Smad3 proteins fused to GST-GFP were coinjected together with the injection control Cy3-labeled BSA into one of the nuclei in dikaryons generated from NIH 3T3 cells and incubated for 1 h at 37°C. Protein localization was detected by GFP autofluorescence and DAPI staining of nuclei served as control. L denotes the linker domain of Smad3. Arrows indicate the injected nuclei in dikaryon cells. (D) Recombinant wild-type Smad3 and Smad3 with changes of its three C-terminal serine residues either to aspartate (3D) or to alanine (3A) were microinjected and analyzed after 1 h of incubation at 37°C as described for panel B.
In order to identify protein domains responsible for this export, various Smad3 deletion mutants were tested in vivo. When GST-Smad3 MH1+Linker was microinjected into the nucleus, the protein was retained in the nucleus after 1 h of incubation (Fig. 1B). However, nuclear injection of GST-Smad3 Linker+MH2 was followed by export to the cytoplasm; in contrast, GST itself was largely retained in the nucleus. Despite the absence of nuclear export of the MH1 and linker domains, it is possible that an NES exists in the MH1 domain whose activity is masked by the strong NLS localized in the same domain (35). In order to rule out such a possibility, we generated dikaryons by fusing cells after polyethylene glycol treatment and microinjected GST-GFP-tagged Smad3 deletions into one of the two nuclei (Fig. 1C). We never observed transfer of GST-GFP-tagged Smad3 MH1 or linker (L) to the uninjected nucleus, indicating that export or shuttling of these domains did not occur and supporting the notion that there is no functional NES in the MH1 or linker domains. In order to exclude the possibility that the microinjected Smad3 MH1 domain failed to be exported because it was anchored in the nucleus by binding to DNA, we tested the mutant Smad3-K74R/R81K, which lacks DNA binding ability because of point mutations of two critical amino acid residues that contact DNA (20). This mutant always remained in the microinjected nucleus of the dikaryon cell. In contrast, microinjection of the isolated MH2 domain of Smad3 always led to efficient cytoplasmic export (Fig. 1C). However, this domain never showed any capacity of import to the second, noninjected nucleus of the dikaryons. Thus, we conclude that the MH2 domain of Smad3 is sufficient to carry robust nuclear export.
Next, to examine whether receptor-mediated phosphorylation of the C-terminal SSVS motif of Smad3 might affect its export, we used two triple-point mutants, Smad3D, in which aspartate replaced the three serine residues to mimic phosphorylation, and Smad3A, a nonphosphorylatable mutant with serine-to-alanine mutations. Flag-Smad3A was exported as efficiently as wild-type Flag-Smad3 protein after nuclear microinjection of NIH 3T3 cells (Fig. 1D). On the other hand, Flag-Smad3D exhibited significantly weaker export to the cytoplasm, and the majority of this protein remained in the injected nuclei. Since Smad3D mimics the activated form of Smad3 that is normally produced after phosphorylation by the type I receptor, these data suggest that receptor-activated Smad3 primarily segregates in the nucleus, as has been reported for Smad2 (21). The fact that Smad3A was more efficiently exported than Smad3D is compatible with the notion that the nuclear export of Smad3 might be enhanced when its C terminus is not phosphorylated, which would support the hypothesis that nuclear Smad3 undergoes dephosphorylation prior to export (10). Obviously, the present data cannot address directly the requirement of Smad3 dephosphorylation for its export.
Nuclear export of Smad3 requires cytosolic factors and the Ran GTPase.
To further study the mechanism of Smad3 nuclear export, we set up an in vitro reverse import assay. For this purpose, we used an established system of digitonin-permeabilized HeLa cells (1). First, GFP-Smad3D was imported into the nucleus in the presence of cytosol. The cells then were washed and further incubated in the presence or absence of cytosol. When cells were incubated with buffer, no obvious export was observed, whereas the addition of cytosol stimulated the export of Smad3, which led to a dramatic decrease in nuclear staining and overall loss of fluorescence due to diffusion of the exported protein from the permeabilized cytoplasm (Fig. 2A). We also observed a similar export of wild-type GFP-Smad3, although the import of this protein to the nucleus was inefficient (data not shown) due to the poor association of nonphoshorylated Smad3 to importin-β1 (14). The export was dependent on energy and temperature, as it was not observed when ATP was depleted by incubation with sodium azide or when permeabilized cells were incubated on ice, respectively (Fig. 2A). Pretreatment of cytosol with LMB prior to incubation could not inhibit the export of GFP-Smad3D. These data suggest that the in vitro export of Smad3 requires cytosolic factors other than CRM1.
To analyze the required cytosolic factors, we performed in vitro reverse import assays with recombinant proteins instead of cytosol. This time, the nuclei of permeabilized HeLa cells were loaded with GFP-Smad3D with the help of importin-β1, Ran-GDP, and the specific importin for Ran, p10, as we have previously shown (14). When cells were washed and further incubated with Ran-GDP and p10 for time periods of 10 min or longer, we observed a measurable decrease of the nuclear GFP-Smad3D, which became more significant after 20 min of incubation (Fig. 2B). When a control protein containing an NLS and an NES sequence (GST-NLS-NES-GFP) was used, similar nuclear decrease was observed. On the other hand, when the cells were incubated with CRM1 in the absence or presence of Ran-GDP and p10, we did not observe any significant export of GFP-Smad3D, whereas the control GST-NLS-NES-GFP protein was exported in the presence of CRM1, Ran-GDP, and p10. Similar to CRM1, exportin-t, a specific exportin for tRNA (2, 17), which served as a negative control, failed to support export of Smad3D. These data suggest that Ran is required for Smad3 export, and a specific exportin for Smad3 must mediate this transport.
To confirm the Ran dependency of Smad3 nuclear export, we investigated the effect of the RanT24N mutant, which exists exclusively in its GDP form because it binds tightly to the guanine exchange factor RCC1 (regulator of chromosome condensation 1), resulting in its inactivation (4). When permeabilized cells were incubated with RanT24N-GDP and p10, we could not observe any decrease of nuclear GFP-Smad3D (Fig. 2C); this mutant also blocked the export of the control GST-NLS-NES-GFP protein. This shows that in vitro nuclear export of Smad3 is dependent on the GTP form of Ran.
An active Ran cycle supports Smad3 export.
To demonstrate the involvement of the GTP form of Ran in vivo, we coinjected RanGAP1, which stimulates the hydrolysis of Ran-GTP, together with GFP-Smad3 into nuclei (Fig. 3A). Microinjection of RanGAP1 inhibited the nuclear export of GFP-Smad3 as well as of the control GST-NES-GFP protein, supporting the conclusion that Ran-GTP is important for the export of Smad3. To further verify the Ran-GTP dependency, we used the hamster tsBN2 cell line, which has a temperature-sensitive point mutation in the Ran guanine nucleotide exchange factor RCC1 (30). When GFP-Smad3 or GFP-Smad3D was microinjected into the nucleus at the permissive temperature (33.5°C), both proteins were exported to the cytoplasm, similar to the control GST-NES-GFP protein (Fig. 3B). In contrast, at the nonpermissive temperature (39.5°C) neither Smad3 nor the control protein was exported. Together, these data establish firmly that Smad3 requires the GTP form of Ran for its export.
In order to verify that inactivation of the Ran cycle in the tsBN2 cells that carry the temperature-sensitive RCC1 mutation could also affect the subcellular distribution of endogenous Smad3, we analyzed the levels of nuclear accumulation of Smad3 and C-terminally phosphorylated Smad3 in tsBN2 cell fractions highly enriched for nuclear proteins (Fig. 3C). Immunoblot analysis for endogenous Smad3 and C-terminally phosphorylated Smad3, using specific antibodies during time course experiments in response to TGF-β1, revealed proper nuclear accumulation of both Smad3 and its phosphorylated form as early as 1 h after TGF-β1 stimulation when cells were grown at the permissive temperature (33.5°C). These nuclear levels declined with time quite significantly, which is consistent with the model of Smad3 export at late time points post-TGF-β treatment. As predicted from all previous experiments, inactivation of RCC1 by shifting the temperature to 39.5°C completely blocked the export of nuclear Smad3 as late as 4 h post-TGF-β stimulation (Fig. 3C and D). Interestingly, under the same conditions, nuclear import of endogenous Smad3 occurred to a large extent, demonstrating that nuclear export is more sensitive to perturbation of RCC1 activity than nuclear import. However, a small reduction of nuclear phospho-Smad3 and total Smad3 levels was observed 8 h poststimulation, even after inactivation of RCC1. This result is consistent with proteasomal degradation of nuclear Smad3 at late points post-TGF-β signaling, as has been demonstrated previously for both Smad2 and Smad3 (11). We conclude that endogenous nuclear Smad3 (and its phosphorylated form) is actively exported to the cytoplasm in a Ran-dependent manner.
Exportin 4 associates with Smad3.
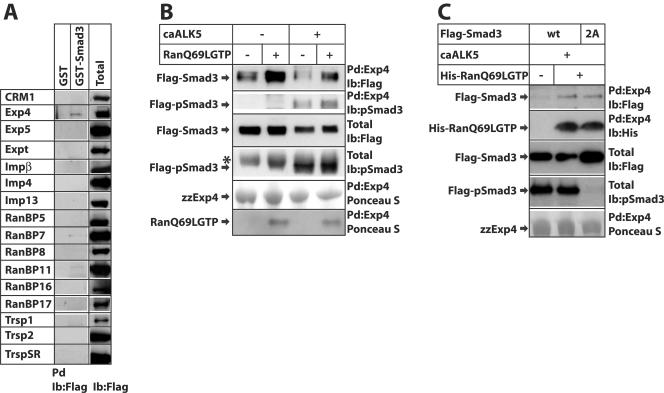
In order to identify a specific transporter for Smad3 nuclear export, we performed a protein-protein interaction screen for putative members of the importin-β family that includes both importins and exportins (7, 19) and looked for proteins that would specifically interact with the MH2 domain of Smad3 in a Ran-dependent manner. For this reason we performed pull-down experiments using 16 family members of Flag-tagged mammalian transportins expressed in 293T cells and GST-Smad3 or GST alone as a negative control, incubated in vitro in the presence of constitutively active RanQ69L-GTP, and screened by immunoblotting with the Flag antibody (Fig. 4A). We could score clear and reproducible association between Smad3 and exportin 4, an exportin previously shown to transport eukaryotic initiation factor eIF-5A from the nucleus to the cytoplasm (18). Occasionally, we could also observe very weak but detectable association of Smad3 with RanBP7 and RanBP11; however, over the course of several repeats of this experiment, we could not obtain reproducible and convincing evidence that these proteins interacted with Smad3 in a specific manner. All other tested transportins failed to show specific interaction with Smad3 in the presence of constitutively active RanQ69L-GTP (Fig. 4A). Importin-β1 served as a control for the specificity of the reaction, as this transportin interacts with phosphorylated Smad3 and Ran-GTP dissociates the complex (14). Accordingly, in the presence of RanQ69L-GTP, we could not observe any significant association of importin-β1 with GST-Smad3 (Fig. 4A).
FIG. 4.
Exportin 4 associates with Smad3 in a Ran-dependent manner. (A) Pull-down assay for the screening of Smad3-interacting transportins. The indicated 16 Flag-tagged transportins were expressed in 293T cells, and total cell extracts of transfected cells were incubated with GST or GST-Smad3 beads in the presence of 2 μM recombinant His-tagged RanQ69L-GTP prior to immunoblotting with anti-Flag antibody. Aliquots of the transfected cell lysate are also immunoblotted with the anti-Flag antibody for reference of transportin expression (Total). (B) Pull-down assays using zzExp4-IgG beads and total cell extracts from transiently transfected 293T cells with N-terminal Flag-tagged full-length Smad3 in the absence (−) or presence (+) of a constitutively active TGF-β type I receptor, ALK5 (caALK5). The association reactions were performed in the absence (−) or presence (+) of 2 μM recombinant purified His-tagged RanQ69L-GTP. Immunoblot analysis of proteins bound to the exportin 4 beads (Pd) and of total cell extracts (Total) was performed using the anti-Flag or the anti-phospho-Smad3 antibodies. Ponceau S staining of the immunoblots revealed the recombinant proteins used. Arrows indicate the relevant protein bands, and an asterisk marks a nonspecific band. (C) Pull-down assays performed as described for panel B, except that the transfected Flag-Smad3 was either full-length wild type (wt) or carried substitutions of the two C-terminal serines to alanines (2A), and the recombinant His-tagged RanQ69L-GTP was detected in the pull-down assays using an antihistidine antibody instead of staining by Ponceau S. Ib, immunoblot.
We then examined whether C-terminal phosphorylation of Smad3 induced by coexpression of the constitutively active TGF-β type I receptor (also known as activin receptor-like kinase 5 [ALK5]) had any effect on the interaction with exportin 4 (Fig. 4B and C). Coexpression of caALK5 led to robust levels of phosphorylated Smad3, as detected by immunoblotting with antibody specific for the C-terminal phosphorylated di-serine motif of Smad3 (Fig. 4B and C). Phosphorylated Smad3 interacted with exportin 4, albeit not as strongly as unphosphorylated Smad3. The nonphosphorylatable Smad3(2A) mutant associated with exportin 4 as efficiently as wild-type Smad3 (Fig. 4C). This suggests that exportin 4 may preferentially associate with unphosphorylated Smad3, which corroborates the findings of in vivo Smad3 export in microinjected cells (Fig. 1D). The association of Smad3 and exportin 4 was strongly enhanced by coincubating the two proteins with the RanQ69L-GTP mutant that is constitutively locked in the active, GTP-bound form (Fig. 4B and C). These results are in agreement with the in vitro and in vivo export data of Fig. 1 and 2 and suggest that Ran-GTP enforces a high-affinity interaction between exportin 4 and Smad3.
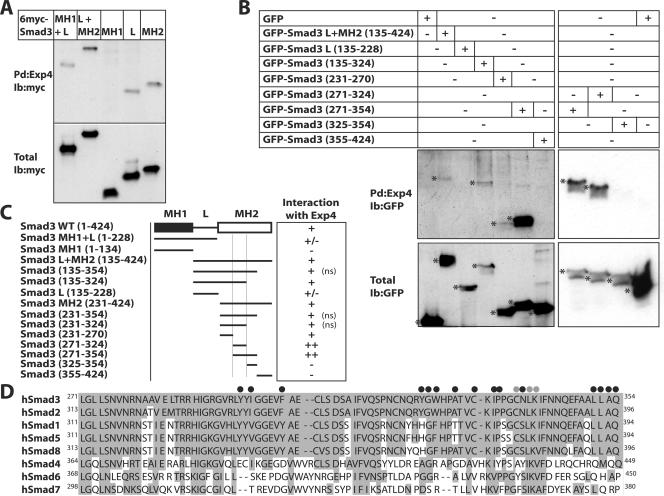
We then mapped the domain of Smad3 that associated with exportin 4 with high affinity and specificity (Fig. 5A). While the MH1 domain of Smad3 failed to bind to exportin 4, both the linker and the MH2 domains showed significant association and a protein fragment consisting of both the linker and the MH2 domain exhibited the strongest interaction. In contrast, a fragment encompassing the MH1 and linker domains of Smad3 showed weaker interaction with exportin 4 compared to the linker domain alone. In order to delineate a minimal peptide motif in Smad3 that was required for interaction with exportin 4, we created a large panel of GFP-Smad3 linker+MH2 domain deletion mutants and tested their interaction with recombinant exportin 4 in the presence of RanQ69L-GTP (Fig. 5B and C). Trimming down the Smad3 MH2 domain into a short peptide spanning amino acids 271 to 354 (Fig. 5C and D) resulted in the strongest interaction observed with exportin 4 in the presence of Ran-GTP. An even shorter peptide, amino acids 271 to 324, also showed the same strong interaction with exportin 4 (Fig. 5B and C). We conclude from this analysis that a core peptide within the center of the Smad3 MH2 domain is required for specific association with exportin 4 in a Ran-dependent manner. The MH2 sequence mapped as the exportin 4 interaction motif is well characterized structurally and functionally and is highly conserved between Smad3 and Smad2 and between the BMP-specific R-Smads (Fig. 5D).
FIG. 5.
Mapping the domain of Smad3 that interacts with exportin 4. (A) Pull-down assays in the presence of RanQ69L-GTP using exportin 4 beads (zzExp4-IgG) and total cell extracts from transiently transfected 293T cells with the indicated mammalian expression vectors for various deletion mutants of Smad3 that were tagged at their N termini with the 6myc epitope. (B) Pull-down assays as described for panel A with deletion mutants of the Smad3 linker (L) and MH2 domain N-terminally tagged with GFP. Asterisks indicate the relevant protein bands in the pull down (Pd) and total cell extract immunoblots. (C) Summary of the interaction domains of Smad3 to exportin 4. The minimal MH2 peptide sequence required for interaction is marked between two thin vertical lines. The interaction table is based on the data of panels A and B and of experiments not shown (ns). (D) Amino acid sequence of the human Smad3 fragment that interacts with exportin 4, residues 271 to 354, and alignment with all other members of the human Smad (hSmad) family. Amino acid conservation within the family is indicated by gray shading. Black circles above the Smad3 sequence mark critical amino acids mapped by crystallography and mutagenesis in Smad2 as responsible for the interaction of Smad2 with the adaptor protein SARA and transcription factors of the FoxH1 family (25, 33). Gray circles mark amino acids which, when mutated in Smad2, block its nuclear export (41). Ib, immunoblot; WT, wild type.
Exportin 4 mediates nuclear export of Smad3 in vitro and in vivo.
In order to demonstrate that exportin 4 indeed can act as a specific exportin for Smad3, we repeated the in vitro reverse import assays of Fig. 2B, but instead of crude cytosolic extracts we used fully purified recombinant proteins (Fig. 6A). In these assays we used shorter incubation periods of 6 min instead of 20 min (see Fig. 2B and C), because at short time intervals (0 to 10 min) the combination of Ran and p10 was not sufficient to export Smad3 efficiently to the cytoplasm (Fig. 5A). It can be readily observed that whereas buffer alone or Ran plus p10 induce weak export of GFP-tagged Smad3D within the 6-min period, inclusion of recombinant exportin 4 led to quantitative export of Smad3D within the same short time period. As a negative control, we repeated the experiment by replacing exportin 4 with CRM1, which fails to support Smad3 export (Fig. 6A) or interact with Smad3 with high affinity (Fig. 4A). In order to analyze the ability of exportin 4 to carry nuclear export of Smad3 in a more quantitative manner, we performed kinetic experiments of in vitro export and counted the number of nuclei that stained positively for GFP-Smad3D (Fig. 6B). During the course of 15 min, transport buffer was ineffective in sustaining Smad3 export. Ran could induce time-dependent export of Smad3, especially during the 10- to 15-min interval, which demonstrates the presence of weak but functional exportin activity in the nuclei of the permeabilized cells. Finally, exportin 4 together with Ran catalyzed rapid and time-dependent export of Smad3 which was evident and significant as early as 5 min. The Smad3 export catalyzed by exportin 4 in vitro was clearly dependent on Ran, since replacing wild-type Ran with its inactive mutant RanT24N completely failed to support Smad3 export despite the presence of exportin 4 (Fig. 6C). Exclusion of Ran from the transport reaction in the presence of exportin 4 also led to undetectable Smad3 export. These results are in full agreement with the in vivo analysis of Smad3 export and demonstrate that Ran is a limiting component in the permeabilized cell system. On the basis of these data, we conclude that exportin 4 is sufficient for the export of Smad3 and cooperates with the Ran GTPase to execute this process.
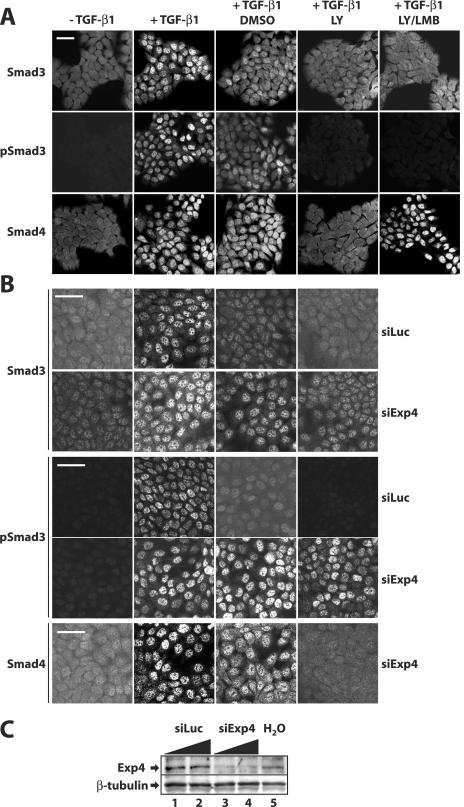
In order to demonstrate that endogenous exportin 4 is necessary for the export of endogenous Smad3, we employed an siRNA knockdown approach using a pool of four RNA oligonucleotides specifically targeting human exportin 4 (see Materials and Methods). In order to study nuclear export of endogenous Smad3, we adapted a protocol based on the immortalized human HaCaT keratinocyte cell system as previously described (24). In HaCaT cells starved from serum for 24 to 48 h, all of the endogenous Smad3 (and Smad4) was distributed primarily in the cytoplasm and weakly in the nucleus (Fig. 7A). Under the same conditions, no specific signal for C-terminally phosphorylated Smad3 (pSmad3) could be detected. Upon stimulation with 2 ng/ml TGF-β1 for 1 h, the majority of endogenous Smad3 (and Smad4) accumulated in the nucleus, while strong nuclear pSmad3 signals were also recorded. After the 1-h stimulation with TGF-β1, the ligand was removed and the cells were incubated in medium in the presence of DMSO for another 2 h (Fig. 7A). During the 2-h interval we could readily observe a gradual decrease of the nuclear Smad3 (or pSmad3 and Smad4) signal with redistribution towards the cytoplasm, representing active nuclear export of these proteins as previously described (24). This active export was even more enhanced when the endogenous TGF-β receptor system was shut down by the use of the type I receptor kinase-specific inhibitor LY580276 (Fig. 7A). In the presence of the inhibitor, HaCaT cells return to the resting state and endogenous Smad3 (and Smad4) redistributes primarily to the cytoplasm, whereas the pSmad3 signal is completely lost. Coincubation of cells with the type I receptor inhibitor LY580276 and the CRM1 inhibitor LMB left the distribution of endogenous Smad3 (and pSmad3) unchanged, while it enforced nuclear accumulation of Smad4 despite the absence of any signal from the TGF-β receptor, as previously described (24). The above system appeared sensitive and robust enough to ask whether the apparent export of endogenous Smad3 was sensitive to knockdown of endogenous exportin 4. As a specificity control, we also measured the distribution of endogenous Smad4. Transfection of HaCaT cells under conditions where more than 90% of the cells were incorporating the control siRNA oligonucleotides against luciferase (siLuc) exhibited the same pattern of endogenous Smad3 distribution as described above (Fig. 7B). The same was true for endogenous pSmad3 and Smad4 (unpublished results). Upon transfection of the exportin 4-specific siRNA, a knockdown on the order of 85% was reproducibly achieved (Fig. 7C) while no obvious change to the nuclear accumulation of Smad3, pSmad3, or Smad4 in response to TGF-β1 could be observed, confirming that knockdown of exportin 4 was not perturbing nonspecifically the overall nucleocytoplasmic shuttling machinery (Fig. 7B). However, after removal of the ligand and especially after inhibition of the type I receptor kinase with LY580276, export of endogenous Smad3 could not be detected and the protein remained constantly in the nucleus. The nuclear pSmad3 also remained constitutively nuclear under the same conditions of exportin 4 knockdown (Fig. 7B), strongly suggesting that exportin 4 is required for the loss of nuclear pSmad3 levels upon shutdown of TGF-β receptor signaling. Under the same knockdown conditions, endogenous Smad4 redistributed to the cytoplasm, further confirming the specificity of the knockdown, which could not perturb the normal export of Smad4 that is catalyzed by CRM1. The nuclear accumulation of endogenous Smad3 and pSmad3 after knockdown of exportin 4 could be observed even 6 or 8 h postremoval of TGF-β1 from the culture medium (unpublished results). These data demonstrate that endogenous exportin 4 is a critical mediator of nuclear export of Smad3 and reinforce the conclusion that Smad3 export is an active process whose presence is revealed robustly upon shutting down the endogenous TGF-β signaling pathway.
FIG. 7.
Endogenous exportin 4 is required for nuclear export of Smad3. (A) HaCaT cells were treated with vehicle (−TGF-β1) or with 2 ng/ml TGF-β1 for 1 h (+TGF-β1) and with 2 ng/ml TGF-β1 for 1 h followed by 2 h of incubation with DMSO or with the specific ALK5 inhibitor LY580276 (LY) alone or in combination with 8 ng/ml LMB. Cells were immunostained for endogenous Smad3, C-terminally phosphorylated Smad3 (pSmad3), with Smad4 as control. (B) HaCaT cells were transiently transfected with 25 nM control (siLuc) or exportin 4-specific (siExp4) for 48 h prior to treatments with compounds as in panel A and immunofluorescence for the indicated proteins. Bars correspond to 10 μm. (C) Immunoblot analysis of the same cell monolayers used in panel B with anti-exportin 4 and anti-β-tubulin (control) antibodies. Cells were transfected with increasing amounts of siRNA oligonucleotides (25 nM and 100 nM; triangles) and treated with vehicle (−) or with 2 ng/ml TGF-β1 for 1 h.
In vivo export of Smad3 requires its interaction with exportin 4.
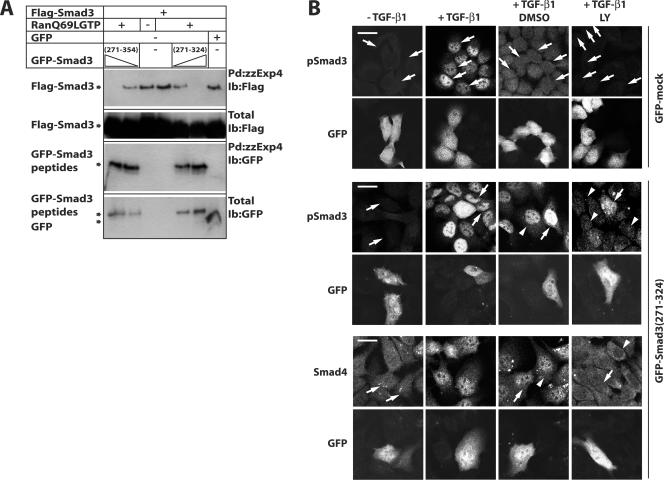
In order to demonstrate that the molecular interaction between exportin 4 and Smad3 was important for the nuclear export of Smad3, we took advantage of the fine mapping of the Smad3 interface with exportin 4 (Fig. 5). We reasoned that transfection of cells with the interface peptide (amino acids 271 to 354) could saturate the binding to endogenous exportin 4, thus limiting this transportin from access to the endogenous Smad3. If this were true, then the peptide should act as a dominant-negative inhibitor and block the export of endogenous Smad3 in the HaCaT cell system described above. We first demonstrated that the interface peptides could competitively inhibit the interaction of Smad3 with exportin 4, using in vitro pull-down assays in the presence of the constitutively active RanQ69L-GTP (Fig. 8A). Indeed, overexpression of GFP-Smad3(271-354) and GFP-Smad3(271-324) dose dependently blocked the in vitro interaction of Smad3 with recombinant exportin 4.
FIG. 8.
Interaction between Smad3 and exportin 4 is crucial for the nuclear export of Smad3. (A) Pull-down assays in the presence of RanQ69L-GTP using exportin 4 beads (zzExp4-IgG) and total cell extracts from transiently transfected 293T cells with Flag-Smad3 (full length) and different doses of GFP-Smad3 fragments (amino acids 271 to 354 and 271 to 324). (B) HaCaT cells were transiently transfected with pEGFP (GFP-mock) or pEGFP-Smad3(271-324) and 24 h later were treated exactly as described in the legend to Fig. 7 prior to immunofluorescence microscopy for endogenous phospho-Smad3 and Smad4 as control. GFP indicates autofluorescence of transfected cells in each field. Bars correspond to 10 μm, white arrows point to transfected nuclei, and white arrowheads point to nontransfected nuclei. Ib, immunoblot; LY, LY580276.
Based on these results, we transiently transfected control GFP or the GFP-Smad3(271-324) peptide in HaCaT cells and then repeated the TGF-β1 stimulation followed by ligand withdrawal and receptor kinase inhibition (Fig. 8B). Transient transfection of HaCaT cells with plasmid DNA was only 15 to 30% efficient, in contrast to the very efficient (>90%) transfection of siRNA oligonucleotides. Despite this limitation, we could readily detect transfected cells based on their GFP autofluorescence and directly compared them to the neighboring nontransfected cells. For convenience, and in order to avoid possible cross-reactivity of our anti-Smad3 polyclonal antibody with the transfected Smad3 peptides, we monitored the levels of endogenous pSmad3 and of Smad4 as a negative control (Fig. 8B). Control GFP transfection did not perturb the normal processes of pSmad3 nuclear accumulation and its subsequent loss from the nucleus. In contrast, transfection of GFP-Smad3(271-324) did not perturb nuclear accumulation of endogenous pSmad3 but characteristically resulted in persistent nuclear residence of this protein, even when the endogenous type I receptor kinase was fully blocked. Under the same conditions, endogenous Smad4 was free to shuttle and redistribute to the cytoplasm, confirming that the Smad3 peptide was acting in a specific manner and did not affect the independent exportin CRM1. We therefore conclude that inhibitory peptides that block the interaction between Smad3 and exportin 4 are effective inhibitors of the active Smad3 nuclear export process. This strongly supports the model that Smad3 nuclear export depends on its physical interaction with the specific exportin 4, identified here for the first time.
DISCUSSION
Despite current attempts to describe the molecular pathways that guide Smad protein nucleocytoplasmic shuttling, many critical steps are not yet understood (21, 43). The nuclear import mechanisms of Smad1, Smad2, Smad3, and Smad4 have already been described (14, 36, 37, 39-42). One established model relies on the transporter importin-β1, which mediates nuclear import of Smad3 independent of importin-α (14, 36) or import of Smad4 via an importin-α/importin-β complex (39). In both Smad3 and Smad4, importins recognize NLSs in the MH1 domain of Smads (39). The second model of carrier-independent nuclear import of Smads is based on the interaction of the Smad MH2 domain with the phenylalanine- and glycine-rich repeats of nucleoporins. This model was originally demonstrated to mediate Smad2 import (40) but recently has also been extended to Smad3 and Smad4 (42). This raises the possibility that Smads utilize multiple mechanisms of nuclear import that rely on structural signals in both of their MH1 and MH2 domains, possibly as a means to control more accurately the need for continuous shuttling of these proteins between the cytoplasm and the nucleus (21, 27). It is currently unknown whether the two current models of Smad nuclear import apply to distinct sets of Smad complexes, such as monomeric Smads, homooligomeric Smads, or heterooligomeric Smads, for example.
Recently, the nuclear export of the BMP-specific Smad1 was shown to be carried out by CRM1 via recognition of multiple NESs (37, 38). However, the nuclear export of the TGF-β R-Smads has remained poorly characterized (27, 43). For these reasons, we have carefully examined the nuclear export of one TGF-β R-Smad, Smad3, whose nuclear import mechanism we previously elucidated (14). Our data demonstrate the ability of Smad3 to be exported from the nucleus via a mechanism that involves Ran and the novel exportin 4. The microinjection approach allowed us to assign an intrinsic export function to recombinant Smad3 (Fig. 1). This activity resides in the MH2 domain (Fig. 1B and C) and is not inhibited by LMB (Fig. 1A) as previously reported (10, 24). The LMB-independent export of Smad3 indicates that it must rely on protein motifs that are distinct from the well-characterized CRM1-dependent NES of Smad4 that resides in a unique segment of its linker region (24, 32) or the CRM1-dependent NES defined in Smad1 (37, 38). Fine mapping of the interaction between Smad3 and exportin 4 resulted in the delineation of a minimal peptide sequence within the Smad3 MH2 domain that is required for this association (Fig. 5). The region spanning amino acids 271 to 324 in Smad3 (Fig. 5D) has been previously analyzed, as it contributes to the proper folding of the MH2 domain in Smad3 but also in Smad2 and Smad4, as revealed by crystallographic studies (3, 28, 34). In addition, this region forms an interface used repeatedly in Smad signaling processes, such as the interaction between Smad2 and its adaptor protein Smad anchor for receptor activation (SARA) or the interaction between Smad2 and forkhead/winged helix transcription factors of the Foxh1 family (25, 26, 33). The exportin 4-interacting peptide is highly conserved among all R-Smads and to a lesser extent among the co-Smad, Smad4, and the inhibitory Smads, Smad6 and Smad7 (Fig. 5D). In favor of the importance of this sequence in processes of Smad protein shuttling, specific mutations of Cys380, Leu382, and Lys383 in Smad2 specifically impair nuclear export of Smad2 (41). These three amino acids are fully conserved among all R-Smads and fall towards the C-terminal border of the peptide motif identified here for Smad3. Unlike CRM1-dependent nuclear export, which utilizes a leucine-rich motif as the NES in the cargo protein, Smad3(271-354) does not have any apparent leucine-rich motif, suggesting that the NES used by exportin 4 may be a three-dimensional motif, similar to the hydrophobic pocket recognized by SARA or the FoxH1 transcription factors.
Our work has provided clear evidence for the role of the Ran GTPase in both the import (14) and export (Fig. 2, 3, and 6) of Smad3. This is in accordance with the established model of import and export of proteins (7). It also underscores the necessity of specific adaptor proteins such as importins and exportins to mediate the distinct translocations of Smad proteins into and out of the nucleus. It is of interest that at least in the case of Smad3, the import-associated function maps to the MH1 domain, whereas the export-associated function maps to the MH2 domain. Establishing whether Ran is a critical cofactor for the shuttling of other Smad proteins, such as Smad1, Smad2, and Smad4, is important, as this may allow the formation of a unified model of Smad nucleocytoplasmic shuttling.
The identification of exportin 4 as a transporter of Smad3 results in this exportin acquiring a second role in nucleocytoplasmic transport. Its first role was identified when exportin 4 was discovered and is the export of a specific cofactor of the translational initiation machinery, eIF-5A (18). In eIF-5A, exportin 4 recognizes an unusual structural motif of the protein that carries a modified lysine residue named hypusine. This posttranslational modification of eIF-5A enhances the recognition of this cargo protein by exportin 4 dramatically (18). Whether such posttranslational modification also occurs on one of the two lysine residues of the MH2(271-354) subdomain of Smad3 currently remains unknown. It also remains to be tested in future studies whether exportin 4 can carry out nuclear export of additional Smad members. This could be suggested based on the high degree of conservation of the MH2(271-354) subdomain among all R-Smads (Fig. 5D). It is worth noting that exportin 4 was the only protein that scored clearly and reproducibly in a positive manner in the Smad3 protein-protein interaction screen that we performed (Fig. 4A). Under these conditions, importin-β1 failed to interact as expected, since Ran-GTP leads to the dissociation of importin-β1-Smad3 complexes (14). However, a few other transportins could show weaker and less reproducible associations with Smad3 in a Ran-GTP-dependent manner. Thus, it is formally possible that additional exportins may cooperate with Smad3 or other Smad proteins to elicit their efficient nuclear export.
The above hypothesis remains open, yet when human cells were depleted from endogenous exportin 4 via siRNA knockdown, the active export of Smad3 was undetectable (Fig. 7). This strongly suggests that exportin 4 plays a primary physiological role in the process of nucleocytoplasmic shuttling of Smad3. Furthermore, since short peptides that represent the Smad3 interface with exportin 4 act as dominant-negative mutants that interfere with proper shuttling of endogenous Smad3, we conclude that the interaction of nuclear Smad3 with exportin 4 in the presence of Ran-GTP is critical for the physiological export of Smad3 (Fig. 8). On the other hand, inhibition of exportin 4 did not affect the nuclear export of Smad4. The utilization of distinct exportins by these two Smad proteins that are known to form functional heterooligomers suggests that their nuclear export follows a dissociation process in the nucleus. This model is compatible with the hypothesis of an unknown nuclear phosphatase that leads to dissociation of R-Smad complexes with Smad4 (10, 41).
Based on the present work, we propose the following model for the regulation of Smad3's subcellular distribution. In unstimulated cells, Smad3 can shuttle in and out of the nucleus, but it primarily resides in the cytoplasm because of its constitutive export by exportin 4 and the presence of cytoplasmic anchor proteins. Ligand stimulation induces the phosphorylation-dependent association of Smad3 to importin-β1 and its import into the nucleus (14, 36). During passage through the nuclear pore, both importin-β1 and Smad3 are capable of interacting with specific nucleoporins (41), which mediates the efficient translocation of the protein complex towards the nucleus. Since phosphorylation counteracts the predominant export activity of Smad3, it is logical that dephosphorylation by cellular phosphatases would restore the constitutive export of Smad3. Such a dephosphorylation mechanism would also inhibit spontaneous reimport of Smad3 to the nucleus in the absence of rephosphorylation by the active type I receptor in the plasma membrane or endocytic vesicles as proposed by Inman et al. and Pierreux et al. (10, 24). Such a mechanism provides a means for the R-Smads to monitor the activation status of the receptor complex and thus propagate multiple rounds of signaling until the receptors stop being activated by TGF-β. An important open question is whether this model applies generally to all cell types responding to TGF-β or whether cells of different developmental origins can modulate their responsiveness to TGF-β superfamily members by regulating the nucleocytoplasmic shuttling of specific Smad proteins. Of equally high importance is the question of whether the nuclear export of Smad3, as described here, directly links to its degradation by proteasomes in the cytoplasm or whether it plays any direct regulatory role on the transcriptional functions of Smad3 in the nucleus. The identification of a new export pathway for Smad3 opens the path for detailed elucidation of all these critical aspects of regulation of this evolutionarily conserved signaling pathway.
Acknowledgments
The technical assistance of P. Lönn, K. Pardali, and M. Shono and the advice of K. Lin on structural details of Smad3 are gratefully acknowledged. We thank A. Shimamoto for various importin family expression vectors, M. Yoshida for providing leptomycin B, J. M. Yingling for the ALK5 inhibitor LY580276, M. Reiss for affinity-purified rabbit anti-phospho-Smad3 antibody, and G. Blobel, D. Görlich, S. Itoh, S. Kuroda, X. Liu, H. F. Lodish, I. W. Mattaj, K. Miyazono, P. ten Dijke, K. Yagami, and N. Yaseen for various bacterial and mammalian expression plasmids and adenoviruses.
This research was partly supported by grants from the Human Frontier Science Program (to A.M. and Y.Y.). A.K. was supported by a postdoctoral fellowship from the Swedish Foundation for International Cooperation in Research and High Education and by a grant from the Ichiro Kanehara Foundation.
REFERENCES
- 1.Adam, S. A., R. Sterne-Marr, and L. Gerace. 1992. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 219**:**97-110. [DOI] [PubMed] [Google Scholar]
- 2.Arts, G. J., S. Kuersten, P. Romby, B. Ehresmann, and I. W. Mattaj. 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17**:**7430-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chacko, B. M., B. Qin, J. J. Correia, S. S. Lam, M. P. de Caestecker, and K. Lin. 2001. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat. Struct. Biol. 8**:**248-253. [DOI] [PubMed] [Google Scholar]
- 4.Dasso, M., T. Seki, Y. Azuma, T. Ohba, and T. Nishimoto. 1994. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 13**:**5732-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng, X.-H., and R. Derynck. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21**:**659-693. [DOI] [PubMed] [Google Scholar]
- 6.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90**:**1051-1060. [DOI] [PubMed] [Google Scholar]
- 7.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15**:**607-660. [DOI] [PubMed] [Google Scholar]
- 8.Hieda, M., T. Tachibana, F. Yokoya, S. Kose, N. Imamoto, and Y. Yoneda. 1999. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J. Cell Biol. 144**:**645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamoto, N., T. Shimamoto, T. Takao, T. Tachibana, S. Kose, M. Matsubae, T. Sekimoto, Y. Shimonishi, and Y. Yoneda. 1995. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 14**:**3617-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inman, G. J., F. J. Nicolas, and C. S. Hill. 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell 10**:**283-294. [DOI] [PubMed] [Google Scholar]
- 11.Izzi, L., and L. Attisano. 2004. Regulation of the TGFβ signalling pathway by ubiquitin-mediated degradation. Oncogene 23**:**2071-2078. [DOI] [PubMed] [Google Scholar]
- 12.Kose, S., N. Imamoto, T. Tachibana, T. Shimamoto, and Y. Yoneda. 1997. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol. 139**:**841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kose, S., N. Imamoto, T. Tachibana, M. Yoshida, and Y. Yoneda. 1999. β-Subunit of nuclear pore-targeting complex (importin-β) can be exported from the nucleus in a Ran-independent manner. J. Biol. Chem. 274**:**3946-3952. [DOI] [PubMed] [Google Scholar]
- 14.Kurisaki, A., S. Kose, Y. Yoneda, C.-H. Heldin, and A. Moustakas. 2001. Transforming growth factor-β induces nuclear import of Smad3 in an importin-β1 and Ran-dependent manner. Mol. Biol. Cell 12**:**1079-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurisaki, K., A. Kurisaki, U. Valcourt, A. A. Terentiev, K. Pardali, P. ten Dijke, C.-H. Heldin, J. Ericsson, and A. Moustakas. 2003. Nuclear factor YY1 inhibits transforming growth factor β- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 23**:**4494-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutay, U., E. Izaurralde, F. R. Bischoff, I. W. Mattaj, and D. Görlich. 1997. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16**:**1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutay, U., G. Lipowsky, E. Izaurralde, F. R. Bischoff, P. Schwarzmaier, E. Hartmann, and D. Görlich. 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1**:**359-369. [DOI] [PubMed] [Google Scholar]
- 18.Lipowsky, G., F. R. Bischoff, P. Schwarzmaier, R. Kraft, S. Kostka, E. Hartmann, U. Kutay, and D. Görlich. 2000. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19**:**4362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67**:**265-306. [DOI] [PubMed] [Google Scholar]
- 20.Morén, A., S. Itoh, A. Moustakas, P. ten Dijke, and C.-H. Heldin. 2000. Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene 19**:**4396-4404. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas, F. J., K. De Bosscher, B. Schmierer, and C. S. Hill. 2004. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 117**:**4113-4125. [DOI] [PubMed] [Google Scholar]
- 22.Pardali, K., A. Kurisaki, A. Morén, P. ten Dijke, D. Kardassis, and A. Moustakas. 2000. Role of Smad proteins and transcription factor Sp1 in p21Waf1/Cip1 regulation by transforming growth factor-β. J. Biol. Chem. 275**:**29244-29256. [DOI] [PubMed] [Google Scholar]
- 23.Peng, S. B., L. Yan, X. Xia, S. A. Watkins, H. B. Brooks, D. Beight, D. K. Herron, M. L. Jones, J. W. Lampe, W. T. McMillen, N. Mort, J. S. Sawyer, and J. M. Yingling. 2005. Kinetic characterization of novel pyrazole TGF-β receptor I kinase inhibitors and their blockade of the epithelial-mesenchymal transition. Biochemistry 44**:**2293-2304. [DOI] [PubMed] [Google Scholar]
- 24.Pierreux, C. E., F. J. Nicolas, and C. S. Hill. 2000. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20**:**9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randall, R. A., S. Germain, G. J. Inman, P. A. Bates, and C. S. Hill. 2002. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J. 21**:**145-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall, R. A., M. Howell, C. S. Page, A. Daly, P. A. Bates, and C. S. Hill. 2004. Recognition of phosphorylated-Smad2-containing complexes by a novel Smad interaction motif. Mol. Cell. Biol. 24**:**1106-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reguly, T., and J. L. Wrana. 2003. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 13**:**216-220. [DOI] [PubMed] [Google Scholar]
- 28.Shi, Y., A. Hata, R. S. Lo, J. Massagué, and N. P. Pavletich. 1997. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 388**:**87-93. [DOI] [PubMed] [Google Scholar]
- 29.Siegel, P. M., and J. Massagué. 2003. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 3**:**807-820. [DOI] [PubMed] [Google Scholar]
- 30.Tachibana, T., N. Imamoto, H. Seino, T. Nishimoto, and Y. Yoneda. 1994. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J. Biol. Chem. 269**:**24542-24545. [PubMed] [Google Scholar]
- 31.Tachibana, T., M. Hieda, T. Sekimoto, and Y. Yoneda. 1996. Exogenously injected nuclear import factor p10/NTF2 inhibits signal-mediated nuclear import and export of proteins in living cells. FEBS Lett. 397**:**177-182. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe, M., N. Masuyama, M. Fukuda, and E. Nishida. 2000. Regulation of intracellular dynamics of Smad4 by its leucine-rich nuclear export signal. EMBO Rep. 1**:**176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, G., Y. G. Chen, B. Ozdamar, C. A. Gyuricza, P. A. Chong, J. L. Wrana, J. Massagué, and Y. Shi. 2000. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 287**:**92-97. [DOI] [PubMed] [Google Scholar]
- 34.Wu, J. W., M. Hu, J. Chai, J. Seoane, M. Huse, C. Li, D. J. Rigotti, S. Kyin, T. W. Muir, R. Fairman, J. Massagué, and Y. Shi. 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-β signaling. Mol. Cell 8**:**1277-1289. [DOI] [PubMed] [Google Scholar]
- 35.Xiao, Z., X. Liu, Y. I. Henis, and H. F. Lodish. 2000. A distinct nuclear localization signal in the N terminus of Smad3 determines its ligand-induced nuclear translocation. Proc. Natl. Acad. Sci. USA 97**:**7853-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, Z., X. Liu, and H. F. Lodish. 2000. Importin β mediates nuclear translocation of Smad 3. J. Biol. Chem. 275**:**23425-23428. [DOI] [PubMed] [Google Scholar]
- 37.Xiao, Z., N. Watson, C. Rodriguez, and H. F. Lodish. 2001. Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J. Biol. Chem. 276**:**39404-39410. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, Z., A. M. Brownawell, I. G. Macara, and H. F. Lodish. 2003. A novel nuclear export signal in Smad1 is essential for its signaling activity. J. Biol. Chem. 278**:**34245-34252. [DOI] [PubMed] [Google Scholar]
- 39.Xiao, Z., R. Latek, and H. F. Lodish. 2003. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 22**:**1057-1069. [DOI] [PubMed] [Google Scholar]
- 40.Xu, L., Y. G. Chen, and J. Massagué. 2000. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nat. Cell Biol. 2**:**559-562. [DOI] [PubMed] [Google Scholar]
- 41.Xu, L., Y. Kang, S. Cöl, and J. Massagué. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10**:**271-282. [DOI] [PubMed] [Google Scholar]
- 42.Xu, L., C. Alarcón, S. Cöl, and J. Massagué. 2003. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J. Biol. Chem. 278**:**42569-42577. [DOI] [PubMed] [Google Scholar]
- 43.Xu, L., and J. Massagué. 2004. Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 5**:**209-219. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., and Y. Xiong. 1999. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3**:**579-591. [DOI] [PubMed] [Google Scholar]