Envelope Glycoprotein Incorporation, Not Shedding of Surface Envelope Glycoprotein (gp120/SU), Is the Primary Determinant of SU Content of Purified Human Immunodeficiency Virus Type 1 and Simian Immunodeficiency Virus (original) (raw)
Abstract
Human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) particles typically contain small amounts of the surface envelope protein (SU), and this is widely believed to be due to shedding of SU from mature virions. We purified proteins from HIV-1 and SIV isolates using procedures which allow quantitative measurements of viral protein content and determination of the ratios of _gag_- and _env_-encoded proteins in virions. All of the HIV-1 and most of the SIV isolates examined contained low levels of envelope proteins, with Gag:Env ratios of approximately 60:1. Based on an estimate of 1,200 to 2,500 Gag molecules per virion, this corresponds to an average of between 21 and 42 SU molecules, or between 7 and 14 trimers, per particle. In contrast, some SIV isolates contained levels of SU at least 10-fold greater than SU from HIV-1 isolates. Quantification of relative amounts of SU and transmembrane envelope protein (TM) provides a means to assess the impact of SU shedding on virion SU content, since such shedding would be expected to result in a molar excess of TM over SU on virions that had shed SU. With one exception, viruses with sufficient SU and TM to allow quantification were found to have approximately equivalent molar amounts of SU and TM. The quantity of SU associated with virions and the SU:TM ratios were not significantly changed during multiple freeze-thaw cycles or purification through sucrose gradients. Exposure of purified HIV-1 and SIV to temperatures of 55°C or greater for 1 h resulted in loss of most of the SU from the virus but retention of TM. Incubation of purified virus with soluble CD4 at 37°C resulted in no appreciable loss of SU from either SIV or HIV-1. These results indicate that the association of SU and TM on the purified virions studied is quite stable. These findings suggest that incorporation of SU-TM complexes into the viral membrane may be the primary factor determining the quantity of SU associated with SIV and HIV-1 virions, rather than shedding of SU from mature virions.
The envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) are encoded by the respective viral env genes and synthesized as precursor proteins (44). In the endoplasmic reticulum, cotranslational glycosylation of the precursor protein generates a 160-kDa protein, where further assembly of the precursor into trimeric structures is presumed to take place (25, 56). Studies of other enveloped viruses, including both influenza virus and vesicular stomatitis virus, have shown that viral glycoproteins assemble into conformationally correct oligomeric structures prior to transport from the endoplasmic reticulum for further processing in the exocytotic pathway (24, 48). Cleavage of the HIV-1 and SIV precursor proteins at a conserved cleavage site by a cellular protease yields a surface (SU) glycoprotein of approximately 120 kDa and a transmembrane (TM) glycoprotein of approximately 41 kDa (gp41) that associate through noncovalent interactions (12, 13, 47). These trimeric structures are then transported to the surface of the cell, where they associate with forming virus particles during the assembly and budding process. Once on the surface of the virus, these envelope trimers are responsible for binding to specific cell surface receptors as well as for fusion events that allow the core proteins and viral genome to enter the cell during the infection process.
The number of envelope trimers per virion and surface density of SU on virions has been a subject of some controversy. Shortly after HIV-1 was isolated, several studies suggested that SU was shed from infected cells and/or from the virus soon after budding. Schneider et al. (46) reported that virus-free culture fluid contained at least 100-fold more SU than virus pelleted from the same volume of supernatant. We previously reported (43) that virus purified by sucrose gradient centrifugation contained little SU. Culture supernatants from HIV-1-infected cells and membranes of these cells contained high levels of SU and were used as sources of SU, rather than purified viruses.
Immunoelectron microscopic analysis using sera from HIV-1-infected patients showed little labeling of mature HIV-1 particles but heavy labeling of virus associated with cell membranes, suggesting that little SU is associated with mature virus. Based on electron microscopic analysis in which surface features on virions were assumed to represent envelope glycoprotein spikes, Gelderblom proposed an organization of spikes or knobs on the surface of virus budding from cells and postulated that an “ideal” well-preserved SIV or HIV particle was coated with 72 knobs or spikes (15). If the spikes represented trimeric structures of TM and SU, it followed that virus particles contained up to 72 × 3, or 216 copies of SU. It was also noted that the “spikes” were much less apparent on free virions, and it was suggested that much of the SU may “shed” from the particle during or shortly after maturation by a process that involves disruption of the noncovalent association between SU and TM.
On the other hand, Layne et al. (30) analyzed the SU content of purified mature HIV-1 and concluded that the Gag-to-Env ratio was about 40:1, with an average particle having ∼30 copies of SU or ∼10 “gp120 knobs” per particle. They also analyzed the rate of SU dissociation from purified virus and concluded that SU on the purified virus was reasonably stable and that the dissociation rate could account for only a small fraction of the “apparent” loss of SU. This seemed to support the suggestion that some of the SU on the viral surface was in a stable state and that a substantial portion of the SU originally associated with the virus was assumed to be labile and dissociated from the particles shortly after budding and/or during purification. Several investigators therefore interpreted these early observations to mean that SU is easily dissociated from the virus particle and that much of the free SU found in culture media from infected cells was at one time virus associated. The alternative hypothesis, that the relatively crude starting materials used for the analyses contained a significant fraction of SU that was not and had never been particle associated, was not evaluated.
We have employed protein purification and analysis techniques that allow quantitative recovery of proteins from purified viruses, including the Gag proteins as well as SU and TM. We have analyzed several strains of HIV-1 and SIV produced from a variety of cell lines and specifically focused on determining the approximate ratios of SU to TM and Gag to SU and TM. Physical parameters that might affect shedding of SU from the virus, such as temperature and centrifugation, were also examined.
MATERIALS AND METHODS
Viruses and reagents.
Unless otherwise noted, all viruses were produced from chronically infected cell lines. Viruses are designated according to the virus strain and cell line in which they were propagated (i.e., virus strain/cell line, cell log number; the AIDS Vaccine Program [AVP], Frederick, Md.): SIVMne/HuT-78 cl.E11S CL#63, SIV NC-MAC/Sup-T1 CL#130, SIV CP-MAC/Sup-T1 CL#131, SIVmac 239/CEMx174 CL#215, SIVsmE660/CEMx174 CL#186, HIV-1 including MN/H9 cl.4 CL#71, IIIB/BC7 (26), NL4-3/CEMx174 CL#151, JR-FL/Sup-T1-CCR5 CL30 CL#202, BAL/Sup-T1-CCR5 CL30 CL#204, and ADA-M/Sup-T1-CCR5 CL30 CL#203.
Dithiothreitol was from Calbiochem, La Jolla, Calif. Acetonitrile and H2O (UV grade) were obtained from EM Science; trifluoroacetic acid (HPLC/Spectra grade) was from Perkin Elmer (Applied Biosystems Division, Warrington, Great Britain). The 8 M guanidine-HCl was purchased from Pierce, Rockford, Ill.
Virus purification.
Virions were purified by sucrose density gradient ultracentrifugation after cells were removed from harvested cell cultures by tangential flow filtration using a 0.65-μm-pore-size membrane (Millipore Prostack) or by clarification using a Beckman J-6 M centrifuge model JS4.2 rotor for 30 min at 4,000 rpm. Batches of up to 30 liters were subjected to continuous-flow ultracentrifugation (Beckman continuous-flow rotor model CF32Ti) at 4°C in a 25 to 50% (wt/vol) sucrose (RNase free) density gradient in TNE (0.01 M Tris-HCl [pH 7.2], 0.1 M NaCl, and 1 mM EDTA in Milli-Q water) buffer. The sample was pumped at 5 liters per h with the centrifuge running at 30,000 rpm, and centrifugation was continued for 30 min after all of the sample was applied to allow banding of the virus. The rotor was decelerated to 3,000 rpm, and the gradient was displaced by pumping 55% sucrose into the rotor at 25 ml/min. UV-absorbing material in the gradient was detected by using a UV monitor (ISCO model UA-5) set at 280 nm (2 absorbance units, full scale) to detect proteins.
Fractions (25 ml each) were collected and analyzed for sucrose density using a refractometer (Bausch and Lomb). The virus-containing fractions, as determined by the UV profile and/or sucrose density (1.1612 to 1.1868 g of sucrose per ml), were pooled, diluted approximately 1:3 with TNE buffer, and centrifuged at 30,000 rpm for 1 h to pellet the virus. The resulting viral pellet was resuspended in TNE buffer to a final 1,000-fold concentration relative to the original volume of cell culture fluid. Aliquots were stored in liquid nitrogen vapor. To analyze components of the sucrose density gradient, a sample of each gradient fraction was diluted to less than 20% sucrose with TNE, centrifuged, and stored at −20°C for subsequent high-pressure liquid chromatography (HPLC) analysis.
Infectivity assays.
Samples were analyzed for the presence of infectious virus by incubating samples with AA2 CL.1 or AA2 CL.5 cells for detection of HIV-1 and SIV, respectively. Samples of viruses (500 μl) were added to 10-ml cultures containing 500,000 viable cells per ml and incubated at 37°C. After overnight incubation, the cells were washed three times and monitored for progeny virus production for the following 3 weeks by determining the concentration of capsid antigen of cell-free culture supernatants (HIV-1 p24CA and SIV p28CA antigen capture kits; AVP, National Cancer Institute [NCI] at Frederick, Frederick, Md.).
Freezing and thawing.
Frozen aliquots of sucrose-banded SIVMne(E11S) and HIV-1 MN (1,000×) were thawed in a 37°C water bath for 5 min. This was considered one freeze-thaw cycle. Additional freeze-thaw cycles were accomplished by placing samples in methanol-dry ice for 10 min and then thawing in a 37°C water bath for 5 min. Following the final thaw, samples were transferred to 72-ml polycarbonate centrifuge tubes containing 50 ml of sterile TNE buffer at 4°C. The tubes were centrifuged at 100,000 × g for 45 min. After careful decanting, supernatant was discarded, and the tubes were inverted on sterile gauze pads for 10 min to drain. Any residual fluid from the shoulders or lips of tubes was removed by using gauze pads and forceps. The pellets were carefully resuspended in 0.5 ml of TNE buffer and analyzed by HPLC and immunoblotting.
Heat treatment of SIVMne(E11S) and HIV-1 MN cl.4.
Sucrose density gradient-purified SIVMne(E11S) and HIV-1 (MN)/H9 cl.4 virus concentrates (1,000×) were diluted 1:10 in either sterile TNE buffer or complete medium (RPMI 1640 with 10% fetal bovine serum [FBS] and antibiotics). The protein concentrations (Bio-Rad DC protein assay using a bovine serum albumin [BSA] standard) of both diluted virus preparations were ∼0.28 mg/ml. Aliquots (1 ml) were dispensed into sterile, 2.0-ml tubes (Sarstedt catalog no. 72.694.006) and placed in an ice-water bath. Sets were then heat treated for 1 h at 50, 55, 60, or 65°C by immersion in a water bath. Sample temperature was monitored by a thermocouple (Honeywell) placed in a control tube containing 1 ml of water that was heated in parallel with the samples. Prior to use, the thermocouple was validated against a National Institute of Standards and Technology traceable calibrated mercury thermometer (serial no. HB/B38781) and found to be within 0.65°C of the thermometer reading at temperatures between 37°C and 70°C. Aliquots were incubated for 1 h after they had reached the selected temperature. Equilibration times were approximately 5 min for each set. After incubation at various temperatures, viral samples were centrifuged at 25,000 rpm for 60 min through a 20% sucrose-TNE pad (layer at the bottom of the tubes). Pellets were resuspended in TNE buffer (final concentration, 1,000-fold) and analyzed for SU content by using HPLC.
Treatment of virions with sCD4.
CHO cell-derived recombinant soluble human CD4 (sCD4), which is reactive with HIV-1 gp120 (K d < 10 nM) and inhibits HIV-1 binding to and infection of CD4+ T cells, was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health (Norbert Schuelke, contributor).
Purified SIVMne(E11S), HIV-1 MN/H9 cl.4, and HIV-1 NL4-3/CEMx174 were incubated for 2 h at 37°C in either the presence or absence of sCD4 (20 μg/ml) and centrifuged through 20% sucrose in a phosphate-buffered saline (PBS) pad at 25,000 rpm for 60 min. Pellets were resuspended in PBS buffer, disrupted in 8 M guanidine-HCl, and analyzed for SU retention by using HPLC and immunoblots.
HPLC separation and analysis of viral proteins.
Viral samples were disrupted in 8 M guanidine-HCl (Pierce, Rockford, Ill.) with or without 50 mM dithiothreitol (Calbiochem, La Jolla, Calif.) and fractionated by HPLC to isolate viral proteins. HPLC was performed at a flow rate of 300 μl/min on a Poros R2/H narrow-bore column (2.1 by 100 mm; Boehringer, Mannheim, Germany), using aqueous acetonitrile-trifluoroacetic acid solvents and a Shimadzu HPLC system equipped with LC-10AD pumps, an SCL-10A system controller, a CTO-10AC oven, an FRC-10A fraction collector, and an SPD-M10AV diode array detector. The gradient of buffer B (0.1% trifluoroacetic acid in acetonitrile) was 10% to 36.5%, 12 min; 36.5% to 37%, 4 min; 37% to 41%, 7 min; 41% to 70%, 12 min; and 70%, 5 min. A temperature of 55°C was maintained during HPLC separation. Peaks were detected by UV absorption at 206 and 280 nm and analyzed by sequencing using an automated Applied Biosystems Inc. 477 protein sequencer, by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and by immunoblot analysis using the enhanced chemiluminescence procedure (Amersham Life Science, Arlington Heights, Ill.). Quantitation of total viral and purified proteins was performed by amino acid analysis using a Hitachi L-8800 amino acid analyzer.
Immunoblot analysis of HIV-1 MN cl.4.
In adjacent gel lanes, three replicate aliquots of viral sample were loaded, and the gel (4 to 20% gradient) was calibrated with preparations of recombinant SU and p24 purified from the same virus by HPLC and quantitated by amino acid analysis. Gel electrophoresis was performed under nonreducing conditions. After proteins were transferred onto an Immobilon-P membrane (Millipore, Bedford, Mass.), SU and p24 were visualized simultaneously with polyclonal antisera generated against HIV-1 SU and monoclonal p24 antisera using enhanced chemiluminescence (Amersham, Arlington, Ill.). Densitometry was performed, and the quantities of SU and p24 in the virus were determined by interpolation onto standard curves produced from scanning of known input amounts of SU and p24 on the same blots. A Scan Ace III from Pacific Image Electronics was used to digitally scan in data in grayscale transparency mode from an immunoblot film in TIFF format. After scanning of membranes, band intensities were quantified using Scion Image analysis software (Scioncorp, Frederick, Md.). No image enhancement options that might have altered the relative intensity ratios of the bands being analyzed were used. Care was taken to use series of dilutions of sample which gave band intensities within the nonsaturated, linear response range of the optical hardware.
RESULTS
Analysis of SIVs. (i) Separation and quantitation of SIV proteins.
Stocks of virus concentrated to 1,000-fold were prepared as described in Materials and Methods prior to analysis. Viral infectivity titers from the starting material (clarified cell supernatant) and the resuspended virus pellets following sucrose banding and centrifugation indicated that there was minimal loss of infectivity (data not shown). To determine the amounts of Gag and Env glycoproteins retained on purified virus and to calculate the ratio of Gag to Env proteins in virus samples, we employed microscale HPLC methods. This approach allows viral proteins to be isolated, characterized, and quantitated in one combined procedure.
After purification, concentrated (1,000-fold) virions were disrupted in 8 M guanidine-HCl and subjected to HPLC separation under nonreducing conditions. For initial studies, we used SIVMne(E11S) (SIVMne/HuT-78 cl.E11S), which was produced from a single cell clone (E11S) obtained from HuT-78 cells infected with wild-type SIVMne and which we have previously shown contains large amounts of envelope glycoproteins (3). Proteins eluted from the HPLC column were detected by UV absorption, collected, and analyzed by SDS gel electrophoresis followed by Coomassie brilliant blue or silver staining, immunoblot analysis, mass spectrometry, and protein sequence analysis.
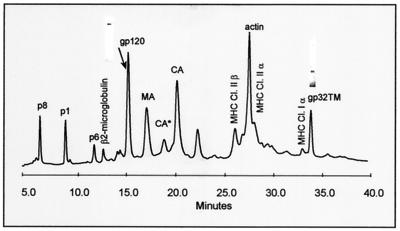
Figure 1 shows the HPLC elution profile of SIVMne(E11S). The viral Gag proteins p8NC, p1, p6, and p17MA and Env proteins SU and TM(gp32) were each eluted as single UV peaks (Fig. 1). The envelope genes of HIV-1 and SIV encode a 41-kDa form of the TM protein. Propagation of SIV in certain human T-cell lines, such as HUT-78, selects for isolates containing a premature stop codon within the cytoplasmic domain of the transmembrane envelope glycoprotein (23, 57). Propagation of SIVMne(E11S) in HUT-78 results in truncation of TM to yield a protein which migrates at approximately 32 kDa. The amount of purified protein in each peak was determined by amino acid analysis, and the molar amounts were calculated from the known amino acid sequence and composition of the protein. Molar ratios of the purified proteins recovered from the HPLC separation were calculated from these data.
FIG. 1.
HPLC analysis of sucrose density gradient-purified SIVMne(E11S). After purification, virions were disrupted in 8 M guanidine-HCl and separated by HPLC. Peaks were detected at 280 nm. Proteins were eluted from the column with an acetonitrile gradient as described in Materials and Methods and identified by SDS-PAGE, immunoblot, mass spectrometry, protein sequencing, and amino acid analysis. Protein purity of SU(gp120) and TM(gp32) peaks is shown as insertions on the HPLC profile. Bands were visualized by Coomassie staining. Two UV-absorbing peaks (labeled CA* and CA) were found to contain highly purified monomeric Gag p28CA protein. Subsequent analysis showed that following reduction with 2-mercaptoethanol, the protein in peak CA* eluted as CA, indicating that CA* contained a form of p28CA with at least one internal disulfide bond.
The molar ratios determined for the Gag proteins (p17MA, p28CA, p8NC, p6, and p1) were ∼1:1, in agreement with earlier observations (19, 20). Quantification of the proteins by amino acid analysis gave a ratio of Gag to Env of ∼6:1 and the estimated molar ratio of SU to TM(gp32) was approximately 1:1. The p6:SU and p6:TM ratios were 5.61:1 (±9%) and 5.58:1 (±4%), respectively. Based on an estimate of Gag molecules per viral particle ranging from 1,200 to 2,500 (5, 10, 30, 42, 49, 51), this Gag-to-Env ratio corresponds to an average of 65 to 140 trimeric complexes of SU (200 to 420 SU molecules) per virion. These data are not inconsistent with previous proposals that there may be 72 envelope trimers per virion (15). The finding of equimolar amounts of SU and TM does not support the suggestion that SU was shed from the SU-TM complex following maturation and during purification, as this would result in a molar deficiency of SU relative to TM.
(ii) Viral proteins in microvesicles.
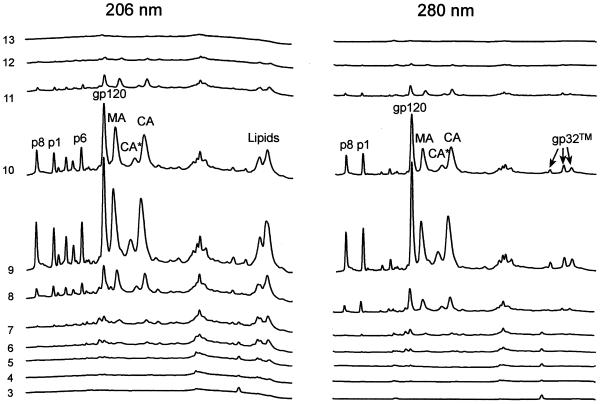
Microvesicles are heterogeneously sized membrane fragments shed from both uninfected and infected cells and are an inevitable contaminant of sucrose gradient-purified retrovirus preparations (4). CP-MAC (Y723C) is an SIV isolate known to express high levels of SU on the surface of infected cells (27-29). We used SIV CP-MAC to investigate whether SU or TM or both were incorporated into microvesicles and might thereby contribute to the measured SU content of purified virus preparations. SIV CP-MAC/Sup-T1 was prepared from infected cell culture supernatants processed as described in Materials and Methods, and fractions across the sucrose density gradient were collected and analyzed by HPLC.
Figure 2 shows that SU and TM were primarily recovered with fractions that also contained Gag viral proteins (p8NC, p1, p17MA, and p28CA) (fractions 8 through 11, predominantly fractions 9 and 10), suggesting that the vast majority of SU(gp120) and TM(gp32) bands with the virus. Equal aliquots from all fractions were analyzed by immunoblot blot with gp120, p28, and CD45 antibodies (data not shown). CD45 is highly expressed on both infected and uninfected cells and is well represented on microvesicles but is not incorporated into virions, and it thus served as a marker for microvesicles (8, 39). Immunoblot analysis revealed that CD45 was mostly present in fractions 6 and 7. In contrast, very little SU and no p28CA antibody-positive bands were found in these fractions, which contained the majority of the CD45. Thus, although microvesicles do contaminate purified virus preparations, the vast majority of the microvesicle-associated marker CD45 was present in fractions that contained little or no gp120, suggesting that SU is associated with virions but not microvesicles (4).
FIG. 2.
HPLC analysis of sucrose density gradient fractions from an SIV CP-MAC purification. Cell culture supernatants were processed as described in Materials and Methods. Fractions were collected from the sucrose density gradient after centrifugation. The percent sucrose was determined for each fraction. A 25-ml sample from each fraction was diluted 1:3 with TNE buffer and centrifuged at 100,000 × g to pellet any virus that may have been present. The pellets were resuspended in 8 M guanidine-HCl and analyzed for protein composition by HPLC under nonreducing conditions.
In addition to assessing the presence of SU glycoprotein in microvesicles, the HPLC profile for SIV CP-MAC also allowed evaluation of Gag-to-SU and SU-to-TM ratios for this virus. The ratios of integrated peak areas for the various Gag proteins to SU for SIV CP-MAC were equivalent to those shown in Fig. 1 for SIVMne(E11S), indicating that SIV CP-MAC also has a Gag-to-SU ratio of ∼6:1. The TM protein of SIV CP-MAC was eluted in three separate peaks, as shown in Fig. 2. Immunoblot analysis of the eluted protein showed monomeric TM protein in the first peak and disulfide-bonded homodimeric forms of TM in the last two peaks. The disulfide-bonded forms are likely due to spontaneous oxidations involving the Cys residue substituted for Tyr at position 723 in this strain. The ratio of the integrated peaks for SU to the combined integrated areas of the three peaks of TM protein in Fig. 2 is equivalent to the SU:TM ratio estimated for SIVMne(E11S) (Fig. 1). Thus, purified SIV CP-MAC, like SIVMne(E11S), has a ratio of SU to TM of ∼1:1 and represents a second example of a mature SIV that does not shed SU from the SU-TM complex during maturation and purification.
(iii) SU and TM content and SU:TM ratios for SIV isolates.
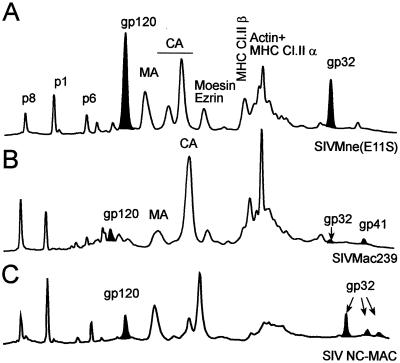
Figure 3 shows results for three different SIV isolates analyzed by the same HPLC method to assess the relative proportions of Gag and Env proteins in preparations of purified virus. Comparison of the peak areas and fraction analysis in the HPLC profiles were used to evaluate the content of the Gag, SU, and TM proteins. These data revealed three different patterns for purified SIV particles, as typified by the examples shown in Fig. 3.
FIG. 3.
Comparison of SIV preparations by HPLC analysis of SIVMne(E11S) (A), SIVmac239 (B), and SIV NC-MAC (C). After purification, virions were disrupted in 8 M guanidine-HCl and separated by HPLC. Peaks were detected at 280 nm. Proteins were eluted from the column with an acetonitrile gradient as described in Materials and Methods and analyzed by SDS-PAGE, immunoblot, mass spectrometry, protein sequencing, and amino acid analysis. The peaks corresponding to gp120 (SU) and the peaks identified as gp32 (TM) are denoted black. For SIVmac239 (panel B), both the gp32 and gp41 forms of TM are present, reflecting ongoing selection of truncated forms in the cultures used to produce the virus analyzed. The combined integrated areas of both peaks were used to calculate SU:TM ratios.
The first profile is characterized by high relative amounts of Env proteins, with Gag-to-Env proteins in ∼6:1 molar ratio and approximately equivalent molar amounts of SU and TM proteins; examples are SIVMne(E11S) and SIV CP-MAC (Fig. 3A). The second pattern (Fig. 3B) shows a low relative amount of envelope glycoproteins, with a Gag:Env ratio of ∼60:1 and both SU and TM proteins at low and approximately comparable molar amounts (Fig. 3B). Examples of SIV isolates with low levels of virion-associated SU and TM are SIVmac239 and SIVsmE660. The third profile is exemplified by SIV NC-MAC (Fig. 3C), which, like SIVMne(E11S) (Fig. 3A), also has a Gag:TM ratio of 6:1. However, SIV NC-MAC has an SU:TM ratio of approximately 1:10, rather than the roughly equimolar ratio seen for SIVMne(E11S). This profile is consistent with a virus that incorporated high relative amounts of SU and TM during assembly and budding but subsequently shed SU, presumably due to relatively lower affinity of the noncovalent interactions between SU and TM. The alternative hypothesis, that SIV NC-MAC represents a virus which may have acquired more TM than SU during assembly and budding, cannot be excluded but seems unlikely based on current understanding of retrovirus assembly. Of note, SIV NC-MAC is the only example we have identified to date that demonstrates this profile of a large amount of TM relative to Gag, but with a significant excess of TM over SU.
Analysis of HIV-1.
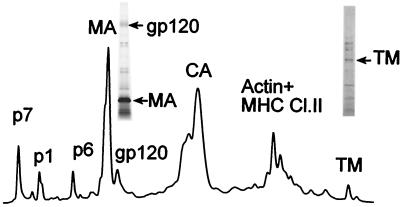
We also used the same approach to analyze multiple different isolates of purified HIV-1 virions, including HIV-1 MN cl.4, HIV-1 IIIB/BC7 (26), HIV-1 NL4-3/CEMx174, HIV-1 JR-FL/Sup-T1-CCR5 CL30, HIV-1 BAL/Sup-T1-CCR5 CL30, and HIV-1 ADA-M/Sup-T1-CCR5 CL30 (33). The representative HPLC elution profile shown in Fig. 4 for purified HIV-1 MN produced from clone 4 cells is typical of all HIV-1 samples analyzed. The Gag proteins (p17MA, p24CA, p7NC, p6, and p1) were identified by amino acid sequence analysis and were recovered in amounts consistent with equivalent molar amounts being present in virions, as reported previously (19). Proteins in various fractions (Fig. 4) were visualized by silver staining, and the presence of SU and TM in fractions was confirmed by immunoblot analysis.
FIG. 4.
HPLC analysis of HIV-1 MN/H9 cl.4. After purification, virions were disrupted in 8 M guanidine-HCl and separated by HPLC. Peaks were detected at 280 nm. Proteins eluted from the column with an acetonitrile gradient were analyzed by SDS-PAGE, immunoblot, mass spectrometry, protein sequencing, and amino acid analysis. After gp120 and gp41 were identified on the HPLC profile by immunoblot, fractions containing SU and TM were analyzed by SDS-PAGE followed with silver staining (insertions on HPLC picture).
Silver staining revealed that fractions containing SU and TM were mixtures of different proteins (Fig. 4, insets). Consequently, direct quantitation of Env proteins by amino acid analysis of these fractions to help determine the Env/Gag ratio was not feasible. However, inspection of the HPLC profile clearly indicated that the amount of TM protein present is quite small, making the corresponding Gag:TM ratio high relative to that in SIVMne(E11S). For multiple technical reasons, the HPLC profile for HIV-1 underestimates the actual amount of TM protein present. Not only does the TM(gp41)-containing peak reflect contributions from other non-TM proteins (Fig. 4), but the calculated extinction coefficient at 280 nm for HIV-1 TM(gp41) (107,090) is nearly twice that of SIV TM(gp32) (61,570). Thus, the TM(gp41) content of HIV-1 is very low compared to the TM(gp32) content of SIVMne(E11S), even lower than would be suggested by visual comparison of the TM peaks in Fig. 1 and 4. In fact, when the 280-nm extinction coefficients are taken into consideration, in order for HIV-1 to have a Gag:TM ratio similar to that of SIVMne(E11S), the area of the TM(gp41) peak would need to be at least 20 times greater than that observed.
Nevertheless, in all HIV-1 strains analyzed, only small quantities of SU and TM were found. The data suggest that the low levels of SU may in large part be determined by the amounts of SU-TM complexes incorporated during assembly. Separate experiments confirmed that the small amounts of TM observed by HPLC were not due to loss of the hydrophobic TM protein on the column, ruling out preferential loss of TM relative to SU and potential artifactual skewing of the SU:TM ratio on this basis (data not shown).
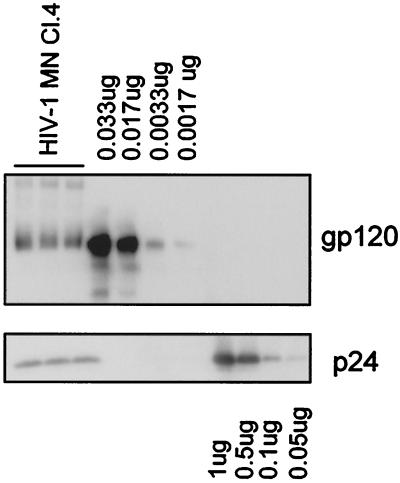
Given the above technical limitations in applying the analytical methods used for evaluation of SIV preparations to the evaluation of HIV-1 samples, semiquantitative immunoblot analysis based on serial dilutions was employed to estimate the Gag:Env ratio in HIV-1 MN/H9 cl.4 virus (Fig. 5). It was estimated that the analyzed virus contained ∼11 ng or 0.09 pmol of SU and ∼140 ng or 5 pmol of p24 per lane. This corresponds to an estimated p24/SU ratio of ∼60:1. Assuming that there are 1,200 to 2,500 Gag molecules in one viral particle, we thus estimate that HIV-1 MN/H9 cl.4 incorporates, on average, approximately 21 to 42 molecules of SU, or 7 to 14 trimeric SU-TM complexes per virion.
FIG. 5.
Analysis of SU and p24CA amounts in HIV-1 MN/H9 cl.4 virus. Virion-associated SU and p24CA levels were analyzed by immunoblot analysis under nonreducing conditions. Bands were visualized with mouse monoclonal antibodies prepared against purified p24 and gp120, followed by enhanced chemiluminescence staining. Band intensities were quantified by densitometry using Scion Image analysis software.
Freezing and thawing of SIVMne(E11S) and HIV-1 MN/H9 cl.4 results in no loss of Env proteins.
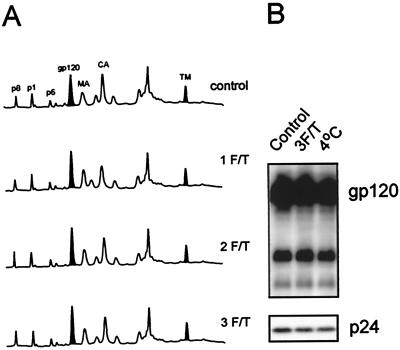
To examine physical parameters which may induce dissociation of SU from virions, purified virus was subjected to multiple freeze-thaw cycles (see Materials and Methods) and examined by HPLC (Fig. 6A) or immunoblot (Fig. 6B). Figure 6A shows an alignment of HPLC elution profiles for SIVMne(E11S) subjected to one, two, and three freeze-thaw cycles compared to a control viral sample. Figure 6B shows a comparison of a control sample of HIV-1 MN/H9 cl.4, a virus sample which was frozen and thawed three times, and a virus sample incubated at 4°C for 1 h. The amounts of pelletable SU and other viral proteins did not decrease as the number of freeze-thaw cycles increased. Taken together, these observations show that freezing and thawing, even after multiple cycles, did not result in loss of SU from either purified SIV or HIV-1.
FIG. 6.
Freezing and thawing of purified viruses does not result in shedding of SU proteins. Three aliquots of sucrose banded SIVMne(E11S) and HIV-1MN cl.4 (both 1,000-fold) were resuspended and thawed in a 37°C waterbath for 5 min. One aliquot was then immersed in an ice-water bath, completing one freezing and thawing (1 F/T) cycle. Another aliquot was frozen in methanol-dry ice for 10 min, thawed in a 37°C water bath for 5 min, and then immersed in an ice-water bath (2 cycles). The third aliquot was subjected to a third cycle of freezing and thawing (3 cycles). All samples were centrifuged at 100,000 × g for 45 min. The pellets were resuspended and analyzed by the HPLC method (A) for SIVMne(E11S) (280 nm) and by immunoblot analysis (B) for HIV-1 MN cl.4. The control sample was thawed just before HPLC or immunoblot analysis.
Effect of sCD4 on SU retention.
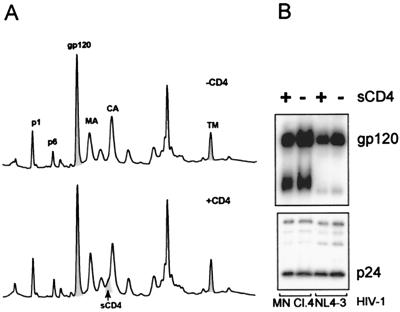
sCD4 has been reported to induce shedding of SU from HIV-1 (6, 14, 18, 36). To determine the susceptibility of SU to sCD4-induced shedding from purified virus, samples of SIVMne(E11S), HIV-1 MN cl.4, and HIV-1 NL4-3 were incubated with sCD4 (20 μg/ml) for 2 h at 37°C. Samples were then pelleted and analyzed by HPLC and immunoblot as previously described. As shown in Fig. 7, the amounts of envelope SU and TM, Gag proteins p1 and p6, MA, CA, and NC proteins recovered in the pelleted virus after treatment with sCD4 were indistinguishable from the amounts recovered before treatment. In order to verify the integrity of the recombinant sCD4, it was tested for bioactivity in an infectivity assay. sCD4 (30 μg/ml) reduced infectivity by at least 3,000-fold (data not shown), indicating that it was biologically active. Immunoblot analysis of the HIV-1 samples before and after treatment with sCD4 (Fig. 7B) revealed that at least 85% of the SU present in the samples before treatment was retained in the pelleted fraction after treatment with sCD4. These data clearly show that prolonged incubation with an excess of sCD4 does not induce extensive dissociation of SU from the purified virus particles, suggesting that the SU-TM interaction is stable. The sCD4 associated with the viral pellet was detected as a shoulder on the p24 peak by silver staining and immunoblot with anti-CD4.
FIG. 7.
SIVMne(E11S), HIV-1MN/H9 cl.4, and HIV-1 NL4-3/CEMx174 were incubated with or without sCD4 for 2 h at 37°C. Viral samples were then pelleted, and SIVMne(E11S) was analyzed by HPLC (A). Peaks were detected at 280 nm. (B) Virion-associated gp120 and p24CA on HIV-1 isolates were analyzed by SDS-PAGE and immunoblot analysis, followed by densitometry.
Heat treatment of SIVMne(E11S) and HIV-1 MN/H9 cl.4.
To investigate the effect of thermal treatment on the stability of virus Env proteins, SIVMne(E11S) and HIV-1 MN/H9 cl.4 were heated and then examined for retention of viral proteins (Fig. 8 and 9, respectively). The samples were thawed for 5 min in a 37°C water bath, incubated for 1 h at 4, 50, 55, 60, and 65°C, and centrifuged through a 20% sucrose-TNE buffer pad. Pelleted virus was analyzed for infectivity and by HPLC. Thermal treatment of both HIV-1 and SIV resulted in a loss of detectable infectivity in the samples incubated at 55°C and higher temperatures. The virus preparations heated at 50°C still retained some residual infectivity (data not shown).
FIG. 8.
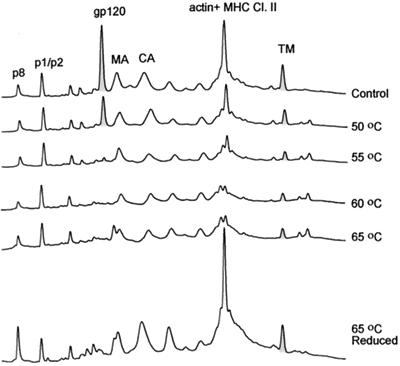
Heat treatment of SIVMne(E11S). SIVMneE11S virus stock (2 ml, 1,000-fold) were thawed for 5 min in a 37°C waterbath, and then five separate 0.3-ml aliquots were each diluted with 0.7 ml of sterile TNE buffer and heated for 1 h at 50, 55, 60, or 65°C; a fifth sample was not heated but incubated at 4°C for 1 h. After incubation, viral samples were centrifuged through a 20% sucrose-TNE buffer pad at 25,000 rpm for 60 min. Pellets were resuspended in TNE buffer and analyzed for gp120 content using HPLC methods under nonreducing and reducing (65°C) conditions. Peaks were detected at 280 nm.
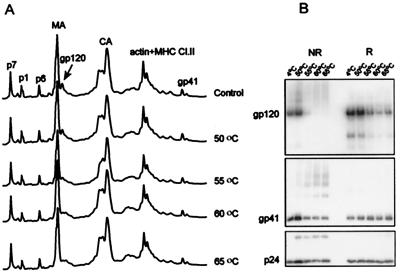
FIG. 9.
Heat treatment of HIV-1 MN/H9 cl.4. Virus was incubated at 4, 50, 55, 60, and 65°C for 1 h, and one sample was subjected to freezing and thawing three times. After incubation at various temperatures, viral samples were centrifuged, and pellets were analyzed for gp120 content using HPLC methods (A) under reducing conditions. Peaks were detected at 280 nm. (B) immunoblot analysis of gp120 retention by heat-treated HIV-1MN/H9 cl.4 virus. Virion-associated gp120 and p24CA levels were analyzed by SDS-PAGE under both reducing (R) and nonreducing (NR) conditions, followed by immunoblot analysis. Bands were visualized with mouse monoclonal antibodies prepared against purified p24 and gp120, followed by enhanced chemiluminescence staining.
Analysis of the HPLC profiles of SIV incubated at different temperatures revealed significant loss of SU protein in virus treated at 55°C and higher. When the sample heated at 65°C was reduced and reanalyzed by HPLC (Fig. 8, bottom HPLC profile), NC, MA, CA, and TM peaks were restored but SU was not, indicating that SU was shed from the virion. Some Gag proteins changed their elution behavior, suggesting that at high temperatures, free Cys residues might oxidize, giving rise to disulfide-bonded forms of proteins with altered chromatographic mobility. The p1 and p6 peaks were unchanged after heating of the viral preparation. These Gag proteins do not contain Cys residues and consequently would not be sensitive to oxidation. Quantitative recovery of the p1, p6, MA, CA, and TM proteins in the viral pellet (Fig. 8, bottom HPLC profile) strongly suggested that the viral lipid membrane remained intact through the heat treatment and that TM is not dissociated from the particles by heating. Comparing the ratio of protein peak areas reveals a significant loss of SU relative to p1, p6, and TM proteins, with the greatest loss occurring in samples heated to 55°C or higher. Samples of HIV-1 MN/H9 cl.4 (Fig. 9) responded to heat treatment with the majority of SU shedding at temperatures of 55°C or greater. Immunoblot analysis with anti-HIV gp120, p24CA, and gp41 antibodies was used in addition to HPLC profiles to further analyze the amount of SU shedding. We conclude that thermal treatment of viral particles induced shedding of SU but not TM protein from the virion surface. These results are in agreement with a study reported by Moore and Klasse for HIV-1 RF (34).
DISCUSSION
Knowledge of the surface density of the SU envelope protein on HIV-1 and SIV may provide insights into basic aspects of the viral life cycle, such as the processes of virion binding and fusion to target cells, assembly, and budding, in addition to having potential practical ramifications in relation to viral neutralization and in the design and development of prototype HIV-1 vaccines. We have developed an analytical approach that allows purification of virions combined with separation and quantitative analysis of individual viral proteins. We used these methods to characterize the relative amounts of different viral proteins present in purified virions.
Previously it has been proposed that HIV-1 virions more or less uniformly incorporate 72 envelope glycoprotein spikes during assembly and budding, with variable but typically extensive subsequent shedding of SU from SU-TM complexes on virions accounting for the relative lack of SU found in preparations of purified, mature viral particles (15). The alternative hypothesis, that the variable but typically small amounts of SU found on virions may reflect the amounts incorporated during assembly and budding, has generally not been considered. Key data that would help discriminate between these models (the amount of virion-associated TM protein and the SU:TM ratio) have not been available heretofore.
Using our approach for quantitative analysis of viral proteins in purified virus preparations, we have previously shown that the different _gag_-encoded proteins from HIV-1 are recovered in equimolar amounts (19, 20), which strongly suggests that the precursor is packaged in the virus prior to processing by the viral protease. In the present study, we used similar protein isolation and analysis techniques to quantitate Gag and Env proteins and the relative amounts of SU and TM envelope proteins in different preparations of purified SIV and HIV-1 virions. After proteolytic cleavage of a gp160 envelope precursor by a cellular protease, SU and TM associate through noncovalent interactions. While we found variable levels of SU on different virions, with a single exception (discussed below), we found no examples of SIV or HIV-1 with levels of TM in excess of SU, a pattern that would suggest shedding of SU. Moreover, the ratio of SU to TM for purified viruses was independent of the ratio of Gag to these envelope proteins. SIV isolates with high levels of envelope proteins [SIVMne(E11S) and SIV CP-MAC] had ratios of Gag to SU of ∼6:1 (Fig. 1, 2, and 3) and the molar ratios of SU to TM were ∼1:1.
Interestingly, all of the SIV isolates having high levels of SU (Gag:SU, ∼6:1) had truncated TM proteins (gp32). Other SIV isolates and all HIV-1 isolates examined had Gag-to-SU ratios of ∼60:1, and these viruses had low levels of both SU and TM, with the TM protein being in the full-length (gp41) form. The low levels of both SU and TM found on these viruses argues against shedding of gp120 as the explanation for the low levels of virion-associated SU found. Any significant loss of SU from the SU-TM complex incorporated into the virions would have been detected as a change in the SU-to-TM ratio. The possibility of shedding the entire SU-TM complex to yield a decrease in SU content without affecting the SU:TM ratio seems highly unlikely, since this would require an energetic process to dislodge the TM transmembrane domain from the viral envelope lipid.
The results of our heat treatment experiments for both SIV and HIV are consistent with dissociation of SU from SU-TM complexes, leaving TM with the particles. These results further support the notion that both gp32 and gp41 TM proteins are firmly anchored in the virion membrane, even when gp120 dissociates. In addition, there is experimental evidence for the association of the intracellular portion of TM with Gag proteins, specifically matrix protein (11, 13, 31). It thus seems far more likely that the SU-TM complex on the virus particle is stable during purification and that the ratio of Gag to SU is primarily determined by the amount of SU-TM complex incorporated during virus assembly.
It has been shown previously that HIV-1 virions contain a small quantity of SU, and in this study we have demonstrated that purified HIV-1 virions from both X4 and R5 isolates have a Gag-to-Env ratio of approximately 60:1. Assuming 1,200 to 2,500 Gag molecules per virion (5, 10, 30, 42, 50, 51), this corresponds to approximately 21 to 42 SU molecules per virus particle, which would result in 7 to 14 envelope trimers per virion. These findings are in broad agreement with a previous report (40) in which the ratio of p24Gag to SU on T-cell line-adapted viruses ranged between 70:1 and 297:1. They used virus with X4 or R5 envelope glycoproteins on a common NL4-3 background and found that R5 enveloped viruses appeared to contain more SU than X4 enveloped viruses. Willey et al. (54, 55) also reported small differences in the Env/Gag ratios of R5 and X4 viruses. There appears to be variation from virus to virus and between experimental systems with regard to differences in levels of SU on R5 and T-cell line-adapted viruses. Our results for R5 envelopes from HIV-1 JR-FL, HIV-1 BAL, and HIV-1 ADA-M propagated in SUPT-1 CCR5 (33) cells showed no significant increase in SU over X4 envelopes from T-cell line-adapted viruses such as HIV-1 IIIB, HIV-1 MN, and HIV-1 NL4-3, which is consistent with the findings of Karlsson et al. (21).
As mentioned above, we did find one SIV isolate with an excess of TM over SU, consistent with the profile expected if there was significant shedding of gp120. LaBranche et al. (27-29) first described SIV NC-MAC and CP-MAC as variants of SIV-MAC (BK28) that differ by 11 amino acid substitutions in the env gene (six in SU and five in TM). Our analysis of purified NC-MAC and CP-MAC showed that they each have a Gag:TM ratio of ∼6:1. CP-MAC had an SU-to-TM ratio of ∼1:1, similar to most other viruses, but NC-MAC had an SU-to-TM ratio of ∼1:10 (Fig. 3). This result shows that NC- and CP-MAC incorporate the same amounts of TM (Gag-to-TM ratio of 6:1) but differ greatly in the amounts of SU associated with the TM protein on the purified virus. This suggests that the amino acid substitutions separating NC- and CP-MAC affect the stability of the SU-TM complex but do not affect the incorporation of TM into the virion. Thus, NC-MAC may be a true case of a virus that sheds SU. It is not clear from the current studies whether the SU of NC-MAC dissociates from TM before or after virus assembly.
From the more general finding that purified HIV-1 and SIV have equivalent ratios of their envelope proteins SU and TM, we conclude that incorporation of TM into the viral membrane is the determining factor governing the amount of SU found on purified virions rather than loss or shedding of SU during the purification process. We further conclude that the amount of SU-TM complex incorporated into the virion lipid bilayer can vary greatly depending upon the viral isolate. In this regard it is interesting that all of the viruses that we have identified as having a Gag-to-SU ratio of ∼6:1 are members of the SIV group with truncated TM proteins (gp32). This observation is consistent with the findings of Littman et al. and Vzorov et al. (52, 53, 57), showing that truncation of the TM cytoplasmic tail of SIV increased the amount of SU copurifying with the virus.
We also experimentally evaluated the stability of the SU-TM complexes on purified virions by subjecting the viral particles to sucrose gradient purification, repeated freeze-thaw cycles, exposure to sCD4, or heating. Neither repeated freezing and thawing of the viruses nor centrifugation induced any significant loss of SU from the viruses (Fig. 6), confirming the relatively stable association of SU with TM on the surface of virions. Several conflicting reports have been published regarding sCD4-induced shedding of SU from HIV (1, 6, 9, 14, 16, 18, 22, 32, 34-36, 40, 41, 46, 50, 54, 55). We found no significant sCD4-induced shedding of SU from either purified HIV-1 or SIV virions at 37°C. Previously published studies on sCD4-induced shedding of SU involved shedding of SU either from cells expressing SU or from virus in culture fluid, and either may be contaminated by the presence of non-virion-associated SU in the culture fluid. It is possible that a small quantity of SU may be loosely associated with viral particles and that purification through a sucrose gradient may remove this SU, resulting in preparations of purified particles with only stable SU-TM complexes. Our studies utilized purified virus as the source of SU, and although the sCD4 was capable of binding to gp120 and preventing infection with HIV-1, it did not induce appreciable shedding of SU from purified virus.
Grovit-Ferbas et al. (17) reported that after heating various HIV-1 preparations at 62°C for 2 min, most of the SU was retained with the virus after centrifugal ultrafiltration through a 300-kDa cutoff device. However, fractionation of the heat-treated virus through a Percoll gradient showed that most of the SU did not cosediment with virus. The authors suggested that perhaps the SU retained with the virus during the ultrafiltration was in the form of SU oligomers or as membrane fragments containing SU. We found that heating of purified HIV-1 or SIV at 55°C or higher resulted in a significant loss of SU from the virus (Fig. 8 and 9). In fact, heat was the only physical procedure we found which readily dissociated SU from purified virus. Our results showed that prolonged heating (1 h) at temperatures of less then 50°C did not result in significant loss of any viral proteins, including SU, from the pelleted virus particles, suggesting that the whole virus, including the SU-TM complex, is stable at moderate temperatures. At higher temperatures (greater than 50°C), SU was shed from the particle, although the TM protein remained with the pelleted virion fraction. This result was consistent with the hypothesis that shedding of SU involved disrupting the noncovalent associations in the SU-TM complex, leaving TM embedded in the viral lipid membrane.
These thermal stability experiments were conducted with HIV-1 (MN) (Fig. 9). Semiquantitative immunoblot analysis showed that the HIV-1 MN virus particle, including the SU-TM complex, was stable at temperatures up to 50°C but readily shed SU at 55°C and higher. Taken together, the results of the thermal stability experiments for both HIV-1 and SIV strongly support the conclusion that SU does not readily dissociate from the SU-TM complex on the virus at moderate temperatures.
In aggregate, our observations provide strong support for the proposition that incorporation of SU-TM complexes into the virion envelope during the process of particle assembly and budding is the primary determinant of SU content of purified mature virions for the viruses we studied. In combination with data demonstrating retention of SU on purified viruses subjected to repeated freezing-thawing and sucrose gradient centrifugation, our results suggest that the association of SU and TM on purified mature virions is quite stable. While SIV NC-MAC provides an example of a virus which appears to have shed a large proportion of its SU complement, the fact that this was the only example of a virus we identified which had an significant excess of TM over SU suggests that such shedding may be less common than proposed previously. It will be of interest to extend the type of analyses described here to additional virus isolates, including viruses produced from primary cells.
Previously, whole inactivated HIV-1 depleted of functional SU has been used in vaccine studies (37, 38). Presentation of native, authentic viral envelope antigens, SU and TM, may be important for induction of optimal responses to vaccination. Native Env structures are postulated to be important in eliciting neutralizing antibodies and appear to be required for efficient cross priming of major histocompatibility complex I-restricted responses by CD8+ T cells in vitro (7). We recently described a novel chemical method that irreversibly inactivates retroviruses while retaining the conformational and functional integrity of the envelope glycoproteins (2, 45). Because they are reagents to ascertain the contribution of native envelope proteins to vaccine efficacy, it is essential that these proteins maintain functional and structural conformation and be retained on the virus during the purification procedures. Retention of SU by these purified viruses also makes these inactivated viral particles ideal platforms for testing molecularly modified SU in enhancing and broadening the immune responses. Understanding the underlying basis for the different levels of SU found on different viruses will be important in being able to design and produce vaccine immunogens with different desired levels of SU. The current studies represent a step in this direction.
Acknowledgments
We thank R. J. Center and B. Moss for the generous gift of the recombinant SU, N. Schuelke for recombinant sCD4, A. J Scarzello for virus infectivity assays, and D. E. Ott for stimulating discussion.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. N01-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. government.
REFERENCES
- 1.Allan, J. S., E. M. Whitehead, K. Strout, M. Short, P. Kanda, T. K. Hart, and P. J. Bugelski. 1992. Strong association of simian immunodeficiency virus (SIVagm) envelope glycoprotein heterodimers: possible role in receptor-mediated activation. AIDS Res. Hum. Retrovir. 8**:**2011-2020. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, L. O., J. W. Bess, E. N. Chertova, J. L. Rossio, M. T. Esser, R. E. Benveniste, L. E. Henderson, and J. D. Lifson. 1998. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res. Hum. Retrovir. 14(Suppl. 3)**:**S311-S319. [PubMed] [Google Scholar]
- 3.Benveniste, R. E., R. W. Hill, L. J. Eron, U. M. Csaikl, W. B. Knott, L. E. Henderson, R. C. Sowder, K. Nagashima, and M. A. Gonda. 1990. Characterization of clones of HIV-1-infected HuT 78 cells defective in gag gene processing and of SIV clones producing large amounts of envelope glycoprotein. J. Med. Primatol. 19**:**351-366. [PubMed] [Google Scholar]
- 4.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230**:**134-144. [DOI] [PubMed] [Google Scholar]
- 5.Bess, J. W., P. J. Powell, H. J. Issaq, L. J. Schumack, M. K. Grimes, L. E. Henderson, and L. O. Arthur. 1992. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J. Virol. 66**:**840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugelski, P. J., H. Ellens, T. K. Hart, and R. L. Kirsh. 1991. Soluble CD4 and dextran sulfate mediate release of gp120 from HIV-1: implications for clinical trials. J. Acquir. Immune Defic. Syndr. 4**:**923-924. [PubMed] [Google Scholar]
- 7.Buseyne, F., S. Le Gall, C. Boccaccio, J. P. Abastado, J. D. Lifson, L. O. Arthur, Y. Riviere, J. M. Heard, and, O. Schwartz. 2001. MHC-I-restricted presentation of HIV-1 virion antigens without viral replication. Nat. Med. 7**:**344-349. [DOI] [PubMed] [Google Scholar]
- 8.Esser, M. T., D. R. Graham, L. V. Coren, C. M. Trubey, J. W. Bess, Jr., L. O. Arthur, D. E. Ott, and J. D. Lifson. 2001. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75**:**6173-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, P. B., G. B. Karlsson, R. A. Dwek, and F. M. Platt. 1996. _N_-Butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with impaired gp120 shedding and gp41 exposure. J. Virol. 70**:**7153-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372**:**359-362. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70**:**341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O., and M. A. Martin. 1995. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270**:**23883-23886. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69**:**1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, Y. K., T. K. Hart, Z. L. Jonak, and P. J. Bugelski. 1993. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J. Virol. 67**:**3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelderblom, H. R. 1991. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS 5**:**617-637. [PubMed] [Google Scholar]
- 16.Groenink, M., J. P. Moore, S. Broersen, and H. Schuitemaker. 1995. Equal levels of gp120 retention and neutralization resistance of phenotypically distinct primary human immunodeficiency virus type 1 variants upon soluble CD4 treatment. J. Virol. 69**:**523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grovit-Ferbas, K., J. F. Hsu, J. Ferbas, V. Gudeman, and I. S. Chen. 2000. Enhanced binding of antibodies to neutralization epitopes following thermal and chemical inactivation of human immunodeficiency virus type 1. J. Virol. 74**:**5802-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart, T. K., R. Kirsh, H. Ellens, R. W. Sweet, D. M. Lambert, S. R. Petteway, Jr., J. Leary, and P. J. Bugelski. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 88**:**2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, L. E., R. E. Benveniste, R. Sowder, T. D. Copeland, A. M. Schultz, and S. Oroszlan. 1988. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne). J. Virol. 62**:**2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, L. E., R. C. Sowder, T. D. Copeland, S. Oroszlan, and R. E. Benveniste. 1990. Gag precursors of HIV and SIV are cleaved into six proteins found in the mature virions. J. Med. Primatol. 19**:**411-419. [PubMed] [Google Scholar]
- 21.Karlsson, G. B., F. Gao, J. Robinson, B. Hahn, and J. Sodroski. 1996. Increased envelope spike density and stability are not required for the neutralization resistance of primary human immunodeficiency viruses. J. Virol. 70**:**6136-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirsh, R., T. K. Hart, H. Ellens, J. Miller, S. A. Petteway, Jr., D. M. Lambert, J. Leary, and P. J. Bugelski. 1990. Morphometric analysis of recombinant soluble CD4-mediated release of the envelope glycoprotein gp120 from HIV-1. AIDS Res. Hum. Retrovir. 6**:**1209-1212. [DOI] [PubMed] [Google Scholar]
- 23.Kodama, T., D. P. Wooley, Y. M. Naidu, H. W. Kestler, 3rd, M. D. Daniel, Y. Li, and R. C. Desrosiers. 1989. Significance of premature stop codons in env of simian immunodeficiency virus. J. Virol. 63**:**4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreis, T. E., and H. F. Lodish. 1986. Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell 46**:**929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393**:**648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBranche, C. C., T. L. Hoffman, J. Romano, B. S. Haggarty, T. G. Edwards, T. J. Matthews, R. W. Doms, and J. A. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73**:**10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, and J. A. Hoxie. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J. Virol. 68**:**7665-7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, and J. A. Hoxie. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J. Virol. 68**:**5509-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, M. Marsh, and J. A. Hoxie. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69**:**5217-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189**:**695-714. [DOI] [PubMed] [Google Scholar]
- 31.Mammano, F., E. Kondo, J. Sodroski, A. Bukovsky, and H. G. Gottlinger. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 69**:**3824-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKeating, J. A., A. McKnight, and J. P. Moore. 1991. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J. Virol. 65**:**852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Means, R. E., T. Matthews, J. A. Hoxie, M. H. Malim, T. Kodama, and R. C. Desrosiers. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75**:**3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, J. P., and P. J. Klasse. 1992. Thermodynamic and kinetic analysis of sCD4 binding to HIV-1 virions and of gp120 dissociation. AIDS Res. Hum. Retrovir. 8**:**443-450. [DOI] [PubMed] [Google Scholar]
- 35.Moore, J. P., J. A. McKeating, Y. X. Huang, A. Ashkenazi, and D. D. Ho. 1992. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J. Virol. 66**:**235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250**:**1139-1142. [DOI] [PubMed] [Google Scholar]
- 37.Moss, R. B., W. Giermakowska, P. Lanza, J. L. Turner, M. R. Wallace, F. C. Jensen, G. Theofan, S. P. Richieri, and D. J. Carlo. 1997. Cross-clade immune responses after immunization with a whole-killed gp120-depleted human immunodeficiency virus type-1 immunogen in incomplete Freund's adjuvant (HIV-1 immunogen, REMUNE) in human immunodeficiency virus type-1 seropositive subjects. Viral Immunol. 10**:**221-228. [DOI] [PubMed] [Google Scholar]
- 38.Moss, R. B., R. J. Trauger, W. K. Giermakowska, J. L. Turner, M. R. Wallace, F. C. Jensen, S. P. Richieri, F. Ferre, A. E. Daigle, C. Duffy, G. Theofan, and D. J. Carlo. 1997. Effect of immunization with an inactivated gp120-depleted HIV-1 immunogen on beta-chemokine and cytokine production in subjects with HIV-1 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 14**:**343-350. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74**:**3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Brien, W. A., S. H. Mao, Y. Cao, and J. P. Moore. 1994. Macrophage-tropic and T-cell line-adapted chimeric strains of human immunodeficiency virus type 1 differ in their susceptibilities to neutralization by soluble CD4 at different temperatures. J. Virol. 68**:**5264-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orloff, S. L., M. S. Kennedy, A. A. Belperron, P. J. Maddon, and J. S. McDougal. 1993. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J. Virol. 67**:**1461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piatak, M., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K.-C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259**:**1749-1754. [DOI] [PubMed] [Google Scholar]
- 43.Pyle, S. W., J. W. Bess, Jr., W. G. Robey, P. J. Fischinger, R. V. Gilden, and L. O. Arthur. 1987. Purification of 120,000 dalton envelope glycoprotein from culture fluids of human immunodeficiency virus (HIV)-infected H9 cells. AIDS Res. Hum. Retrovir. 3**:**387-400. [DOI] [PubMed] [Google Scholar]
- 44.Robey, W. G., B. Safai, S. Oroszlan, L. O. Arthur, M. A. Gonda, R. C. Gallo, and P. J. Fischinger. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228**:**593-595. [DOI] [PubMed] [Google Scholar]
- 45.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72**:**7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider, J., O. Kaaden, T. D. Copeland, S. Oroszlan, and G. Hunsmann. 1986. Shedding and interspecies type sero-reactivity of the envelope glycopolypeptide gp120 of the human immunodeficiency virus. J. Gen. Virol. 67**:**2533-2538. [DOI] [PubMed] [Google Scholar]
- 47.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-335. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 48.Tatu, U., C. Hammond, and A. Helenius. 1995. Folding and oligomerization of influenza hemagglutinin in the ER and the intermediate compartment. EMBO J. 14**:**1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372**:**363-365. [DOI] [PubMed] [Google Scholar]
- 50.Thali, M., C. Furman, E. Helseth, H. Repke, and J. Sodroski. 1992. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J. Virol. 66**:**5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogt, V. M., and M. N. Simon. 1999. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73**:**7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vzorov, A. N., and R. W. Compans. 1996. Assembly and release of SIV env proteins with full-length or truncated cytoplasmic domains. Virology 221**:**22-33. [DOI] [PubMed] [Google Scholar]
- 53.Vzorov, A. N., D. Lea-Fox, and R. W. Compans. 1999. Immunogenicity of full-length and truncated SIV envelope proteins. Viral Immunol. 12**:**205-215. [DOI] [PubMed] [Google Scholar]
- 54.Willey, R. L., M. A. Martin, and K. W. Peden. 1994. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J. Virol. 68**:**1029-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willey, R. L., T. S. Theodore, and M. A. Martin. 1994. Amino acid substitutions in the human immunodeficiency virus type 1 gp120 V3 loop that change viral tropism also alter physical and functional properties of the virion envelope. J. Virol. 68**:**4409-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393**:**705-711. [DOI] [PubMed] [Google Scholar]
- 57.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J. Virol. 67**:**2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]