RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain (original) (raw)
Abstract
microRNAs (miRNAs) are small (~22 nucleotide) regulatory RNAs which play fundamental roles in many biological processes. Recent studies have shown that the expression of many miRNAs is altered in various human tumors and some miRNAs may function as oncogenes or tumor suppressor genes. However, with the exception of glioblastoma multiforme, the expression of miRNAs in brain tumors is unknown. Furthermore, methods to profile miRNAs from formalin-fixed, paraffin-embedded (FFPE) archival tissues or to study their cellular and subcellular localization in FFPE tissues have been lacking. Here we report the coordinated miRNA expression analysis from the tissue level to the subcellular level, using the RAKE (RNA-primed, array-based, Klenow Enzyme) miRNA microarray platform in conjunction with Locked Nucleic Acid (LNA)-based in situ hybridization (LNA-ISH) on archival FFPE human brains and oligodendroglial tumors. The ability to profile miRNAs from archival tissues at the tissue level, by RAKE microarrays, and at the cellular level by LNA-ISH, will accelerate studies of miRNAs in human diseases.
Keywords: MicroRNA, miRNA, RAKE, in situ hybridization, brain tumors, Locked Nucleic Acid
INTRODUCTION
MicroRNAs (miRNAs) are a predominant subtype of small (~22 nucleotide) regulatory RNAs (for reviews, see Nelson et al. 2003; Ambros 2004; Bartel 2004; Meister and Tuschl 2004; Wienholds and Plasterk 2005). miRNAs bind directly to Argonaute (Ago) proteins and form effector complexes known as RNA-induced silencing complexes (RISCs) or miRNPs (Hammond et al. 2001; Mourelatos et al. 2002). Ago proteins are a large family of ~95 kDa proteins that are found in most organisms; humans contain four Ago proteins (Ago1–Ago4) (Carmell et al. 2002). miRNAs, in the form of miRNPs or RISCs, exert their influence through complementary base-pairing with specific target mRNAs, leading in turn to target mRNA degradation or translational repression (for reviews, see Meister and Tuschl 2004; Murchison and Hannon 2004; Filipowicz et al. 2005; Zamore and Haley 2005). Bioinformatics predict that the number of human miRNAs exceeds 500 and may approach 1000 (Bentwich et al. 2005; Berezikov et al. 2005), thus comprising ~5% of the transcriptome. Human miRNAs, in turn, are predicted to regulate numerous mRNA targets (Lewis et al. 2003, 2005; John et al. 2004; Kiriakidou et al. 2004; Krek et al. 2005; Lim et al. 2005; Xie et al. 2005); recent estimates suggest that one-third of human mRNAs may be regulated by miRNAs (Lewis et al. 2005; Xie et al. 2005). An increasing body of research has linked miRNAs with human cancers (for reviews, see McManus 2003; Caldas and Brenton 2005; Croce and Calin 2005; Meltzer 2005; Wienholds and Plasterk 2005). However, pathogenetic mechanisms are yet to be demonstrated conclusively.
RAKE miRNA profiling of archival human brain
Expression profiling of miRNAs in cancers has been reported only with RNA isolated from fresh tissues. Furthermore, there have been no reports of in situ hybridization for human miRNAs. We previously developed the RNA-primed, array-based, Klenow Enzyme (RAKE) micro-array platform, which involves on-slide application of the Klenow fragment of DNA polymerase to extend unmodified miRNAs hybridized to immobilized DNA probes (Nelson et al. 2004). A major advantage of RAKE is that it requires no sample RNA manipulation prior to hybridization. We have also demonstrated that miRNAs survive formalin fixation and paraffin embedding and can be readily isolated from formalin-fixed, paraffin-embedded (FFPE) tissues (Nelson et al. 2004). In this study, we used RAKE to profile miRNAs from normal human adult and fetal brains and from reactive astrocytosis and oligodendroglial tumors (materials and methods are described in Supplementary Material, which can be found at http://www.med.upenn.edu/camb/faculty/ggr/mourelatos.html). The cases analyzed included four grade II oligodendrogliomas, 16 grade III oligodendrogliomas, nine fetal brains, six adult temporal lobe cortices, one adult normal hippocampus, three adult hippocampi with mesial temporal sclerosis, and two adult cerebella (Supplementary Table 1).
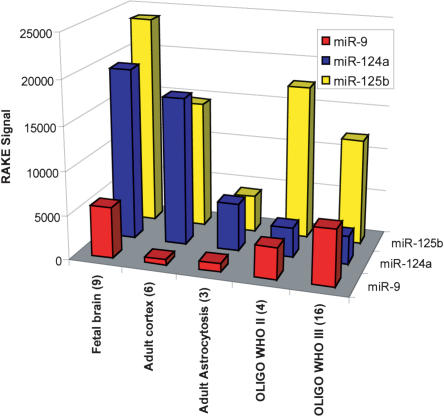
The microarray data showed that different tissue types had characteristic miRNA expression profiles (Supplementary Fig. 1; Supplementary Table 2). Using a hierarchical clustering algorithm (using the Spotfire software and the following standard clustering criteria: weighted; correlation; input rank; hierarchical), all tumor samples clustered separately from all nontumor tissues (Supplementary Fig. 1). Of special interest were miRNAs expressed relatively specifically in brain, and/or which showed informative differences across the tissue types (Fig. 1 ▶; Supplementary Fig. 1). By RAKE, the expression of miR-125b was high in fetal and adult brains and in oligodendroglial tumors (Fig. 1 ▶; Supplementary Fig. 1). miR-124a was also highly expressed in fetal and adult brain, but its expression was very low in oligodendrogliomas (Fig. 1 ▶; Supplementary Fig. 1). miR-9 showed increased expression in fetal brain and neoplastic tissue but low expression in adult brain (Fig. 1 ▶; Supplementary Fig. 1). The current generation of RAKE microarray has six replicates for each miRNA; in the case of miR-9, there is a duplicate of these six replicates, as additional technical controls. The two miR-9 replicate groups showed a very high correlation with each other across all the cases sampled (Supplementary Fig. 1; Supplemental Table 2). Previous studies have shown that miR-125b is enriched in brain (Sempere et al. 2004), while miR-124a and miR-9 are almost exclusively expressed in brain (Lagos-Quintana et al. 2002; Sempere et al. 2004), with the exception of miR-124a, which is also expressed in pancreatic islet cells (Poy et al. 2004). Other miRNAs showed increased expression in fetal brain and oligodendrogliomas (including miR-15, -16, -20), but these are ubiquitously expressed and are not specific to brain (Supplementary Fig. 1; Supplementary Table 2). As was found previously (Nelson et al. 2004), Northern blots correlated well with the RAKE microarray results (Supplementary Fig. 2).
FIGURE 1.
miRNA expression levels of human brain-specific or brain-enriched miRNAs. RAKE signal for each miRNA is the average of the signal obtained from all listed samples, with the number of cases for each sample shown in parentheses.
LNA-ISH for miRNAs in archival human tissues
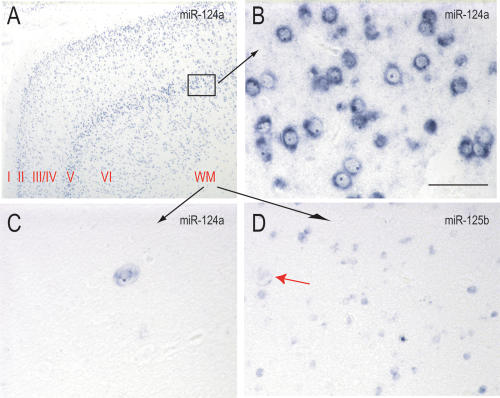
We next applied an in situ hybridization method to visualize the cellular and subcellular distribution of miRNAs in FFPE tissues. For this, we used LNA-modified oligonucleotide probes because they display markedly increased hybridization affinities toward complementary RNAs, compared to traditional RNA or DNA based probes (Vester and Wengel 2004). This property allows for very stringent hybridization conditions, increasing the specificity and sensitivity of miRNA detection (Valoczi et al. 2004; Vester and Wengel 2004). LNA probes have been used successfully to visualize miRNAs in whole-mount preparations of zebrafish (Wienholds et al. 2005). We evaluated four different miRNAs using LNA-ISH: miR-124a, miR-125b, miR-9 (these three miRNAs had particularly high levels of expression in the brain), and miR-122a. miR-122a is expressed only in liver (Lagos-Quintana et al. 2002; Sempere et al. 2004) and was used as a negative control for the brain sections. We first analyzed the expression of these miRNAs in adult brain. The probe for miR-124a stained exclusively neurons, including dentate granule cells of the hippocampus and cortical neurons (Fig. 2A–C ▶; Supplementary Fig. 3A,B). The staining pattern of sections from the temporal lobe was particularly striking and revealed the hexalaminar arrangement of cortical neurons (Fig. 2A ▶), but it also highlighted ectopic neurons in the white matter (Fig. 2C ▶). On higher magnification, the staining was predominantly cytoplasmic (Fig. 2B ▶). In contrast, the probe for miR-125b stained neurons and glial cells (Fig. 2D ▶; Supplementary Fig. 3D,E). Staining of glial cells was particularly evident in white matter (Fig. 2D ▶). miR-9 staining was prominent in the dentate granule cells of the hippocampus (Supplementary Fig. 3G), but it was very low in the neurons of adult cortex (Supplementary Fig. 3H). The probe for the liver-specific miR-122a showed intense staining in sections from human liver (Supplementary Fig. 3L) but very low staining in sections from brain (Supplementary Fig. 3J,K), which represented background. Sections of human liver stained with probes for miR-124a, miR-125b, and miR-9 also showed very low diffuse staining that we consider background (Supplementary Fig. 3C,F,I).
FIGURE 2.
Cellular and subcellular distribution of miRNAs in human adult cortex. ISH with LNA probes against miR-124a and miR-125b were performed in human temporal isocortex. (A) Low-power view of miR-124a staining; cortical layers are indicated; (WM) white matter. (B) High-power view of cortical layer V. (C) High-power view of white matter. (D) High-power view of white matter stained with miR-125b; red arrow shows ectopic neuron. All photomicrographs were obtained and processed with identical software and hardware settings. Scale bar for B_–_D = 100 microns.
Next, we studied the expression of miR-9, miR-124a, and miR-125b in human fetal brains by LNA-ISH. The miR-9 probe stained intensely the cells of the germinal matrix (Supplementary Fig. 4A), while staining of the cerebral cortex was much less intense (Supplementary Fig. 4B). In contrast, the miR-124a probe stained intensely the cerebral cortex (Supplementary Fig. 4D) but showed weaker staining of the germinal matrix (Supplementary Fig. 4C). The miR-125b probe stained both the germinal matrix and cerebral cortex with the same intensity (Supplementary Fig. 4E, F). These findings demonstrate that miR-9 is predominantly expressed in the proliferating neuroblasts and glioblasts of the germinal matrix while miR-124a is predominantly expressed in the mature neurons of the cerebral cortex.
LNA-ISH for miRNAs in oligodendrogliomas
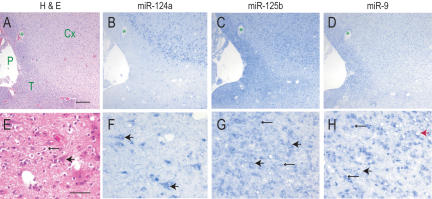
We then studied the staining pattern of oligodendroglial tumors and representative photomicrographs are shown in Figure 3 ▶. We prepared near-serial sections derived from an oligodendroglioma that was infiltrating cerebral cortex and also demonstrating subpial tumor spread (Fig. 3A,E ▶) and stained them by LNA-ISH for miR-124a, miR-125b, and miR-9. miR-124a was negative in tumor cells, but was strongly positive in neurons that were overrun by infiltrating glioma cells (Fig. 3B,F ▶). miR-125b was positive in both the normal and neoplastic glial cells and in neurons (Fig. 3C,G ▶). miR-9 showed very strong staining in tumor cells and much weaker staining of neurons (Fig. 3D,H ▶).
FIGURE 3.
Expression of miRNAs in oligodendrogliomas. ISH of indicated miRNAs of oligodendroglial (WHO grade II) brain tumors. (A_–_D) Depicts near-serial sections of the same tumor, as do (E_–_H). The green star in (A_–_D) depicts the same blood vessel. (H&E) hematoxylin and eosin stains; (P) pia; (T) subpial spread of tumor; (Cx) cortex. All photomicrographs were obtained and processed with identical software and hardware settings. Scale bar for A_–_D = 250 microns; E_–_H = 60 microns. Large arrowheads indicate neurons; small arrowheads indicated tumor cells. Red arrow in H shows a neuron surrounded by miR-9-positive tumor cells.
We also studied whether there was a correlation between the staining intensity of miR-9 and the grade of oligodendrogliomas and representative findings are shown in Supplementary Figure 5. The staining intensity of miR-9 in tumor cells was similar between grade II (Supplementary Fig. 5A) and grade III oligodendrogliomas (Supplementary Fig. 5C). In addition, we found no correlation between miR-9 staining of the tumor cells and proliferation index, as assessed by immunohistochemistry for the Ki-67/MIB-1 proliferation marker (Supplementary Fig. 5B,D).
Allelic losses of chromosome 1p do not alter miRNA expression in oligodendrogliomas
Allelic losses of chromosome 1p are frequently found in oligodendrogliomas (in 80%–90% of grade II, and 50%–70% of grade III oligodendrogliomas), and they typically involve deletions of the entire 1p region (Reifenberger and Louis 2003). Deletion of the 1p chromosomal region in tumor cells correlates with improved response to chemotherapy in the context of oligodendrogliomas (Buckner 2003; Fallon et al. 2004). Three of the four genes that code for Agos (Ago4, Ago1, and Ago3), the proteins that bind to miRNAs and mediate miRNA function, are localized on chromosome 1p34. We compared the miRNA profiles of four grade III oligodendrogliomas, two with and two without chromosome 1p deletions (chromosomal deletions were detected by FISH analysis of tumor cells using 1p probes). The miRNA repertoire and expression levels were very similar between all samples, indicating that allelic losses of three Ago genes do not substantially alter miRNAs in oligodendrogliomas (Supplementary Fig. 6). Moreover, the staining patterns of miR-9, miR-124a, and miR-125b, as revealed by ISH, between these tumors were also similar (data not shown).
SUMMARY
miRNAs were profiled for the first time using complementary tissue and cellular techniques on human archival FFPE biopsy- and autopsy-derived tissues. The RAKE microarray revealed particular miRNAs that are expressed in normal human brain and in oligodendrogliomas. In situ hybridization was applied successfully to refine the data obtained by RAKE and to localize particular miRNAs. These findings indicate the remarkable stability of miRNAs in archival human tissues. The importance of using in situ hybridization to complement tissue-level studies is well illustrated by the contrasting expression patterns of miR-124a and miR-9 in human brain and oligodendrogliomas. At the tissue level (by RAKE microarray), miR-124a is high in normal brain and lower in oligodendrogliomas. One might hypothesize that decreased levels of miR-124a might correlate with neoplastic transformation. However, in situ hybridization provides direct evidence against this hypothesis because miR-124a is expressed in neurons only and not in tumor cells. The low levels of miR-124a expression by microarrays in tissues derived from oligodendrogliomas is due to these residual neurons. By contrast, microarray data show that miR-9 expression is increased in oligodendrogliomas versus normal adult brain. This could be due to miR-9 being normally expressed mainly in glial cells, and the increase in gliomas being due only to an increase of the density of cells of glial lineage. However, in situ hybridization shows that miR-9 is expressed in low levels in adult neurons, but in very high levels in proliferating neuroblasts and glioblasts in fetal brain and also in tumor cells. miR-9 is a brain-specific miRNA that is 100% conserved between known mammalian, zebrafish, chicken, Drosophila, and Anopheles species, with a somewhat lower degree of conservation between those species and Caenorhabditis elegans. In humans, copies of miR-9 genes reside on chromosomes 1, 5, and 15; those on chromosomes 1 and 5 are present within the introns of known protein-encoding genes. In zebrafish, miR-9 is expressed in proliferating brain, spinal cord, and eyes, beginning between 16–24 h post-fertilization (Wienholds et al. 2005). In flies, miR-9 is required for cellularization (Leaman et al. 2005). Bioinformatics have predicted numerous mRNA targets for miR-9, suggesting that miR-9 may regulate multiple pathways (Lewis et al. 2003, 2005; Kiriakidou et al. 2004). The apparent maturation of neuro-blasts, migrating from the germinal matrix (relatively high miR-9 expression, low miR-124a) to the fetal cerebral cortex (relatively high miR-124a expression, low miR-9) shows that there may be a reciprocal relationship between these two miRNAs. Further studies are required to uncover the function of miR-9 in neurodevelopment and neoplasia. We expect that the application of RAKE and LNA-ISH in archival tissues will greatly accelerate studies of miRNA in numerous human diseases.
Acknowledgments
We thank J.-C. Oberholtzer and members of our lab for helpful discussions. We thank Exiqon Inc. for generously providing the LNA probes. This work was supported by NIH grants K08-NS050110 to P.T.N. and R01-GM0720777, P30-CA016520, P30-AG10124, P30-HD026979 to Z.M.
REFERENCES
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431**:** 350–355. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116**:** 281–297. [DOI] [PubMed] [Google Scholar]
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37**:** 766–770. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H., and Cuppen, E. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120**:** 21–24. [DOI] [PubMed] [Google Scholar]
- Buckner, J.C. 2003. Factors influencing survival in high-grade gliomas. Semin. Oncol. 30**:** 10–14. [DOI] [PubMed] [Google Scholar]
- Caldas, C. and Brenton, J.D. 2005. Sizing up miRNAs as cancer genes. Nat. Med. 11**:** 712–714. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stemcell maintenance, and tumorigenesis. Genes & Dev. 16**:** 2733–2742. [DOI] [PubMed] [Google Scholar]
- Croce, C.M. and Calin, G.A. 2005. miRNAs, cancer, and stem cell division. Cell 122**:** 6–7. [DOI] [PubMed] [Google Scholar]
- Fallon, K.B., Palmer, C.A., Roth, K.A., Nabors, L.B., Wang, W., Carpenter, M., Banerjee, R., Forsyth, P., Rich, K., and Perry, A. 2004. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J. Neuropathol. Exp. Neurol. 63**:** 314–322. [DOI] [PubMed] [Google Scholar]
- Filipowicz, W., Jaskiewicz, L., Kolb, F.A., and Pillai, R.S. 2005. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 15**:** 331–341. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293**:** 1146–1150. [DOI] [PubMed] [Google Scholar]
- John, B., Enright, A.J., Aravin, A., Tuschl, T., Sander, C., and Marks, D.S. 2004. Human microRNA targets. PLoS Biol. 2**:** e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou, M., Nelson, P.T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., and Hatzigeorgiou, A. 2004. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 18**:** 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, A., Grun, D., Poy, M.N., Wolf, R., Rosenberg, L., Epstein, E.J., MacMenamin, P., da Piedade, I., Gunsalus, K.C., Stoffel, M., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37**:** 495–500. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12**:** 735–739. [DOI] [PubMed] [Google Scholar]
- Leaman, D., Chen, P.Y., Fak, J., Yalcin, A., Pearce, M., Unnerstall, U., Marks, D.S., Sander, C., Tuschl, T., and Gaul, U. 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121**:** 1097–1108. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115**:** 787–798. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120**:** 15–20. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433**:** 769–773. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. 2003. MicroRNAs and cancer. Semin. Cancer Biol. 13**:** 253–258. [DOI] [PubMed] [Google Scholar]
- Meister, G. and Tuschl, T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431**:** 343–349. [DOI] [PubMed] [Google Scholar]
- Meltzer, P.S. 2005. Cancer genomics: Small RNAs with big impacts. Nature 435**:** 745–746. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous micro-RNAs. Genes & Dev. 16**:** 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison, E.P. and Hannon, G.J. 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16**:** 223–229. [DOI] [PubMed] [Google Scholar]
- Nelson, P., Kiriakidou, M., Sharma, A., Maniataki, E., and Mourelatos, Z. 2003. The microRNA world: Small is mighty. Trends Biochem. Sci. 28**:** 534–540. [DOI] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., and Mourelatos, Z. 2004. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 1**:** 155–161. [DOI] [PubMed] [Google Scholar]
- Poy, M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432**:** 226–230. [DOI] [PubMed] [Google Scholar]
- Reifenberger, G. and Louis, D.N. 2003. Oligodendroglioma: Toward molecular definitions in diagnostic neuro-oncology. J. Neuropathol. Exp. Neurol. 62**:** 111–126. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5**:** R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi, A., Hornyik, C., Varga, N., Burgyan, J., Kauppinen, S., and Havelda, Z. 2004. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32**:** e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester, B. and Wengel, J. 2004. LNA (locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry 43**:** 13233–13241. [DOI] [PubMed] [Google Scholar]
- Wienholds, E. and Plasterk, R.H. 2005. MicroRNA function in animal development. FEBS Lett 579**:** 5911–5922. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., Kloosterman, W.P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, R.H., Kauppinen, S., and Plasterk, R.H. 2005. MicroRNA expression in zebrafish embryonic development. Science 309**:** 310–311. [DOI] [PubMed] [Google Scholar]
- Xie, X., Lu, J., Kulbokas, E.J., Golub, T.R., Mootha, V., Lindblad-Toh, K., Lander, E.S., and Kellis, M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434**:** 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D. and Haley, B. 2005. Ribognome: The big world of small RNAs. Science 309**:** 1519–1524. [DOI] [PubMed] [Google Scholar]