Novel thermostable Y-family polymerases: applications for the PCR amplification of damaged or ancient DNAs (original) (raw)
Abstract
For many years, Taq polymerase has served as the stalwart enzyme in the PCR amplification of DNA. However, a major limitation of Taq is its inability to amplify damaged DNA, thereby restricting its usefulness in forensic applications. In contrast, Y-family DNA polymerases, such as Dpo4 from Sulfolobus solfataricus, can traverse a wide variety of DNA lesions. Here, we report the identification and characterization of five novel thermostable Dpo4-like enzymes from Acidianus infernus, Sulfolobus shibatae, Sulfolobus tengchongensis, Stygiolobus azoricus and Sulfurisphaera ohwakuensis, as well as two recombinant chimeras that have enhanced enzymatic properties compared with the naturally occurring polymerases. The Dpo4-like polymerases are moderately processive, can substitute for Taq in PCR and can bypass DNA lesions that normally block Taq. Such properties make the Dpo4-like enzymes ideally suited for the PCR amplification of damaged DNA samples. Indeed, by using a blend of Taq and Dpo4-like enzymes, we obtained a PCR amplicon from ultraviolet-irradiated DNA that was largely unamplifyable with Taq alone. The inclusion of thermostable Dpo4-like polymerases in PCRs, therefore, augments the recovery and analysis of lesion-containing DNA samples, such as those commonly found in forensic or ancient DNA molecular applications.
INTRODUCTION
The Y-family of DNA polymerases consists of phylogenetically separated UmuC, DinB, Rev1 and Rad30 subfamilies (1). UmuC-like proteins are only found in prokaryotes, while Rev1 and Rad30 (polη and polι) orthologs exist exclusively in eukaryotes. DinB-like proteins are, however, found in all three kingdoms of life. In prokaryotes, DinB-like proteins are typified by Escherichia coli polIV; in eukaryotes, by polκ; and in archaea, by Dbh and Dpo4. The Dbh (DinB homolog) protein was originally identified from Sulfolobus acidocaldarius using a degenerate PCR approach designed to obtain UmuC homologs (2), while Dpo4 (DNA polymerase IV) was identified by searching the completed genome of Sulfolobus solfataricus (3). Both Dbh and Dpo4 have been characterized biochemically and have been shown to be thermostable Y-family DNA polymerases. While Dbh exhibits poor processivity and poor lesion bypass activity (4,5), Dpo4 is significantly more robust on both undamaged and damaged templates (3,6). The crystal structures of both Dbh and Dpo4 have been solved, revealing that they are topologically similar to classical polymerases in that they resemble a right hand and possess ‘fingers’, ‘palm’ and ‘thumb’ sub-domains (7–9). However, they also possess an additional domain that has been termed the ‘little finger’ (LF) (9). The LF domain holds the polymerase on to the primer-template and studies of Dpo4/Dbh chimeras in which the LF domains were interchanged, revealed that the LF domain is the major factor determining the overall processivity of the enzyme (6).
Similar to many Y-family polymerases, Dpo4 is capable of replicating past several types of DNA lesions, including abasic sites (3,10), a cis–syn cyclobutane pyrimidine dimer (CPD) (3,11), and to a lesser degree pyrimidine 6-4 pyrimidone (6-4PP) at a dithymine site (3), _cis_-Platin (_cis_-Pt) guanine adducts (3) and _N_-acetyl-2-aminofluorene (AAF) guanine adducts (3). Indeed, crystal structures of Dpo4 in the process of bypassing an abasic site (12), a CPD (13) and a Benzo[_a_]pyrene-adenine lesion (14) have served as a paradigm for lesion bypass by the Y-family polymerases and have provided insights into the molecular mechanisms of translesion replication.
In contrast to Y-family polymerases, polymerases that belong to the A- or B-family possess a smaller and constricted active site that cannot accommodate bulky or non-coding adducts. As a consequence, such lesions are often kinetic blocks to continuing replication. Indeed, a major limitation in forensic science or the analysis of ancient DNAs is the ability of conventional PCR enzymes, such as Taq, to amplify damaged DNA (15,16). One way to potentially circumvent this problem would be to utilize a Y-family polymerase to replicate the damaged DNA template, so as to generate one or more lesion-free copies of the template that could be subsequently amplified by a higher fidelity PCR enzyme. In the first steps toward achieving this goal, we report here a quick and simple strategy for the identification of thermostable Y-family polymerases. We have overproduced, purified and characterized five novel Y-family polymerases, as well as two recombinant chimeras with enhanced enzymatic properties, and we provide ‘proof of principle’ that the inclusion of a thermostable Y-family DNA polymerase in the PCR amplification of damaged DNA facilitates the recovery of amplicons previously unobtainable with Taq polymerase alone.
MATERIALS AND METHODS
Identification and cloning of novel dpo4-like genes
Based on protein alignments of S.solfataricus Dpo4, S.acidocaldarius Dbh and a hypothetical Dpo4-like protein identified within the genome of Sulfolobus tokodaii, the degenerate PCR primers, dpo4_FRIDegen1 and dpo4_RXhDegen1, were designed and used to amplify an ∼300 bp region of _dpo4_-like genes from the genomes of Acidianus infernus (Ain) (DSM 3191), Sulfolobus shibatae (Ssh) (DSM 5389), Sulfolobus tengchongensis (Ste) (17), Stygiolobus azoricus (Saz) (DSM 6296) and Sulfurisphaera ohwakuensis (Soh) (DSM 12321). The sequence of dpo4_FRIDegen1 is 5′-CCG GAA TTC YWY RTI GAY WTB GAY WVI TTY TWY GC-3′ and is based on the conserved amino acid sequence (FH)(VI)D (MF)D(YCS) F(FY)A. The sequence of dpo4_RXhDegen1 is 5′-GCC GCT CGA GTC IAD RWA IGC YTC RTC IAY ISW IRY-3′ and is based on the conserved amino acid sequence (AVI)S(IV)DEA(YF)(LI)(DE). The PCR products were ‘TA’ cloned into pGEM-T (Promega) and sequenced. All subsequent PCR products were also cloned into pGEM-T except where noted. Additional regions of the _dpo4_-like genes were amplified using the degenerate PCR primers, dpo4_RXhDegen5 (5′-GCC GCT CGA GYT TRT WRT AIS HYT CIT YDA TIG C-3′ [AI(NED)E(AS)Y(NY)K)] and dpo4_RXhDegen6 (5′-GCC GCT CGA GRT AIG CIB KIT CIW TIS WIA TIC C-3′ [GIS(IK)(DE)(NTR)AY]), in conjunction with gene-specific primers (Ai_FRI276 (5′-CCG GAA TTC AAT AAT GTA CGG AAT ACT CTC T-3′), Sa_FRI267 (5′-CCG GAA TTC TTC TAA TAG GAT TAT GAA CGG A-3′), So_FRI283 (5′-CCG GAA TTC CAT AAT CAG TAG CTA TTC GG-3′) and Ss_FRI235 (5′-CCG GAA TTC GCC CAT GAG AAA GAA AGT GT-3′). The sequence of the 5′ ends of the S.azoricus and S.shibatae genes were determined by inverse PCR using the primer pairs Sa_FRI142(5′-CCG GAA TTC GCT ACA GGC AAT TAC GAG G-3′) and Sa_RXh74 (5′-GCC GCT CGA GTA AAC GCA GAC AAT TAC CG-3′) and the primer pairs Ss_FRI138 (5′-CCG GAA TTC GTG GCT ACT GCG AAC TAC-3′) and Ss_RXh96 (5′-GCC GCT CGA GCA CTA TCC TCA AAT CTA CCA-3′). The sequence of the 5′ and 3′ ends of A.infernus, S.ohwakuensis genes as well as the 3′ ends of S.azoricus and S.shibatae genes were isolated by performing PCR utilizing degenerate primers designed to the conserved upstream ribokinase gene and conserved downstream hypothetical gene; ribo_FRIDegen1 [5′-CCG GAA TTC GAY ACI ACI GGI GCI GGI GAY-3′ [DTTGAGD]) and hypo_FXhDegen1a (5′-GCC GCT CGA GTA YGA RGA YGT IGA RGG IGG3′ [YEDVEGG]) in combination with gene-specific primers. The gene-specific primers used are Ai_FRI772 (5′-CCG GAA TTC AAC TCA GGA GAT ATT AAA GAG A-3′), Ai_RXh43 (5′-GCC GCT CGA GAG TTC TGG GTT AAG GAC TTC-3′), Sa_FRI762 (5′-CCG GAA TTC ATT ACC GTA TAA TAC GAG AAA G-3′), So_FRI783 (5′-CCG GAA TTC AAA GTG ATA GAA CCT TAT CTG-3′), So_RXh76 (5′-GCC GCT CGA GCA GAA TAA ACA CAG ACT ATA AG-3′) and Ss_FRI696 (5′-CCG GAA TTC AGA GAT GAG TAT AAC GAA CCT-3′). Isolation of the 5′ and 3′ ends of the _S.tengchongensis dpo4_-like genes was accomplished by performing PCR using the degenerate PCR primer pairs ribo_FRIDegen1/dpo4_RXhDegen1 and dpo4_FRIDegen4 (5′-CCG GAA TTC RYI WSI RTI GAY GAR GCI TWY HTI GA-3′ [(AVI)S(IV)DEA(YF)(LI)(DE)])/hypo_FXhDegen1a.
The full-length nucleotide sequence of the polIV-like genes can be located in the following GenBank accession numbers: DQ124670 (A.infernus); DQ124673 (S.shibatae); DQ124669 (S.tengchongensis); DQ124671 (S.azoricus); and DQ124672 (S.ohwakuensis).
The full-length dpo4_-like genes were then PCR amplified using gene-specific primers and cloned or sub-cloned into pET22b+ (Novagen) from NdeI or FauI to BamHI for expression in RW382, a Δ_umuDC derivative of BL21(λDE3) (Novagen) (18). The primers sets are Ai_FNde10 (5′-TTA ATG TTA AAT GCA T_AT G_AT TGT AC-3′) (the initiation ATG is italicized)/Ai_RBam1088 (5′-GTA CGG TGA GAT GGA TCC TTC-3′), Sa_FFau7 (5′-TTA CAC TCC AAG TGC CCG CTG TAT ATG ATT G-3′)/Sa_RBam1078 (5′-TAT CGA AGT ATG GAT CCG TAA ATT TC-3′), So_FNde5 (5′-AGC CTA AGG TAA AAT GCA T_AT G_AT-3′)/So_RBam1078 (5′-GGA AGA ATG GAT CCT ACT AAA GTG-3′), Ss_FNde5 (5′-GAA ACC AAA TGT TAT ATG CAT_ATG_ AT-3′)/Ss_RBam1089 (5′-CTA TTT GTC GGA TCC TAA GGA AAT AG-3′) and Ste_FNde5 (5′-AAA AAC CAA AAG TTA TAT GCA T_AT G_AT-3′)/Ste_RBam1089 (5′-TTA CCT CAA GGA TCC TAA GGA AAT TG-3′). These clonings created plasmids overexpressing Ain _dpo4_-like pol (pJM534), Saz _dpo4_-like pol (pJM520), Soh _dpo4_-like (pJM521) Ssh _dpo4_-like pol (pJM522), and Ste _dpo4_-like pol (pJM551) respectively.
Construction of chimeric enzymes
Chimeric _dpo4_-like genes were constructed by introducing a novel MscI restriction site into the Ste _dpo4_-like and the Ain _dpo4_-like genes in pJM551 and pJM534, just prior to the region encoding the little finger (LF) domain, using the QuickChange Mutagenesis kit (Stratagene, La Jolla, CA) to create pJM563 and pJM564 respectively. The Ain _dpo4_-like LF domain was replaced in pJM564 by subcloning the Ste _dpo4_-like LF domain out of pJM563 from MscI to BamHI to create the Ain/Ste _dpo4_-like chimeric gene in pJM580. In a similar way, the Sso LF domain from the previously described plasmid p1947 (6) was sub-cloned from MscI to BamHI into pJM564 to create the Ain/Sso _dpo4_-like chimeric gene in pJM590. The full-length nucleotide sequence of the polIV chimeras can be located in the following GenBank accession numbers: DQ124674 (A.infernus with the LF from Sso Dpo4) and DQ124675 (A.infernus with the LF from Ste Dpo4).
Purification of Dpo4-like enzymes
Purification of the new Dpo4-like and chimeric enzymes was accomplished essentially as previously reported (3) except the protocol was simplified as outlined below. One or two liter LB-ampicillin cultures of the RW382 strain harboring the various Dpo4-like expression plasmids were grown at 37°C overnight. In the case of Ste Dpo4, Saz Dpo4, Soh Dpo4 and the two chimeric Dpo4s, AinLFSso and AinLFSte, the LB-ampicillin cultures also contained 0.01 mM isopropyl-β-d-thiogalactopyranoside. Cleared lysates in buffer A (75 mM NaCl, 10 mM KHPO4 pH 7, 0.1 mM EDTA, 1 mM DTT) were heat treated for 10 min at 70°C and the denatured E.coli proteins were removed by centrifugation at 21000 g for 20 min. The soluble supernatant was applied to a Hydroxyapatite Bio-Gel HTP Gel column (HP) (BioRad, Hercules, CA) and bound proteins were eluted with a 10–1000 mM linear gradient of KHPO4. Fractions containing the Dpo4-like proteins from the HAP column were desalted on a HiPrep 26/10 desalting column (Amersham Pharmacia Biotech, Piscataway, NJ) using a 20 mM NaCl, 20 mM Tris, pH 7.5, 0.1 mM EDTA and 1 mM DTT wash and subsequently applied to a SP Sepharose HP column (Sph) (Amersham Pharmacia Biotech). Bound proteins were eluted with a 20–1000 mM linear gradient of NaCl. Fractions containing the purified Dpo4-like polymerases were concentrated and stored at −80°C with the addition of 10% glycerol. The Saz and Soh Dpo4-like proteins were applied to one additional column; a HiLoad 26/60 Superdex 75 column (Amersham Pharmacia Biotech), washed with 100 mM NaCl, 20 mM Tris, pH 7.5, 0.1 mM EDTA and 1 mM DTT to ensure the purity of these two samples.
In vitro primer extension assays
The undamaged (HTU50) and the abasic site-containing (HTX50) synthetic oligonucleotides used in the in vitro primer extension assays were synthesized by Lofstrand Laboratories (Gaithersburg, MD) using standard techniques and were PAGE purified prior to use. The synthetic abasic site (dSpacer) was purchased from Glen Research (Sterling, VA) and incorporated into the oligonucleotide template using standard techniques by Lofstrand Laboratories. The cis–syn CPD-containing oligonucleotide (HMTT50) was synthesized by Phoenix Biotechnologies (Huntsville, AL) and has been described previously (6). The 5-hydroxy-5-methylhydantoin-containing oligonucleotide (ODN) was synthesized as described previously (19). Single-stranded M13mp18 DNA was purchased from New England Biolabs (Beverly, MA). The sequence of each primer-template pair is given in the legend of the respective figures. Primers were labeled with [γ-32P]ATP using T4 polynucleotide kinase by Lofstrand Laboratories. Radiolabeled primers were annealed to the unlabeled templates at a ratio of 1:2 or 1:1.4, in the case of M13mp18, by heating the primer and template together at 95°C in annealing buffer (0.4 M Tris–HCl, pH 8.0, 50 mM MgCl2, 5 µg/ml BSA and 14.2 mM β-mercaptoethanol) and allowing the mixture to slowly cool to room temperature. Standard 10 µl reactions contained 40 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 100 µM each dNTP (or a single dNTP, where noted), 10 mM DTT, 250 µg/ml BSA, 2.5% glycerol and 10 nM primer-template DNA. Reactions were carried out at 60 or 37°C, in the case of the hydantoin-containing template, with varying concentrations of enzyme and for varying lengths of time which are specified in the legend of the respective figures. Reactions were terminated by the addition of 10 µl of 95% formamide, 10 mM EDTA, heated to 95°C for 5 min and briefly chilled on ice. Aliquots containing 5 µl of the samples were subjected to electrophoresis through polyacrylamide, 8 M urea gels and replication products were visualized by PhosphorImager analysis.
PCRs on undamaged DNA
For the undamaged plasmid DNA template, 50 µl PCRs were set up containing standard ThermoPol reaction buffer [20 mM Tris–HCl, pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100] (New England Biolabs), 10 ng of circular pJM548 (Ste _dpo4_-like in pGEM-T), 200 µM of each ultrapure dNTP and 1 µM of each primer; Ste_FNde5 and Ste_RBam1089 whose sequences are provided above. Aliquots containing 2.5 U of Taq DNA polymerase (New England Biolabs) were used as a positive control. Each of the Dpo4 enzymes was added to a concentration of 200 nM, except the Ssh Dpo4-like enzyme, which was added to a concentration of 400 nM. Reactions were heated to 85°C for 3 min and then treated with 35 cycles of 85°C for 30 s, 56°C for 1 min and 60°C for 4 min. Products were separated on a 0.9% agarose gel stained with ethidium bromide.
Genomic DNA sample preparation
K562 human genomic DNA (Promega Corporation, Madison, WI) was diluted in sterile water to a concentration of 100 ng/µl in a 1.5 ml microcentrifuge tube (Fisher Scientific, Norcross, GA) and exposed to UVC in a Stratalinker 1800 (Stratagene, La Jolla, CA) at a flux rate of 0.197 J/cm2/min.
PCR amplification and detection of Alu sequences
PCR amplification of the Alu insert was carried out with 2 ng of genomic DNA. The 25 µl reaction contained 1× Reaction Buffer (40 mM Tris–HCl, pH 8.0, 10 mM DTT, 60 mM KCl and 2.5% glycerol), 2.5 µM dNTPs, 2.5 U Taq Gold DNA Polymerase (Applied Biosystems, Foster City, CA), 3 mM MgCl2, 20 pmol each of the forward and reverse primers, and 100 nM Dpo4, if applicable. Primer sequences were as follows: forward, 5′-GCG GTG GCT CAC GCC T-3′ and reverse, 5′-GGA GTC TCG CTC TGT CG-3′ (20). The forward primer was labeled at its 5′ end with the reactive fluorescent dye 6-carboxyfluorescein (6-FAM). The forward primer binds a site near the Alu transcription start site. This sequence is identical in both the Major and the Precise Alu subfamily consensus (21), which comprise ∼99% of Alu repeats in the human genome (20). The cycling conditions were: initial denaturation at 95°C for 11 min; 17 cycles: 94°C for 10 s, 56°C for 30 s, 72°C for 30 s; final extension 72°C for 5 min. For some experiments a smaller Alu fragment was amplified: the 25 µl reaction was carried out with 2 ng genomic DNA, 2.5 µM dNTPs, 3.25 mM MgCl2, 10 µg non-acetylated BSA, 2.5 U AmpliTaq DNA Polymerase (Applied Biosystems), and 20 pmol each of the forward and reverse primers in 1× Buffer D3 (40 mM Tris–HCl, pH 8.0, 10 mM DTT, 6 mM KCl, 2.5% glycerol), and 100 nM Ste or Ain, if applicable. Cycling conditions were as follows: 85°C for 1 min; 22 cycles: 85°C for 30 s, 56°C for 1 min, 60°C for 5 min; 60°C for 5 min.
Amplified fragments were detected using the ABI Prism 310 capillary electrophoresis (CE) system (Applied Biosystems). A 1.5 µl aliquot of each amplified sample was added to 24 µl Hi-Di formamide (Applied Biosystems) and 1 µl of GeneScan 500 TAMRA internal lane standard (Applied Biosystems). Tubes were heated at 95°C for 3 min and snap cooled on ice for at least 3 min. Samples were injected onto the CE using the GS STR module (5 s injection, 15 kV, 60°C, run time 28 min, Filter Set C) module. Samples were subject to laser-induced fluorescence and analyzed with GeneScan 3.1.2 software (Applied Biosystems).
RESULTS AND DISCUSSION
Identification of novel Dpo4-like genes
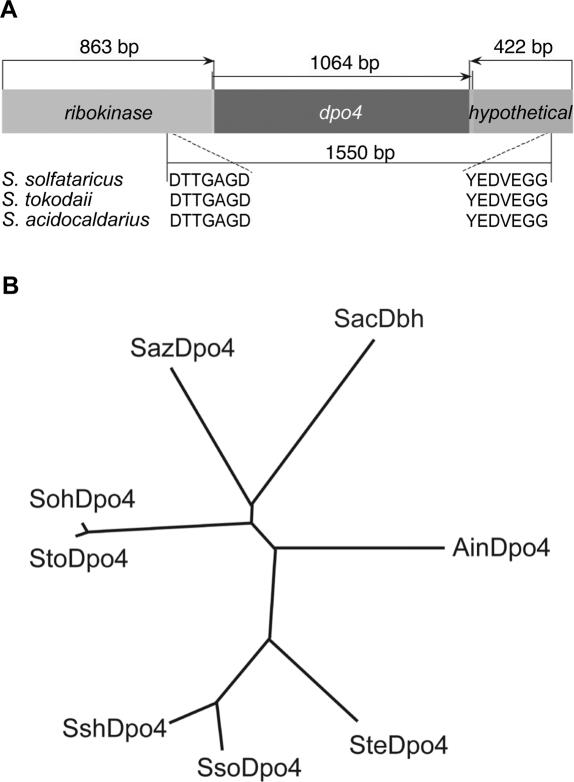
We utilized the amino acid sequences of S.acidocaldarius Dbh (Sac) and S.solfataricus Dpo4 (Sso), as well as a hypothetical Dpo4-like protein encoded within the S.tokodaii genome (GenBank Accession number NC 003106) to design degenerate PCR primers to identify additional genes encoding polIV-like enzymes from other members of the Sulfolobaceae family. Five new polIV homologs were identified from A.infernus (Ain), S.shibatae (Ssh), S.tengchongensis (Ste), S.azoricus (Saz) and S.ohwakuensis (Soh). Once a fragment of a polIV-like gene was amplified, cloned and sequenced, we took advantage of the conserved genomic architecture surrounding the known polIV-like genes to develop a strategy to quickly obtain full sequence of the new genes (Figure 1A). Degenerate PCR primers to the upstream ribokinase gene and the downstream hypothetical gene were designed and in combination with either _dpo4_-like gene-specific or degenerate PCR primers, the ends of each of the new _dpo4_-like genes were cloned and sequenced. Subsequently, PCR primers to the 5′ and 3′ ends of each of the new genes were used to amplify the full-length dpo4-like gene.
Figure 1.
Identification, cloning, and phylogenetic analysis of novel Dpo4-like orthologs. (A) Illustration of the cloning strategy for identification and isolation of full-length _dpo4_-like orthologs. In some cases, degenerate PCR primers based on the Dpo4 and Dbh amino acid sequences were used to amplify a section a _dpo4_-like ortholog. Subsequently, degenerate PCR primers based on the amino acid sequences of the conserved upstream ribokinase (DTTGAGD) and downstream hypothetical (YEDVEGG) genes in combination with dpo4 ortholog gene-specific primers were used to amplify the beginning and the ending of each gene. In the case of the Ste _dpo4_-like gene, degenerate PCR primers based on the Dpo4 and Dbh amino acid sequences were used instead of gene-specific primers to isolate the ends of the gene. Once the sequence of the 5′ and 3′ of each gene was identified, primers were designed to amplify and clone the full-length genes. (B) Unrooted phylogenetic tree of Dpo4-like polymerases. A protein alignment was performed using the Clustal X program (version 1.62b) (27). This protein alignment was used to calculate the tree using the Draw N-J Tree algorithm. The unrooted tree was drawn from the calculated tree using the TreeView program (version 1.6) (28). The derivation of each protein is designated as follows: S.acidocaldarius (SacDbh), S.solfataricus Dpo4 (SsoDpo4), S.tokodaii (StoDpo4), S.shibatae (SshDpo4), S.tengchongensis (SteDpo4), A.infernus (AinDpo4), S.azoricus (SazDpo4) and S.ohwakuensis (SohDpo4).
Dbh and Dpo4 are from closely related organisms, but have very different biochemical properties (6). We therefore reasoned that determining the phylogenetic relationship of the new polIV-like polymerases to Dpo4 or Dbh might be predictive of their general enzymatic activities. Figure 1B shows an unrooted phylogenetic tree of S.acidocaldarius Dbh, S.solfataricus Dpo4, S.tokodaii Dpo4-like and the five novel enzymes. This analysis revealed that the orthologs most closely related to S.solfataricus Dpo4 are, in order of relatedness, S.shibatae Dpo4-like, S.tengchongensis Dpo4-like and A.infernus Dpo4-like, and as we show below, these polymerases possess enzymatic properties somewhat similar to Dpo4. The S.azoricus and S.ohwakuensis polIV-like genes are more distantly related to Dpo4 and have properties somewhat similar to Dbh. Thus, a simple phylogenetic analysis appears to be a good indicator of the overall enzymatic properties of the novel archaeal Y-family polymerases.
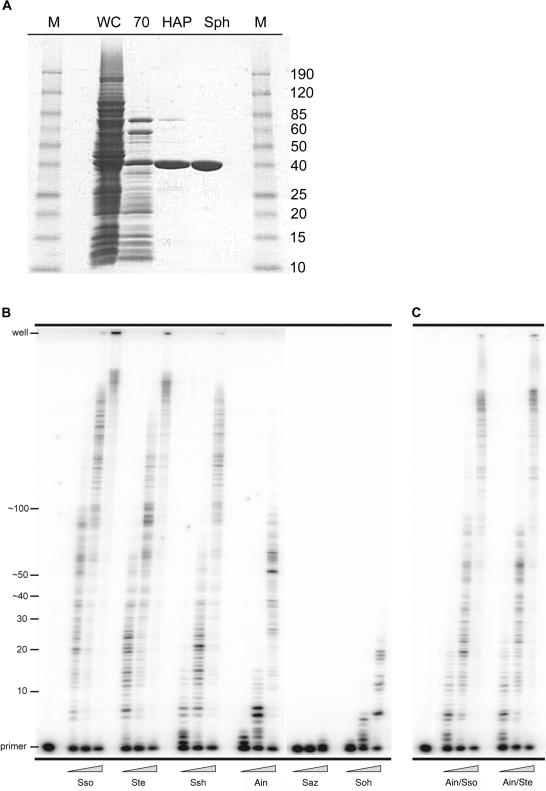
Overexpression, purification and processivity of novel Dpo4-like polymerases
In order to characterize the biochemical properties of the new polIV-like enzymes, they were overexpressed in an E.coli strain, RW382, lacking umuDC (polV) (22) and purified utilizing a similar, but somewhat simplified scheme, to that was previously reported (Figure 2A) (6). Such an approach allowed us to readily purify anywhere between 1 and 5 mg of each polymerase from just 1–2 l of E.coli cell culture. Once the polymerases were purified to >95% homogeneity, we initially characterized them for their relative degree of processivity. The primer extension assays were performed at 60°C, with various concentrations of each enzyme and utilized a primer annealed to a circular 7.2 kb M13mp18 template (Figure 2B). Under these conditions, the Saz and Soh polIV-like proteins exhibited weak polymerase activity. Indeed, even at the highest enzyme concentration, the Saz and Soh enzymes added only few nucleotides to the primer. As a consequence, we did not characterize the Saz and Soh enzymes further. In contrast, the Ssh Dpo4-like and the Ste Dpo4-like enzymes exhibited a similar level of polymerase activity and processivity to Sso Dpo4, demonstrating that they are robust thermostable polymerases. Intriguingly, while the Ain Dpo4-like enzyme showed a somewhat similar level of primer utilization compared with Dpo4, Ssh Dpo4-like or Ste Dpo4-like, the size of the replication products was substantially shorter, suggesting that the Ain Dpo4-like polymerase is less processive than the other three enzymes. As noted above, however, the LF domain of the polymerase has been shown to be the major determinant of polymerase processivity (6). In an attempt to increase the processivity of the Ain Dpo4-like enzyme, we interchanged the LF domain of the Ain Dpo4-like enzyme with the LF domain of either Sso Dpo4 or Ste Dpo4-like polymerase to create the Ain/Sso Dpo4-like and the Ain/Ste Dpo4-like chimeras. As anticipated, the processivity of each chimera was significantly enhanced relative to the native Ain Dpo4-like enzyme (Figure 2C).
Figure 2.
Purification and processivity of Dpo4-like proteins. (A) Scheme for the purification of Dpo4-like proteins. Whole-cell extracts (WC) were clarified and then heat treated at 70°C for 10 min. Denatured E.coli proteins were removed from the extracts by centrifugation (70). Subsequently, the soluble extracts were passed over a Hydroxyapatite Bio-Gel HTP Gel column (HAP) and then a SP Sepharose HP column (Sph). The Saz and Soh Dpo4-like proteins were purified over one additional column, a HiLoad 26/60 Superdex 75 column (data not shown). M designates the Benchmark prestained protein size marker (Invitrogen). (B) M13mp18 primer extension reactions using the previously identified S.solfataricus Dpo4 (Sso) and the five newly identified polymerases; S.tengchongensis (Ste), S.shibatae (Ssh), A.infernus (Ain), S.azoricus (Saz) and S.ohwakuensis (Soh). Primer extension of primer M13HTP (5′-CCT TAG AAT CCT TGA AAA CAT AGC GA-3′) annealed to M13mp18 from base pairs 4101 to 4126 was performed at 60°C for 5 min utilizing 10 nM of the M13HTP/M13mp18 primer-template and increasing concentrations of the Dpo4-like enzymes; from left to right 0.2, 2.0 and 20 nM. Replication products were separated on 12%/8 M urea polyacrylamide gels and visualized by PhosphorImager analysis. Primer location and number of nucleotides added are indicated to the left of the gel. (C) M13mp18 primer extension reactions using the ‘little finger’ domain chimeric polymerases; A.infernus/S.solfataricus (Ain/Sso) and A.infernus/S.tengchongensis (Ain/Ste).
Ability of Dpo4-like enzymes to facilitate PCR
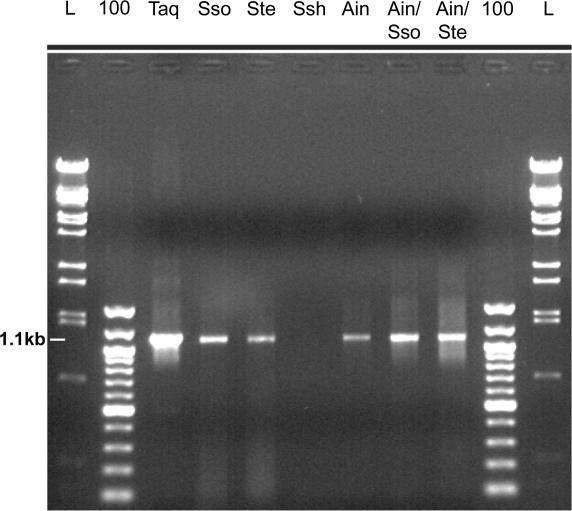
Although the thermostable Dpo4-like enzymes are typically less processive than replicative polymerases, they are nevertheless capable of replicating hundreds of bases in the M13mp18 primer extension assay and we have previously shown that Dpo4 could effectively amplify an ∼1300 bp pre-denatured linear target DNA molecule at high template concentrations (∼400 ng) in a PCR (3). The ability of Dpo4 and our new Dpo4-like enzymes to substitute for Taq in PCRs was reexamined under more optimal reaction conditions. As shown in Figure 3, Dpo4-like enzymes can efficiently amplify an ∼1100 bp target DNA in a ‘closed-tube reaction’ when the denaturation temperature is lowered from the typical ≥90 to 85°C. Using this lower denaturation temperature, we could also eliminate the pre-denaturation step, amplify non-linear DNA, reduce the amount of DNA to about 10 ng and reduce the primer concentration from 30 µM of each primer used previously, to 1 µM. Under these conditions, it appears that the Ain/Sso Dpo4-like and Ain/Ste Dpo4-like chimeras are the most robust PCR enzymes belonging to the Y-family examined to date. Further optimization of PCR amplification by Dpo4-like polymerases is certainly possible, e.g. by altering the reaction buffer components such as salts, protein stabilizers or reagents that lower melting temperature.
Figure 3.
PCR amplification of the 1.1 kb _S.tengchongensis Dpo4_-like gene by Dpo4-like polymerases. A total of 2.5 U of Taq or 200 nM (400 nM of Ssh) of each Dpo4-like polymerase was added to the PCRs. Two microliters of the 50 µl PCRs were loaded onto a 0.9% agarose gel and the PCR products separated by electrophoresis. The enzymes employed in the PCRs are specified above each lane. The position of the 1.1 kb PCR product is indicated on the left. L and 100 designates the lambda BstE II and 100 bp markers, respectively.
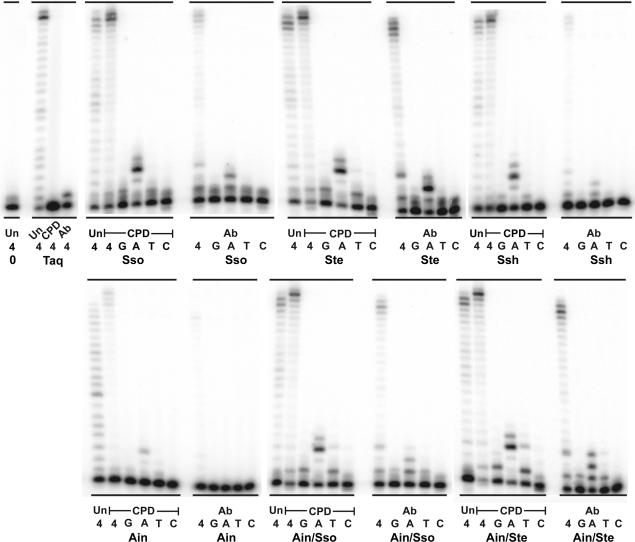
Translesion synthesis activity of the novel Dpo4-like enzymes
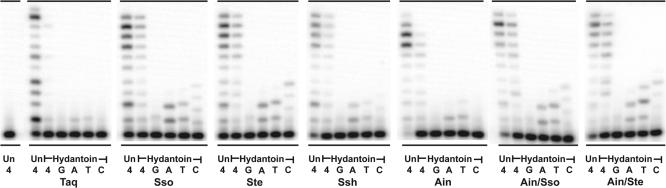
The major biological function of Y-family polymerases is thought to be the synthesis of damaged DNA refractory to replication by the cell's conventional replicases. We therefore compared the lesion bypass capabilities of our new enzymes with that of Dpo4 in a primer extension assay using DNA templates that contain lesions such as a cis_–_syn CPD, an abasic site (Ab) (Figure 4) or a 5-hydroxy-5-methylhydantoin (5-OH-5-MeHyd) (hyd) (Figure 5). These lesions arise endogenously as a result of exposure to ultraviolet (UV) radiation in the case of CPDs, or due to the action of base-excision repair (BER) mechanism(s) or spontaneous hydrolysis of the _N_-glycosyl bond in the case of abasic sites (23). In addition, exogenous DNA, such as forensic or ancient DNA (aDNA) samples, can suffer the same types of insults due to exposure to environmental factors including UV-light, heat, humidity and other genotoxic substances. Indeed, hydantoin derivatives of pyrimidine bases, which are the most prevalent lesions encountered in aDNA (24), can be produced via several oxidative processes as a result of prolonged environmental exposure (25). Using approximately equal activities of Taq and the Dpo4-like polymerases, we found that all three lesions block Taq polymerase (Figures 4 and 5). However, in dramatic contrast, all of the Dpo4-like enzymes not only preferentially incorporated the correct nucleotides opposite each lesion, but they also promoted full lesion bypass, albeit with varying efficiencies (Figures 4 and 5). Of particular note, is the fact that the Ain/Sso and Ain/Ste Dpo4-like chimeras appear to be the most efficient of the novel enzymes at bypassing all three DNA lesions.
Figure 4.
Primer extension on CPD and abasic site-containing templates. Primer SSHTP2 (5′-GCG GTG TAG AGA CGA GTG CGG AG-3′) was annealed to the undamaged template, HTU50 (Un) (5′-CTC TCA CAA GCA GCC AGG CAT TCT CCG CAC TCG TCT CTA CAC CGC TCC GC-3′); the CPD-containing template, HMTT50 (CPD) (5′-CTC TCA CAA GCA GCC AGG CAT TCT CCG CAC TCG TCT CTA CAC CGC TCC GC-3′) (where the T-T CPD is underlined); or the abasic site-containing template, HTX50 (Abasic) (5′-CTC TCA CAA GCA GCC AGG CAT XCT CCG CAC TCG TCT CTA CAC CGC TCC GC-3′), (where X denotes the position of the abasic site). Primer extension assays utilized 10 nM of the three different primer/templates. The reactions containing the undamaged template (HTU50) was incubated at 60°C for 3 min, whereas the reactions containing the CPD (HMTT50) and abasic site (HTX50) templates were incubated at 60°C for 10 min. The concentrations of each enzyme utilized for the undamaged template (HTU50) reactions is as follows: Taq, 0.0025 U; Sso, 0.1 nM; Ste, 0.33 nM; Ssh, 0.4 nM; Ain, 0.2 nM; Ain/Sso, 0.5 nM; Ain/Ste, 0.5 nM. The concentrations of each enzyme utilized for the CPD-containing template (HMTT50) reactions was 50 times higher than that used for the undamaged template and is as follows: Taq, 0.125 U; Sso, 5 nM; Ste, 16.5 nM; Ssh, 20 nM; Ain, 10 nM; Ain/Sso, 25 nM; Ain/Ste, 25 nM. The concentrations of each enzyme utilized for the abasic site-containing template (HMTT50) reactions was five times higher than used for the undamaged template and is as follows: Taq, 0.0125 U; Sso, 0.5 nM; Ste, 1.65 nM; Ssh, 2 nM; Ain, 1 nM; Ain/Sso, 2.5 nM; Ain/Ste, 2.5 nM. Reactions were initiated by the addition of 100 µM of all four dNTPs (4) or 100 µM of individual dNTPs (G, A, T, C) (indicated below each lane). Replication products were separated on12%/8 M urea polyacrylamide gels and visualized by PhosphorImager analysis.
Figure 5.
Primer extension on the hydantoin-containing template. Primer SSHydP (5′-AGA TCA GTC ACG-3′) was annealed to the undamaged template, HydU22 (Un) (5′-CAC TTC GGA TCG TGA CTG ATC T-3′), and the 5-hydroxy-5-methylhydantoin-containing template, (5′-CAC TTC GGA HCG TGA CTG ATC T-3′), (where H indicates the position of the Hydantoin). Primer extension assays utilized 10 nM of these primer/templates and the reactions were incubated at 37°C for 5 min. The concentrations of each enzyme utilized for the undamaged template (HTU50) and hydantoin-containing template reactions is as follows: Taq, 0.0083 U; Sso, 0.33 nM; Ste, 0.75 nM; Ssh, 1 nM; Ain, 0.5 nM; Ain/Sso, 1.25 nM; Ain/Ste, 1.25 nM. Reactions were initiated by the addition of 100 µM of all four dNTPs (4) or 100 µM of individual dNTPs (G, A, T, C) (indicated below each lane). Replication products were separated on 18%/8 M urea polyacrylamide gels and visualized by PhosphorImager analysis.
Dpo4-like enzymes and their ability to PCR amplify damaged DNA templates
The ability to detect DNA polymorphisms using molecular genetic techniques has revolutionized the forensic analysis of biological evidence. DNA typing now plays a critical role within the criminal justice system, but one of the limiting factors with the technology is that DNA isolated from biological stains recovered from the crime scene is sometimes so damaged as to be intractable to analysis. Currently no methods exist to rectify such damaged genomic DNA. The development of a method to restore and recover DNA profiles from damaged DNA would thus represent a significant advance with widespread applications in forensic casework and mass disaster investigations.
We reasoned, therefore, that a potential solution to this problem would be to perform the PCR of damaged DNA in the presence of a thermostable Y-family polymerase. To specifically test this hypothesis, human genomic DNA was irradiated with UVC light at a flux rate of 0.197 J/cm2/min for 6 min. Exposure of naked DNA in this manner causes a multitude of lesions including significant quantities of CPDs and (6-4) photoproducts (26). Genetic profile recovery from the UVC damaged substrate was assessed by amplification of Alu short interspersed elements comprising ∼10% of the genome that contain a limited number of bipyrimidine sites known to form photoproducts (20). The ubiquity of Alu sequences throughout the genome thus renders the repair substrate a de facto whole genome scan of UVC-induced DNA damage and its repair.
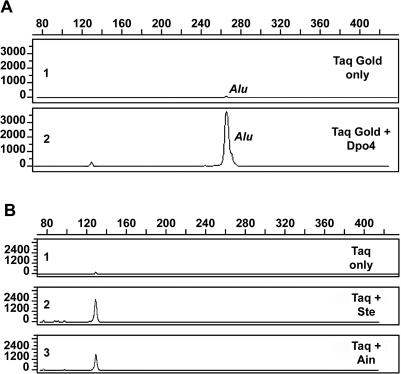
Taq Gold alone gave an Alu signal that was barely detectable (Figure 6A, top panel). Remarkably, a blend of Taq with 100 nM Sso Dpo4 in the PCR permitted the recovery of an Alu signal from the damaged genomic DNA substrate (Figure 6A, bottom panel). In four replicate experiments, there was a 36- to 45-fold enrichment of Alu sequences recovered from the damaged template by the Taq/Dpo4 blend. Alternative PCR conditions were employed with AmpliTaq to test the other Dpo4-like thermostable Y-family enzymes described above and these included a lower denaturation temperature (85°C), an increased extension time to permit efficient polymerase switching at damaged sites, and amplifying a smaller Alu fragment. Using these conditions in replicate experiments, Ste and Ain Dpo4-like enzymes produced a 14- to 18-fold and a 10- to 15-fold enrichment of Alu sequences from the damaged template, respectively (Figure 6B). As controls, the Taq Gold or AmpliTaq concentrations were doubled in the absence of the thermostable Y-family enzyme and this resulted in an average 1.5-fold increase in Alu signal. No further improvement in signal intensity was obtained with additional increases in AmpliTaq or Taq Gold concentration (data not shown). Thus, the Alu enrichment observed in these polymerase blend experiments is consistent with bona fide lesion bypass and not an increase in PCR efficiency mediated by the presence of more DNA polymerase. An amplification negative control was run with each experiment in which all PCR reactants, including the Dpo4 enzyme, were present with the exception of the human DNA template. All such amplification negative controls indicated the absence of human DNA contamination in any of the PCR reactants, including the Dpo4-like enzyme preparations. Sequencing of the repaired substrates was not conducted, since it would not provide confirmation that repair of UV-damaged DNA had taken place. Some of the original starting molecules will have been bypassed, some will remain undamaged and some will be damaged but be permissive of lesion bypass. If this population was sequenced, it is likely that the vast majority would possess that wild-type sequence, including even some that were repaired and possess the wild-type sequence. Thus, the chances of obtaining a few sequences that differ from the wild-type are remote and, if sequenced subsequent to cloning, could be confused with the normal PCR mutational processes.
Figure 6.
PCR amplification of UV-damaged DNA. (A) Human genomic DNA (K562) was damaged by exposure to UVC at a flux rate of 0.197 J/cm2/min. Two nanograms of damaged genomic DNA were amplified with primers specific for the Major and Precise Alu subfamilies (21). The PCR contained, inter alia, 2.5 U Taq Gold DNA Polymerase, 20 pmol each of the forward and reverse primers and, if applicable, 100 nM Dpo4. The forward primer was labeled at its 5′ end with the reactive fluorescent dye 6-FAM. Amplified fragments were separated by capillary electrophoresis and detected by laser-induced fluorescence of the incorporated dye-labeled primer. The electropherograms depict Alu element amplification products of the UVC damaged genomic substrate with Taq Gold DNA polymerase alone (1) or with a cocktail of Taq DNA polymerase and 100 nM Dpo4 (2). (B) Experimental details are the same as (A) except that the DNA was exposed to UVC at the same flux rate for 30 min and PCR was performed at 85°C, as noted in Materials and Methods. Results show the Alu element products obtained with AmpliTaq DNA polymerase alone (1) or with a cocktail of Taq DNA polymerase and 100 nM Ste (2) or Ain (3).
Experiments are currently in progress to assess the success of this same strategy for the recovery of locus specific microsatellite marker profiles from damaged blood, semen and other biological stains, with Dpo4 and the Ain/Sso and Ain/Ste chimeras. Possible improvements in the efficiency of the repair of genomic DNA templates possessing multiple types of DNA lesions include the development of an in vitro system that combines translesion synthesis and base-excision repair.
CONCLUSIONS
Here, we report a quick and simple method for the identification of polIV-like polymerases from Archaea. Due to their thermostable nature, large quantities of the recombinant enzymes can be readily purified from E.coli. Of the five novel enzymes reported here, three were Dpo4-like in their processivity and lesion bypassing properties, while two were Dbh-like and exhibited poor processivity and lesion bypass. Second generation recombinant enzymes, with improved enzymatic properties, were simply generated by interchanging the LF domain of the respective polymerase. Last, but most significantly, we have shown that by using a thermostable Y-family polymerase in a blend with a conventional PCR enzyme like Taq, we can PCR amplify damaged DNA samples. This has immediate applications in forensic science and the analysis of ancient DNA samples, which are often degraded and are not easily amplified by Taq alone.
Acknowledgments
This work was supported by funds from the NIH/NICHD Intramural Research Program (J.P.M. and R.W.), the EU Marie Curie training and mobility program, project number MRTN-CT 2003-505086 ‘CLUSTOXDNA (J.C.)’, and Office of Justice Programs, National Institute of Justice, Department of Justice, Award Number 2002-IJ-CX-K001 (J.B.). Points of view in this document are those of the authors and do not necessarily represent the official position of the US Department of Justice; the authors would also like to thank Dr Li Huang, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100080, People's Republic of China for the generous gift of the S.tengchongensis genomic DNA. Funding to pay the Open Access publication charges for this article was provided by the NIH/NICHD Intramural Research Program.
Conflict of interest statement. J.P.M and R.W are co-inventors on the International Patent Application No. PCT/US2005/017941 by the National Institutes of Health, which was filed on May 20, 2005.
REFERENCES
- 1.Ohmori H., Friedberg E.C., Fuchs R.P.P., Goodman M.F., Hanaoka F., Hinkle D., Kunkel T.A., Lawrence C.W., Livneh Z., Nohmi T., et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 2.Kulaeva O.I., Koonin E.V., McDonald J.P., Randall S.K., Rabinovich N., Connaughton J.F., Levine A.S., Woodgate R. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res. 1996;357:245–253. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 3.Boudsocq F., Iwai S., Hanaoka F., Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DNA polymerase with lesion-bypass properties akin to eukaryotic polη. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruz P., Pisani F.M., Shimizu M., Yamada M., Hayashi I., Morikawa K., Nohmi T. Synthetic activity of Sso DNA polymerase Y1, an archaeal DinB-like DNA polymerase, is stimulated by processivity factors proliferating cell nuclear antigen and replication factor C. J. Biol. Chem. 2001;276:47394–47401. doi: 10.1074/jbc.M107213200. [DOI] [PubMed] [Google Scholar]
- 5.Potapova O., Grindley N.D., Joyce C.M. The mutational specificity of the dbh lesion bypass polymerase and its implications. J. Biol. Chem. 2002;277:28157–28166. doi: 10.1074/jbc.M202607200. [DOI] [PubMed] [Google Scholar]
- 6.Boudsocq F., Kokoska R.J., Plosky B.S., Vaisman A., Ling H., Kunkel T.A., Yang W., Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low-fidelity synthesis and translesion replication. J. Biol. Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 7.Zhou B., Pata J.D., Steitz T.A. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 8.Silvian L.F., Toth E.A., Pham P., Goodman M.F., Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nature Struct. Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 9.Ling H., Boudsocq F., Woodgate R., Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 10.Kokoska R.J., McCulloch S.D., Kunkel T.A. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 11.McCulloch S.D., Kokoska R.J., Masutani C., Iwai S., Hanaoka F., Kunkel T.A. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 12.Ling H., Boudsocq F., Woodgate R., Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 13.Ling H., Boudsocq F., Plosky B.S., Woodgate R., Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 14.Ling H., Sayer J.M., Plosky B.S., Yagi H., Boudsocq F., Woodgate R., Jerina D.M., Yang W. Crystal structure of a Benzo[a]pyrene Diol Epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl Acad. Sci. USA. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellinger R.E., Thoma F. Taq DNA polymerase blockage at pyrimidine dimers. Nucleic Acids Res. 1996;24:1578–1579. doi: 10.1093/nar/24.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sikorsky J.A., Primerano D.A., Fenger T.W., Denvir J. Effect of DNA damage on PCR amplification efficiency with the relative threshold cycle method. Biochem. Biophys. Res. Commun. 2004;323:823–830. doi: 10.1016/j.bbrc.2004.08.168. [DOI] [PubMed] [Google Scholar]
- 17.Xiang X., Dong X., Huang L. Sulfolobus tengchongensis sp. nov., a novel thermoacidophilic archaeon isolated from a hot spring in Tengchong, China. Extremophiles. 2003;7:493–498. doi: 10.1007/s00792-003-0355-2. [DOI] [PubMed] [Google Scholar]
- 18.McDonald J.P., Frank E.G., Levine A.S., Woodgate R. Intermolecular cleavage of the UmuD-like mutagenesis proteins. Proc. Natl Acad. Sci. USA. 1998;95:1478–1483. doi: 10.1073/pnas.95.4.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasparutto D., Ait-Abbas M., Jaquinod M., Boiteux S., Cadet J. Repair and coding properties of 5-hydroxy-5-methylhydantoin nucleosides inserted into DNA oligomers. Chem. Res. Toxicol. 2000;13:575–584. doi: 10.1021/tx000005+. [DOI] [PubMed] [Google Scholar]
- 20.Englander E.W., Howard B.H. Alu-mediated detection of DNA damage in the human genome. Mutat. Res. 1997;385:31–39. doi: 10.1016/s0921-8777(97)00036-0. [DOI] [PubMed] [Google Scholar]
- 21.Kariya Y., Kato K., Hayashizaki Y., Himeno S., Tarui S., Matsubara K. Revision of consensus sequence of human Alu repeats—a review. Gene. 1987;53:1–10. doi: 10.1016/0378-1119(87)90087-4. [DOI] [PubMed] [Google Scholar]
- 22.McDonald J.P., Maury E.E., Levine A.S., Woodgate R. Regulation of UmuD cleavage: role of the amino-terminal tail. J. Mol. Biol. 1998;282:721–730. doi: 10.1006/jmbi.1998.2044. [DOI] [PubMed] [Google Scholar]
- 23.Friedberg E.C., Walker G.C., Siede W. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology; 1995. [Google Scholar]
- 24.Hoss M., Jaruga P., Zastawny T.H., Dizdaroglu M., Paabo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadet J. DNA damage caused by oxidation, deamination, ultraviolet radiation and photoexcited psoralens. IARC Sci. Publ. 1994:245–276. [PubMed] [Google Scholar]
- 26.Hall A., Ballantyne J. Characterization of UVC-induced DNA damage in bloodstains: forensic implications. Anal. Bioanal. Chem. 2004;380:72–83. doi: 10.1007/s00216-004-2681-3. [DOI] [PubMed] [Google Scholar]
- 27.Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 28.Page R.D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]