A Spectrum of PCSK9 Alleles Contributes to Plasma Levels of Low-Density Lipoprotein Cholesterol (original) (raw)
Abstract
Selected missense mutations in the proprotein convertase subtilisin/kexin type 9 serine protease gene (PCSK9) cause autosomal dominant hypercholesterolemia, whereas nonsense mutations in the same gene are associated with low plasma levels of low-density lipoprotein cholesterol (LDL-C). Here, DNA sequencing and chip-based oligonucleotide hybridization were used to determine whether other sequence variations in PCSK9 contribute to differences in LDL-C levels. The coding regions of PCSK9 were sequenced in the blacks and whites from the Dallas Heart Study (_n_=3,543) who had the lowest (<5th percentile) and highest (>95th percentile) plasma levels of LDL-C. Of the 17 missense variants identified, 3 (R46L, L253F, and A443T) were significantly and reproducibly associated with lower plasma levels of LDL-C (reductions ranging from 3.5% to 30%). None of the low–LDL-C variants were associated with increased hepatic triglyceride content, as measured by proton magnetic resonance spectroscopy. This finding is most consistent with the reduction in LDL-C being caused primarily by accelerating LDL clearance, rather than by reduced lipoprotein production. Association studies with 93 noncoding single-nucleotide polymorphisms (SNPs) at the PCSK9 locus identified 3 SNPs associated with modest differences in plasma LDL-C levels. Thus, a spectrum of sequence variations ranging in frequency (from 0.2% to 34%) and magnitude of effect (from a 3% increase to a 49% decrease) contribute to interindividual differences in LDL-C levels. These findings reveal that PCSK9 activity is a major determinant of plasma levels of LDL-C in humans and make it an attractive therapeutic target for LDL-C lowering.
Elevated levels of plasma low-density lipoprotein cholesterol (LDL-C) are a major risk factor for the development and progression of atherosclerosis, which can result in coronary artery disease and stroke. Plasma levels of LDL-C vary over a threefold range in the population, and both family and twin studies have consistently shown that ∼50% of the variation is genetic in etiology (Rao et al. 1982; Heller et al. 1993). Although multiple genetic defects have been identified that cause rare Mendelian forms of severe hypercholesterolemia or hypocholesterolemia, the sequence variations in the genome accounting for most of the variation in plasma cholesterol levels in the general population have not been determined.
LDLs are formed in the circulation as a metabolic product of very-low-density lipoprotein (VLDL) and are cleared from the blood by LDL receptor (LDLR)–mediated endocytosis in the liver. Disruption of the LDLR pathway is associated with severe hypercholesterolemia. Inactivating mutations in LDLR (MIM 606945) or in the LDLR-binding region of apolipoprotein B-100 (apoB-100) cause severe autosomal dominant hypercholesterolemia (MIM 144010) (Innerarity et al. 1990; Goldstein et al. 2001), and mutations in ARH, an adaptor protein required for LDLR endocytosis, cause autosomal recessive hypercholesterolemia (MIM 603813) (Garcia et al. 2001). No common sequence variations in the genes defective in Mendelian forms of hypercholesterolemia (or hypocholesterolemia) have been convincingly and reproducibly shown to contribute significantly to interindividual differences in plasma levels of cholesterol in the general population.
Recently, selected missense mutations in PCSK9 were found to cause severe hypercholesterolemia (Abifadel et al. 2003; Leren 2004; Timms et al. 2004). PCSK9 encodes a 692-aa glycoprotein that is a member of the subtilisin/kexin type 9 serine protease subfamily of proprotein convertases. PCSK9 contains a signal sequence and prodomain at its N-terminus, followed by a catalytic domain and cysteine-rich carboxy-terminal domain (fig. 1) (Naureckiene et al. 2003; Seidah et al. 2003; Benjannet et al. 2004). PCSK9 undergoes autocatalytic cleavage in the endoplasmic reticulum, releasing the aminoterminal prodomain (Naureckiene et al. 2003; Seidah et al. 2003; Benjannet et al. 2004). The prodomain remains associated with the processed form of PCSK9 as it transits through the secretory pathway (Seidah et al. 2003). PCSK9 is expressed most abundantly in the liver, kidney, and small intestine (Seidah et al. 2003). Hepatic levels of PCSK9 mRNA change inversely with cholesterol feeding in mice (Maxwell et al. 2003) and are elevated in mice overexpressing constitutively active forms of the cholesterol regulatory transcription factors SREBP-1a and SREBP-2 (Horton et al. 2003).
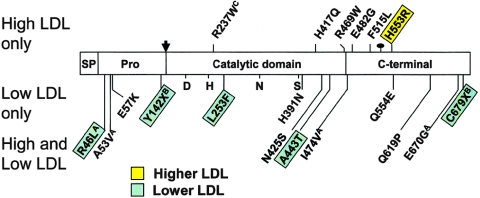
Figure 1.
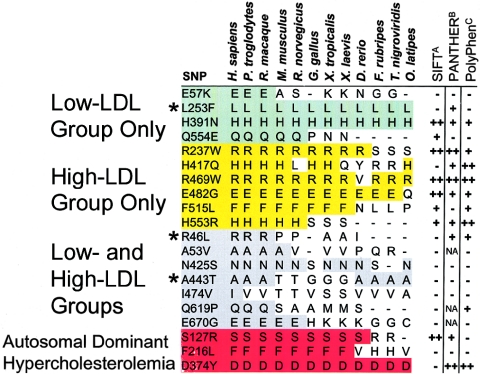
Nonsynonymous sequence variations in PCSK9 identified in the DHS subjects. PCSK9 contains a 30-aa signal peptide (SP) followed by a prodomain (Pro). The proconvertase undergoes autocatalytic processing to release the 14-kDa prodomain peptide from the amino-terminus (Benjannet et al. 2004). The catalytic domain, which contains the catalytic triad of aspartate (D), histidine (H), and serine (S) as well as a highly conserved asparagine (N), is followed by a carboxy-terminal domain, which contains an _N_-linked glycosylation site. The sites of the nonsynonymous mutations identified in only the high-LDL subjects, in only the low-LDL subjects, or in both groups of the DHS sample are shown. The mutations that are significantly associated with an increase (yellow) or decrease (green) in plasma LDL-C levels in the DHS samples are indicated. Mutations identified initially by Abifadel et al. (2003), Cohen et al. (2005), or Benjannet et al. (2004) are indicated by a superscript A, B, or C, respectively.
The physiological substrate(s) of PCSK9 is not known, but high-level expression of recombinant PCSK9 in the livers of mice results in a pronounced reduction in LDLR protein level and hypercholesterolemia without any associated changes in LDLR mRNA level (Benjannet et al. 2004; Maxwell and Breslow 2004; Park et al. 2004). These data suggest that missense mutations in PCSK9 cause hypercholesterolemia by a gain-of-function mechanism and promote the degradation of LDLRs in hepatocytes (Benjannet et al. 2004; Maxwell and Breslow 2004; Park et al. 2004). An alternative hypothesis is that PCSK9 affects plasma LDL-C levels by altering the rate of secretion of apoB-100 in an LDLR-independent manner (Benjannet et al. 2004; Ouguerram et al. 2004; Sun et al. 2005). Metabolic studies measuring VLDL–apoB-100 synthesis in two individuals with a missense mutation in PCSK9 found a significant increase in VLDL production (Ouguerram et al. 2004), whereas conflicting results have been obtained with regard to the effect of PCSK9 expression on apoB-100 synthesis in the livers of mice or in cultured hepatocytes isolated from mice expressing recombinant PCSK9 (Park et al. 2004; Lalanne et al. 2005). Additional studies will be required to determine whether increased synthesis of apoB-containing lipoproteins by the liver contributes to the hypercholesterolemia associated with missense mutations in PCSK9.
Mutations in PCSK9 result not only in severe hypercholesterolemia but also in hypocholesterolemia (Cohen et al. 2005). Two different nonsense mutations in PCSK9 are associated with a 40% reduction in mean plasma levels of LDL-C (Cohen et al. 2005), presumably because of a loss of PCSK9 function resulting in elevated hepatic LDLR activity. Mice lacking PCSK9 have markedly increased hepatic LDLR levels (Rashid et al. 2005). Thus, selected missense mutations in PCSK9 produce hypercholesterolemia due to a gain of function, resulting in reduced hepatic levels of LDLR, whereas inactivating mutations in the same gene cause hypocholesterolemia due to a loss of function, resulting in an increased level of LDLR in the liver.
Since mutations in PCSK9 can cause severe dominant hypercholesterolemia (Abifadel et al. 2003; Leren 2004; Timms et al. 2004) as well as significantly reduced plasma levels of LDL-C (Cohen et al. 2005), other investigators have queried whether other more common sequence variations in PCSK9 contribute to variations in plasma levels of LDL-C. Shioji et al. (2004) identified two SNPs, and Chen et al. (2005) identified a haplotype associated with differences in plasma LDL-C level, but no systematic examination of the relationship between sequence variations in PCSK9 and plasma levels of LDL-C has been reported.
The aim of the present study was to characterize the spectrum of sequence variations in PCSK9 and to investigate the contributions of these variations to differences in plasma levels of LDL-C in the general population. In addition, we investigated whether functional sequence variations in PCSK9 associated with low plasma levels of LDL-C are also associated with increased hepatic triglyceride content (HTGC), as would be expected if defects in PCSK9 cause reduced production of apoB-containing lipoproteins by the liver.
Subjects and Methods
Subjects
Informed consent, blood samples, and clinical evaluations were obtained for all participating subjects by institutional review board–approved protocols. The study population included all participants in the Dallas Heart Study (DHS) from whom fasting venous blood samples were obtained (_n_=3,543). The DHS sample is a multiethnic, probability-based sample of Dallas County, weighted to include 50% blacks, in which ethnicity was self-assigned in accordance with United States census categories (Victor et al. 2004). In the present study, “blacks” and “whites” refer to individuals who self-identified as non-Hispanic black and non-Hispanic white, respectively. The blood samples were maintained at 4°C until the plasma and serum were separated, aliquoted, and stored at −80°C. Genomic DNA was isolated from the leukocytes with Pure Gene (Gentra Systems). Significant associations were replicated in a sample of blacks from Maywood, Cook County, Illinois (Cohen et al. 2005).
Measurement of Plasma LDL-C Levels and Determination of HTGC
Plasma LDL-C concentrations were determined using commercial enzymatic reagents. HTGC was determined by proton nuclear magnetic resonance spectroscopy as described elsewhere (Browning et al. 2004; Szczepaniak et al. 2004). Hepatic steatosis was defined as HTGC >5.5% (Browning et al. 2004).
PCR and DNA Sequencing
Within each ethnic- and sex-specific group, plasma LDL-C levels were adjusted for the effects of age by linear regression and for the effects of lipid-lowering medications by the assumption of a 25% decrease in plasma LDL-C level. For sequencing, we selected the 5% of individuals with the highest and lowest adjusted plasma LDL-C levels in each ethnic- and sex-specific group whose fasting plasma levels of triglycerides were <250 mg/dl. The 5th and 95th percentiles, respectively, of the adjusted plasma LDL-C levels were 49 mg/dl and 177 mg/dl for black men (_n_=770), 56 mg/dl and 172 mg/dl for black women (_n_=1,051), 61 mg/dl and 171 mg/dl for white men (_n_=499), and 57 mg/dl and 168 mg/dl for white women (_n_=544).
The exons and flanking intronic sequences of PCSK9 were amplified by PCR and treated with recombinant exonuclease I and shrimp alkaline phosphatase (Exo-Sap [USB]). Both strands of each product were sequenced on an ABI 3730 automated sequencer with BigDye terminator cycle sequencing reagents (Applied Biosystems). The sequences of the oligonucleotides used for sequencing are available on request.
Assay of Mutations
Specific 5′-nucleotidase assays for all nonsynonymous sequence variations identified by sequencing in the present study were developed using the TaqMan system (Applied Biosystems). The assays were performed on an HT7900 Real-Time PCR system with probes and reagents purchased from Applied Biosystems. All other SNPs were genotyped by PCR-based amplification of genomic DNA, followed by hybridization to high-density oligonucleotide arrays (Perlegen Sciences).
Statistical Analysis
We used the statistical software package R for all statistical analyses (The R Development Core Team 2004; The R Project for Statistical Computing). The observed genotype frequencies were tested for deviation from Hardy-Weinberg equilibrium by using χ2 tests. Tests for association were performed in the DHS sample and were then validated by replication in the Cook County sample. Individuals taking lipid-lowering drugs were excluded from all tests for association with plasma LDL-C levels. Lipoprotein levels were adjusted for age and sex by linear regression and were tested for association with each sequence variant by using one-way analysis of variance, to compare the mean values of the three genotype groups. Genotypes represented by fewer than six individuals were omitted from the analysis. Blacks and whites in the DHS sample were analyzed separately. Sequence variants found to be significantly associated with plasma LDL-C levels at a nominal significance threshold of 0.05 in the DHS blacks were tested for association in the Cook County sample by using unpaired t tests. Since the direction of effect was already established in the DHS sample, associations in the Cook County sample were examined using one-sided tests. Sequence variants that achieved a nominal significance threshold of 0.05 in the DHS and Cook County samples and that showed the same direction of effect in the two samples were considered to be statistically significant.
Haplotype Analysis of PCSK9
We estimated haplotype blocks in each ethnic group, using the expectation-maximization algorithm (as implemented in Haploview [Barrett et al. 2005]) and the Gabriel et al. (2002) definition of haplotype blocks, and we calculated the association of each haplotype block with LDL-C values by using the haplo.stats package (Mayo Foundation for Medical Education and Research). The results are presented as P values based on the global statistic for the association of haplotype blocks with LDL-C level. Testing was performed at the nominal significance threshold of 0.05.
Results
Nonsynonymous Sequence Variations in PCSK9
To identify sequence variations in PCSK9 associated with hyper- or hypocholesterolemia in the general population, we sequenced the exons and flanking intronic regions of PCSK9 in four groups of individuals (black men, black women, white men, and white women) from the DHS sample who had either very low (<5th percentile) or very high (>95th percentile) plasma levels of LDL-C. The DHS sample is a multiethnic (52% non-Hispanic black, 29% non-Hispanic white, 17% Hispanic, and 2% other ethnicities) probability-based population sample of Dallas County for which ethnicity was self-assigned (Victor et al. 2004). The low- and high-LDL groups each included 39 (of 770) black men, 50 (of 1,051) black women, 25 (of 499) white men, and 27 (of 544) white women. A total of 6.1% of the sample population were taking cholesterol-lowering agents (6.0% of blacks and 7.6% of whites). Since the mean reduction in plasma LDL-C level in individuals taking a statin is ∼25% (Sacks and Katan 2002), we estimated the untreated plasma LDL-C level of those individuals taking a cholesterol-lowering agent by adding 33% to their levels measured while taking treatment. A total of 35 subjects who were taking lipid-lowering agents were included among the 141 individuals in the high-LDL group (26 of 89 blacks and 9 of 52 whites).
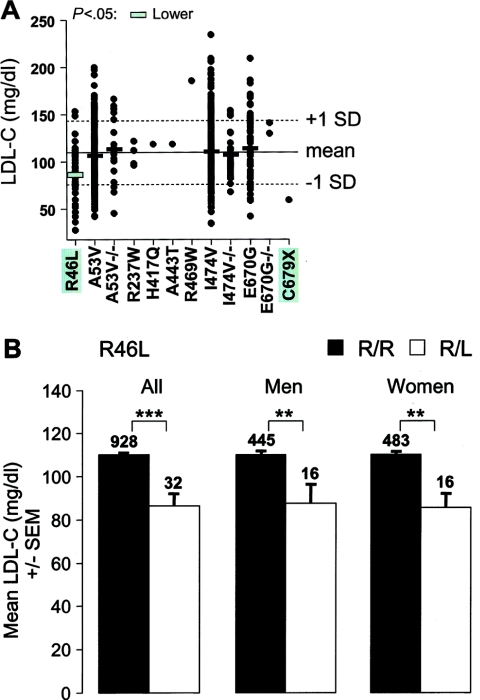
DNA sequencing revealed 19 nonsynonymous sequence variations in PCSK9, including 17 missense mutations and 2 nonsense mutations (fig. 1). Six mutations (four missense mutations and two nonsense) were identified in only the low-LDL subjects, and six were found in only the high-LDL subjects. Thus, the overall prevalence of PCSK9 mutations was similar in the groups with low and high LDL-C. We have shown elsewhere that both of the nonsense mutations in PCSK9 are associated with low plasma levels of LDL-C (Cohen et al. 2005). To determine whether any of the missense mutations identified were associated with plasma LDL-C levels, each mutation was assayed in the entire DHS sample with fluorogenic 5′-nucleotidase (TaqMan) assays (table 1). In the DHS sample, all 19 nonsynonymous mutations were present in blacks, whereas only 9 were found in whites. Accordingly, the two populations were analyzed separately by using one-way analysis of variance. Only one of the nonsynonymous sequence variations, R46L, was significantly associated with plasma LDL-C levels in the white subjects (P<10-4) (fig. 2A). In the white men and white women, heterozygotes for the minor allele (L46) had significantly lower plasma levels of LDL-C (fig. 2B).
Table 1.
Nonsynonymous Sequence Variations in PCSK9 Identified in Blacks and Whites in the DHS Sample Who Had Plasma LDL-C Levels <5th or >95th Percentile
| Minor-Allele Frequencya in DHS(%) | ||||
|---|---|---|---|---|
| Subjects andNucleotide Substitution | Amino AcidSubstitution | Blacks(_n_=1,822) | Whites(_n_=1,045) | Hispanics(_n_=601) |
| Low-LDL subjects only: | ||||
| c.169G→A | E57K | .33 | … | … |
| c.426C→G | Y142Xb | .19 | … | … |
| c.757C→T | L253F | .25 | … | .084 |
| c.1171C→A | H391N | .41 | … | .083 |
| c.1660C→G | Q554E | .30 | … | .084 |
| c.2037C→A | C679Xb | .72 | .048 | .084 |
| Total | 2.2 | .048 | .33 | |
| High-LDL subjects only: | ||||
| c.709C→T | R237Wb | .028 | .24 | .084 |
| c.1251C→A | H417Q | .30 | .048 | … |
| c.1405C→T | R469W | .75 | .049 | .084 |
| c.1445A→G | E482G | .11 | … | … |
| c.1545T→G | F515L | .028 | … | … |
| c.1658A→G | H553R | 1.3 | … | … |
| Total | 2.5 | .33 | .17 | |
| Both low- and high-LDL subjects: | ||||
| c.137G→T | R46Lb | .28 | 1.6 | .75 |
| c.158C→T | A53Vb | 1.8 | 13 | 4.6 |
| c.1274A→G | N425S | 1.9 | … | .17 |
| c.1327G→A | A443T | 9.4 | .048 | 1.3 |
| c.1420A→G | I474Vb | 22 | 18 | 9.7 |
| c.1856A→C | Q619P | 1.5 | … | … |
| c.2009A→G | E670Gb | 26 | 3.6 | 4.2 |
| Total | 62 | 35 | 20 |
Figure 2.
Plasma LDL-C levels in whites in the DHS sample. A, Subjects were genotyped by specific 5′-nucleotidase assay. The mean age- and sex-adjusted plasma LDL-C level of all whites in the DHS sample is indicated by the solid horizontal line (±1 SD is indicated by the dotted lines). Each point in the plot represents the LDL-C level of an individual who is heterozygous or homozygous for the rare allele (−/−) of the indicated sequence variation. Horizontal bars, Mean plasma LDL-C levels for each sequence variant (omitted if n<6; see the “Subjects and Methods” section). The green-shaded bar indicates a significantly lower plasma LDL-C level (P<10-4, by one-way analysis of variance). B, Mean (± SEM) of the plasma LDL-C levels for the R46L variation. R46L is significantly associated with decreased plasma LDL-C level in the DHS whites (left set of bars), white men (center set of bars), and white women (right set of bars). The number of subjects in each group is given above the bars. **P<.01, ***P<10-4; by two-sided t test.
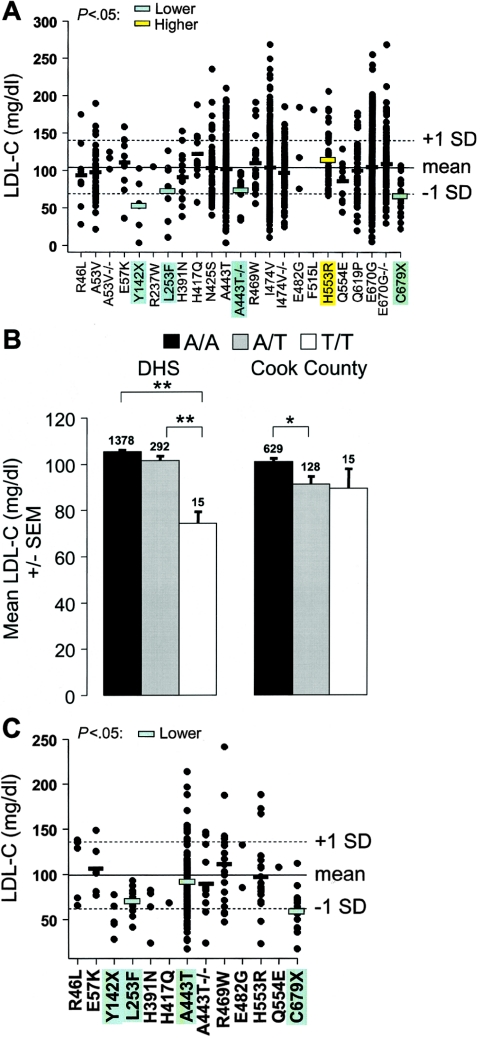
Among blacks, three missense mutations (L253F, A443T, and H553R) were associated with plasma LDL-C levels (fig. 3A). The L253F and A443T mutations were associated with low LDL-C levels: heterozygotes for the F253 allele had a 30% reduction in mean plasma LDL-C levels (L/L 104.6 mg/dl; L/F 73.2 mg/dl), whereas homozygotes for the T443 variant had a 29% reduction in plasma levels of LDL-C (fig. 3A and 3B). The H553R mutation was associated with increased plasma levels of LDL-C (P<.05). All three mutations were either very rare (A443T) or absent (L235F and H553R) among whites. The R46L sequence variation, which was significantly associated with LDL-C levels in whites, was much less common in blacks (minor-allele frequency 0.28%), and the decrease in LDL-C levels among heterozygotes did not achieve statistical significance in this population (mean plasma levels: R/R 104.5 mg/dl; R/L 93.8 mg/dl; _P_=.35).
Figure 3.
Plasma LDL-C levels in blacks in the DHS (A) and Cook County (C) samples. The mean (solid lines) and SD (dashed lines) of plasma LDL-C levels adjusted for age and sex are shown. Horizontal bars, Mean plasma LDL-C levels for each sequence variant (omitted if n<6). Significantly lower or higher plasma LDL-C levels are indicated by green- or yellow-shaded bars, respectively. B, Mean (± SEM) of the plasma LDL-C level at the A443T variation in the DHS blacks (left set of bars) and Cook County subjects (right set of bars). The number of subjects in each group is given above the bars. *P<.05, **P<.01; by pairwise two-sided t tests with Holm correction for multiple comparisons.
To test the reproducibility of these findings, we assayed the nonsynonymous PCSK9 sequence variations that were significantly associated with plasma LDL-C levels and all nonsynonymous sequence variants that were found only in the low-LDL group or only in the high-LDL group in 850 blacks from Cook County (fig. 3C). The mean plasma LDL-C level of the Cook County sample was slightly lower than that of the DHS sample, perhaps reflecting the younger ages and lower BMIs (calculated as weight in kilograms divided by the square of height in meters) of the Cook County participants (table 2). Our power to detect associations was lower in the Cook County sample than in the DHS blacks because of the smaller sample size. Nevertheless, the L235F and A443T sequence variations were both significantly associated with low LDL-C levels in the Cook County sample. The H553R variation was not associated with LDL-C levels in this sample. This could reflect limited statistical power to detect modest effects of rare alleles, but the finding that the mean plasma LDL-C level was slightly lower, rather than higher, in the H553R heterozygotes than in the H553 homozygotes suggests that this is unlikely.
Table 2.
Demographic Characteristics of the Blacks in the DHS and Cook County Samples[Note]
| All Blacks | Black Men | Black Women | ||||
|---|---|---|---|---|---|---|
| Characteristic | DHS | Cook County | DHS | Cook County | DHS | Cook County |
| Total no. of subjects | 1,822 | 850 | 771 | 523 | 1,051 | 315 |
| No. (%) of men | 771 (42) | 523 (62)a | 771 (100) | 523 (100) | NA | NA |
| Age (years) | 45 ± 10 | 42 ± 7a | 45.1 ± 10 | 43 ± 7a | 45 ± 10 | 41 ± 7a |
| BMI | 32.0 ± 8.1 | 26.6 ± 7.3a | 29.6 ± 7.1 | 25.2 ± 5.9a | 33.7 ± 8.8 | 28.8 ± 9.1a |
| Cholesterol (mg/dl) | 177 ± 39 | 172 ± 38b | 177 ± 39 | 170 ± 37b | 178 ± 39 | 175 ± 39 |
| Triglyceride (mg/dl)c | 88 (59) | 85 (62) | 92 (72) | 85 (64) | 83 (50) | 82 (59) |
| LDL-C (mg/dl) | 104 ± 36 | 99 ± 37a | 104 ± 37 | 96.8 ± 36a | 105 ± 35 | 103 ± 39 |
| HDL-C (mg/dl) | 52 ± 15 | 52 ± 15 | 50 ± 16 | 52 ± 16d | 54 ± 15 | 52 ± 15d |
| VLDL-C (mg/dl)c | 17 (12) | 17 (12) | 18 (14) | 17 (13) | 17 (10) | 16 (12) |
Mutations in PCSK9 Are Not Associated with Increased HTGC
Mutations that cause low plasma levels of LDL-C due to reduced hepatic VLDL secretion are associated with accumulation of triglyceride in the liver (Schonfeld et al. 2003). If mutations in PCSK9 cause a reduction in plasma LDL-C levels by reducing the rate of VLDL secretion, it would be anticipated that subjects with hypocholesterolemia due to mutations in PCSK9 would have hepatic steatosis. We examined the relationship between the LDL-lowering variants in PCSK9 (R46L in whites and Y142X [Cohen et al. 2005], L253F, A443T, and C679X [Cohen et al. 2005] in blacks) and the HTGC as determined using proton magnetic resonance spectroscopy (Browning et al. 2004; Szczepaniak et al. 2004) in 747 whites and 1,090 blacks in the DHS sample. The HTGC was not measured in the one DHS white individual who had a PCSK9 nonsense mutation. The “LDL-lowering variants” group includes the DHS whites with R46L (_n_=26) and the DHS blacks with a nonsense mutation (_n_=20), L253F (_n_=3), or A443T (heterozygotes and homozygotes, _n_=185).
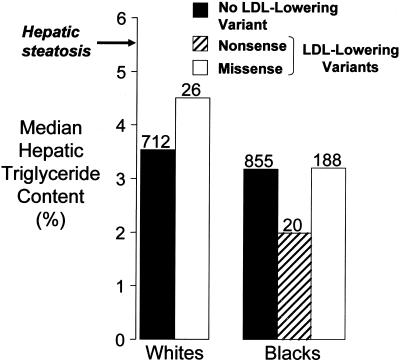
In the whites, the median HTGC did not differ significantly between the subjects with the LDL-lowering variant R46L, and the subjects with no LDL-lowering variant (4.5% vs. 3.5%; _P_=.4, by two-sided Wilcoxon rank sum test) (fig. 4). In the blacks, the median HTGC did not differ significantly between the subjects with an LDL-lowering nonsense mutation and the subjects with no LDL-lowering variant (2.0% vs. 3.2%; _P_=.2) or between the subjects with an LDL-lowering missense variant and the subjects with no LDL-lowering variant (3.2% in both groups). Essentially identical results were obtained when individuals with diabetes or impaired glucose tolerance were excluded (data not shown).
Figure 4.
Median HTGC in whites and blacks with nonsynonymous sequence variations in PCSK9 associated with low plasma levels of LDL-C. HTGC was measured using proton magnetic resonance spectroscopy as described elsewhere (Browning et al. 2004). The median HTGC of the whites with or without an LDL-lowering variation (R46L) and the median HTGC of the blacks with or without either a nonsense mutation (Y142X or C679X) or a missense sequence variant (L253F or A443T) associated with a lower level of LDL-C were compared (by two-sided Wilcoxon rank sum test). The number of subjects in each group is given above the bars. The threshold for hepatic steatosis, defined as HTGC >5.5% (Browning et al. 2004), is indicated (arrow).
Evolutionary Conservation of Nonsynonymous Sequence Variations in PCSK9
To assess the relationship between the evolutionary conservation and functional effects of the nonsynonymous sequence variations identified, we aligned the human PCSK9 amino acid sequence with the orthologous sequences from other mammals, birds, amphibians, and fish (fig. 5). The three PCSK9 mutations associated with Mendelian hypercholesterolemia were at highly conserved amino acid residues. Of the 17 missense sequence variations identified in the present study, 12 changed amino acid residues that are completely conserved in primates and mice, and 8 were at amino acid residues conserved from humans to frogs. Only one (L253F) of these eight was associated with plasma LDL-C levels. The other two mutations associated with plasma LDL-C levels (R46L and A443T) were at amino acid residues conserved only among primates. One sequence variant (F515L) at a highly conserved residue was identified in only a single hypercholesterolemic black subject, so its functional significance could not be determined. These data indicate that the extent of evolutionary sequence conservation did not reliably predict the impact of sequence variation on protein function.
Figure 5.
Evolutionary sequence conservation and predicted functional effects of missense sequence variations in PCSK9. Conservation for each variation found in the low-LDL subjects only (green), the high-LDL subjects only (yellow), or both groups (gray) of the DHS sample and in patients with autosomal dominant hypercholesterolemia (red) (Abifadel et al. 2003; Leren 2004; Timms et al. 2004) is highlighted. An asterisk (*) indicates a SNP significantly associated with plasma LDL-C level. Sequences are shown for Homo sapiens, Pan troglodytes, Rhesus macaque, Mus musculus, Rattus norvegicus, Gallus gallus, Xenopus tropicalis, Xenopus laevis, Danio rerio, Fugu rubripes, Tetraodon nigroviridis, and Oryzias latipes. The predicted effect of each amino acid sequence variation on protein function is indicated (right columns). ASIFT v.2: − = tolerated; + = deleterious (low-confidence prediction); ++ = deleterious. BPANTHER v.5.0: − = unlikely functional effect; + = possible deleterious functional effect; ++ = high probability of deleterious functional effect; NA = not modeled by the PANTHER hidden Markov model (family sequence alignment absent or poor). CPolyPhen: − = benign; + = possibly damaging; ++ = probably damaging (Sunyaev et al. 2001; Ng and Henikoff 2003; Thomas et al. 2003).
We also examined the utility of three computer-based algorithms for predicting the functional effects of the nonsynonymous variations: PolyPhen (polymorphism phenotyping) (Sunyaev et al. 2001), SIFT (sorting intolerant from tolerant) (Ng and Henikoff 2003), and PANTHER (protein analysis through evolutionary relationships) (Thomas et al. 2003). SIFT and PANTHER use sequence similarity to predict whether an amino acid substitution affects protein function, whereas PolyPhen uses sequence similarity as well as structural information for its predictions. Each of these algorithms predicted that approximately one-half of the PCSK9 missense sequence variations detected in the DHS sample have deleterious effects on PCSK9 function (41%, 57%, and 53% for SIFT, PANTHER, and PolyPhen, respectively) (fig. 5). SIFT failed to correctly identify any of the three missense sequence variations associated with plasma levels of LDL-C and also failed to predict two of the mutations in PCSK9 that cause dominant hypercholesterolemia (Abifadel et al. 2003; Leren 2004; Timms et al. 2004). PANTHER predicted two of the three functional sequence variations as well as two of the three disease-causing mutations; however, this program also incorrectly predicted that six of the missense substitutions would be functional. Finally, PolyPhen correctly predicted just one of the three missense polymorphisms and mutations that were associated with plasma levels of LDL-C.
Noncoding Sequence Variations in PCSK9
To investigate more comprehensively the contributions of synonymous and noncoding sequence variations in PCSK9 to interindividual differences in plasma LDL-C level in the general population, we genotyped the black and white subjects in the DHS sample for 94 SNPs that span the entire PCSK9 locus, extending ∼10 kb upstream and downstream of the 5′ and 3′ UTRs of PCSK9. One of these SNPs is a synonymous substitution in exon 9 (V460V); the remainder are located outside the PCSK9 coding sequence. For each ethnic group, we analyzed all SNPs with a minor-allele frequency of at least 1% (75 SNPs in the whites and 89 SNPs in the blacks).
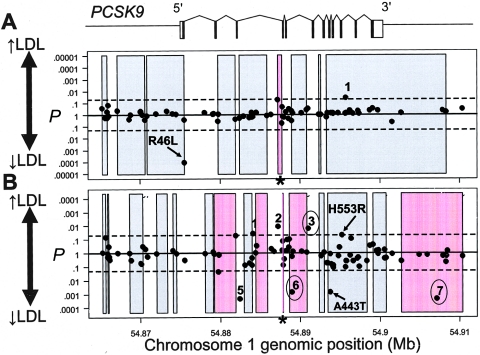
Analysis of variance indicated that one SNP was significantly associated with plasma levels of LDL-C in whites, and seven were significantly associated with plasma levels of LDL-C in blacks (fig. 6). Six other SNPs met the nominal significance threshold of 0.05 but were excluded from further consideration because the mean plasma LDL-C levels of heterozygotes were higher or lower than the mean plasma LDL-C levels of homozygotes for both the common and rare alleles.
Figure 6.
Haplotype block structure across PCSK9 and P values for significance of association of common SNPs with plasma LDL-C level in whites (A) and blacks (B) in the DHS sample. SNPs and haplotype blocks are plotted along the _X_-axis according to genomic position. The P value for each SNP is plotted above or below the midline according to whether the mean plasma LDL-C level of the heterozygous genotype is higher (above midline) or lower (below midline) than the mean plasma LDL-C level of the homozygous common genotype. Coding SNPs significantly associated with plasma LDL-C level are indicated (arrows), and the corresponding amino acid sequence variations are given. Arabic numerals indicate the noncoding SNPs significantly associated with plasma LDL-C level in whites (1) or blacks (1–7). SNPs that were also significant in the Cook County sample are circled. The extents of haplotype blocks are indicated (shaded rectangles). Haplotype blocks significantly associated with plasma LDL-C level are shaded in pink. P values for haplotype blocks are based on the global statistic. Asterisks (*) indicate haplotype blocks that were significantly associated with plasma LDL-C level after exclusion of all SNPs that were individually associated with plasma LDL-C level. Dashed lines, _P_=.05. The genomic structure of PCSK9 is shown schematically above the plots. Genomic positions are based on National Center for Biotechnology Information (NCBI) build 34 of the human genome.
The single noncoding SNP that was associated with LDL-C levels in whites was rare (minor-allele frequency 1.7%) and was not associated with LDL-C levels in blacks (data not shown). Therefore, this allele was not evaluated further. For each of the seven noncoding SNPs that were significantly associated with plasma LDL-C levels in blacks, we tested for replication in the Cook County subjects (table 3). The genotype frequencies of all seven SNPs were similar in the two populations. Three of the seven noncoding SNPs were also significantly associated with plasma LDL-C levels in the Cook County subjects (SNPs 3, 6, and 7 in fig. 6B). SNP 7 is located downstream of the PCSK9 3′ UTR, in the last intron of an adjacent gene, USP24, which is transcribed in the opposite direction from PCSK9 and encodes a ubiquitin carboxyl-terminal hydrolase. No diseaseassociated mutations in USP24 have been reported to date. SNP 7 is in linkage disequilibrium with the SNP corresponding to A443T (_r_2=0.5; _D_′=0.85). The effects of these noncoding SNPs on plasma LDL-C level were much smaller than those of the coding sequence variations (2.6% increase to 4.1% decrease) (table 4). None of these three noncoding SNPs were located in a region of sequence conservation between human and mouse PCSK9, as determined by VISTA alignment (Couronne et al. 2003).
Table 3.
Association between Sequence Variations in PCSK9 and Plasma LDL-C Levels in Blacks[Note]
| Subjects and Variants | DHS Blacksa(_n_=1,711) | Cook County Sampleb(_n_=780) |
|---|---|---|
| Low-LDL subjects only: | ||
| E57K | .60 | .637 |
| Y142X | .00017 | NA |
| L253F | .021 | .00010 |
| H391N | .156 | NA |
| Q554E | .103 | NA |
| C679X | 4.3 × 10 −8 | 1.3 × 10 −5 |
| High-LDL subjects only: | ||
| R237W | NA | − |
| H417Q | .106 | NA |
| R469W | .47 | .158 |
| E482G | NA | NA |
| F515L | NA | − |
| H553R | .0476 | .59 |
| Low- and high-LDL subjects: | ||
| R46L | .345 | NA |
| A53V | .298 | ND |
| N425S | .845 | ND |
| A443T | .0014 | .0027 |
| I474V | .125 | ND |
| Q619P | .387 | ND |
| E670G | .244 | ND |
| Decreased LDL-C: | ||
| 54879790_4427356c | .042 | .45 |
| 54882467_4342503c | .000431 | .93 |
| 54888986_1033060c | .00134 | .0015 |
| 54907116_1033098c | .000462 | .027 |
| Increased LDL-C: | ||
| 54884133_4319928c | .037 | .13 |
| 54887262_1033043c | .011 | .057 |
| 54891027_4814882c | .016 | .014 |
Table 4.
Magnitude of Effect of the PCSK9 Sequence Variations Reproducibly Associated with Plasma LDL-C Levels in the DHS Sample[Note]
| Effect,Sequence Variation,and Genotype | No. ofIndividuals | Mean ± SDLDL-C(mg/dl) | Difference inMean LDL-Ca(mg/dl) |
|---|---|---|---|
| Decreased LDL-C: | |||
| 54875565_R46Lb: | |||
| G/G | 928 | 110.1 ± 33.31 | |
| G/T | 32 | 86.61 ± 30.52 | 23.5 (21.3) |
| 54882140_Y142Xc: | |||
| C/C | 1,691 | 104.7 ± 35.73 | |
| C/G | 7 | 53.84 ± 32.89 | 50.9 (48.6) |
| 54888340_L253Fc: | |||
| C/C | 1,692 | 104.6 ± 35.92 | |
| C/T | 7 | 73.17 ± 41.28 | 31.4 (30.0) |
| 54893773_A443Tc: | |||
| G/G | 1,378 | 105.3 ± 36.15 | |
| G/A | 292 | 101.6 ± 33.92 | 3.7 (3.5) |
| A/A | 15 | 74.46 ± 18.48 | 30.8 (29.2) |
| 54899133_C679Xc: | |||
| C/C | 1,672 | 105 ± 35.82 | |
| C/A | 26 | 66.27 ± 20.84 | 38.7 (36.9) |
| 54888986_1033060c: | |||
| A/A | 1,021 | 106.7 ± 36.78 | |
| A/T | 535 | 102.3 ± 34.69 | 4.4 (4.1) |
| T/T | 71 | 92.8 ± 31.14 | 13.9 (13.0) |
| 54907116_1033098c: | |||
| G/G | 1,218 | 105.7 ± 36.67 | |
| G/A | 383 | 102.4 ± 33.92 | 3.3 (3.1) |
| A/A | 21 | 76.46 ± 25.68 | 29.2 (27.6) |
| Increased LDL-C: | |||
| 54891027_4814882c: | |||
| G/G | 704 | 102.4 ± 34.55 | |
| G/A | 750 | 105.1 ± 36.61 | 2.7 (2.6) |
| A/A | 173 | 111 ± 38.60 | 8.6 (8.4) |
Haplotype Blocks in PCSK9
Next, we queried whether combinations of closely linked sequence variations in PCSK9 contributed to plasma levels of LDL-C. We used all SNPs with a minor-allele frequency of at least 1% to construct haplotype blocks across the PCSK9 locus and then calculated a global P value for association of each haplotype block with plasma LDL-C level (fig. 6). Of the nine estimated haplotype blocks in the DHS whites, one was significantly associated with plasma LDL-C level (fig. 6A). Neither of the two sequence variations (R46L and SNP 1033079) that were significantly associated with plasma LDL-C level in the DHS whites is located within this 518-bp haplotype block. Of the 15 estimated haplotype blocks in the DHS blacks, 5 were significantly associated with plasma LDL-C levels (fig. 6B). Two of the PCSK9 sequence variations that were significantly associated with plasma LDL-C level in both the DHS blacks and the Cook County subjects (SNPs 6 and 7 in fig. 6B) were located within one of the five haplotype blocks associated with plasma levels of LDL-C.
To determine whether the observed associations between the haplotype blocks and plasma LDL-C levels were driven by the SNPs that were individually associated with plasma LDL-C levels (see above), we excluded these SNPs from the analysis and retested for association of the haplotype blocks with plasma LDL-C level. In the whites, the exclusion of the significant SNPs did not affect the association of block 6 with plasma LDL-C level (fig. 6A). In the blacks, however, only one of the five haplotype blocks—block 10, which contained just two SNPs (1033044 and 1033045), located 58 bp apart—remained associated (fig. 6B). This block did not overlap with block 6 in the whites, and neither of these two haplotype blocks was located in a region of sequence conservation between human and mouse PCSK9 (Bray et al. 2003; Couronne et al. 2003).
Discussion
The major finding of this study is that a spectrum of alleles of PCSK9 are associated with a range of effects on plasma levels of LDL-C. We identified three missense variations and two noncoding sequence variants that were significantly and reproducibly associated with lower plasma levels of LDL-C. These sequence variants most likely lower LDL-C levels by impairing PCSK9 activity, since nonsense mutations in PCSK9 are associated with low plasma levels of LDL-C (Cohen et al. 2005). In addition, we identified a single noncoding SNP associated reproducibly with a modest (∼3%) but statistically significant increase in LDL-C. The effects of the PCSK9 variants on lipoprotein metabolism were extremely specific; individuals with LDL-lowering PCSK9 alleles did not have significant alterations in plasma levels of other lipids or lipoproteins (table 5) and did not have increased HTGC. These findings are consistent with the sequence variants associated with low plasma levels of LDL-C impairing PCSK9 activity and lowering LDL-C levels by accelerating LDL clearance rather than by reducing the secretion of VLDL from the liver. The spectrum of PCSK9 alleles associated with LDL-C spanned a wide range of allele frequencies (0.2%–34%) and magnitude of phenotypic effects (∼3% increase to 49% reduction in LDL-C) (table 4). Thus, unlike the other genes associated with Mendelian forms of hypercholesterolemia (LDLR, APOB, ARH, ABCG5, and ABCG8), common sequence variants in PCSK9 also contribute significantly to interindividual variation in plasma levels of LDL-C. Taken together, our results suggest that the genetic architecture of plasma LDL-C levels is complex, with multiple common and rare alleles with small and large phenotypic effects contributing significantly to plasma LDL-C levels in the population.
Table 5.
Demographic Characteristics of DHS Blacks With and Without LDL-Lowering Nonsynonymous Variations in PCSK9[Note]
| LDL-LoweringNonsynonymous SequenceVariationsa | ||
|---|---|---|
| Characteristic | Blacks Without | Blacks With |
| Total no. of subjects | 1,423 | 354 |
| No. (%) of men | 601 (42) | 150 (42) |
| Age (years) | 44.9 ± 10.3 | 44.1 ± 10.1 |
| BMI | 32 ± 8.12 | 31.8 ± 7.73 |
| No. (%) of diabetics | 286 (20) | 70 (20) |
| Cholesterol (mg/dl) | 179 ± 39.3 | 172 ± 36.1b |
| Triglyceride (mg/dl)c | 88 (58) | 91 (60) |
| LDL-C (mg/dl) | 106 ± 35.9 | 97.3 ± 34.5d |
| HDL-C (mg/dl) | 52 ± 14.9 | 53.4 ± 16.3 |
| VLDL-C (mg/dl)c | 17 (11) | 18 (12) |
| HTGC (%)c | 3.2 (3.2) | 3.2 (3.0) |
Five of the six PCSK9 alleles that were reproducibly associated with LDL-C in this study caused a reduction in plasma LDL-C levels. Only a single missense mutation that might cause autosomal dominant hypercholesterolemia was identified in this study; one black with an LDL-C of 184 mg/dl had a phenylalanine→leucine substitution at residue 515. We are currently examining the family of this individual to determine whether the sequence variation cosegregates with hypercholesterolemia. None of the missense mutations in PCSK9 that have been associated with autosomal dominant hypercholesterolemia (S127R, F216L, and D374Y) (Abifadel et al. 2003; Leren 2004; Timms et al. 2004) were identified in the DHS sample. One common noncoding sequence variant was associated with a small but reproducible increase in plasma LDL-C levels. Thus, most PCSK9 alleles that contribute to interindividual variation in plasma LDL-C levels in whites and blacks act by reducing PCSK9 activity.
Comprehensive evaluation of both coding and noncoding variation in PCSK9 required statistical analysis of >100 sequence variants (17 nonsynonymous, 1 synonymous, and 93 noncoding). Nonetheless, the genetic associations reported here are unlikely to be a fortuitous consequence of multiple testing. Of the 106 sequence variants tested in blacks in the DHS sample, 5 would be expected to be significantly associated with LDL-C at a nominal significance threshold of 0.05 simply by chance, and none of these would be expected to achieve this significance threshold in a replicate analysis of an independent sample. In contrast, 10 of the 106 sequence variants assayed in blacks in the DHS sample achieved the nominal significance threshold of 0.05, and 5 of these also achieved a P value <.05 in the Cook County sample. For all five sequence variants, the direction of effect was the same in both populations.
We failed to confirm any of the previously reported associations between sequence variations in PCSK9 and differences in plasma levels of LDL-C (Shioji et al. 2004; Chen et al. 2005). Two common SNPs, IVS1−161C→T and I474V, associated with decreased plasma levels of LDL-C in a Japanense sample were not associated with reduced plasma levels of LDL-C in the DHS sample (Shioji et al. 2004). Nor was any association found between plasma levels of LDL-C and a common E670G substitution whose corresponding allele was part of a haplotype associated with increased plasma levels of LDL-C in hypercholesterolemic subjects from Houston (10% nonwhite) (Chen et al. 2005).
The molecular mechanism by which sequence variations in PCSK9 affect plasma LDL-C levels remains controversial. Studies in genetically manipulated mice suggest that PCSK9 affects plasma LDL-C levels by altering hepatic LDLR levels: overexpression of wild-type PCSK9 in mouse liver by adenovirus-mediated transgenesis is associated with a marked reduction in hepatic LDLRs and a corresponding increase in plasma cholesterol levels (Dubuc et al. 2004; Park et al. 2004), whereas deletion of PCSK9 results in an increase in LDLR protein levels (Rashid et al. 2005). It has also been proposed that mutations in PCSK9 affect plasma LDL-C levels by altering the rate of secretion of VLDL from the liver (Benjannet et al. 2004; Ouguerram et al. 2004; Sun et al. 2005). If the hypercholesterolemia associated with selected missense mutations in PCSK9 is due to increased VLDL synthesis, then the hypocholesterolemia associated with the nonsense mutations is predicted to result in decreased VLDL secretion, as occurs in two Mendelian forms of hypocholesterolemia: hypobetalipoproteinemia and abetalipoproteinemia. Both of these diseases are characterized by an accumulation of triglyceride in the liver due to the reduction in VLDL secretion (Schonfeld et al. 2003). In the present study, we did not detect any significant difference in the median HTGC or in the prevalence of hepatic steatosis between the subjects with and the subjects without an LDL-lowering mutation in PCSK9, in either ethnic group. This finding suggests that impaired VLDL secretion is not the major mechanism by which loss-of-function mutations in PCSK9 decrease plasma LDL-C levels.
The mechanisms by which the sequence variations identified in this study affect PCSK9 activity are not known. One of the missense mutations associated with plasma levels of LDL-C (L235F) affected a residue that was completely conserved among all species examined, from humans to fish. Surprisingly, the residues that were altered by the other two functional missense mutations (R46L and A443T) were poorly conserved. Codon 46 specifies a hydrophobic amino acid in most species (proline in rodents, alanine in frogs, and isoleucine in zebrafish). Similarly, the residue affected by the A443T substitution is threonine in rodents and glycine in birds and frogs. Since no biochemical assay for PCSK9 is available, we cannot determine whether these substitutions directly affect protein activity or whether they affect the expression level of the protein. It is also possible that these sequence variants do not directly affect PCSK9 activity but are in linkage disequilibrium with other noncoding polymorphisms that affect PCSK9 expression.
The identification of a wide spectrum of amino acid sequence variations in PCSK9, coupled with the ability to test for association of these variations with a phenotype directly related to PCSK9 function in a large population sample, provided an opportunity to assess the utility of computer-based prediction algorithms. None of the three algorithms predicted any of the missense variations significantly associated with plasma LDL-C level to have a deleterious effect with high probability; moreover, each of the three functional sequence variations was predicted by at least one of the algorithms to be benign, and A443T was predicted by all three algorithms to be benign. This latter prediction by all three programs is likely a result of the presence of T443 in rodent PCSK9. This might be explained by differences in the effect of this mutation between humans and rodents or by the co-occurrence of one or more compensating mutations (Kondrashov et al. 2002). In addition, 64% of the sequence variations that were not associated with plasma LDL-C levels were predicted by at least one of the algorithms to be deleterious. Thus, in silico prediction methods were poor predictors of functional variation in PCSK9. It is possible that the number of orthologous sequences used in this analysis did not provide adequate power to assess the spectrum of amino acids permitted at each position in the protein. In this case, the performance of these algorithms could potentially be improved by greatly increasing the number of reference sequences included in the alignment. Alternatively, the poor performance of the algorithms may reflect intrinsic limitations of in silico prediction. This problem may be particularly pertinent for sequence variations that have more-subtle effects on protein function.
The results of this study have important implications for the genetic architecture of quantitative traits and for strategies to identify functional sequence variants. First, our data provide further support for the importance of rare sequence variants with large phenotypic effects. Among blacks, 3% of the population carries a PCSK9 allele that is associated with a mean reduction in plasma LDL-C levels of ∼35%. The LDL-lowering effect of these rare PCSK9 alleles is comparable to the effects achieved by 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors (statins). Thus, carriers of these mutations should have substantial protection from coronary heart disease. Second, our data also support a role for common PCSK9 variants (both coding and noncoding) in determining plasma levels of LDL-C. The effects of these variants were much smaller than the effects associated with the rare variants; the plasma LDL-C levels among individuals with one of the common LDL-lowering PCSK9 alleles differed by <5% from the levels of those without the allele. Finally, although >100 sequence variants in PCSK9 were analyzed in this study, only a small number were systematically associated with plasma LDL-C levels. For each of the functional sequence variants identified, several closely linked SNPs did not show any significant association with plasma LDL-C levels (fig. 6). Haplotype blocks constructed using multiple highly informative SNPs also failed to reveal association with plasma LDL-C levels unless the putatively functional SNP was included in the analysis. These results suggest that very dense SNP analysis may be required for genetic association studies and that analysis of common sequence variants is unlikely to reveal the effects of rare alleles in the population.
Acknowledgments
We thank the DHS Investigators (Victor et al. 2004) for providing the clinical material for this study. We thank Tommy Hyatt, Sijing Niu, and Kevin Vo for excellent technical assistance. This work was supported by grants from the Donald W. Reynolds Foundation, the W. M. Keck Foundation, the National Institutes of Health (HL 20948), The Veterans Affairs Merit Award Grant, The Perot Family Fund, and the Le Ducq Foundation.
Web Resources
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for LDLR, autosomal dominant hypercholesterolemia, and autosomal recessive hypercholesterolemia)
- PANTHER, https://panther.appliedbiosystems.com/
- PolyPhen, http://www.bork.embl-heidelberg.de/PolyPhen/
- Sorting Intolerant From Tolerant (SIFT), http://blocks.fhcrc.org/sift/SIFT.html
- The R Project for Statistical Computing, http://www.r-project.org/
References
- Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, et al (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34:154–156 10.1038/ng1161 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chretien M, Prat A, Seidah NG (2004) NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the LDLR and LDL-cholesterol. J Biol Chem 279:48865–48875 10.1074/jbc.M409699200 [DOI] [PubMed] [Google Scholar]
- Bray N, Dubchak I, Pachter L (2003) AVID: a global alignment program. Genome Res 13:97–102 10.1101/gr.789803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH (2004) Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 40:1387–1395 10.1002/hep.20466 [DOI] [PubMed] [Google Scholar]
- Chen SN, Ballantyne CM, Gotto AM Jr, Tan Y, Willerson JT, Marian AJ (2005) A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am Coll Cardiol 45:1611–1619 10.1016/j.jacc.2005.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37:161–165 10.1038/ng1509 [DOI] [PubMed] [Google Scholar]
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I (2003) Strategies and tools for whole-genome alignments. Genome Res 13:73–80 10.1101/gr.762503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, Prat A (2004) Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol 24:1454–1459 10.1161/01.ATV.0000134621.14315.43 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N, Barnes R, Cohen JC, Hobbs HH (2001) Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science 292:1394–1398 10.1126/science.1060458 [DOI] [PubMed] [Google Scholar]
- Goldstein J, Hobbs H, Brown M (2001) Familial hypercholesterolemia. In: Scriver C, Beaudet A, Sly W, Valle D (eds) The metabolic and molecular bases of inherited disease. Vol II. McGraw Hill, New York, pp 2863–2913 [Google Scholar]
- Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE (1993) Genetic and environmental influences on serum lipid levels in twins. N Engl J Med 328:1150–1156 10.1056/NEJM199304223281603 [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100:12027–12032 10.1073/pnas.1534923100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innerarity TL, Mahley RW, Weisgraber KH, Bersot TP, Krauss RM, Vega GL, Grundy SM, Friedl W, Davignon J, McCarthy BJ (1990) Familial defective apolipoprotein B-100: a mutation of apolipoprotein B that causes hypercholesterolemia. J Lipid Res 31:1337–1349 [PubMed] [Google Scholar]
- Kondrashov AS, Sunyaev S, Kondrashov FA (2002) DobzhanskyMuller incompatibilities in protein evolution. Proc Natl Acad Sci USA 99:14878–14883 10.1073/pnas.232565499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne F, Lambert G, Amar MJ, Chetiveaux M, Zair Y, Jarnoux AL, Ouguerram K, Friburg J, Seidah NG, Brewer HB Jr, Krempf M, Costet P (2005) Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells. J Lipid Res 46:1312–1319 10.1194/jlr.M400396-JLR200 [DOI] [PubMed] [Google Scholar]
- Leren TP (2004) Mutations in the PCSK9 gene in Norwegian subjects with autosomal dominant hypercholesterolemia. Clin Genet 65:419–422 10.1111/j.0009-9163.2004.0238.x [DOI] [PubMed] [Google Scholar]
- Maxwell KN, Breslow JL (2004) Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA 101:7100–7105 10.1073/pnas.0402133101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL (2003) Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res 44:2109–2119 10.1194/jlr.M300203-JLR200 [DOI] [PubMed] [Google Scholar]
- Naureckiene S, Ma L, Sreekumar K, Purandare U, Lo CF, Huang Y, Chiang LW, Grenier JM, Ozenberger BA, Jacobsen JS, Kennedy JD, DiStefano PS, Wood A, Bingham B (2003) Functional characterization of Narc 1, a novel proteinase related to proteinase K. Arch Biochem Biophys 420:55–67 10.1016/j.abb.2003.09.011 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouguerram K, Chetiveaux M, Zair Y, Costet P, Abifadel M, Varret M, Boileau C, Magot T, Krempf M (2004) Apolipoprotein B100 metabolism in autosomal-dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler Thromb Vasc Biol 24:1448–1453 10.1161/01.ATV.0000133684.77013.88 [DOI] [PubMed] [Google Scholar]
- Park SW, Moon YA, Horton JD (2004) Post-transcriptional regulation of LDL receptor protein by proprotein convertase subtilisin/kexin type 9a (PCSK9) in mouse liver. J Biol Chem 279:50630–50638 10.1074/jbc.M410077200 [DOI] [PubMed] [Google Scholar]
- Rao DC, Laskarzewski PM, Morrison JA, Khoury P, Kelly K, Wette R, Russell J, Glueck CJ (1982) The Cincinnati Lipid Research Clinic family study: cultural and biological determinants of lipids and lipoprotein concentrations. Am J Hum Genet 34:888–903 [PMC free article] [PubMed] [Google Scholar]
- Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD (2005) Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci USA 102:5374–5379 10.1073/pnas.0501652102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks FM, Katan M (2002) Randomized clinical trials on the effects of dietary fat and carbohydrate on plasma lipoproteins and cardiovascular disease. Am J Med Suppl 9B 113:13S–24S [DOI] [PubMed] [Google Scholar]
- Schonfeld G, Patterson BW, Yablonskiy DA, Tanoli TS, Averna M, Elias N, Yue P, Ackerman J (2003) Fatty liver in familial hypobetalipoproteinemia: triglyceride assembly into VLDL particles is affected by the extent of hepatic steatosis. J Lipid Res 44:470–478 10.1194/jlr.M200342-JLR200 [DOI] [PubMed] [Google Scholar]
- Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M (2003) The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. PNAS 100:928–933 10.1073/pnas.0335507100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioji K, Mannami T, Kokubo Y, Inamoto N, Takagi S, Goto Y, Nonogi H, Iwai N (2004) Genetic variants in PCSK9 affect the cholesterol level in Japanese. J Hum Genet 49:109–114 10.1007/s10038-003-0114-3 [DOI] [PubMed] [Google Scholar]
- Sun XM, Eden ER, Tosi I, Neuwirth CK, Wile D, Naoumova RP, Soutar AK (2005) Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet 14:1161–1169 10.1093/hmg/ddi128 [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W 3rd, Kondrashov AS, Bork P (2001) Prediction of deleterious human alleles. Hum Mol Genet 10:591–597 10.1093/hmg/10.6.591 [DOI] [PubMed] [Google Scholar]
- Szczepaniak LS, Nurenberg P, Leonard D, Browning JD, Reingold JS, Grundy S, Hobbs HH, Dobbins RL (2004) Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab 288:E462–E468 10.1152/ajpendo.00064.2004 [DOI] [PubMed] [Google Scholar]
- The R Development Core Team (2004) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O (2003) PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31:334–341 10.1093/nar/gkg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms KM, Wagner S, Samuels ME, Forbey K, Goldfine H, Jammulapati S, Skolnick MH, Hopkins PN, Hunt SC, Shattuck DM (2004) A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum Genet 114:349–353 10.1007/s00439-003-1071-9 [DOI] [PubMed] [Google Scholar]
- Victor RG, Haley RW, Willett DL, Peshock RM, Vaeth PC, Leonard D, Basit M, Cooper RS, Iannacchione VG, Visscher WA, Staab JM, Hobbs HH (2004) The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol 93:1473–1480 10.1016/j.amjcard.2004.02.058 [DOI] [PubMed] [Google Scholar]