Subtelomeric proteins negatively regulate telomere elongation in budding yeast (original) (raw)
Abstract
The Tbf1 and Reb1 proteins are present in yeast subtelomeric regions. We establish in this work that they inhibit telomerase-dependent lengthening of telomere. For example, tethering the N-terminal domain of Tbf1 and Reb1 in a subtelomeric region shortens that telomere proportionally to the number of domains bound. We further identified a 90 amino-acid long sequence within the N-terminal domain of Tbf1 that is necessary but not sufficient for its length regulation properties. The role of the subtelomeric factors in telomere length regulation is antagonized by TEL1 and does not correlate with a global telomere derepression. We show that the absence of TEL1 induces an alteration in the structure of telomeric chromatin, as defined biochemically by an increased susceptibility to nucleases and a greater heterogeneity of products. We propose that the absence of TEL1 modifies the organization of the telomeres, which allows Tbf1 and Reb1 to _cis_-inhibit telomerase. The involvement of subtelomeric factors in telomere length regulation provides a possible mechanism for the chromosome-specific length setting observed at yeast and human telomeres.
Keywords: Reb1, subtelomeric repeat, Tbf1, telomerase, telomere
Introduction
Telomeres prevent chromosome termini from being recognized as double-strand breaks and play a regulatory role in cell proliferation and survival (Blackburn, 2000b; Lundblad, 2000). They are also essential for various chromosome functions, including the regulation of gene expression, chromosome positioning and dynamics, recombination as well as for proper mitotic and meiotic divisions (Zakian, 1995; Chan and Blackburn, 2004).
Telomeres are generally composed of short tandem repeated DNA sequences (Blackburn, 1991), which are folded into nucleoprotein complexes that are reorganized during the cell cycle (Brunori et al, 2005). In budding and fission yeasts, these end-complexes, called telosomes, are characterized by their resistance to nuclease digestion in a pattern that is distinct from the bulk of nucleosomal chromatin (Wright et al, 1992; Cooper et al, 1998). The budding yeast end-complex requires an array of the essential repressor/activator protein 1 (Rap1) (Gilson and Gasser, 1995), which serves as a nucleation site for the assembly and spreading within the subtelomeric regions of a silent chromatin state (Perrod and Gasser, 2003). The natural yeast subtelomeric chromatin appears to juxtapose active and silent regions, reflecting a chromosome-specific combination of insulator and silencer elements (Fourel et al, 1999; Pryde and Louis, 1999). In vertebrates, telomeric nucleosomes present a heterochromatin-like profile of histone modifications (Garcia-Cao et al, 2004). In all organisms studied, telomeres are associated with a complex network of non-histone factors including chromatin, DNA repair and telomere-specific components (Brunori et al, 2005). At a higher level of organization, telomeres can fold into looped structures, which are likely to be critical for distinguishing natural chromosome ends from accidental double-strand breaks (Griffith et al, 1999). Finally, in lower eukaryotes such as yeast, telomeres are found as multitelomeric clusters, often located near the nuclear periphery (Gilson et al, 1993; Palladino et al, 1993; Gotta et al, 1996). The fact that telomeres combine both heterochromatin and telomere-specific features is consistent with their capacity to exert position effects (Gottschling et al, 1990) and with the involvement of epigenetic mechanisms in the maintenance of their structure (Sherman and Pillus, 1997; Surralles et al, 1999; Maillet et al, 2001; Sadaie et al, 2003; Garcia-Cao et al, 2004; Perrini et al, 2004; Joseph et al, 2005).
In many organisms, telomerase, a specialized reverse transcriptase, compensates for the systematic shortening of telomeric DNA that occurs during the conventional replication of chromosomal DNA termini (Blackburn, 2000a). This enzyme uses an internal RNA template to add specific repeats to the terminal 3′ overhang. The action of telomerase is cell-cycle regulated, occurring preferentially in late S/G2 but not G1 phase (Diede and Gottschling, 1999; Marcand et al, 2000). A switch between telomere extendible states, for example, accessible to telomerase, and non-extendible states determines the probability for the enzyme to act during one cell cycle (Teixeira et al, 2004).
The in vivo activity of telomerase is inhibited by specific components of the telomeric chromatin (Smogorzewska and de Lange, 2004). In Saccharomyces cerevisiae, the accessibility of a productive telomerase to its substrate is restricted proportionally to the number of Rap1–Rif1–Rif2 complexes assembled at the telomere, thus creating a negative feedback loop that determines the mean number of telomeric repeat sequences (Marcand et al, 1997; Cooper et al, 1998; Kanoh and Ishikawa, 2001; Levy and Blackburn, 2004; Teixeira et al, 2004). This mode of telomere length regulation was called a ‘counting mechanism'. In cells carrying mutations that impair the Rap1-dependent counting, telomeres lengthen in an uncontrolled way (Kyrion et al, 1992; Wotton and Shore, 1997). Conversely, increasing the number of Rap1 or Rif1/Rif2 proteins bound in the immediate subtelomeric region induces a _cis_-shortening of the telomere relative to the number of bound proteins (Marcand et al, 1997; Ray and Runge, 1999b; Levy and Blackburn, 2004).
In cells lacking TEL1 (a gene encoding a PI3-kinase involved in DNA damage response and telomere integrity), the mean telomere length is shorter than in wild-type cells and appears relatively insensitive to mutations that impair the Rap1-counting pathway (Craven and Petes, 1999; Ray and Runge, 1999a). In _tel1_Δ cells, the rate of elongation of a telomere that has been specifically shortened progressively declines upon the return to the equilibrium length (Brevet et al, 2003). Hence, a factor different from the Rap1–Rif1–Rif2 complex should limit telomere lengthening in cis. This process might be more potent in _tel1_Δ cells than in wild-type cells, thus explaining their shorter mean length. Such a notion is in agreement with the fact that the lack of TEL1 does not directly affect telomerase activity, but appears to modify the structure of the end-complex in such a way that telomerase accessibility is impaired (Chan et al, 2001).
A distinctive feature of the Rap1-independent counting mechanism of telomere length regulation is its capacity to accommodate both yeast and vertebrate telomeric sequences. For example, in the absence of TEL1, vertebrate repeats inserted at the internal side of a single telomere are taken into account for telomere length regulation (Brevet et al, 2003). In tlc1-h cells, also known as humanized yeast (Henning et al, 1998), a telomere formed only by vertebrate repeats can be maintained at a regulated mean length (Alexander and Zakian, 2003; Brevet et al, 2003). It is noteworthy that, in tlc1-h cells containing a functional TEL1 gene, the number of vertebrate repeats at a chromosome end is determined in a Rap1-independent manner, indicating that TEL1 does not completely abolish the ability of vertebrate repeats to regulate telomere length. Overall, one can predict in budding yeast the existence of factors distinct from the Rap1–Rif1–Rif2 complex that are capable of binding yeast and vertebrate telomeric repeats and that inhibit in cis telomerase function.
In this study, we searched in budding yeast for factor(s) that upon their association to vertebrate telomeric repeats limit telomere elongation. We showed that the binding of Tbf1 to vertebrate repeats, a subtelomeric protein previously reported to act as a transcriptional insulator, shortens telomere length. Strikingly, we also discovered that Reb1, another insulator protein present in yeast subtelomeric regions, behaves similar to Tbf1 in terms of telomere length regulation.
Results
Subtelomeric DNA binding sites for Tbf1 or Reb1 are taken into account for length regulation in tel1_Δ_ cells
We have previously described the ability of an array of 10 vertebrate telomeric repeats, inserted at a newly formed _URA3_-tagged truncated 7L telomere beyond the ADH4 gene (named hereafter Tel 7Ltr), to be taken into account for telomere length regulation in _tel1_Δ but not TEL1 cells. Their presence leads to a _cis_-shortening of Tel 7Ltr (Brevet et al, 2003). Here, we investigate the nature of the factor(s) responsible for this effect. As Tbf1 binds to vertebrate repeats and DNA binding sites for Tbf1 are often combined with those for Reb1 in natural yeast subtelomeric regions (Fourel et al, 1999; Koering et al, 2000), we asked whether a DNA sequence containing three binding sites for either Tbf1 or Reb1 (named (Tbf1)3 and (Reb1)3) could recapitulate the _cis_-shortening activity of the vertebrate repeats. The sequence of these Tbf1 or Reb1 sites are designed in accordance with the results of a selection procedure using random oligonucleotide pools (Koering et al, 2000) and their specificity has been previously confirmed (Fourel et al, 1999). We note that the (Tbf1)3 and (Reb1)3 sequences do not exactly match the Tbf1 and Reb1 sites found in natural subtelomeric regions. These sites were inserted next to the distal TG1–3 repeats at Tel 7Ltr. As a negative control, in the same location we also inserted a portion of the RAD52 gene, which was previously shown to be neutral with regard to telomere length regulation (named NR) (Brevet et al, 2003) (Figure 1A).
Figure 1.
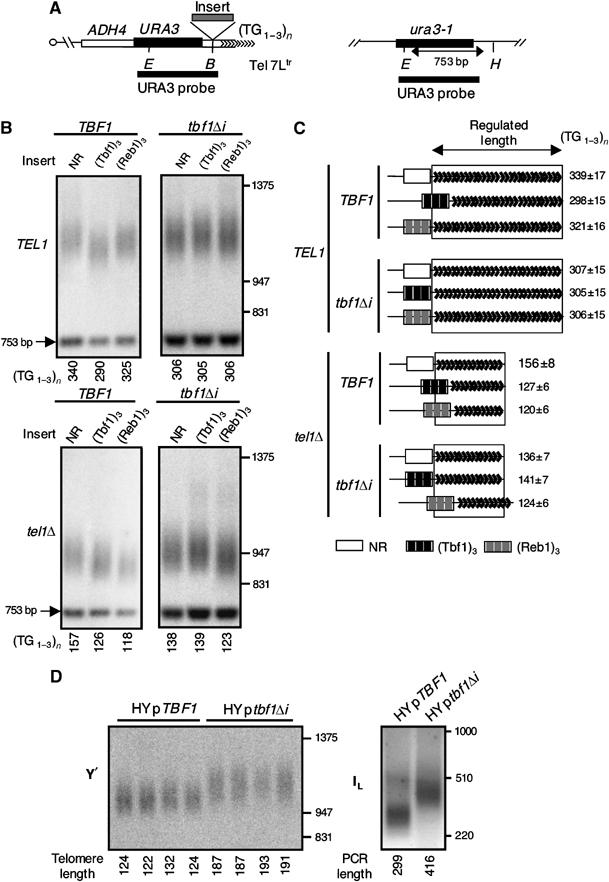
The role of Tbf1 or Reb1 in telomere length regulation. (A) Structure of Tel 7Ltr and the endogenous ura3-1 locus. _B_=_Bam_HI; _E_=_Eco_RV, _H_=_Hin_dIII. (B) Examples of Southern blots used to measure the length of Tel 7Ltr containing either the NR control sequence, (TG1–3)23 or (T2AG3)10 insert in different genetic settings, as indicated. The genomic DNA was digested with _Hin_dIII and _Eco_RV and hybridized with the URA3 probe shown in panel A. The reference band of 753 bp emanates from the ura3-1 locus (see panel A) and the smear corresponds to the terminal restriction fragment that contains the distal yeast repeats. The calculated length of the distal (TG1–3)n is indicated below each lane. In order to obtain the length of the distal telomeric repeats, 800 nt was subtracted from the size of the measured terminal fragment, as shown below each lane. (C) Schematic representation of the results. The mean length corresponding to the average of at least three independent cultures is indicated on the right, together with the standard deviation. For an identical genetic setting, the part of the telomere corresponding to the distal repeats of Tel 7Ltr containing an NR control insert is boxed. As the NR sequence is not taken into account for length regulation, the boxed sequence is named the ‘regulated length'. Thus, in Tel 7Ltr containing the yeast- or vertebrate-type inserts, the part of the chromosome end that is not taken into account for length regulation appears outside the box of the ‘regulated length'. (D) _Xho_I Southern analysis of Y′ telomeres (left part) and PCR analysis of Tel 1L (right part) of humanized yeasts containing a plasmid copy of either TBF1 (pTBF1) or tbf1_Δ_i (ptbf1Δi).
In TEL1 cells, there is shortening (roughly 40 nt) of the distal repeats for the (Tbf1)3-containing telomere but not for those with either (Reb1)3 or the control NR sequence (Figure 1B and C). The _tel1_Δ cells revealed a _cis_-shortening effect of the (Reb1)3 insert, which is now similar to that of (Tbf1)3 (Figure 1B and C). As expected, the NR control sequence does not lead to a _cis_-shortening in this context. The shortening effect of the non-telomeric Tbf1 and Reb1 sites used in these experiments appears to reflect the property of the sites found in natural subtelomeric regions. Indeed, natural sequences from the X and Y′ subtelomeric elements containing multiple Tbf1 and Reb1 sites also lead to telomere shortening when inserted at Tel 7Ltr (data not shown). Overall, these results show that an array of DNA binding sites for Reb1 or Tbf1 behaves similar to vertebrate telomere repeats, being taken into account for length regulation in _tel1_Δ cells. Only Tbf1 binding sites appear to lead to _cis_-shortening in the presence of TEL1.
Identification of an allele of TBF1 that alleviates the Tbf1-dependent telomere shortening
It is conceivable that additional proteins binding to vertebrate repeats or (Tbf1)3, or some special features of these sequences themselves, and not the binding of Tbf1, were responsible for the length regulation effect. Therefore, we repeated the above experiments in cells expressing a viable allele of TBF1 named tbf1_Δ_i. This allele encodes a mutated form of Tbf1 lacking a sequence of 90 amino acids (Tbf1I) located within the transcriptional insulation domain of the protein (Tbf1N) (Fourel et al, 2001) (Figure 2A). Upon expression of Tbf1Δi instead of Tbf1, the (Tbf1)3 element no longer downregulates telomere length in either TEL1 or _tel1_Δ cells (Figure 1B and C). Importantly, the shortening effect of (Reb1)3 in _tel1_Δ cells is only slightly affected in tbf1_Δ_i cells. These results establish the specificity of TBF1 toward the effect of (Tbf1)3 on telomere length regulation.
Figure 2.
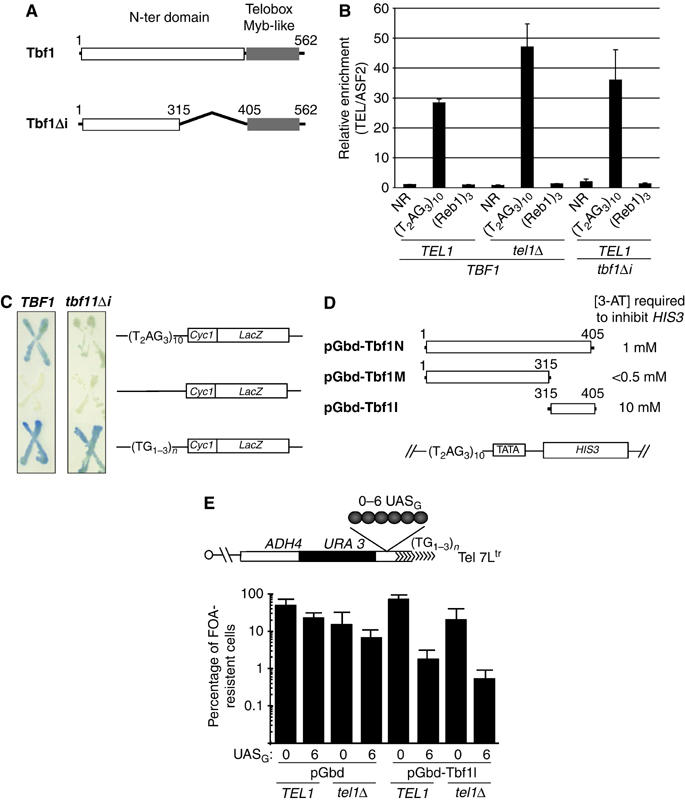
Characterization of the tbf1_Δ_i allele. (A) Representation of Tbf1 and Tbf1Δi proteins. (B) The binding of Tbf1 was assayed by ChIP analysis followed by real-time PCR. The results are expressed as the enrichment of the immunoprecipitated Tel 7Ltr DNA to that of an ASF2 DNA (see Materials and methods). The average value for at least three experiments is shown as well as the estimated standard deviation. (C) β-Galactosidase assay performed by a X-gal coloration of patches of cells corresponding to the indicated genetic setting and containing the plasmids pSX178, pSXHuTel2 and pSXYTel carrying the CYC1-lacZ reporter construct shown on the right (see Koering et al, 2000). (D) Transactivation assays for several hybrid proteins containing different parts of TBF1. The threshold of 3-AT concentration required to inhibit the HIS3 reporter gene is indicated for each protein. (E) Antisilencing assay using the tethering of Gbd-Tbf1I between a telomere and a URA3 reporter gene. As the FOA compound is toxic for cells expressing URA, the percentage of FOA-resistant cells reflects URA3 repression.
Chromatin immunoprecipitation (ChIP) experiments show that the Tbf1Δi protein binds in vivo to vertebrate repeats similar to Tbf1 (Figure 2B). Neither Tbf1 nor Tbf1Δi binds to the (Reb1)3 insert used in Figure 1, which is consistent with the idea that the effect of this sequence on telomere length is Tbf1 independent.
In addition to its role in length regulation, the Tbf1I domain is involved in the transcriptional properties of Tbf1: namely, (i) it is necessary for the full transactivation activity of Tbf1, as revealed by a reduced capacity of vertebrate repeats to transactivate the expression of a UAS-less CYC1″LacZ reporter gene in tbf1_Δ_i cells as compared to TBF1 cells (Figure 2C); (ii) it exhibits a potent transactivation activity when fused to the Gal4 DNA-binding domain (amino acids 1–147, named Gbd) in a _HIS3_-based reporter assay (Figure 2D); (iii) it prevents silencing when tethered at an immediate subtelomeric region, in both TEL1 and _tel1_Δ cells (Figure 2E).
We then asked whether the terminally located Tbf1 proteins present at humanized yeast telomeres can also play a role in length regulation. Although we have been unable to replace TBF1 by tbf1_Δ_i in humanized yeast, the presence of both TBF1 and tbf1_Δ_i reveals a dominant effect of the mutant allele. This leads to lengthening of Y′-type, Tel 7Ltr and 1L telomeres (Figure 1D and data not shown). These results indicate that Tbf1 is a negative regulator of telomere length in the context of the humanized yeast and confirm the important role of the Tbf1I domain in this regulation.
Telomere length is decreased proportionally to the number of Gbd-Tbf1 and Gbd-Reb1 tethered proteins
We then asked whether the number of Tbf1 or Reb1 proteins assembled at a single subtelomere determines the length of the terminal repeats, as in the Rap1-dependent counting mechanism of telomere length regulation. For this, we used a tethering procedure previously described to assay the counting effect of Rap1 (Marcand et al, 1997). Specifically, Gbd-Tbf1N, GbdTbf1I and Gbd-Reb1N hybrids are expressed in a series of strains with increasing numbers of binding sites for Gal4 (two, four and six UASG) inserted at Tel 7Ltr (Figure 3A). As control, we used the Gbd-Rap1 hybrid containing the C-terminus of Rap1 (amino acids 653–827) (Marcand et al, 1997). The length of Tel 7Ltr in cells expressing these various hybrids was normalized to that in cells expressing Gbd alone. Because we noticed some telomere length variations in different clones derived from the transformation with the plasmid encoding the fusion proteins, we measured telomere length in at least five independent transformants grown for more than 50 generations. Importantly, we systematically determined the number of UASG sequences inserted at Tel 7Ltr by PCR analysis in all the clones tested for telomere length. We never observed a modification in the number of UASG in any of the genetic settings used in these experiments (data not shown).
Figure 3.
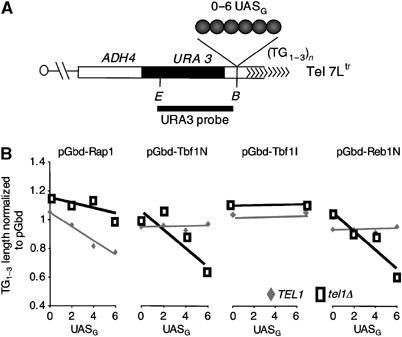
Tethering at an immediate subtelomeric location of Tbf1 and Reb1 N-terminal domains reduces telomere length. (A) Schematic representation of Tel 7Ltr and UASG sites. (B) The graphs represent, as a function of the number of UASG, the ratio of the mean length calculated from cells expressing the hybrid indicated above each graph to that from cells expressing the pGbd plasmid. The gray line represents experiments performed in TEL1 cells, and the black line, in _tel1_Δ cells.
As expected, in cells expressing a Gbd-Rap1 hybrid, the length of the targeted telomere is reduced proportionally to the number of UASG (Figure 3B). In _tel1_Δ cells expressing Gbd-Rap1, this reduction is much less pronounced, confirming that the counting effect of the C-terminal part of Rap1 is partially TEL1 dependent (Brevet et al, 2003). In TEL1 cells, the expression of Gbd hybrids with Tbf1N, Tbf1I or Reb1N does not significantly change the length of the terminal repeats, indicating that Reb1 and Tbf1 do not influence telomere length in TEL1 cells (Figure 3B). By contrast, the expression of Gbd-Tbf1N and Gbd-Reb1N in _tel1_Δ cells leads to an incremental shortening of the terminal repeats proportional to the number of UASG, to the same extent as the shortening induced by Gbd-Rap1 in TEL1 cells (Figure 3B). Importantly, the expression of Gbd-Tbf1I does not lead to telomere shortening (Figure 3B). This result, together with the impaired capacity of Tbf1 DNA binding site to shorten telomere in tbf1_Δ_i cells (Figure 1), shows that Tbf1I is necessary but not sufficient to downregulate telomere length. The length of the Y′ telomeres in _tel1_Δ cells remains unchanged upon expression of the various Gbd hybrid proteins, showing that the effect of the hybrids is specific of the targeted Tel 7Ltr and does not correspond to a trans effect such as the decreased expression of the telomerase (data not shown). Overall, in _tel1_Δ cells, telomere length regulation becomes sensitive to the number of Tbf1 and Reb1 proteins bound at a subtelomeric location.
The cis-shortening due to Tbf1 can be uncoupled from telomeric position effect, short mean telomere length and MEC1-dependent function
We investigated the possibility that the _cis_-shortening induced by Tbf1 results from the derepression of the subtelomeric URA3 gene. In _sir3_Δ cells, there is a complete loss of telomeric repression (Aparicio et al, 1991) and consequently a normal expression of URA3 at Tel7Ltr. Cells carrying a deletion of yKU70 display a partial derepression albeit far more than that seen in _tel1_Δ cells (Boulton and Jackson, 1998; Gravel et al, 1998; Laroche et al, 1998; Nugent et al, 1998). In _sir3_Δ and _yku70_Δ cells, we observed that the insertion at Tel7Ltr of a stretch of vertebrate repeats, containing several Tbf1 binding sites, leads to a shortening of 19 and 12 nt, respectively (Figure 4B). This is consistent with the mild effect of (Tbf1)3 in TEL1 cells (see Figure 1). Importantly, this shortening effect is significantly less as compared to the potent effect of the vertebrate repeats in _tel1_Δ cells (−76 bp; Figure 4B). These results indicate that the role of Tbf1 in telomere length regulation can be uncoupled from a global telomere derepression.
Figure 4.
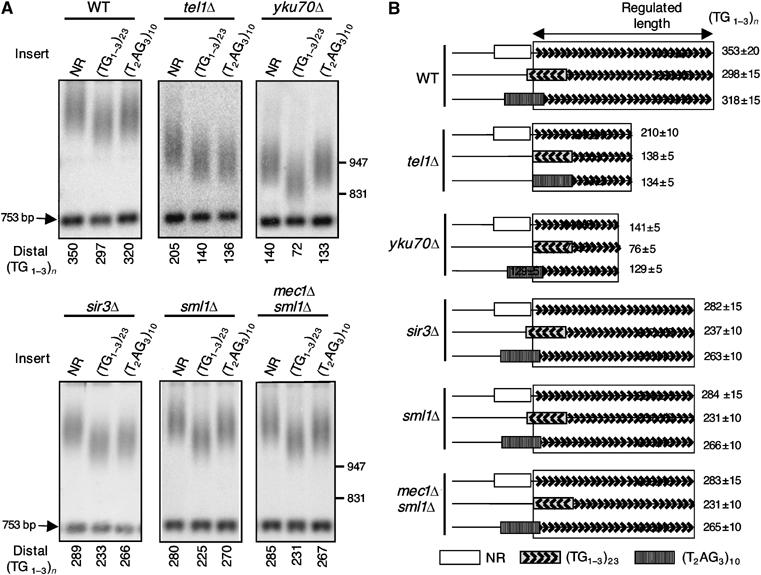
Absence of relationships between SIR3, yKU70 and MEC1 and the _cis_-shortening induced by a subtelomeric stretch of vertebrate telomeric repeats. (A, B) Southern blots and schematic representation of the results as described in Figure 1.
As TEL1 exhibits overlapping functions with its paralog MEC1 in DNA-damage checkpoint and telomere regulation (Morrow et al, 1995), we asked whether _mec1_Δ could behave as _tel1_Δ, reinforcing the ability of vertebrate repeats to regulate telomere length. As the lethality owing to _mec1_Δ can be rescued by the deletion of SML1, we compared the effect of the vertebrate repeats in _sml1_Δ and _sml1_Δ _mec1_Δ cells (Figure 4B). In both cases, the vertebrate repeats are partially taken into account for length regulation but not at the level observed in _tel1_Δ cells (Figure 4B). Hence, _MEC1_-dependent functions do not counteract the ability of Tbf1 binding sites to act as telomere length regulators.
In conclusion, neither silencing alleviation (_sir3_Δ and _yku70_Δ cells) nor short mean telomere length (_yku70_Δ cells) nor checkpoint alteration (_mec1_Δ _sml1_Δ cells) is sufficient to recapitulate the effect of the _tel1_Δ genetic background regarding the ability of the vertebrate repeats to be taken into account for telomere length regulation.
The absence of TEL1 leads to a more accessible conformation of the telomeric end-complex
We next looked for changes in chromatin and telomere structure that could account for the telomere length regulatory role of Tbf1. Using DNase I and micrococcal nuclease (MNase) sensitivity analyses, we failed to detect any major subtelomeric chromatin change associated with the presence of subtelomeric Tbf1 binding sites, in both TEL1 and _tel1_Δ cells (Supplementary Figure 1). Then, we investigated whether the absence of TEL1 modifies the structure of the end-complex. DNA was isolated from DNase I- and MNase-treated cell lysates and hybridized with a TG1–3 or a Y′ probe (Figure 5). The digestion rate of the TG 1–3 DNA is similar in TEL1 and _tel1_Δ lysates, as indicated by the similar decrease of total TG 1–3 signal during the time course of the nuclease digestion (the measurements of total signals are not shown), indicating that the comparison was performed within a similar linear range for the degradation of total and TG 1–3 DNA.
Figure 5.
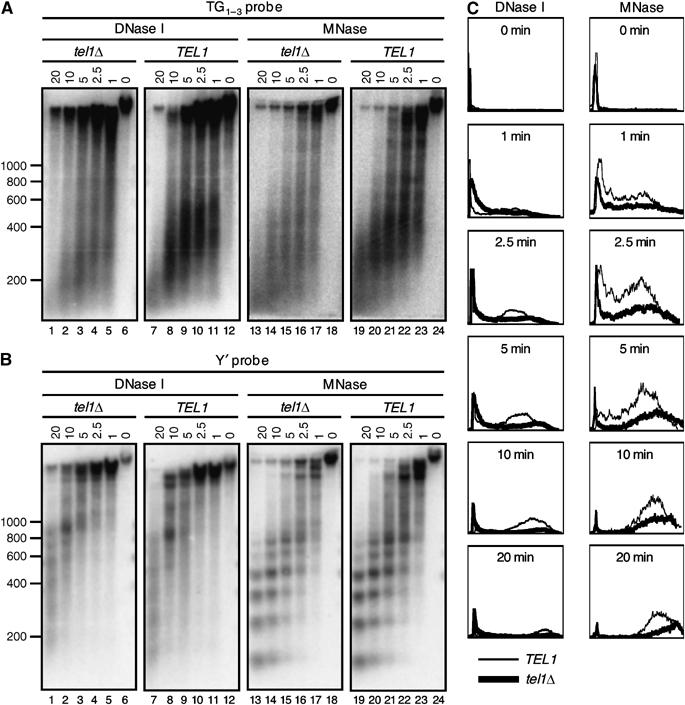
Structure of the telomeric end-complex and subtelomeric chromatin in TEL1 and _tel1_Δ cells. (A) DNA was isolated from fresh cell lysates treated with DNase I or MNase and then analyzed by Southern blot with a TG1–3 probe. (B) DNA was isolated from fresh cell lysates treated with DNase I or MNase and then analyzed by Southern blot with a Y′ probe. (C) Analysis of the profiles of the TG1–3 hybridizing DNA. The thick line represents the _tel1_Δ samples. The thin line represents the TEL1 samples.
The Southern blot analysis with a TG1–3 probe is shown in Figure 5A. In TEL1 cells, both DNase I and MNase produce a broad peak (between 550 and 300 bp) of TG1–3 interacting DNA (Figure 5A, lanes 7–12 and 19–24). Under our experimental conditions, this DNA is resistant to nuclease treatment for about 10 min, which likely corresponds to the telosome structure described in earlier studies (Wright et al, 1992). In _tel1_Δ cells, both nucleases produce TG1–3 DNA peaks, which have higher mobility (corresponding to the overall shorter telomeres in this strain) and are broader (between 500 and 200 bp) as compared to the TEL1 samples (Figure 5A, compare lanes 1–6 to 7–12 and 13–18 to 19–24). Importantly, the signals in the TG1–3 peaks of the _tel1_Δ samples are weaker despite the approximately equal intensity of the non-degraded TG1–3 DNA at the top of the corresponding lanes. For example, after 5 min of DNase I treatment of _tel1_Δ lysate (Figure 5A, lane 3), 43% of the TG1–3 signal is in the end-complex peak, whereas in the TEL1 lysate this signal represents 74% (Figure 5A, lane 9). A graphical comparison of the differences in the TG 1–3 signals at each time point is presented in Figure 5C. The images extend the notion of a broader and lower intensity TG1–3 DNA peaks in the _tel1_Δ versus TEL1 samples. Together, these results show a higher sensitivity of telomere DNA to nucleases in _tel1_Δ cells and suggest a more accessible conformation of the end-complex. The higher sensitivity of telomeres to nuclease digestion in _tel1_Δ is not a mere consequence of the shorter mean telomere length in this strain. In _yku70_Δ cells, the mean telomere length is comparable to _tel1_Δ, yet the _yku70_Δ telomeres have a nuclease resistance comparable to that of WT, rather than _tel1_Δ cells (Supplementary Figure 2). We also compared the patterns of degradation of the Y′ subtelomeric DNA in _tel1_Δ and TEL1 cells. The data are shown in Figure 5B. In contrast to telomere DNA, the Y′ regions in both cells display little differences in sensitivity to both nucleases (Figure 5B, compare lanes 1–6 to 7–12 and 13–18 to 19–24), arguing that the difference in the end-complex between _tel1_Δ and TEL1 cells does not extend into the subtelomeric regions of the chromosomes. This digestion pattern further supports the notion that the nucleosomal structure of subtelomeric DNA in _tel1_Δ cells is intact, as revealed by the analysis of Tel 7Ltr (Supplementary Figure 1).
Overall, the results with MNase and DNase I indicate a more accessible conformation of the end-complex in the absence of TEL1. This altered organization of telomeric structure is likely to be related to the ability of Tbf1 to affect telomere length regulation.
The cis-shortening effect of Tbf1 requires telomerase expression
To determine whether the effects of Tbf1 were dependent on telomerase, we constructed diploid strains (EST2/_est2_Δ TEL1/_tel1_Δ) containing a Tel 7Ltr telomere with or without Tbf1 sites ((T2AG3)10). The diploid strains were sporulated and the rate of telomere shortening at the Tel 7Ltr telomere was measured in cell cultures established from different spores. As expected, in cultures from spores lacking EST2, the median telomere length decreased linearly (Figure 6A). The degradation rates were similar in cultures from spores lacking EST2 and from spores lacking EST2 and TEL1, at Tel7Ltr or Tel7Ltr-(T2AG3)10 (Figure 6B). Therefore, _tel1_Δ does not exacerbate the rate of telomeric DNA degradation caused by the loss of telomerase, even in the presence of subtelomeric Tbf1 sites. It is noteworthy that the presence of Tbf1 sites appears to reduce telomere length heterogeneity with increasing generations (Figure 6A), as well as slightly reducing the degradation rate of the Tel 7Ltr (Figure 6B). A decrease in the degradation rate should lead to telomere lengthening in telomerase-positive cells and thus cannot account for the telomere shortening induced by Tbf1 in telomerase-positive cells. Overall, these data are consistent with the physical presence of Tbf1 at telomeres impairing the telomerase-mediated telomere elongation pathway. As the tethering of Tbf1 acts only on its proximal telomere but not on the other telomeres, we conclude that Tbf1 _cis_-inhibits telomerase without affecting the overall level of telomerase expression.
Figure 6.
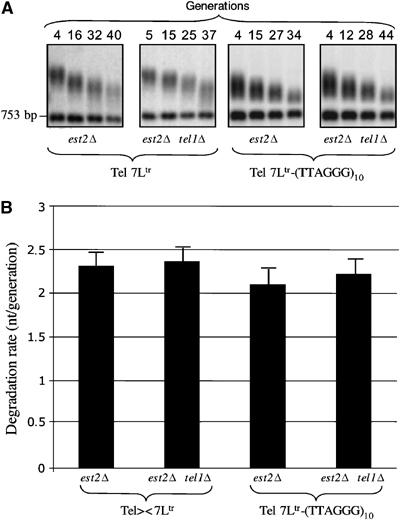
Tbf1 shortening effect requires telomerase expression. (A) Representative Southern blots used to calculate the degradation rates shown in panel B. Spores were grown in a rich liquid medium. Genomic DNAs were digested with _Eco_RV and _Hin_dIII and probed with URA3. (B) Telomere length was plotted versus generations and telomeric degradation rates were calculated from linear regression curves. The values represent the average of at least two independent experiments.
Discussion
In this work, we demonstrate that the subtelomeric proteins Tbf1 and Reb1 act as negative regulators of telomere length (Figure 1). This activity can be revealed at engineered telomeres containing subtelomeric Tbf1 sites and at telomeres of humanized yeast, which contain terminally located Tbf1 binding sites. In particular, we show that the number of Tbf1 or Reb1 N-terminal domains present in a subtelomeric region is ‘counted' to determine the overall length of the telomere (Figure 3). The ability of Tbf1 and Reb1 to regulate telomere length appears maximal in the absence of the TEL1 gene, which encodes a kinase involved in the Rap1-dependent telomere length regulation pathway. We were unable to detect any change in the phosphorylation status of Tbf1 between TEL1 and _tel1_Δ cells (A-S Berthiau and E Gilson, unpublished data). Confirming these results in another telomere setting, the tethering of Tbf1 and Reb1 at a remote subtelomeric location (>23 kb) of Tel 6R triggers telomere shortening of the proximal telomere in _tel1_Δ but not TEL1 cells (Hediger et al, 2006). Importantly, we found that the presence of subtelomeric Tbf1 sites in telomerase-negative cells does not exacerbate telomere shortening, showing that the _cis_-shortening effect of Tbf1 requires telomerase expression. Based on these results, we conclude that Tbf1 and Reb1 negatively regulate telomere length by _cis_-inhibiting telomerase in a Tel1- and Rap1-independent manner. As telomerase activity is linked to conventional replication (Chakhparonian and Wellinger, 2003), Tbf1 and Reb1 could interfere with replication. However, the telomeric replication fork pause appears to be independent of these proteins (Makovets et al, 2004).
The cis-shortening effect of Tbf1 and Reb1 does not result from transcription activation
We identify a 90 amino-acid long sequence within the N-terminal domain of Tbf1, which is necessary but not sufficient for the length regulation properties of the protein. This portion of Tbf1, named the Tbf1I domain, exhibits both antisilencing and transactivation activities (Figure 2). This, together with the fact that the N-terminal domain of Reb1 also acts as a transcriptional activator and as an inhibitor of telomere elongation, raises the possibility that the subtelomeric factors couple transcription to telomere length regulation, as previously reported for a forced transcription into the telomeric tract that slightly shortens telomere length (Sandell et al, 1994). However, the telomere _cis_-shortening induced by Tbf1 and Reb1 cannot be explained by transcription or derepression of the telomeric region: (i) the Tbf1I domain fused to the DNA binding domain of Gal4 acts as a transcriptional insulator and as a much more potent transactivator than a similar construct containing the whole N-terminal domain of Tbf1 (Figure 2), whereas it is unable to downregulate telomere length in _tel1_Δ cells by itself (Figure 3); (ii) a deletion of SIR3, leading to a total derepression of the subtelomeric region, neither recapitulates nor favors the _cis_-shortening effect of Tbf1 or Reb1 (Figure 4); (iii) Tbf1 binding does not alleviate silencing initiation but prevents the spreading of the silent state away from the telomere (Fourel et al, 1999), suggesting that the telomere is still in a silent state; (iv) by sequence inspection, the DNA binding sites for Tbf1 and Reb1 are not associated with TATA boxes; (v) the remote tethering (>23 kb) of Tbf1 or Reb1 at Tel 6R does not induce the derepression of a Tel 6R-associated reporter gene but leads to telomere shortening in _tel1_Δ cells (Hediger et al, 2006). Therefore, we conclude that the effects of Tbf1 and Reb1 on telomere length regulation do not simply result from transcription activation.
Tbf1 and Reb1 alter the nuclear organization of the telomere: a link to telomerase regulation
We noticed that in _tel1_Δ cells, the telomere end-complex appears less structured as demonstrated by its higher sensitivity to nuclease digestion (Figure 5). This structural alteration seems to be restricted at the very terminal part of the chromosome, as the chromatin structure of the immediate subtelomeric regions is not modified by the absence of TEL1 (Figure 5 and Supplementary Figure 1). It does not seem that the shortening of the telomere per se is the cause, as similarly short telomeres in _yku70_Δ cells do not show this characteristic alteration of the telomere structure (Supplementary Figure 2). The fact that Tel1 has a direct bearing on the structure of the end-complex is in agreement with its direct association with yeast telomeres (Takata et al, 2004) and with the fact that the TEL1 ortholog ATM is involved in the higher-order structure of mammalian telomeres (Pandita, 2001).
The less structured end-complexes present in _tel1_Δ yeasts could render the telomeres more sensitive to the action of additional factors involved in length regulation or to alterations in the subnuclear telomere organization. In agreement with the latter possibility, Tbf1 and Reb1 act as antianchoring factors in _tel1_Δ but not TEL1 cells, pulling the telomere away from the nuclear periphery (Hediger et al, 2006). Consequently, a tempting model is that Tbf1 and Reb1 counteract telomerase by moving the telomere away from a telomerase-competent compartment (Hediger et al, 2006).
Subtelomeric proteins as telomere length regulators
Our results suggest that length regulation at natural telomeres depends on the array of Tbf1 and Reb1 sites adjacent to the terminal repeats of many yeast telomeres. This could account for the telomere-specific sensitivity toward TEL1 shown in yeast (Craven and Petes, 1999) and, if similar mechanisms exist in mammalian cells, for the chromosome-specific length profile of human telomeres (Graakjaer et al, 2004). The antagonism between TEL1 and the telomeric role of subtelomeric factors suggests a new link between DNA damage checkpoint and telomere length control. For instance, TEL1 activation upon DNA damage might modulate the contribution of subtelomeric regions to telomere length regulation. In both TEL1 and _tel1_Δ backgrounds, we did not detect any significant difference between TBF1 and tbf1_Δ_i cells in the mean length of Y′ telomeres or of individual X-linked telomeres (A-S Berthiau and E Gilson, unpublished observations). However, the capacity of subtelomeric factors to regulate telomere length is likely to have been underestimated in the tbf1_Δ_i setting, as this allele leads to only a partial loss of function and does not interfere with the activity of Reb1, leaving open the possibility that redundant effects exist between Tbf1, Reb1 and possibly other subtelomeric factors.
Implications for telomere evolution
It is worth noting that Tbf1 belongs to a family of proteins possessing a very similar and evolutionarily related Myb-like DNA binding domain, including the authentic telomeric proteins TRF1 and TRF2 (Bilaud et al, 1996; Teixeira and Gilson, 2005). Interestingly, TRF1 and TRF2 also alter the telomerase pathway, in a seemingly hRAP1-independent manner for TRF1 (Ancelin et al, 2002; Loayza and De Lange, 2003) and through the binding of hRAP1 for TRF2 (Li and de Lange, 2003). What determines the relative contribution of each of these pathways at individual telomeres or between different cell types is not known. Interestingly, ATM, the human ortholog of TEL1, is defective in the cancer-prone disorder ataxia telangiectasia (A-T) and influences telomere length as well as higher-order organization of chromosome ends (Pandita, 2001). Furthermore, ATM is known to interact with both TRF1 (Kishi et al, 2001) and TRF2 (Karlseder et al, 2004). As the deletion of TEL1 converts yeast telomeres from a Rap1-dependent to a Rap1-independent mechanism of length regulation, telomere length defects in A-T patients might result from a modified balance between the TRF1 and TRF2 pathways.
In the light of the work presented here, Tbf1 in yeast should be considered not only as an evolutionary remnant of an earlier telomere-binding protein but also as a functional telomeric factor. This, together with the involvement in telomere length regulation of the subtelomeric protein Reb1 and with the role of the subtelomeric regions in modulating telomeric silencing (Fourel et al, 1999) and nuclear telomere positioning (Hediger et al, 2006), led us to propose that the subtelomeric regions are a functional part of telomeres. Given that Tbf1 and Reb1 behave as essential regulatory factors for gene expression at non-telomeric loci and that their transactivation domains are also involved in length regulation, these subtelomeric proteins might have been conserved through essential non-telomeric functions, while preserving their capacity to regulate telomere structure.
Materials and methods
Strains and constructs
The engineer Tel 7Ltr telomeres were previously described (Marcand et al, 1997; Fourel et al, 1999; Brevet et al, 2003). The pRS314-tbf1Δi plasmid results from the replacement of an _Eco_RI–_Sac_I fragment of pRS314-TBF1 (Brigati et al, 1993) by a PCR fragment generated by Tbf1a (5′-AGTATCGAATTCAGAAAACCAAGCG) and Tbf1b (5′-AAAATTGAGCTCCTGTTCATTTAAGCG) digested with _Eco_RI and _Sac_I. In order to shuffle to tbf1_Δ_i allele, first the genomic TBF1 gene was disrupted by a HIS3 insertion in W303-1A derivatives containing pRS316-TBF1 and then the pRS314-tbf1Δi plasmid was introduced and cells having lost pRS316-TBF1 were screened.
Assays for telomeric structure and composition
Telomere blotting and telomere length measurement were performed as described by Brevet et al (2003). The humanized telomere from Tel1L was PCR amplified according to a previously described method (Forstemann et al, 2000).
ChIP was performed, as described previously (Schramke et al, 2001), with a polyclonal antibody designed against the N-terminal domain of TBF1 (Bilaud et al, 1996). Real-time quantitative PCR was realized, as described by Brevet et al (2003).
Analysis of chromatin structure
Spheroplasts were suspended in nuclease buffer (30 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10 mM CaCl2, 100 mM NaCl, 0.1 mg/ml BSA, 2 μg/ml pepstatin, 2 μg/ml aprotinin, 2 μg/ml leupeptin). MNase (Worthington) or DNase I (Sigma) was added at 50 or 12.5 U/ml final concentration, respectively. Aliquots were removed after 0, 1, 2.5, 5, 10 and 20 min and immediately mixed with STOP solution (3.5 M NaCl, 1.4% SDS, 100 mM EDTA, 100 mM EGTA).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank S Gasser and F Hediger for communicating their results before publication, as well as T Teixeira, F Hediger, S Gasser and A Taddei for their critical reading of the manuscript. This work was supported by grants from the Ligue Nationale contre le Cancer to EG and VG (équipes labellisées) and from the CIHR (grant number 12616) to RW and AB. KY is supported through a sabbatical leave from the Universtity of Guelph. ASB is supported by fellowships from the Ligue Nationale contre le Cancer and the Association pour la Recherche sur le Cancer.
References
- Alexander MK, Zakian VA (2003) Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. EMBO J 22: 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E (2002) Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol 22: 3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res 24: 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH (1991) Structure and function of telomeres. Nature 18: 569–573 [DOI] [PubMed] [Google Scholar]
- Blackburn EH (2000a) The end of the (DNA) line. Nat Struct Biol 7: 847–850 [DOI] [PubMed] [Google Scholar]
- Blackburn EH (2000b) Telomere states and cell fates. Nature 408: 53–56 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17: 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevet V, Berthiau AS, Civitelli L, Donini P, Schramke V, Geli V, Ascenzioni F, Gilson E (2003) The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J 22: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigati C, Kurtz S, Balderes D, Vidali G, Shore D (1993) An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol 13: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunori M, Luciano P, Gilson E, Geli V (2005) The telomerase cycle: normal and pathological aspects. J Mol Med 83: 244–257 [DOI] [PubMed] [Google Scholar]
- Chakhparonian M, Wellinger RJ (2003) Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet 19: 439–446 [DOI] [PubMed] [Google Scholar]
- Chan SR, Blackburn EH (2004) Telomeres and telomerase. Philos Trans R Soc London B 359: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Chang J, Prescott J, Blackburn EH (2001) Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol 11: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination [see comments]. Nature 392: 828–831 [DOI] [PubMed] [Google Scholar]
- Craven RJ, Petes TD (1999) Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152: 1531–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Hoss M, Lingner J (2000) Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res 28: 2690–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Carmine Simmen K, Müller K, Li R, Mermod N, Gilson E (2001) An activation-independent role of transcription factors in insulator function. EMBO Rep 2: 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Gilson E, Gasser SM (1995) Repressor activator protein 1 and its ligands: organising chromatin domains. Nucl Acids Mol Biol 9: 308–327 [Google Scholar]
- Gilson E, Laroche T, Gasser S (1993) Telomeres and the functional architecture of the nucleus. Trends Cell Biol 3: 128–134 [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Shertan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild type S. cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Graakjaer J, Pascoe L, Der-Sarkissian H, Thomas G, Kolvraa S, Christensen K, Londono-Vallejo JA (2004) The relative lengths of individual telomeres are defined in the zygote and strictly maintained during life. Aging Cell 3: 97–102 [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop [see comments]. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Hediger F, Berthiau AS, van Houwe G, Gilson E, Gasser SM (2006) Subtelomeric factors antagonize telomere anchoring and tel1-independent telomere length regulation. EMBOJ [E-pub ahead of print: 9 February 2006; doi:10.1038/sj.emboj.7600976] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning KA, Moskowitz N, Ashlock MA, Liu PP (1998) Humanizing the yeast telomerase template. Proc Natl Acad Sci USA 95: 5667–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph I, Jia D, Lustig AJ (2005) Ndj1p-dependent epigenetic resetting of telomere size in yeast meiosis. Curr Biol 15: 231–237 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, Lange Td T (2004) The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol 2: E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S, Zhou XZ, Ziv Y, Khoo C, Hill DE, Shiloh Y, Lu KP (2001) Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J Biol Chem 276: 29282–29291 [DOI] [PubMed] [Google Scholar]
- Koering CE, Fourel G, Binet-Brasselet E, Laroche T, Klein F, Gilson E (2000) Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res 28: 2519–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol 12: 5159–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol 8: 653–656 [DOI] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, de Lange T (2003) Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell 14: 5060–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, De Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Lundblad V (2000) DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat Res 451: 227–240 [DOI] [PubMed] [Google Scholar]
- Maillet L, Gaden F, Brevet V, Fourel G, Martin SG, Dubrana K, Gasser SM, Gilson E (2001) Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep 2: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH (2004) Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol 24: 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E (2000) Cell cycle restriction of telomere elongation. Curr Biol 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P (1995) TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82: 831–840 [DOI] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol 8: 657–660 [DOI] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM (1993) SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555 [DOI] [PubMed] [Google Scholar]
- Pandita TK (2001) The role of ATM in telomere structure and function. Radiat Res 156: 642–647 [DOI] [PubMed] [Google Scholar]
- Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S (2004) HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell 15: 467–476 [DOI] [PubMed] [Google Scholar]
- Perrod S, Gasser SM (2003) Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell Mol Life Sci 60: 2303–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Runge KW (1999a) Varying the number of telomere-bound proteins does not alter telomere length in tel1Delta cells. Proc Natl Acad Sci USA 96: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Runge KW (1999b) The yeast telomere length counting machinery is sensitive to sequences at the telomere–nontelomere junction. Mol Cell Biol 19: 31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Naito T, Ishikawa F (2003) Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev 17: 2271–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Gottschling DE, Zakian VA (1994) Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci USA 91: 12061–12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V, Neecke H, Brevet V, Corda Y, Lucchini G, Longhese MP, Gilson E, Geli V (2001) The set1Delta mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Genes Dev 15: 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman JM, Pillus L (1997) An uncertain silence [see comments]. Trends Genet 13: 308–313 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Surralles J, Hande MP, Marcos R, Lansdorp PM (1999) Accelerated telomere shortening in the human inactive X chromosome. Am J Hum Genet 65: 1617–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Kanoh Y, Gunge N, Shirahige K, Matsuura A (2004) Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol Cell 14: 515–522 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E (2005) Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res 13: 535–548 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in S. cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA (1992) Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes and Dev 6: 197–210 [DOI] [PubMed] [Google Scholar]
- Zakian VA (1995) Telomeres: beginning to understand the end. Science 270: 1601–1607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2