Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: A model for latency (original) (raw)
Abstract
Human cytomegalovirus (HCMV) resides latently in hematopoietic cells of the bone marrow. Although viral genomes can be found in CD14+ monocytes and CD34+ progenitor cells, the primary reservoir for latent cytomegalovirus is unknown. We analyzed human hematopoietic subpopulations infected in vitro with a recombinant virus that expresses a green fluorescent protein marker gene. Although many hematopoietic cell subsets were infected in vitro, CD14+ monocytes and various CD34+ subpopulations were infected with the greatest efficiency. We have developed an in vitro system in which to study HCMV infection and latency in CD34+ cells cultured with irradiated stromal cells. Marker gene expression was substantially reduced by 4 days postinfection, and infectious virus was not made during the culture period. However, viral DNA sequences were maintained in infected CD34+ cells for >20 days in culture, and, importantly, virus replication could be reactivated by coculture with human fibroblasts. Using an HCMV gene array, we examined HCMV gene expression in CD34+ cells. The pattern of viral gene expression was distinct from that observed during productive or nonproductive infections. Some of these expressed viral genes may function in latency and are targets for further analysis. Altered gene expression in hematopoietic progenitors may be indicative of the nature and outcome of HCMV infection.
Human cytomegalovirus (HCMV) is a ubiquitous β-herpes virus that maintains a lifelong relationship with its host by way of a latent infection. HCMV infection in healthy individuals is typically asymptomatic, whereas a primary infection or reactivation of latent HCMV may cause life-threatening disease in immunologically immature or compromised individuals, including neonates, AIDS patients, and transplant patients (1). The mechanisms governing the establishment and maintenance of latency and reactivation of HCMV from latency are unknown.
HCMV infects a variety of cells types, including hematopoietic and stromal cells of the bone marrow, endothelial cells, epithelial cells, fibroblasts, neuronal cells, and smooth muscle cells (2–5). The bone marrow is a site of HCMV latency (6), but the primary cellular reservoir harboring latent virus within the bone marrow is unknown. Latent viral genomes are detected in CD14+ monocytes and CD33+ myeloid precursor cells (7, 8). Differentiation of CD14+ monocytes into macrophages yields a permissive environment for viral replication and infectious virus is produced (9, 10). HCMV also infects CD34+ hematopoietic progenitor populations (11–15), and viral DNA sequences can be detected in CD34+ cells from healthy seropositive individuals (6, 15), suggesting that a primitive cell population serves as a renewable primary reservoir for latent HCMV.
Two in vitro systems have been developed as models for HCMV latency. Nelson and colleagues (9, 16) have used allogenic stimulation to study HCMV reactivation in monocytes that harbor viral genomes. CD4+ and CD8+ T lymphocytes and the cytokines, IFN-γ, and tumor necrosis factor-α facilitated viral reactivation (16, 17). Mocarski and colleagues (8, 18) have studied HCMV latency in granulocyte–macrophage progenitors expressing CD33 and dendritic cell markers, and identified several HCMV transcripts expressed during latency following in vivo or in vitro infection (18, 19).
Because HCMV DNA is found in CD34+ bone marrow cells (6, 15), it is likely that the virus establishes and maintains latency in hematopoietic cells that are more primitive and longer-lived than cells used in earlier in vitro latency studies. Studying HCMV infection of hematopoietic progenitors in vitro is challenging, given the difficulty of maintaining progenitors in an undifferentiated state in culture. We have developed an in vitro model to study HCMV latency in primitive hematopoietic progenitor populations. Primary CD34+ hematopoietic progenitors are infected immediately after isolation and cultured in long-term bone marrow culture above the immortalized murine stromal cell layer, AFT024 (20). The stromal cells maintain human progenitor cell phenotype and support their proliferation (21–23). During the culture period, viral DNA is maintained in the absence of substantial virus production. The program of HCMV gene expression in CD34+ cells during long-term bone marrow culture is markedly different from productively infected fibroblasts. Importantly, viral replication can be reactivated by coculture on permissive fibroblasts. Our model offers a unique system in which to study HCMV latency, and identifies molecular targets that may contribute to latency.
Materials and Methods
Viruses and Cells.
AD_sub_UL21.5 and TL_sub_UL21.5 were engineered by introducing a marker cassette into the wild-type AD169 and Toledo strains of HCMV, respectively, replacing the UL21.5 coding region (AD169 sequence position 27499–27503). The marker cassette contained the SV40 early promoter controlling the expression of a bicistronic mRNA consisting of an upstream ORF encoding GFP, an internal ribosomal entry site, and a downstream ORF encoding puromycin resistance. Recombinant viruses were constructed by homologous recombination within transfected human fibroblasts as described (24). Virions were purified by density gradient centrifugation through a d-sorbitol cushion as described (25). Virions were resuspended in Iscove-modified Dulbecco medium (IMDM) supplemented with 2% FBS, and virus was stored at −80°C. Before infection, virus was thawed, sonicated, and centrifuged to pellet debris. Infectious virus yields were assayed on human fibroblasts.
Bone marrow cells were obtained from waste materials used during bone marrow harvest procedures from healthy donors at Wake Forest University Baptist Medical Center via a protocol approved by the Institutional Review Board. Umbilical cord blood was obtained from the National Disease Research Interchange. Mononuclear cells were obtained from bone marrow or cord blood by using Ficoll/Paque Plus (Amersham Pharmacia Biosciences) density gradient centrifugation. CD34+ cells were isolated by magnetic cell separation (Miltenyi Biotec, Auburn, CA). Hematopoietic cells (106 cells per ml) were exposed to virus for 12–20 h in IMDM supplemented with 10% BIT serum substitute (StemCell Technologies, Vancouver), 2 mM l-glutamine, 20 ng/ml human low-density lipoprotein (Sigma), and 50 μM 2-mercaptoethanol. Following infection, cells were washed for 1 min in citrate buffer (40 mM Na citrate/10 mM KCl/135 mM NaCl, pH 3.0) to inactivate unabsorbed virus (26).
Cultures of CD34+ cells were established as described (22). Briefly, murine AFT024 cells were plated at a density of 4 × 105 cells per ml in 6-well polystyrene plates coated with 0.1% gelatin (StemCell Technologies) and irradiated (20 Gy, Cesium 137 source, Gammacell 40, Nordion International, Kamata, ON, Canada). CD34+ cells were plated in collagen-coated transwells with a 0.4-μm microporous filter (Transwell-COL, Costar) above irradiated AFT024 cells in RPMI medium 1640 (GIBCO/BRL) supplemented with 20% FBS, 100 μmol/liter 2-mercaptoethanol, and 10 ng/ml each of stem cell factor, Flt-3 ligand, and IL-7 (R & D Systems).
Murine AFT024 cells were maintained in DMEM supplemented with 10% FBS and 50 μM 2-mercaptoethanol at 32°C. Primary human foreskin fibroblasts were maintained in DMEM containing 10% FBS at 37°C.
Flow Cytometry.
Intracellular staining for the pp65 virion protein was performed by fixing and permeablizing cells with Fix and Perm (Caltag, South San Francisco, CA), and then reacting with an anti-pp65 mouse monoclonal antibody followed by a goat anti-mouse secondary IgG conjugated to allophycocyanin (APC; BD Biosciences). Cells were analyzed for the expression of GFP and staining for pp65 by using a FACSort flow cytometer (BD Biosciences Immunocytometry Systems).
Immunophenotyping assays were performed by labeling infected cell populations with fluorescently conjugated monoclonal antibodies (BD PharMingen and BD Immunocytometry Systems) and analyzing GFP+-infected cells in subsets with a FACSort flow cytometer (BD Biosciences Immunocytometry Systems).
Infected CD34+ cells were isolated from infected CD34- enriched populations by labeling samples with a phycoerythrin (PE)-conjugated CD34-specific monoclonal antibody, and then sorting cells that were PE+ and GFP+ by using a FACSVantage flow cytometer (BD Biosciences Immunocytometry Systems). A purity of ≥95% was typically obtained.
Detection of Viral Nucleic Acids.
To isolate DNA, cells were lysed by incubating cells at 50°C in lysis buffer (200 mM Tris⋅HCl, pH 8.0/50 mM EDTA/0.5% SDS/100 μg of proteinase K per ml) for 2 h. DNA was extracted with phenol-chloroform, precipitated, and quantified. The UL5 and actin sequences present in recombinant viruses was detected by PCR amplification.
An HCMV cDNA array (27), together with a series of cellular and control cDNAs, was printed in 50% DMSO on GAPS II amino-silane-coated glass slides (Corning) by using an GMS 417 Arrayer (Genetic MicroSystems, Woburn, MA). Each cDNA was printed in triplicate on different sections of the slide. RNA was isolated from hematopoietic cells by using the Absolutely RNA Microprep kit (Stratagene) and isolated from human fibroblasts by using Trizol LS (Invitrogen). RNA isolated using Trizol was treated with DNaseI by using the DNA-free kit (Ambion, Austin, TX), and polyadenylated transcripts were purified using MicroPoly(A) Pure (Ambion). Equal amounts of RNA were linearly amplified using MessageAmp (Ambion), and the amplified product was primed by using the CyScribe Direct Labeling Kit (Amersham Pharmacia Biosciences) to incorporate Cy3-dUTP or Cy5-dUTP into a cDNA. Labeled cDNA was hybridized to arrays as described (28), and data were analyzed using a GenePix 4000B scanner and GENEPIX PRO VERSION 3.0 software (Axon Instruments). The mean intensities for triplicate spots were averaged. Positive signals were identified by subtracting the mean values for the local background plus two standard deviations of the background and the averaged mean intensity of negative controls from the mean intensity of each spot. Comparisons among arrays were made by normalizing values to the average mean intensity of 18 cellular housekeeping genes included on the array. A portion of the cDNA prepared for arrays was analyzed by RT-PCR to confirm the presence or absence of mRNAs.
Results
Immunophenotype of Hematopoietic Cells Infected by HCMV in Vitro.
The primary cellular reservoir in the bone marrow for latent HCMV is unknown. To identify subsets of bone marrow cells that can be infected in vitro with HCMV, we assayed the infection of a variety of hematopoietic subsets with AD_sub_UL21.5 or TL_sub_UL21.5. These viruses are derived from two strains of HCMV and carry a GFP marker in place of the viral UL21.5 gene, which is dispensable for virus growth in cultured fibroblasts (data not shown).
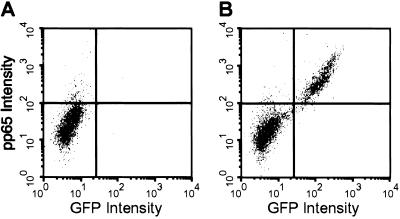
The hematopoietic compartment is comprised of a heterogeneous mixture of cells, and, consequently, GFP expression from the viral genome may depend on the physiological state of each particular cell subset. If GFP is not expressed in all infected subsets, then some infected cells would be missed in our analysis. To determine whether GFP is a reliable marker for infection, we analyzed mononuclear cells infected with TL_sub_UL21.5 by flow cytometry for the presence of both GFP and the viral protein pp65, a virion protein that is delivered to the cell on viral entry. At 10 h postinfection, very few mock-infected cells stained positively for the markers, whereas >98% of the infected cells staining positively for pp65 also expressed GFP (Fig. 1, upper right quadrants). Approximately 2% of the cells stained positively for pp65 in the absence of GFP expression in both the mock and infected populations (Fig. 1, upper left quadrants), indicating that this is background staining. We conclude that GFP expressed from TL_sub_UL21.5 is an appropriate marker for viral infection of primary mononuclear cells.
Fig 1.
GFP is a reliable marker for TL_sub_UL21.5 infection of hematopoietic cells. Mononuclear cells enriched for CD34+ cells (≈60%) were mock-infected (A) or infected with TL_sub_UL21.5 at a multiplicity of 5 plaque-forming units (pfu) per cell (B). At 10 h postinfection, cells were analyzed for expression of GFP and staining for pp65 by flow cytometry. Data from a representative experiment are shown.
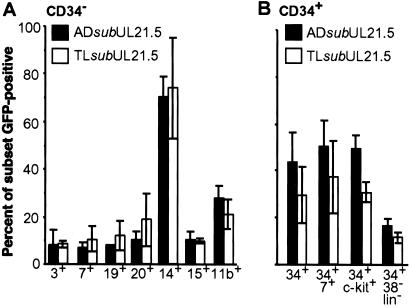
To determine the immunophenotype of cells infected in vitro, populations enriched or depleted for the CD34+ marker were infected with AD_sub_UL21.5 or TL_sub_UL21.5, and cells were stained with fluorescently conjugated monoclonal antibodies to identify various hematopoietic subsets. Flow cytometry was used to determine the percentage of each subpopulation expressing GFP. Lineage-specific subsets within the CD34-depleted fraction of cells were infected with low efficiency (typically <20%), with the exception of CD14+ monocytes (>70%; Fig. 2A). Progenitor populations within the CD34-enriched population were infected at intermediate efficiencies (20–60%; Fig. 2B). Phenotypically primitive CD34+/CD38−/lineage-negative cells were infected, but at relatively low efficiencies (<20%). Precursor T cells expressing CD34 and CD7 were more efficiently infected than CD7+ T cells from the CD34-depleted fraction (compare A and B of Fig. 2). Not surprisingly, the differentiation state appears to affect the relative efficiency by which cells are infected. No difference was observed in the ability of the two HCMV strains to infect these hematopoietic cell populations. From these data we conclude that many subsets of cells can be infected within the hematopoietic compartment, ranging from lineage-specific cells to primitive progenitor cells. We have focused the remainder of our analyses on infection of CD34+ progenitor populations.
Fig 2.
Immunophenotypes of hematopoietic cells infected with AD_sub_UL21.5 or TL_sub_UL21.5. Mononuclear cells were infected at a multiplicity of 5 pfu per cell. At 16–18 h postinfection, cells were labeled with fluorescently conjugated antibodies to distinguish hematopoietic subsets. The percentage of each subset infected (GFP+) was determined by flow cytometry. (A) Analysis of CD34-depleted cells. CD3 and CD7 cell surface markers identify T lymphocytes; CD19 and CD20 identify B lymphocytes; CD14 identifies monocytes; CD15 identifies neutrophils; and CD11b identifies predominantly granulocytes and monocytes. (B) Analysis of CD34-enriched cells containing progenitor cells. CD34+/CD7+ cells and CD34+/c-_kit_+ cells are subsets of progenitor cells. CD34+/CD38−/lineage-negative cells represent a rare progenitor subset containing stem cells. Each bar represents an average of three to five independent experiments. Standard errors are indicated.
Evidence for HCMV Latency After Infection of CD34+ Cells in Vitro.
A model for HCMV latency in primary hematopoietic progenitor cells must meet three criteria. First, ex vivo culture conditions must maintain progenitor populations in a state closely reflecting their in vivo state. Hematopoietic progenitor differentiation is exceptionally sensitive to ex vivo stimuli. Altering the natural balance of differentiation could dramatically effect the outcome of an infection and diminish the model's relevance. Second, viral genomes must be maintained in infected progenitor populations in the absence of viral replication and gene expression characteristic of a productive infection. Third, viral replication must be reactivated in the presence of a stimulus. HCMV-infected CD34+ hematopoietic cells maintained in long-term bone marrow culture satisfy all of these criteria.
The latency model employs AFT024 murine stromal cells (20, 21), which maintain the phenotype and function of primary human hematopoietic progenitor populations and support progenitor proliferation (22, 23). CD34+ progenitor populations cultured above an AFT024 monolayer for up to 28 days can reconstitute the blood system of nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice in competitive repopulation studies (22), demonstrating that these stromal cells can maintain progenitor populations, including stem cells.
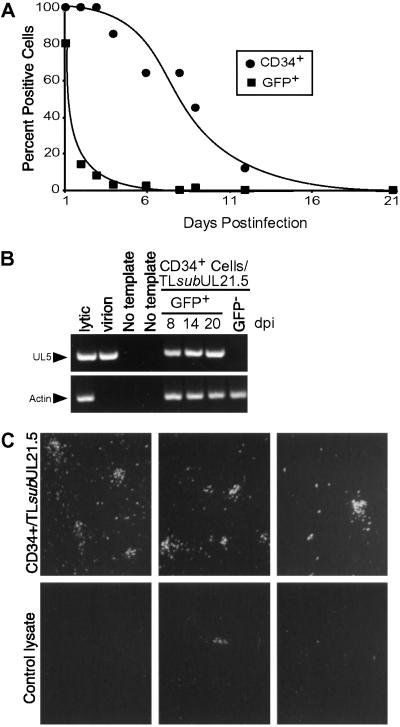
For HCMV latency studies, CD34-enriched mononuclear cells from bone marrow or cord blood were infected with TL_sub_UL21.5 immediately following isolation, so that ex vivo influences were minimized and progenitor cells resembled their in vivo state at the time of infection. Following infection, extracellular virus was inactivated (26), and infected CD34+ cells (GFP+/CD34+) were isolated by flow cytometry and cultured in transwells above an irradiated AFT024 monolayer for up to 21 days. We measured the ability of this stromal layer to maintain progenitor populations by monitoring the cell surface expression of the CD34 progenitor marker (Fig. 3A). Although CD34+ expression was diminished with time, high levels of CD34 were expressed on the cell surface of >50% of infected CD34+ progenitors for up to 8 days in culture. HCMV infection did not alter CD34 cell surface expression from that observed in mock-infected cell populations (data not shown). By contrast, GFP expression was transient. It was maximal at 1 day postinfection, dramatically reduced by 2 days, and undetectable by 6 days (Fig. 3A). Despite the rapid loss in GFP marker expression, DNA sequences from the viral genome could be detected by PCR amplification for 20 days, the last time tested, whereas no viral sequences were detected in GFP− cells from infected populations (Fig. 3B).
Fig 3.
In vitro model for HCMV latency. Mononuclear cells were infected with TL_sub_UL21.5 at a multiplicity of 5 pfu per cell. Infected CD34+ cells were isolated at 16–20 h postinfection and put into long-term bone marrow culture. (A) Time course of CD34 and GFP expression during long-term bone marrow culture. (B) Detection of UL5 and actin sequences from infected cells during long-term bone marrow culture by PCR. (C) Reactivation of HCMV replication in infected hematopoietic cells after coculture with fibroblasts. Fibroblasts were cocultured with infected CD34+ cells from long-term bone marrow cultures (Upper) or with a lysate prepared from an equivalent number of cells (Lower), and reactivation was monitored by GFP fluorescence. Representative data are shown from three independent experiments.
At 10 days postinfection, a time when GFP marker expression was not detected, infected cells were transferred from culture above AFT024 cells and cultured with permissive human fibroblasts to analyze the ability of infected CD34+ cells to support reactivation of viral replication. Within 14 days after initiation of the cocultures, numerous GFP+ fibroblasts were detected (Fig. 3C Upper). These results indicate that virus replication reactivated in infected hematopoietic cells and subsequently infected the fibroblast monolayer. In addition to foci of GFP+-infected fibroblasts, single infected cells were evident. It is not known whether single GFP+ cells represent spread from primary infections of fibroblasts or asynchronous reactivation of viral replication in hematopoietic cells. To distinguish virus production during the culture period with stromal cells from that resulting from reactivation in the presence of fibroblasts, an equivalent number of cells were lysed in their medium by sonication and transferred to fibroblast cultures. Green cells or plaques developed at an ≈200-fold lower frequency than in the reactivation experiments (Fig. 3C Lower), arguing that reactivation of viral replication occurred in the CD34+/fibroblast cocultures. Presumably, infrequent and sporadic reactivation of viral replication occurs during the in vitro latency period. Alternatively, a small number of the CD34+ cells were permissive for viral replication. The latter proposal is attractive, because the CD34+ population is heterogeneous. Yet another possibility, given the low level of viral replication that occurred in fibroblast cultures receiving a lysate of infected CD34+ cells, is that a few cells escaped lysis and were able to reactivate. Taken together, these findings strongly support the view that HCMV establishes a latent infection in CD34+ cells that can be reactivated.
CD34+ Cells Infected with HCMV Exhibit Distinct Patterns of Viral Gene Expression.
The HCMV gene array developed in our lab (27) consists of PCR products corresponding to the 203 known ORFs for the AD169 strain of HCMV (29). The array has been extended to include 19 additional ORFs identified in clinical isolates (30), negative controls, and 80 cellular cDNAs that include housekeeping genes to be used as normalization controls and genes whose expression is modified by HCMV infection in fibroblasts (31, 32).
For array experiments, CD34+ cells were infected and maintained in long-term bone marrow culture. RNA from 10,000–20,000 infected cells was isolated at various times following HCMV infection, and linearly amplified. Fluorescently labeled cDNA probes were synthesized and hybridized to the HCMV arrays. CD34+ cells infected with HCMV expressed a large number of genes during a time course of infection in long-term bone marrow culture. Table 1 lists the mean intensity values for triplicate array spots after background subtraction and normalization for one representative infection. Only the genes that were positive from the hybridization are listed. HCMV genes were not expressed in CD34+ cells in the ordered cascade typical of a lytic infection in fibroblasts, and very few late genes were expressed even at very late times of infection. Indeed, during the 8-day time course, only a subset of mRNAs, representing less than half of the known HCMV genes, were expressed in CD34+ cells. By contrast, mRNAs encoding most HCMV genes were detectable late after infection of fibroblasts (Fig. 4). No viral gene expression was detected on arrays hybridized with cDNA from CD34+ cells that were either mock-infected or infected with UV-inactivated virus (data not shown).
Table 1.
HCMV gene expression in CD34+ cells
| Gene | Signal intensities at dpi | Gene | Signal intensities at dpi | ||||
|---|---|---|---|---|---|---|---|
| 1 | 5 | 8 | 1 | 5 | 8 | ||
| T/IRL2 | 2491.9 | 2107.6 | UL76 | 537.5 | 188.2 | ||
| T/IRL3 | 344.0 | 2107.0 | 2746.2 | UL77 | 2.8 | ||
| T/IRL4 | 1555.7 | 1486.2 | UL78 | 31.7 | |||
| T/IRL5 | 50.8 | 31.7 | UL80 | 57.3 | |||
| T/IRL6 | 135.6 | 49.7 | UL81 | 141.0 | 129.8 | 963.8 | |
| T/IRL7 | 3.3 | 125.0 | 47.2 | UL84 | 52.0 | 71.0 | |
| UL4 | 44.1 | 70.7 | UL87 | 34.9 | 81.4 | ||
| UL5 | 232.7 | 527.7 | 541.8 | UL98 | 24.6 | ||
| UL13 | 515.0 | UL99 | 35.0 | 52.3 | 140.3 | ||
| UL17 | 96.3 | 311.6 | 32.8 | UL105 | 27.5 | 71.0 | |
| UL21 | 27.7 | UL108 | 33.8 | 39.3 | |||
| UL23 | 32.5 | UL110 | 394.2 | 564.6 | |||
| UL25 | 34.0 | UL111 | 17.2 | ||||
| UL26 | 22.0 | UL122 | 87.3 | 38.2 | |||
| UL30 | 76.1 | 276.7 | UL123 | 1110.0 | |||
| UL32 | 111.7 | 48.7 | 485.1 | UL124 | 32.7 | ||
| UL34 | 0.6 | UL125 | 3.0 | ||||
| UL38 | 34.7 | 21.9 | 6.9 | UL128 | 25.8 | ||
| UL39 | 52.7 | 379.0 | 162.6 | UL131 | 17.6 | ||
| UL40 | 31.0 | 70.7 | 179.3 | UL132 | 334.0 | 56.7 | 609.7 |
| UL41 | 6.5 | UL133 | 32.7 | 32.8 | |||
| UL44 | 7.6 | UL135 | 6.9 | ||||
| UL61 | 258.0 | 76.0 | UL138 | 251.7 | 45.3 | 9.2 | |
| UL63 | 14.3 | UL141 | 64.0 | 164.7 | |||
| UL64 | 1164.8 | 436.4 | UL145 | 233.7 | 164.9 | 177.0 | |
| UL65 | 54.0 | UL150 | 506.3 | 259.1 | 584.8 | ||
| UL66 | 40.2 | US6 | 200.6 | ||||
| UL67 | 408.7 | 160.2 | US8 | 63.1 | |||
| UL68 | 3.3 | 1214.1 | 283.2 | US10 | 45.0 | ||
| UL69 | 2.4 | US11 | 25.2 | ||||
| UL70 | 32.7 | 838.2 | 239.1 | US12 | 118.3 | 88.6 | |
| UL72 | 4.2 | US28 | 50.3 | ||||
| UL73 | 9.1 | US32 | 14.8 | ||||
| UL75 | 4.9 | US34 | 13.0 | 0.1 | 0.3 |
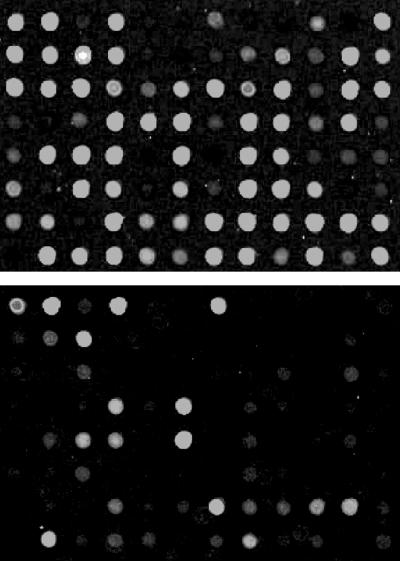
Fig 4.
Gene arrays reveal cell-type-specific differences in HCMV RNA expression. Representative array segments with cDNAs corresponding to HCMV genes UL1 through UL79 are displayed. Fibroblasts were infected at a multiplicity of 5 pfu per cell, and viral RNAs were analyzed at 72 h postinfection (Upper). CD34+ cells were infected at the 10 pfu per cell and RNAs were analyzed at 5 days postinfection (Lower).
In multiple experiments, the range of HCMV mRNAs expressed in CD34+ cells did not vary greatly (data not shown). However, the intensity (i.e., apparent level of expression) and kinetics of expression were somewhat variable. The variability between experiments might result from differences in the nature or composition of the CD34+ cell population among donors. Given the heterogeneous nature of the CD34+ cell population, any subset of cells within the CD34+ population might express only a subset of the genes expressed in the entire CD34+ population. Detection of these genes may be less variable in highly purified subsets of CD34+ populations.
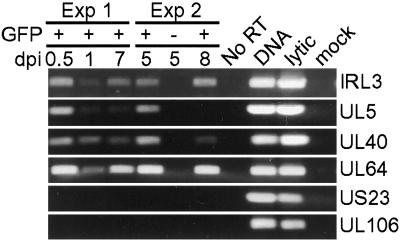
The expression of several HCMV genes was confirmed using RT-PCR (Fig. 5). cDNAs from two independent experiments were amplified by PCR using primers specific to the IRL3, UL5, UL40, UL64, UL106, and US23. Expression of IRL3, UL5, UL40, and UL64 was detected by both array and RT-PCR analysis, whereas expression of UL106 and US23 was not detected by either method. These results validate the patterns of gene expression determined using the viral array.
Fig 5.
HCMV gene expression in CD34+ cells was confirmed by RT-PCR. cDNA prepared from two independent experiments was analyzed for the expression of IRL3, UL5, UL40, UL64, UL106, and US23. IRL3, UL5, UL40, and UL64 were detected in CD34+ cells by array analysis. UL106 and US23 were not detected by array analysis. Controls included GFP− CD34+ cells in experiment 2, an RNA sample that received no reverse transcriptase during cDNA amplification, DNA from a lytic infection, and cDNA from a lytic and mock infection.
Discussion
The mechanisms by which HCMV establishes and maintains latency and reactivates from latency in the bone marrow are not understood. We have developed an in vitro model system for studying HCMV latency in primary human hematopoietic progenitor cells. Our model employs AFT024 stromal cells (20–23) to maintain CD34+ cells in culture, and it meets the key criteria for a latent system. After viral infection, viral genomes were maintained (Fig. 3B) in the absence of viral gene expression characteristic of a productive infection (Fig. 4 and Table 1) and without producing a substantial virus yield (Fig. 3C Lower). Importantly, virus replication was reactivated after extended periods of culture (Fig. 3C Upper). Successful reactivation argues that infection of CD34+ cells in long-term bone marrow culture results in a latent infection and not an abortive or nonproductive infection. Consistent with this interpretation, the pattern of HCMV gene expression in CD34+ cells was markedly different from that observed in nonproductively infected HeLa or Rat-1 cells (data not shown). We conclude that the pattern of HCMV gene expression seen in primary CD34+ cells is distinct and may include genes that contribute to latency.
The efficient reactivation we have observed on coculture of infected CD34+ cells with fibroblasts (Fig. 3C Upper) argues that a substantial portion of the CD34+ population can support a latent infection. A low level of infectious virus was produced before coculturing infected CD34+ cells with fibroblasts (Fig. 3C Lower), consistent with an earlier report that a small portion of mononuclear cells infected with HCMV can support a productive infection (12). The CD34+ population is heterogeneous and comprised of cells ranging from primitive progenitors to cells in the early stages of commitment to a lineage. It is likely that the low level of virus produced within the long-term bone marrow culture results from a productive infection or reactivation in more differentiated cells. The more differentiated cells may preexist in the CD34+ population or may arise in vitro. This hypothesis is consistent with the evidence that reactivation depends on the differentiation state of the cell, and that cytokines can facilitate reactivation of a latent infection (9, 16, 17, 33).
The transcriptional milieu of a cell changes dramatically with differentiation. The nonpermissive state for HCMV replication associated with immature CD34+ cell populations may be the result of chromatin modifications that silence transcription from the viral genome. Indeed, the transcription factor, YY1, has been shown to associate with the major immediate early promoter (MIEP) and has an associated histone deacetylase activity (34, 35). The MIEP has been shown to be deacetylated in the nonpermissive Ntera2 cell line, but becomes acetylated when these cells are differentiated into the permissive neuronal phenotype (34, 35). In our model for latency, GFP was expressed from the viral chromosome at early times after infection but was undetectable by 6 days postinfection. By contrast, GFP is expressed continuously during a lytic infection. Deacetylation could explain the silencing of GFP marker expression from the viral genome. Methylation of the viral genome might also play a role in modulating latency because differential methylation of the EBV genome appears to regulate the program of latency (36–38).
A substantial number of HCMV mRNAs are expressed during infection of CD34+ cells, and some were present after the longest period in culture that was tested (Fig. 4 and Table 1). Because our array experiment measures steady-state levels of viral mRNAs, it is not possible to know whether the viral mRNAs detected late in the time course were still actively transcribed, or whether they might be stable mRNAs whose genes were no longer being transcribed. Nevertheless, some of the viral gene products encoded by mRNAs expressed in our model system may be important to establishing or maintaining a latent infection.
It is possible that a subset of CD34+ cells might express only a portion of the viral genes that we have identified in the entire CD34+ population. Consistent with this hypothesis, distinct patterns of gene expression were observed in CD34-depleted populations compared with CD34+ populations, and we have not detected viral genes expressed in CD14+ monocytes (data not shown). Unique patterns of gene expression might define the nature and outcome of HCMV infection. Consequently, refined analysis of HCMV gene expression in well defined subsets of CD34+ cells will be essential to identifying viral genes that may contribute to latency. It also will be important to determine which of the genes expressed in our latent model are expressed during latency in a natural infection.
It has been argued that the reservoir for latent HCMV is not a primitive progenitor that gives rise to all hematopoietic lineages because HCMV DNA is not readily detected in all blood cells (39). However, we hypothesize that there must be a primitive reservoir to maintain HCMV for the lifetime of the host. The lack of HCMV in all lineages could be explained if infection blocked or preferentially promoted some but not all of the multiple downstream differentiation pathways. Uninfected stem cells could compensate for the infected progenitor's defect in healthy individuals. The ability of some HCMV strains to induce severe myelosuppression following bone marrow transplantation indicates that HCMV could block the differentiation pathways of some or all hematopoietic lineages (40–42). As precedent for this hypothesis, EBV infection activates resting B cells into a proliferative lymphoblastic state to promote differentiation into a memory B cell where EBV can reside latently for the lifetime of the host (43). Alternatively, HCMV genomes might be lost from some cell lineages during lineage commitment, or the virus could reside latently in a long-lived myeloid progenitor such that HCMV is found in cells of the myeloid and not the lymphoid lineage. Although HCMV DNA has been most commonly reported in monocytes (7, 9), other reports have found viral DNA in lymphoid cells (4, 44). Consequently, it is possible that latent HCMV resides in a primitive hematopoietic progenitor. The finding that HCMV DNA sequences are detected in CD34+ cells of seropositive individuals (6, 15) is consistent with the hypothesis that HCMV resides in a progenitor capable of giving rise to many blood cell lineages.
Acknowledgments
We thank I. Lemischka and K. Moore for the generous gift of AFT024 cells; S. Tavazoie for use of an array printer and advice in array data analysis; A. Beavis and B. Grimes for flow cytometry expertise; W. Bresnahan for construction of AD_sub_UL21.5; C. Marion and M. Trader for ensuring timely delivery of bone marrow; and C. Kulesza, A. Flood, and E. Murphy for critical reading of the manuscript. This work was supported by National Cancer Institute Grant CA87661. F.D.G. is a Leukemia and Lymphoma Society Fellow.
Abbreviations
- HCMV, human cytomegalovirus
- pfu, plaque-forming unit
References
- 1.Britt W. J. & Alford, C. A. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia), Vol. 2, pp. 2493–2524. [Google Scholar]
- 2.Sinzger C., Grefte, A., Plachter, B., Gouw, A. S., The, T. H. & Jahn, G. (1995) J. Gen. Virol. 76**,** 741-750. [DOI] [PubMed] [Google Scholar]
- 3.Soderberg C., Larsson, S., Bergstedt-Lindqvist, S. & Moller, E. (1993) J. Virol. 67**,** 3166-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrier R. D., Nelson, J. A. & Oldstone, M. B. (1985) Science 230**,** 1048-1051. [DOI] [PubMed] [Google Scholar]
- 5.Boeckh M., Hoy, C. & Torok-Storb, B. (1998) Clin. Infect. Dis. 26**,** 209-210. [DOI] [PubMed] [Google Scholar]
- 6.Mendelson M., Monard, S., Sissons, P. & Sinclair, J. (1996) J. Gen. Virol. 77**,** 3099-3102. [DOI] [PubMed] [Google Scholar]
- 7.Taylor-Wiedeman J., Sissons, J. G., Borysiewicz, L. K. & Sinclair, J. H. (1991) J. Gen. Virol. 72**,** 2059-2064. [DOI] [PubMed] [Google Scholar]
- 8.Hahn G., Jores, R. & Mocarski, E. S. (1998) Proc. Natl. Acad. Sci. USA 95**,** 3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soderberg-Naucler C., Fish, K. N. & Nelson, J. A. (1997) Cell 91**,** 119-126. [DOI] [PubMed] [Google Scholar]
- 10.Taylor-Wiedeman J., Sissons, P. & Sinclair, J. (1994) J. Virol. 68**,** 1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minton E. J., Tysoe, C., Sinclair, J. H. & Sissons, J. G. (1994) J. Virol. 68**,** 4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maciejewski J. P., Bruening, E. E., Donahue, R. E., Mocarski, E. S., Young, N. S. & St. Jeor, S. C. (1992) Blood 80**,** 170-178. [PubMed] [Google Scholar]
- 13.Movassagh M., Gozlan, J., Senechal, B., Baillou, C., Petit, J. C. & Lemoine, F. M. (1996) Blood 88**,** 1277-1283. [PubMed] [Google Scholar]
- 14.Sindre H., Tjoonnfjord, G. E., Rollag, H., Ranneberg-Nilsen, T., Veiby, O. P., Beck, S., Degre, M. & Hestdal, K. (1996) Blood 88**,** 4526-4533. [PubMed] [Google Scholar]
- 15.von Laer D., Meyer-Koenig, U., Serr, A., Finke, J., Kanz, L., Fauser, A. A., Neumann-Haefelin, D., Brugger, W. & Hufert, F. T. (1995) Blood 86**,** 4086-4090. [PubMed] [Google Scholar]
- 16.Soderberg-Naucler C., Streblow, D. N., Fish, K. N., Allan-Yorke, J., Smith, P. P. & Nelson, J. A. (2001) J. Virol. 75**,** 7543-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soderberg-Naucler C., Fish, K. N. & Nelson, J. A. (1997) J. Clin. Invest. 100**,** 3154-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo K., Kaneshima, H. & Mocarski, E. S. (1994) Proc. Natl. Acad. Sci. USA 91**,** 11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo K., Xu, J. & Mocarski, E. S. (1996) Proc. Natl. Acad. Sci. USA 93**,** 11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wineman J., Moore, K., Lemischka, I. & Muller-Sieburg, C. (1996) Blood 87**,** 4082-4090. [PubMed] [Google Scholar]
- 21.Moore K. A., Ema, H. & Lemischka, I. R. (1997) Blood 89**,** 4337-4347. [PubMed] [Google Scholar]
- 22.Lewis I. D., Almeida-Porada, G., Du, J., Lemischka, I. R., Moore, K. A., Zanjani, E. D. & Verfaillie, C. M. (2001) Blood 97**,** 3441-3449. [DOI] [PubMed] [Google Scholar]
- 23.Nolta J. A., Thiemann, F. T., Arakawa-Hoyt, J., Dao, M. A., Barsky, L. W., Moore, K. A., Lemischka, I. R. & Crooks, G. M. (2002) Leukemia 16**,** 352-361. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch A. J. & Shenk, T. (1999) J. Virol. 73**,** 404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stinski M. F. (1976) J. Virol. 19**,** 594-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tirabassi R. S. & Enquist, L. W. (1998) J. Virol. 72**,** 4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bresnahan W. A. & Shenk, T. (2000) Science 288**,** 2373-2376. [DOI] [PubMed] [Google Scholar]
- 28.Hegde P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J. E., Snesrud, E., Lee, N. & Quackenbush, J. (2000) BioTechniques 29**,** 548-556. [DOI] [PubMed] [Google Scholar]
- 29.Chee M. S., Bankier, A. T., Beck, S., Bohni, R., Brown, C. M., Cerny, R., Horsnell, T., Hutchison, C. A., III, Kouzarides, T., Martignetti, J. A., et al. (1990) Curr. Top. Microbiol. Immunol. 154**,** 125-169. [DOI] [PubMed] [Google Scholar]
- 30.Cha T. A., Tom, E., Kemble, G. W., Duke, G. M., Mocarski, E. S. & Spaete, R. R. (1996) J. Virol. 70**,** 78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H., Cong, J. P., Mamtora, G., Gingeras, T. & Shenk, T. (1998) Proc. Natl. Acad. Sci. USA 95**,** 14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browne E. P., Wing, B., Coleman, D. & Shenk, T. (2001) J. Virol. 75**,** 12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hummel M., Zhang, Z., Yan, S., DePlaen, I., Golia, P., Varghese, T., Thomas, G. & Abecassis, M. I. (2001) J. Virol. 75**,** 4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas M. J. & Seto, E. (1999) Gene 236**,** 197-208. [DOI] [PubMed] [Google Scholar]
- 35.Osborne A., Zhang, H., Yang, W. M., Seto, E. & Blanck, G. (2001) Mol. Cell. Biol. 21**,** 6495-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulson E., Fingeroth, J., Yates, J. & Speck, S. (2002) Virology 299**,** 109-121. [DOI] [PubMed] [Google Scholar]
- 37.Paulson E. J. & Speck, S. H. (1999) J. Virol. 73**,** 9959-9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babcock G. J., Hochberg, D. & Thorley-Lawson, A. D. (2000) Immunity 13**,** 497-506. [DOI] [PubMed] [Google Scholar]
- 39.Jarvis M. & Nelson, J. (2002) Curr. Opin. Microbiol. 5**,** 403-407. [DOI] [PubMed] [Google Scholar]
- 40.Simmons P., Kaushansky, K. & Torok-Storb, B. (1990) Proc. Natl. Acad. Sci. USA 87**,** 1386-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torok-Storb B., Boeckh, M., Hoy, C., Leisenring, W., Myerson, D. & Gooley, T. (1997) Blood 90**,** 2097-2102. [PubMed] [Google Scholar]
- 42.Torok-Storb B., Bolles, L., Iwata, M., Doney, K., Sale, G. E., Gooley, T. A. & Storb, R. (2001) Blood 98**,** 891-892. [DOI] [PubMed] [Google Scholar]
- 43.Thorley-Lawson D. A. & Babcock, G. J. (1999) Life Sci. 65**,** 1433-1453. [DOI] [PubMed] [Google Scholar]
- 44.von Laer D., Serr, A., Meyer-Konig, U., Kirste, G., Hufert, F. T. & Haller, O. (1995) J. Infect. Dis. 172**,** 365-370. [DOI] [PubMed] [Google Scholar]