RNA Polymerase II Elongation Factors of Saccharomyces cerevisiae: a Targeted Proteomics Approach (original) (raw)
Abstract
To physically characterize the web of interactions connecting the Saccharomyces cerevisiae proteins suspected to be RNA polymerase II (RNAPII) elongation factors, subunits of Spt4/Spt5 and Spt16/Pob3 (corresponding to human DSIF and FACT), Spt6, TFIIF (Tfg1, -2, and -3), TFIIS, Rtf1, and Elongator (Elp1, -2, -3, -4, -5, and -6) were affinity purified under conditions designed to minimize loss of associated polypeptides and then identified by mass spectrometry. Spt16/Pob3 was discovered to associate with three distinct complexes: histones; Chd1/casein kinase II (CKII); and Rtf1, Paf1, Ctr9, Cdc73, and a previously uncharacterized protein, Leo1. Rtf1 and Chd1 have previously been implicated in the control of elongation, and the sensitivity to 6-azauracil of strains lacking Paf1, Cdc73, or Leo1 suggested that these proteins are involved in elongation by RNAPII as well. Confirmation came from chromatin immunoprecipitation (ChIP) assays demonstrating that all components of this complex, including Leo1, cross-linked to the promoter, coding region, and 3′ end of the ADH1 gene. In contrast, the three subunits of TFIIF cross-linked only to the promoter-containing fragment of ADH1. Spt6 interacted with the uncharacterized, essential protein Iws1 (interacts with Spt6), and Spt5 interacted either with Spt4 or with a truncated form of Spt6. ChIP on Spt6 and the novel protein Iws1 resulted in the cross-linking of both proteins to all three regions of the ADH1 gene, suggesting that Iws1 is likely an Spt6-interacting elongation factor. Spt5, Spt6, and Iws1 are phosphorylated on consensus CKII sites in vivo, conceivably by the Chd1/CKII associated with Spt16/Pob3. All the elongation factors but Elongator copurified with RNAPII.
The synthesis of functional mRNA precursors by eukaryotic RNA polymerase II (RNAPII) is a multistage process consisting of three major steps: initiation, elongation, and termination. Although much has been learned about steps involved in initiation, including formation of the initiation complex, initiation of the RNA chain, and promoter clearance, much less is known about later stages in transcription, especially chain elongation. It is known, however, that the transition from transcriptional initiation to elongation is associated with a change in the factors that are associated with RNAPII.
Whereas general transcription factors are required for promoter-directed transcription initiation, and the multisubunit Srb/Mediator complex is associated with the RNAPII holoenzyme that binds to promoters in the yeast Saccharomyces cerevisiae, RNAPII can elongate the transcript unaided. However, several factors have been discovered that stimulate elongation on nonchromatin, “naked” DNA templates (e.g., TFIIS and TFIIF) (32, 47). Recently, other factors have been identified that are thought to affect RNAPII transcript elongation via effects on chromatin. These include the yeast factors Spt4 and Spt5 (corresponding to human 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole [DRB] sensitivity-inducing factor [DSIF], which is required for activation of elongation by human immunodeficiency virus Tat) (46), Elongator (30), Spt6 (15), and Spt16/Pob3 (corresponding to human FACT [facilitates chromatin transcription]) (28). Elongator copurified with the elongating form of RNAPII, and it was shown to bind preferentially to the hyperphosphorylated form of RNAPII in vitro (30). The Elp3 subunit of Elongator has histone acetyltransferase (HAT) activity in vitro (48), suggesting that the HAT activity of Elp3 may assist RNAPII during transcription elongation on a chromatin template. Human FACT stimulates transcriptional elongation on a chromatin template in a partially defined in vitro system and has been shown to interact with nucleosomes and histone H2A/H2B dimers (28, 29). Recently, FACT has also been identified in a complex containing human casein kinase II (CKII) that phosphorylates the tumor suppressor protein p53 (18).
Purification of proteins interacting with and affecting the activity of RNAPII has generally involved either conventional chromatography or RNAPII affinity chromatography, a procedure that identifies human TFIIF (RAP30/RAP74) and TFIIS (38). However, conventional or affinity purifications usually require either ion exchangers and hydrophobic columns (in the former case) or high salt concentrations (in the latter case) to reduce nonspecific protein binding to the column matrix. These conditions could potentially disrupt interactions that stabilize some protein complexes. To circumvent these problems, affinity tags that are fused to selected target proteins have been widely used to purify protein complexes. In most cases, however, it has again been necessary to use high salt concentrations to reduce nonspecific binding of proteins to solid supports.
Recently, a tandem affinity purification (TAP) scheme, in which two adjacent tags (calmodulin-binding peptide and Staphylococcus aureus protein A) separated by a tobacco etch virus (TEV) protease site were fused to the protein of interest, was described (33). The use of two tags and, consequently, two independent purification steps allowed protein purification to be carried out under native conditions at near-physiological ionic strengths while still maintaining a low background (defined as the protein yield from an extract derived from an untagged strain). Under these circumstances, protein complexes containing proteins that are weakly associated in vivo have a greater chance of remaining associated during the purification procedure.
Most of the RNAPII elongation factors of S. cerevisiae are poorly characterized. Moreover, conventional purifications of human proteins previously implicated in transcriptional elongation may have resulted in the loss of one or more associated proteins. In order to systematically characterize the yeast transcriptional elongation factors, we devised a modification of the TAP procedure (33) in which dilution of the yeast extract during the purification procedure was minimized and high salt concentrations were avoided. In most cases, purification using this procedure unveiled new protein-protein interactions and sometimes new polypeptides involved in transcriptional elongation in S. cerevisiae. Furthermore, the data revealed that the salt concentrations used during purification were critical in certain cases.
MATERIALS AND METHODS
Yeast strain construction.
All of the strains used in this study (Table 1) are congenic with W303-1A (42). Tags were fused to genes with a PCR-based one-step in vivo tagging strategy (3) using the TAP tag vector pBS1479 (33). Transformed cells (11) were plated onto synthetic medium lacking tryptophan. Integration of the tag-encoding DNA was confirmed by PCR and then Western blotting with nonspecific rabbit immunoglobulin to recognize the protein A component of the TAP tag. Strains with the following genes replaced by a kanamycin resistance cassette were obtained from Research Genetics (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html):CHD1, DST1, PAF1, CDC73, RTF1, and LEO1. Sensitivity to 6-azauracil was tested by plating strains harboring pRS316 (37) onto plates lacking uracil and containing 25 μg of 6-azauracil/ml.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1A | MATaura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 | 39 |
| YCK245 | As W303-1A but trp1 Δ | 23 |
| YNJK1 | As W303-1A, but trp1 Δ _elp1_-TAP::TRP1 | 23 |
| YNJK2 | As W303-1A, but trp1 Δ _elp3_-TAP::TRP1 | 23 |
| YNJK3 | As W303-1A, but trp1 Δ _spt4_-TAP::TRP1 | This study |
| YNJK4 | As W303-1A, but trp1 Δ _spt5_-TAP::TRP1 | This study |
| YNJK6 | As W303-1A, but trp1 Δ _spt6_-TAP::TRP1 | This study |
| YNJK7 | As W303-1A, but trp1 Δ _spt16_-TAP::TRP1 | This study |
| YNJK8 | As W303-1A, but trp1 Δ _pob3_-TAP::TRP1 | This study |
| YNJK9 | As W303-1A, but trp1 Δ _dst1_-TAP::TRP1 | This study |
| YNJK11 | As W303-1A, but trp1 Δ _iws1_-TAP::TRP1 | This study |
| YNJK12 | As W303-1A, but trp1 Δ _rpb3_-TAP::TRP1 | This study |
| YNJK15 | As W303-1A, but trp1 Δ _tfg1_-TAP::TRP1 | This study |
| YNJK16 | As W303-1A, but trp1 Δ _tfg2_-TAP::TRP1 | This study |
| YNJK17 | As W303-1A, but trp1 Δ _tfg3_-TAP::TRP1 | This study |
| YNJK23 | As W303-1A, but trp1 Δ _chd1_-TAP::TRP1 | This study |
| YNJK44 | As W303-1A, but trp1 Δ _elp2_-TAP::TRP1 | 23 |
| YNJK45 | As W303-1A, but trp1 Δ _elp4_-TAP::TRP1 | 23 |
| YNJK46 | As W303-1A, but trp1 Δ _elp5_-TAP::TRP1 | 23 |
| YNJK74 | As W303-1A, but trp1 Δ _rtf1_-TAP::TRP1 | This study |
| YNJK78 | As W303-1A, but trp1 Δ _leo1_-TAP::TRP1 | This study |
| YNJK81 | As W303-1A, but trp1 Δ _cdc73_-TAP::TRP1 | This study |
| YNJK91 | As W303-1A, but trp1 Δ _paf1_-TAP::TRP1 | This study |
| YNJK130 | As W303-1A, but trp1 Δ _ctr9_-TAP::TRP1 | This study |
Protein purification.
Tagged complexes implicated in transcriptional elongation were purified on immunoglobulin G (IgG) and calmodulin columns from extracts of yeast cells (3 liters) grown in yeast extract-peptone-dextrose (YEPD) medium to an optical density at 600 nm (OD600) of 1.0 to 1.5. Cell pellets (7 to 10 g) were frozen in liquid nitrogen and lysed by grinding with dry ice in a Krups coffee grinder (model 203-70). An equal volume of YEB (250 mM KCl, 100 mM HEPES-KOH [pH 7.9], 1 mM EDTA, 2.5 mM dithiothreitol [DTT]) was added, and following centrifugation in a Beckman 70 Ti rotor at 4°C for 2 h at 34,000 rpm, the supernatant was dialyzed against IPP buffer (10 mM Tris-Cl [pH 7.9], 0.1% Triton X-100, 0.5 mM DTT, 0.2 mM EDTA, 20% glycerol containing 100, 125, 150, or 200 mM NaCl). After dialysis, the extract was again centrifuged in a 70 Ti rotor at 4°C for 30 min at 34,000 rpm, and the supernatant was mixed for 3 h with 200 μl of IgG-Sepharose (Pharmacia) equilibrated with IPP buffer. Following binding, the IgG-Sepharose was washed with 1 ml of IPP buffer followed by 400 μl of TEV protease cleavage buffer (50 mM Tris-Cl [pH 7.9], 1 mM DTT, and 0.1% Triton X-100, containing 200, 150, 125, or 100 mM NaCl). The beads were then incubated overnight at 4°C with 100 U of TEV protease (Life Technologies) in 200 μl of TEV cleavage buffer. The eluate was combined with a 200-μl wash with TEV cleavage buffer. To this was added 200 μl of calmodulin binding buffer (10 mM Tris-Cl [pH 7.9], 10 mM β-mercaptoethanol, 2 mM CaCl2, and 0.1% Triton X-100 containing 200, 150, 125, or 100 mM NaCl) and 200 μl of calmodulin beads (Pharmacia) equilibrated with the same buffer. After binding for 1 to 2 h at 4°C, the calmodulin beads were washed with 200 μl of calmodulin binding buffer and 200 μl of calmodulin wash buffer (10 mM Tris-Cl [pH 7.9], 10 mM β-mercaptoethanol, 0.1 mM CaCl2, and 0.1% Triton X-100, containing 200, 150, 125, or 100 mM NaCl). The purified protein complexes were eluted from the calmodulin beads with 5 times with 100 μl of calmodulin elution buffer (10 mM Tris-Cl [pH 7.9], 10 mM β-mercaptoethanol, 3 mM EGTA [pH 8.0], and 0.1% Triton X-100, containing 200, 150, 125, or 100 mM NaCl), and the eluted protein was lyophilized or concentrated by precipitation with acetone or trichloroacetic acid (TCA). Purified proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gels containing 10% polyacrylamide (except where indicated otherwise), and the proteins were visualized by silver staining. Western blotting was performed using standard techniques with monoclonal antibody 8WG16 (43).
Protein identification.
Protein bands were reduced, alkylated, and subjected to in-gel tryptic digestion, and the peptides were then purified and identified by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry with a PerSeptive DE STR instrument (26). For tandem mass spectrometry, the TCA-precipitated protein was resuspended in 100 mM NH4HCO3/1 mM CaCl2 buffer, pH 8.5, and digested overnight at 37°C with 2 μl of immobilized Poros trypsin beads (PerSeptive). The entire digest was fractionated as described previously (10) on a 7.5-cm (inner diameter, 100 μm) reverse-phase C18 capillary column attached in-line to a Finnigan LCQ-Deca ion trap mass spectrometer by ramping a linear gradient from 2 to 60% solvent B in 90 min. Solvent A consisted of 5% acetonitrile, 0.5% acetic acid, and 0.02% heptafluorobutanoic acid (HFBA), and solvent B consisted of acetonitrile-water (80:20) containing 0.5% acetic acid and 0.02% HFBA. The flow rate at the tip of the needle was set to 300 nl/min by programming the high-performance liquid chromatography pump and using a split line. The mass spectrometer cycled through four scans as the gradient progressed. The first was a full mass scan followed by successive tandem mass scans of the three most intense ions. A dynamic exclusion list was used to limit collection of tandem mass spectra for peptides that eluted over a long period of time. All tandem mass spectra were searched by using the SEQUEST computer algorithm against the complete yeast protein database (6/2,000). Each high-scoring peptide sequence was manually compared with the corresponding tandem mass spectrum to ensure that the match was correct.
ChIP.
Chromatin immunoprecipitation (ChIP) and PCRs were performed as described previously (22). All yeast strains were grown at 30°C to an OD595 of 0.4 to 0.6. Sequences for the ADH1 3′ untranslated region primer pair are as follows: ADH11231, ACCGGCATGCCGAGCAAATGCCTG; ADH11400, CCCAACTGAAGGCTAGGCTGTGG.
RESULTS
Purification and identification of the yeast elongation factors.
We used single-step transformation with PCR products and a TRP1 selective marker to place TAP tags containing a calmodulin-binding peptide and S. aureus protein A, separated by a TEV protease cleavage site, at the C termini of TFIIS, the three subunits of TFIIF, all six known subunits of Elongator (23), Spt4, Spt5, Spt6, Spt16, and Pob3. To confirm the success of each tagging procedure, Western blotting was performed on extracts derived from the tagged strains, making use of an irrelevant immunoglobulin to recognize the protein A component of the TAP tag. Each tagged protein, together with any associated proteins, was then purified on IgG and calmodulin columns and analyzed by SDS-PAGE followed by staining with silver. Control purifications were always carried out in parallel by using extracts derived from an otherwise congenic strain that contained no tagged protein. The protein bands absent from the control preparation and corresponding to the tagged protein and any associated proteins were then identified by MALDI-TOF and ion trap mass spectrometry. Proteins that copurified with the tagged proteins were then themselves tagged, purified, and analyzed so as to confirm their presence in the complex and create a picture of the interaction relationships among all the putative elongation factors of S. cerevisiae.
Spt5 is a phosphoprotein that associates with Spt4 or Spt6.
Human DSIF and NELF (negative elongation factor) have previously been identified and purified as inhibitory elongation factors that make transcription sensitive to DRB (46, 49). DSIF consists of 160- and 14-kDa subunits and can also stimulate transcriptional elongation under certain conditions. The yeast homologues of the DSIF subunits, Spt4 and Spt5, also form a complex, and mutations in the genes encoding these proteins result in sensitivity to 6-azauracil (15), often considered to be an important criterion for mutations in yeast genes that encode elongation factors (2, 9). More recently, the Drosophila homologue of Spt5 was shown to colocalize with stalled, hypophosphorylated RNAPII on Drosophila heat shock genes in the absence of induction and with hyperphosphorylated, actively elongating RNAPII during salivary gland development, also implying that Spt5 is an elongation factor (1, 17).
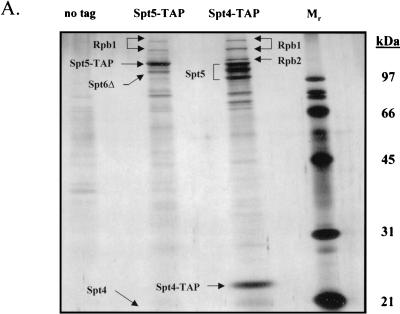
Purification of tagged Spt4 from yeast extracts in the presence of 100 mM NaCl resulted in copurification of three forms of Spt5 as well as a substoichiometric amount of RNAPII (Fig. 1A). The three electrophoretically distinct forms of Spt5 were consistently observed, but they are probably caused by C-terminal degradation, because the tagging and purification of Spt5 resulted in the isolation of only one form of tagged Spt5 (Fig. 1A). Copurifying with the tagged Spt5 were Spt4, a substoichiometric amount of RNAPII, and, surprisingly, a truncated form of Spt6. The small untagged Spt4 polypeptide ran off the gel shown in Fig. 1A, but the presence of Spt4 in the sample applied to the gel was confirmed by tandem mass spectrometry. These results implied that Spt5 can associate either with Spt4 or with a modified form of Spt6. Analysis of the complexes containing tagged Spt4 or Spt5 by use of tandem mass spectrometry served to identify a phosphorylation site in Spt5 on serine 188 in the sequence SDDE, which is the start of a consensus CKII phosphorylation site [(S/T)-(D/E)-X-(D/E)] (see Fig. 2B) (12).
FIG. 1.
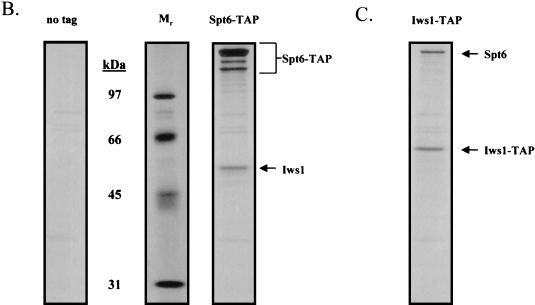
Isolation of protein complexes containing Spt4, Spt5, and Spt6. (A) TAPs of the Spt4/Spt5 complex were carried out on strains containing either no tagged protein or TAP-tagged versions of Spt4 or Spt5. Protein complexes were purified in the presence of 100 mM NaCl and were then analyzed by SDS-PAGE and silver staining. Spt4, Spt5, subunits of RNAPII, and a truncated form of Spt6 were identified by trypsin digestion and MALDI-TOF mass spectrometry. In the purification of tagged Spt5, the presence of Spt4 was identified by subjecting an aliquot of the eluate from the final column directly to trypsin digestion and tandem mass spectrometry. (B) TAP of Spt6. Purification using an extract containing a TAP-tagged version of Spt6 in 100 mM NaCl resulted in isolation of three forms of the tagged protein and a previously uncharacterized protein encoded by ORF YPR133c, whose gene product we have called Iws1 (interacts with Spt6). (C) TAP of Iws1. Tagging and isolation of Iws1 in 100 mM salt resulted in copurification of a stoichiometric amount of one form of Spt6. Because both Spt6-TAP and Iws1-TAP were run on the same gel, it could be concluded that Iws1 copurified with only the slowest-migrating form of Spt6.
FIG. 2.
Detection of phosphorylation on putative CKII sites in Spt5, Spt6, and Iws1. (A) Identification of phosphorylation sites in Spt6. Tandem mass spectrometry was used to identify three tryptic fragments in the N-terminal region of Spt6 that were phosphorylated on serine residues (serines 95 [SEDD], 135 [SESD], and 207 [SDED]). Each phosphorylated serine residue in Spt6 was at the beginning of a consensus CKII site. (B) Phosphorylation of a conserved serine present in both Spt5 and Iws1. Mass spectrometry successfully detected a phosphorylation site on serine 89 (SDDD) of Iws1 as well as on serine 188 (SDDE) of Spt5; each of these occurs at the start of a putative CKII site. The acidic N-terminal regions of Spt5 and Iws1 have sequence similarity, and their phosphorylated serine residues align perfectly. The regions compared have 23% identity and 40% similarity.
Spt6 is a phosphoprotein that interacts with the uncharacterized phosphoprotein Iws1.
SPT6 was originally identified along with SPT4 and SPT5 in a screen for suppressors of _cis_- and _trans_-acting mutations that affected promoter function (15, 41). More recently, Drosophila Spt6 was shown to colocalize with hyperphosphorylated, actively elongating RNAPII on polytene chromosomes (1, 17).
Tagging and purification of yeast Spt6 resulted in copurification of substoichiometric amounts of a previously uncharacterized protein, encoded by the essential open reading frame (ORF) YPR133c, which we have called Iws1 (interacts with Spt6) (Fig. 1B). Conversely, tagging and purification of Iws1 resulted in the isolation of Spt6, further confirming that this interaction occurs in vivo (Fig. 1C). There were three electrophoretically distinct forms of tagged Spt6, but only the form with the lowest electrophoretic mobility copurified with tagged Iws1. It is not clear whether the higher-mobility forms of tagged Spt6 are a consequence of posttranslational modification or in vitro or in vivo degradation.
Tandem mass spectrometry was used to identify three tryptic fragments in the N-terminal region of Spt6 that were phosphorylated on serine residues (serines 95 [SEDD], 135 [SESD], and 207 [SDED]) (Fig. 2A). As was the case for Spt5, each phosphorylated serine residue in Spt6 was at the beginning of a consensus CKII site. Many more putative CKII sites exist in the N-terminal region of Spt6, but tandem mass spectrometry did not detect phosphorylation at these sites. We did detect a phosphorylation site on serine 89 (SDDD) of Iws1, which is also a putative CKII site. Interestingly, the acidic N-terminal regions of Spt5 and Iws1 have sequence similarity (Fig. 2B), and surprisingly, their phosphorylated serine residues align perfectly, perhaps explaining why Spt6 can interact with both of them.
Spt16/Pob3 associates with multiple protein complexes.
Human FACT was purified as a factor that overcame a nucleosomal block to elongation by RNAPII in vitro and is composed of human homologues of yeast Spt16/Cdc68 and Pob3 (28, 29). FACT interacts with nucleosomes and histone H2A-H2B dimers and might, therefore, promote nucleosome disruption during transcriptional elongation. Moreover, the yeast SPT16 gene interacts genetically with the genes encoding the yeast elongation factors TFIIS and Spt4 (41).
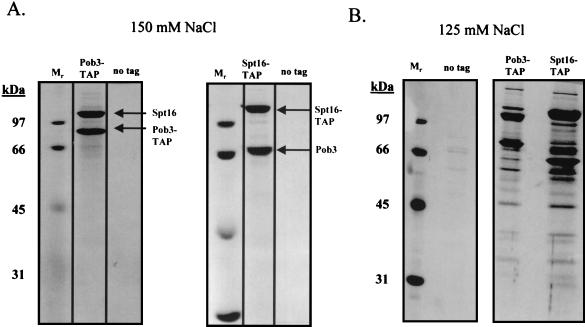
When the yeast counterparts of FACT, Spt16 and Pob3, were tagged and subsequently purified from yeast extracts in buffers containing 150 mM NaCl, large and approximately stoichiometric amounts of Spt16 and Pob3 were isolated and no other proteins were detected by silver staining (Fig. 3A). However, when the NaCl concentration used throughout the purification was reduced to 125 mM, many other proteins copurified with Spt16 and Pob3 in substoichiometric amounts (Fig. 3B). When the NaCl concentration was further reduced to 100 mM (Fig. 3C and D), the yields of the proteins associated with Spt16 and Pob3 increased substantially. In each case, none of the Spt16/Pob3-associated proteins were detected in parallel purifications carried out with extracts derived from an untagged strain (Fig. 3A, B, and C).
FIG. 3.
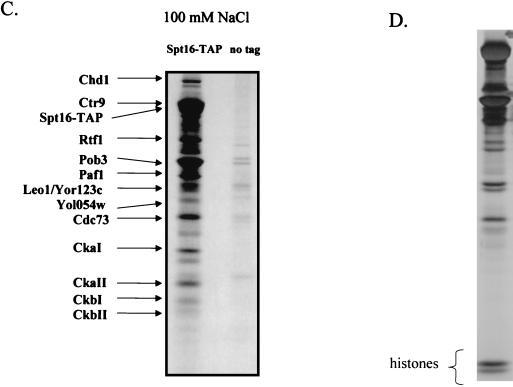
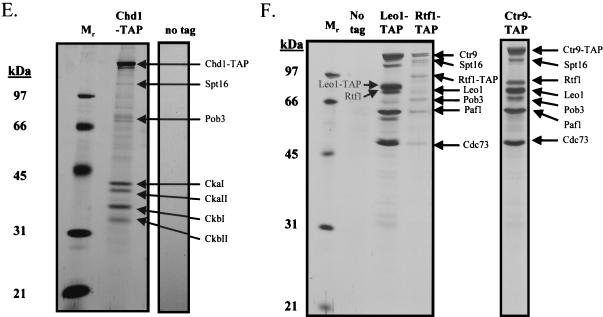
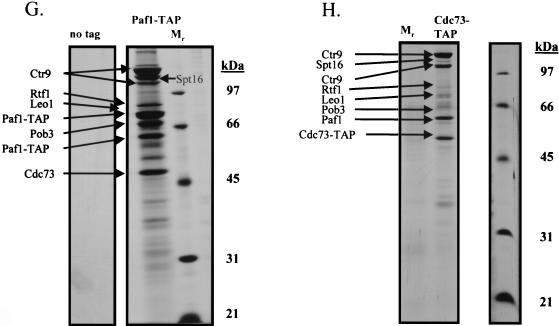
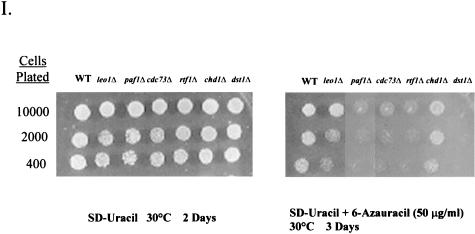
Spt16/Pob3 associates with several protein complexes. (A) TAPs of Spt16 and Pob3 from TAP-tagged strains in buffers containing 150 mM NaCl. Purified preparations were analyzed by SDS-PAGE and silver staining. Spt16 and Pob3 were identified by mass spectrometry. (B) Purification of Spt16/Pob3 complexes in buffers containing 125 mM NaCl. Analysis by SDS-PAGE and silver staining revealed that a number of other polypeptides copurified with tagged Spt16 and Pob3. (C and D) Tagged Spt16 was purified in buffers containing 100 mM NaCl, resulting in an increased yield of proteins associated with Spt16/Pob3. The purified preparation was electrophoresed on gels containing 10% (C) or 15% (D) polyacrylamide. The additional proteins identified by mass spectrometry were Chd1, Ctr9, Paf1, Cdc73, Rtf1, Leo1, Yol054w, all four subunits of CKII, and histones. (E) TAP of tagged Chd1 in the presence of 100 mM salt. The purified protein was analyzed by SDS-PAGE, silver staining, and mass spectrometry. Tagged Chd1 copurified with seemingly stoichiometric amounts of the four subunits of CKII (CkaI, CkaII, CkbI, and CkbII) as well as substoichiometric amounts of Spt16 and Pob3. (F through H) TAPs of Leo1, Rtf1, and Ctr9 (F), Paf1 (G), and Cdc73 (H) (all in the presence of 100 mM NaCl) resulted in isolation of complexes containing Rtf1, Leo1, Ctr9, Paf1, and Cdc73, as well as Spt16 and Pob3, but no histones. (I) Sensitivities of deletion strains to 6-azauracil. Strains containing the rtf1Δ, paf1Δ, cdc73Δ, leo1Δ, chd1Δ, or dst1Δ deletion, as well as plasmid pRS316, were plated on SD-uracil medium with or without the drug 6-azauracil (50 μg/ml) and were grown at 30°C for 2 to 4 days. WT, wild type.
Tandem and MALDI-TOF mass spectrometry were used to identify the following proteins that copurified with Spt16 and Pob3: Chd1, Ctr9, Paf1, Cdc73, Rtf1, CkaI, CkaII, CkbI, CkbII, histones, and proteins encoded by two uncharacterized ORFs, YOL054w and YOR123c (also known as LEO1) (25). Interestingly, Paf1 and Cdc73, as well as Ctr9, have previously been identified as components of a complex that binds to RNAPII (21, 36). Chd1 is a member of the _c_hromodomain-helicase-DNA-binding family and has been shown to be involved in ATP-dependent nucleosome remodeling (44). Recently, another component of the complex, Rtf1, was implicated genetically in transcriptional elongation (8).
The plethora of proteins we identified in association with Spt16 and Pob3 raised the possibility that several distinct protein complexes were associated with Spt16 and Pob3. To resolve these complexes, we tagged and purified from yeast extracts the various proteins that were associated with Spt16 and Pob3. Tagging of Chd1 resulted in copurification of approximately stoichiometric amounts of the four subunits of CKII (CkaI, CkaII, CkbI, and CkbII), as well as substoichiometric amounts of Spt16 and Pob3 (Fig. 3E), but none of the other polypeptides that had copurified with Spt16 and Pob3. Tagging and purification of Ctr9 resulted in the coisolation of Rtf1, Leo1, Paf1, and Cdc73, as well as Spt16 and Pob3, but neither the histones nor CKII and Chd1 (Fig. 3F). However, tagging and purification of histone H3 did result in the copurification of Spt16 and Pob3 (data not shown). These results indicated that there are at least three separable complexes that associate with Spt16 and Pob3: histones and nucleosomes, Chd1/CKII, and the complex containing Ctr9, Paf1, Cdc73, Rtf1, and Leo1. Confirmation came from experiments in which we labeled and purified Leo1 and Rtf1 (Fig. 3F), Paf1 (Fig. 3G), and Cdc73 (Fig. 3H). In each case, we observed all of the untagged components of this subcomplex, as well as amounts of Spt16 and Pob3 that varied from near-stoichiometric to clearly substoichiometric. In certain cases, there were two distinct forms of Paf1, Cdc73, or Ctr9, although we have not yet determined whether these are consequences of degradation, processing, or posttranslational modification. Although Yol054w had copurified with tagged Spt16 and Pob3, we did not observe copurification of Yol054w with tagged Paf1, Ctr9, Cdc73, Rtf1, or Leo1, suggesting that Yol054w may be directly associated with Spt16 or Pob3.
To determine if the polypeptides copurifying with Spt16/Pob3 might be involved in transcriptional elongation in vivo, strains harboring knockouts of genes encoding some of the nonessential subunits of the Spt16/Pob3 complex were tested for sensitivity to 6-azauracil. The drug 6-azauracil results in reductions in UTP and GTP concentrations in yeast cells, leads to induction of the PUR5 gene (35), and has been shown to affect the elongation efficiency of RNAPII (9). Therefore, sensitivity to this compound has been used as an indicator for involvement in transcriptional elongation. Studies using 6-azauracil have helped to show that genes like DST1, which encodes TFIIS, encode factors that have a role in transcriptional elongation (2). When dst1Δ was used as a control, deletions in rtf1, paf1, cdc73, and leo1, but not chd1, resulted in significant or marked sensitivity to 50 μg of 6-azauracil/ml (Fig. 3I).
Purification of TFIIF and TFIIS.
TFIIF is a general transcription factor that interacts with RNAPII and can recruit RNAPII to a preinitiation complex containing TFIID and TFIIB (14). TFIIF also participates in promoter clearance and can associate with RNAPII in vitro during chain elongation, enhancing its polymerization rate and processivity (7, 32). It is thought that TFIIF acts during the early stages of elongation and prevents abortive transcription, perhaps by increasing the rate of nucleotide addition.
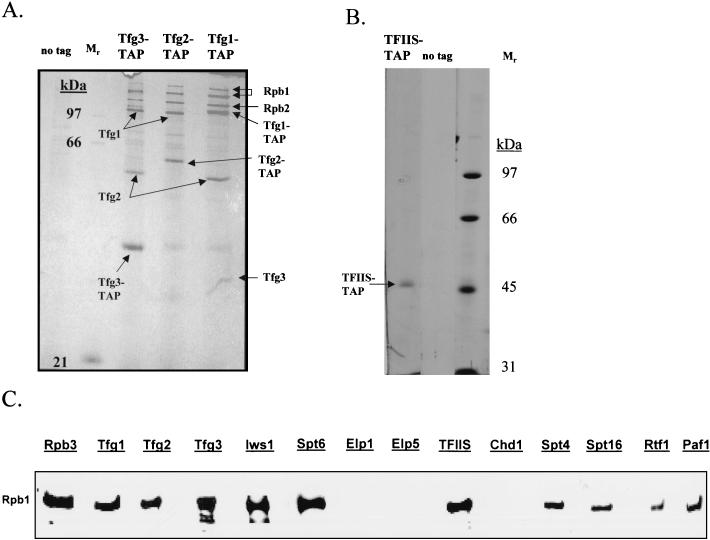
In yeast, TFIIF is comprised of the three subunits Tfg1, Tfg2, and Tfg3. When each subunit was tagged and purified from yeast extracts in buffers containing 150 mM NaCl, a complex containing all three subunits of TFIIF was isolated along with a nearly stoichiometric amount of RNAPII (Fig. 4A).
FIG. 4.
TAPs of TFIIF and TFIIS and detection of RNAPII interactions by Western blot analysis. (A) Purification of TFIIF by using TAP-tagged versions of Tfg1, Tfg2, and Tfg3. Purified proteins were analyzed by SDS-PAGE, silver staining, and mass spectrometry. Purification of any of the three subunits of TFIIF in buffers containing 150 mM NaCl results in isolation of all three components of TFIIF as well as a substoichiometric amount of RNAPII. (B) Tagging and purification of TFIIS. Purification of TFIIS (encoded by the DST1 gene) in buffers containing 100 mM NaCl resulted in the isolation of predominantly TFIIS. Further development of the silver stain revealed only background proteins and none that had copurified with TFIIS. (C) Western blot analysis of transcription elongation factors purified in the presence of 100 mM NaCl by using monoclonal antibody 8WG16, which binds to the CTD of the largest subunit of RNAPII, Rpb1. As a positive control, RNAPII was purified from a strain with a TAP tag on its third-largest subunit, Rpb3. Equal volumes of the column eluates from the various purifications were loaded onto the gel, except that approximately 10 times less of the Rpb3 purification was loaded. RNAPII, in the presence of 100 mM NaCl, copurified with TFIIF (Tfg1, -2, and -3), Spt4/Spt5, Spt6/Iws1, TFIIS, Spt16, and Paf1/Rtf1, but not with Elongator (Elp1 and Elp5) or Chd1.
At certain arrest sites encoded in the template DNA, RNAPII slides backwards, resulting in loss of contact between the 3′ end of the nascent transcript and the catalytic center of the polymerase (34, 45, 47). The transcriptional elongation factor, TFIIS, facilitates escape of arrested RNAPII from such a pause site by promoting cleavage of the nascent transcript by the catalytic center of RNAPII, followed by reextension of the RNA from the newly created 3′ end. When TFIIS was tagged and purified from yeast extracts, a predominantly free form of the protein was isolated (Fig. 4B), even though TFIIS is known to interact directly with RNAPII (38).
Isolation of a holo-Elongator complex.
Because the three-subunit Elongator complex was originally found associated with the elongating form of RNAPII and was shown to preferentially bind to the hyperphosphorylated form of RNAPII in vitro (30), it was hypothesized that Elongator replaces the mediator during the switch from transcription initiation to elongation. By using extracts from individual strains containing tagged versions of the three known Elongator subunits, Elp1, Elp2, and Elp3, a six-subunit yeast holo-Elongator complex containing three additional polypeptides, Elp4, Elp5, and Elp6, was subsequently isolated (23). When purification was carried out at higher salt concentrations, Elongator could be separated into two subcomplexes, one consisting of Elp1, -2, and -3 and the other consisting of Elp4, -5, and -6 (23). Interestingly, no RNAPII detectable by silver staining was present in our Elongator preparations, even when microgram quantities of Elongator were loaded onto the gel (23) (data not shown).
Copurification of the yeast elongation factors with RNAPII.
Human TFIIS, TFIIF, and Spt5 are known to bind directly to RNAPII (32, 38, 46). As well, Ctr9, Paf1, and Cdc73 were identified as components of a yeast protein complex that binds to RNAPII (21, 36), and Elongator copurified with chromatin-bound elongating RNAPII (30). Enough RNAPII copurified with tagged TFIIF, Spt4, and Spt5 for us to detect its two largest subunits, Rpb1 and Rpb2, by silver staining and identify them by mass spectrometry (Fig. 1A and 4A). We used the more sensitive Western blot technique with a monoclonal antibody directed against Rpb1 to determine which yeast elongation factors were associated more weakly with RNAPII (Fig. 4C). Purification of RNAPII itself from a strain containing a TAP tag on Rpb3, the third-largest subunit of RNAPII, showed that the soluble yeast extracts used for our purifications indeed contained RNAPII recognized by monoclonal antibody 8WG16 (Fig. 4C). All of the tagged elongation factor subunits copurified with at least some RNAPII except Chd1 and Elongator. Chd1 may not copurify with RNAPII simply because its association with RNAPII is too indirect: it is weakly associated with Spt16/Pob3, which in turn is weakly associated with Paf1/Ctr9/Cdc73/Rtf1/Leo1, which in turn is weakly associated with RNAPII (see Fig. 6). Remarkably, no detectable RNAPII copurified with Elongator when either Elp1 or Elp5 was tagged and purified, even when as much as 1 μg of purified Elongator was loaded onto the gel (Fig. 4C), and similar results were obtained with tagged Elp2 and Elp3 (data not shown). In other experiments (data not shown), we used antibodies specific for the phosphorylated C-terminal domain (CTD) on Rpb1. Although purification of tagged Rpb3 revealed that our extracts contained some phosphorylated Rpb1, phosphorylated Rpb1, like the mostly unphosphorylated Rpb1 recognized by the antibody (8WG16) used in Fig. 4C, did not copurify with Elongator tagged on Elp1 or Elp5.
FIG. 6.
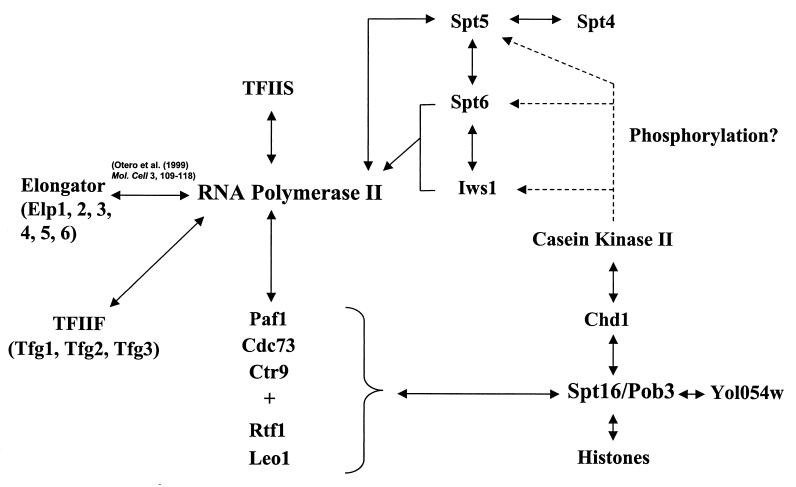
Comprehensive interaction map for the RNAPII elongation factors. For details, see the text.
ChIP assays.
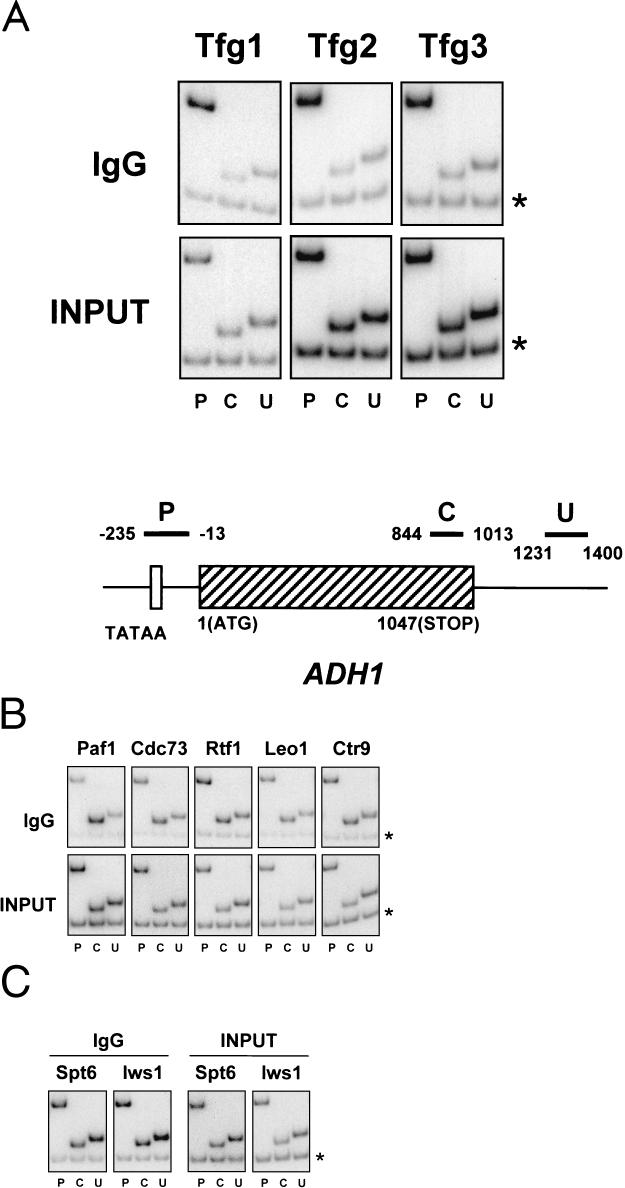
Analysis of the in vivo distribution of the novel elongation factors described here along a transcribed gene was carried out by ChIP assays in which the proteins were cross-linked to DNA in vivo by use of formaldehyde (22). Yeast cells were then lysed, and following isolation and shearing of chromatin, the presence of the proteins near specific DNA sequences was monitored by immunoprecipitation with rabbit immunoglobulin G-agarose. Finally, reversal of the cross-links was carried out, followed by PCR analyses of the coprecipitated DNA. The gene investigated was ADH1, encoding alcohol dehydrogenase, and three sets of primer pairs were used to analyze the gene: one directed against the promoter region, one directed further downstream within the coding region, and finally, one used to amplify a region corresponding to DNA in the 3′ untranslated region.
All three subunits of TFIIF, namely, Tfg1, Tfg2, and Tfg3, cross-linked to the promoter region of ADH1 but not to the coding region or 3′ untranslated region (Fig. 5A). These results suggested that, whereas TFIIF is a stable component of the initiation complex and may play a role in early elongation, it does not remain stably associated with elongating RNAPII in vivo. In contrast, all components of the Paf1 complex were found to cross-link specifically to all three regions of the ADH1 gene that were tested, including the novel, uncharacterized protein Leo1 (Fig. 5B); that is, all three fragments of the ADH1 gene were enriched by immunoprecipitation relative to a DNA fragment from a nontranscribed region, no matter which subunit of the Paf1/Ctr9/Cdc73/Rtf1/Leo1 complex was tagged. Similarly, the ChIP patterns of Spt6 and Iws1 were almost identical, and both displayed the same degree of cross-linking to all three regions of the ADH1 gene (Fig. 5C). Interestingly, ChIP assays demonstrated that Elongator subunits did not cross-link to any part of the ADH1 gene or to a number of other genes that were tested (our unpublished data).
FIG. 5.
ChIP assays with TFIIF, Paf1/Leo1/Ctr9/Cdc73/Rtf1, and Spt6/Iws1. ChIP was performed with strains containing TAP-tagged versions of the TFIIF subunits Tfg1, Tfg2, and Tfg3 (A), Paf1, Cdc73, Rtf1, Leo1, and Ctr9 (B), or Spt6 and Iws1 (C). To monitor the presence of each protein along the ADH1 gene, chromatin was immunoprecipitated with rabbit IgG-agarose and PCR amplified with primer pairs recognizing promoter (P), coding (C), and 3′ untranslated (U) regions (see schematic in panel A). Each PCR contained a second primer pair that amplified a region of chromosome V devoid of ORFs (marked by asterisks), thus providing an internal control for background. Input, signal from chromatin before immunoprecipitation. Primer pairs used are as follows: for P, ADH1-235 and ADH1-13; for C, ADH1844 and ADH11013; for U, ADH11231 and ADH11400; and for the nontranscribed region, Intergenic V-1 and Intergenic V-2.
DISCUSSION
At the outset of this study, a number of genes were known or suspected to encode elongation factors for RNAPII in S. cerevisiae. TFIIS had been shown to stimulate elongation by yeast RNAPII in vitro (34, 45, 47). TFIIF from human and Drosophila cells had been shown to stimulate the rate of elongation by their respective RNAPII enzymes in vitro (4, 32), and we have found that the same is true for purified yeast TFIIF (our unpublished data). Nevertheless, we found that TFIIF is stably associated only with the promoter region and not with the downstream transcribed region of the ADH1 gene in vivo, and similar observations were reported recently by Pokholok et al. (31). Therefore, it is not clear that TFIIF behaves as an elongation factor in vivo in spite of its in vitro activity. Spt4 and Spt5 are yeast homologues of the two subunits of DSIF, a human factor with both positive and negative effects on chain elongation by human RNAPII in vitro (46), and purified yeast Spt4/Spt5 also stimulates elongation by yeast RNAPII in vitro (our unpublished data). The genetic properties of the yeast SPT6 gene are similar to those of SPT4 and SPT5 (15, 41). Moreover, the Drosophila homologues of Spt5 and Spt6 appear to be associated with Drosophila elongation complexes on polytene chromosomes (1, 17). Spt16 and Pob3 are yeast homologues of the two subunits of human FACT, a factor that stimulates elongation by human RNAPII on a chromatin template in a partially reconstituted system in vitro (28, 29). Genetic studies on the yeast RTF1 gene have implicated Rtf1 as a possible RNAPII elongation factor in vivo (8). Finally, biochemical copurification experiments indicated that Elongator is associated with elongating RNA polymerase II in vivo (30).
Collectively, these previously published studies implicated 13 polypeptides as having possible roles in chain elongation by yeast RNAPII. However, only TFIIS, TFIIF, and Elongator had ever been purified; moreover, none of the yeast elongation factors had been purified under conditions designed to minimize subunit loss during the purification procedure, raising the possibility that there were intersubunit associations and new elongation factor subunits that had yet to be discovered. Based on the experiments we have described here and other experiments (N. J. Krogan and J. F. Greenblatt, unpublished data), we believe that complexes containing proteins that are weakly associated in vivo have a greater chance of remaining associated during purification when dilution is minimized and purification is carried out using near-physiological ionic strengths. The web of interactions among the RNAPII elongation factors that we uncovered by these procedures is illustrated in Fig. 6.
Most importantly, these procedures enabled us to discover that Spt16/Pob3 associates with 14 other polypeptides, namely, Rtf1 and the core histones, as well as Chd1, the four subunits of yeast CKII, a three-protein complex (Paf1/Ctr9/Cdc73) known to associate with RNAPII, and one or two proteins of unknown function, Leo1 and possibly Yol054w. A relatively small change in the NaCl concentration used for the purification, from 150 to 125 or 100 mM, enabled us to identify all these Spt16/Pob3-associated proteins rather than just Spt16 and Pob3. Tagging each of the other proteins associated with Spt16/Pob3 revealed that the Spt16/Pob3 complex associates with three distinct protein complexes: the core histones, Chd1/CKII, and Rtf1/Paf1/Ctr9/Cdc73/Leo1 (see Fig. 6). It is not clear whether these three protein complexes associate simultaneously with Spt16/Pob3. Simultaneous association would imply that Spt16/Pob3 is a scaffold for the coassembly of all these factors.
The interaction of Spt16/Pob3 with Chd1 and the core histones is in accord with similar findings for human FACT (19, 28, 29). Kelley et al. (19) showed that the human homologues of Pob3 and Chd1 interact physically in vitro and in vivo. As well, a chd1Δ mutation suppressed the growth defect conferred by a particular pob3 point mutation (8) and may also result in resistance to 6-azauracil in some strain backgrounds (although not particularly in the one we have used here), implicating Chd1 in transcriptional elongation (44). FACT was also observed to copurify with human CKII in a complex that phosphorylated p53 (18), but it is not clear whether human CHD1 was also part of this complex. The results we obtained by tagging and purifying Chd1 imply that yeast Chd1 is directly associated with CKII (see Fig. 6). One possibility is that Chd1 controls elongation by controlling the phosphorylation of an elongation factor by CKII. Definitive proof that Chd1 is an elongation factor awaits experiments showing that it can directly affect elongation by RNAPII in vivo or in vitro.
It was recently shown that Paf1 (RNAPII-associated factor), Cdc73, and Ctr9 form a complex that associates with RNAPII (21, 36). However, the roles of these proteins in transcription by RNAPII were unclear. The observation that combining a PAF1 deletion with deletion of the mediator subunit SRB5 is lethal was used to argue that the Paf1 complex is involved in initiation (36). However, we found that Paf1, Cdc73, and Ctr9 are associated with Spt16/Pob3 in a complex that also contains Rtf1 and Leo1, and so it is likely that this complex mediates the interaction between Spt16/Pob3 and RNAPII. Rtf1 (restores TBP function) was originally shown to affect the function of the TATA-binding protein (TBP) (40). However, recent work has suggested that it is likely to play a role in elongation by RNAPII, since a strain with a deletion of RTF1 is sensitive to 6-azauracil, a phenotype associated with defects in transcriptional elongation (8). Also, a deletion of RTF1 has been shown to genetically interact with point mutations in the essential genes SPT16 and POB3. In view of the genetically defined role of Rtf1 in elongation and the association of Paf1/Ctr9/Cdc73 with RNAPII, it is very likely that the Rtf1/Paf1/Ctr9/Cdc73/Leo1 complex has an important role in the behavior of Spt16/Pob3 as an elongation factor. Consistent with this, we found that paf1Δ, leo1Δ, and cdc73Δ strains are sensitive to 6-azauracil, implicating the products of these genes in transcriptional elongation in vivo. Further confirmation that Paf1/Cdc73/Ctr9/Rtf1/Leo1 is involved in elongation came from ChIP experiments showing that these proteins are associated with the transcribed regions of ADH1 and other genes (our data and reference 31) and from recently published genetic and coimmunoprecipitation experiments examining the roles of these proteins (27, 39). However, in contrast to this work, neither study was able to detect a stable physical association between the Paf1 complex and Spt16/Pob3. Nevertheless, definitive proof that Paf1, Cdc73, Ctr9, Rtf1, and Leo1 are elongation factors awaits experiments showing directly that they have elongation activity in vivo or in vitro.
Tagging and purification of Rtf1, Paf1, Ctr9, or Cdc73 resulted in copurification of the same set of polypeptides that included these proteins as well as Leo1, Spt16, and Pob3, confirming that all are truly associated with Spt16/Pob3. Since it is possible that the presence of the TAP tag on a protein may impair its function in vivo, strains harboring individual tagged components of the Paf1 complex were tested for sensitivity to 6-azauracil. The strains containing the tagged proteins did not confer any significant sensitivity to the drug, implying the tag is not very detrimental to the function of the complex (data not shown). It can similarly be inferred that C-terminal tags on Spt16, Pob3, Spt5, Spt6, Iws1, Tfg1, and Tfg2 are not very detrimental, because these proteins are all encoded by essential genes, and tagged haploid strains are viable and grow well in all cases. Although the uncharacterized polypeptide Yol054w was detected when tagged Spt16 was purified in buffers containing 100 mM NaCl, it was not observed to copurify with any of the other tagged proteins. Efforts to tag Yol054w were unsuccessful, and so we cannot conclude that Yol054w is a bona fide subunit of any of these complexes. It is possible, however, that Yol054w is directly associated with Spt16 or Pob3 (see Fig. 6).
When all three subunits of TFIIF (Tfg1, Tfg2, and Tfg3) were tagged and purified in buffers containing 150 mM NaCl, the three proteins copurified with a near-stoichiometric amount of RNAPII in each case. This showed that TFIIF interacts with RNAPII in vivo, as suggested by experiments in many systems (7, 38) which demonstrated that TFIIF interacts with RNAPII in vitro. However, Tfg3 has also been reported to be associated with a number of other complexes, including the transcription initiation factor TFIID and the chromatin-modifying complexes SWI/SNF and NuA3 (5, 13, 16). When an extract from a Tfg3-tagged strain was used to purify the smallest subunit of TFIIF in buffers containing 75 to 100 mM NaCl, other polypeptides were detected by silver staining of the SDS-polyacrylamide gel, and these may correspond to subunits of these other factors (Krogan and Greenblatt, unpublished).
The Elongator complex reported to copurify with RNAPII was originally shown to have three subunits, Elp1, Elp2, and Elp3, but is now known to have three additional subunits, Elp4 (YPL101w), Elp5 (YHR187w), and Elp6 (YMR312w), which can be separated from Elp1/Elp2/Elp3 in buffers containing 200 mM NaCl (23, 30). Genetic and DNA microarray analysis of strains containing deletions of the Elongator subunits revealed that all are likely to be components of the same functional complex. Interestingly, Western blot analysis showed that no detectable RNAPII copurified with tagged Elp1, Elp2, Elp3, or Elp5. Given our observation that purified Elongator, unlike purified TFIIF, also did not stimulate elongation by RNAPII in vitro (our unpublished data), this result implies either that Elongator, unlike TFIIS, TFIIF, Spt4/Spt5, Spt6/Iws1, and the Spt16/Pob3 complex, does not truly associate with RNAPII or else that Elongator associates with RNAPII only once RNAPII is engaged in transcription and then perhaps only on some genes. In our hands, deletion of the genes encoding subunits of Elongator does not confer resistance or sensitivity to 6-azauracil (23), suggesting that Elongator may not generally be an elongation factor. Yeast Elongator subunits are mainly localized in the cytosol (24, 31), and ChIP experiments did not reveal association of Elongator subunits with the transcribed region of the ADH1 gene (our unpublished data). However, human Elongator has been shown to weakly stimulate transcription by human RNAPII in vitro (20), and one of its subunits has been shown to move from the cytosol to the nucleus in response to interleukin-6 treatment (6). Therefore, Elongator may regulate the transcription of specific genes in response to signaling pathways. This would be consistent with microarray experiments indicating that Elongator affects the expression of only a small set of yeast genes (23).
Tagging and purification of Spt4 resulted in the copurification of Spt5. When purification was carried out in buffers containing 100 mM NaCl, Spt4/Spt5 copurified with RNAPII and was detectable by silver staining. Similarly, tagged Spt5 copurified with Spt4 and RNAPII. These findings are in accord with previous observations that a mutated form of RPB2 genetically interacts with SPT5 and that Spt5 coimmunoprecipitates with Rpb1 (15), the largest subunit of RNAPII. Similarly, DSIF, the human counterpart of Spt4/Spt5, physically interacts with human RNAPII (46).
Tagged Spt5 also copurified with a truncated form of Spt6. A physical interaction between Spt5 and Spt6 was detected in one previously published study (41) but not in a follow-up study (15). Our tagging and purification of Spt6 did not result in the copurification of Spt5, probably because the form of Spt6 that copurified with tagged Spt5 is missing its C-terminal region and we have used C-terminal tags exclusively. However, we did discover a previously uncharacterized 40-kDa polypeptide, Iws1, associated with Spt6. When Iws1 was tagged and purified, it copurified with Spt6, confirming that this is a bona fide interaction. Western blotting demonstrated that Spt6-Iws1, like Spt4/Spt5, copurifies with RNAPII. Conversely, purification of tagged RNAPII resulted in copurification of Spt4, Spt5, Iws1, and Spt6, confirming these associations (data not shown). Our ChIP experiments showing that Iws1 is associated with the transcribed region of the ADH1 gene, together with our observations that Iws1 is associated with Spt6 and RNAPII, suggest that Iws1, like Spt6, is an RNAPII elongation factor. Proof that Spt6 and Iws1 are elongation factors awaits experiments showing that these proteins influence elongation by RNAPII in vivo and in vitro.
Interestingly, the sequence of the acidic N-terminal region of Iws1 displays similarity to the N-terminal region of Spt5. Moreover, tandem mass spectrometry revealed that a conserved serine residue in a consensus CKII phosphorylation site in this region was phosphorylated in both Spt5 and Iws1. Three phosphorylated serines detected by mass spectrometry in the N-terminal region of Spt6 are also located in consensus CKII sites (12). Therefore, phosphorylation by CKII may be important for the association of Spt5 and Iws1 with Spt6 or may have a regulatory role in the activities of Spt5, Spt6, and Iws1. One interesting possibility is that the CKII associated with Chd1 in the Spt16/Pob3 elongation factor complex may be involved in the phosphorylation of Spt5, Spt6, and Iws1 (see Fig. 6).
Our targeted proteomics approach has led to the description of a web of interactions, comprising 25 polypeptides (31 if Elongator is included), among the putative transcriptional elongation factors of S. cerevisiae. Construction of this interaction web was made possible by our use of affinity tagging and purification for each of the polypeptides, followed by electrophoresis, silver staining, and mass spectrometry. Determining which interactions are direct and which are indirect would not have been possible if we had simply relied on the sensitive Western blot technique. In some cases, (for example, TFIIS, TFIIF, and Spt4/Spt5), our results confirmed known or suspected interactions. In other cases, tagging and purification of a known elongation factor resulted in the discovery of novel information regarding that factor. In particular, Iws1, Paf1, Cdc73, Ctr9, and Leo1 were not previously suspected to be RNAPII elongation factors, and Paf1, Cdc73, Ctr9, Leo1, and Rtf1 were not known to associate with Spt16/Pob3. It is intriguing that there are probably so many elongation factors for RNAPII. It is not known whether all the RNAPII elongation factors can associate simultaneously with the elongating transcription complex, nor is it known whether a given factor accompanies RNAPII all the way from its initiation site to its termination site. The nonessentiality in yeast of many of the elongation factor polypeptides may imply that some of them are functionally redundant. However, it is also possible that some of them specifically regulate genes whose expression is not essential under laboratory conditions. The yeast genetic system as well as ChIP assays may make it possible to resolve some of these issues.
Acknowledgments
We thank D. B. Jansma for critical reading of the manuscript.
N.J.K. was supported by a PGS-B Scholarship Award from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Doctoral Fellowship from the Canadian Institute of Health Research (CIHR). This research was supported by grants to J.F.G. from the Canadian Institutes of Health Research and the National Cancer Institute of Canada with funds from the Canadian Cancer Society. A.S. is a scholar of the Leukemia & Lymphoma Society.
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14**:**2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archambault, J., F. Lacroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12**:**4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bähler, J., J. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14**:**943-951. [DOI] [PubMed] [Google Scholar]
- 4.Bengal, E., O. Flores, A. Krauskopf, D. Reinberg, and Y. Aloni. 1991. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol. Cell. Biol. 11**:**1195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16**:**3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collum, R. G., S. Brutsaert, G. Lee, and C. Schindler. 2000. A Stat3-interacting protein (StIP1) regulates cytokine signal transduction. Proc. Natl. Acad. Sci. USA 97**:**10120-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25**:**375-380. [DOI] [PubMed] [Google Scholar]
- 8.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156**:**535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22**:**9-11. [DOI] [PubMed] [Google Scholar]
- 10.Gatlin, C. L., G. R. Kleeman, L. G. Hays, A. J. Link, and J. R. Yates III. 1998. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microscopy and nanospray mass spectrometry. Anal. Biochem. 263**:**93-101. [DOI] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5**:**255-269. [Google Scholar]
- 12.Glover, C. V., III. 1998. On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 59**:**95-133. [DOI] [PubMed] [Google Scholar]
- 13.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25**:**59-63. [DOI] [PubMed] [Google Scholar]
- 14.Greenblatt, J. F. 1998. RNA polymerase II holoenzyme and transcriptional regulation. Curr. Opin. Cell Biol. 9**:**310-319. [DOI] [PubMed] [Google Scholar]
- 15.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12**:**357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14**:**1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14**:**2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller, D. M., X. Zeng, Y. Wang, Q. H. Zhang, M. Kapoor, H. Shu, R. Goodman, G. Lozano, Y. Zhao, and H. Lu. 2001. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7**:**283-292. [DOI] [PubMed] [Google Scholar]
- 19.Kelley, D. E., D. G. Stokes, and R. P. Perry. 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108**:**10-25. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. H., W. Lane, and D. Reinberg. 2002. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 99**:**1241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch, C., P. Wollman, M. Dahl, and F. Lottspeich. 1999. A role for Ctr9 and Paf1 in the regulation of G1 cyclin expression in yeast. Nucleic Acids Res. 27**:**2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14**:**2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogan, N. J., and J. F. Greenblatt. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21**:**8203-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16**:**707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magdolen, V., P. Lang, G. Mages, H. Hermann, and W. Bandlow. 1994. The gene LEO1 on yeast chromosome XV encodes a non-essential, extremely hydrophilic protein. Biochim. Biophys. Acta 1218**:**205-209. [DOI] [PubMed] [Google Scholar]
- 26.Mann, M., P. Hojrup, and P. Roepstorff. 1993. Use of mass spectrometry molecular weight information to identify proteins in sequence databases. Biol. Mass. Spectrom. 22**:**338-345. [DOI] [PubMed] [Google Scholar]
- 27.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22**:**1970-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92**:**105-116. [DOI] [PubMed] [Google Scholar]
- 29.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400**:**284-288. [DOI] [PubMed] [Google Scholar]
- 30.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. G. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3**:**109-118. [DOI] [PubMed] [Google Scholar]
- 31.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9**:**799-809. [DOI] [PubMed] [Google Scholar]
- 32.Price, D. H., A. E. Sluder, and A. L. Greenleaf. 1989. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 9**:**1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17**:**1030-1032. [DOI] [PubMed] [Google Scholar]
- 34.Rudd, M. D., M. G. Izban, and D. S. Luse. 1994. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc. Natl. Acad. Sci. USA 91**:**8057-8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20**:**7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17**:**1160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122**:**19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sopta, M., R. W. Carthew, and J. Greenblatt. 1985. Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 260**:**10353-10360. [PubMed] [Google Scholar]
- 39.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21**:**1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stolinski, L. A., D. M. Eisenmann, and K. M. Arndt. 1997. Identification of Rtf1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17**:**4490-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson, M. S., and F. Winston. 1992. SPT4, SPT5 and SPT6 interactions: effect on transcription and viability in Saccharomyces cerevisiae. Genetics 132**:**325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56**:**619-630. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, N. E., D. B. Aronson, and R. R. Burgess. 1990. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem. 265**:**7069-7077. [PubMed] [Google Scholar]
- 44.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromodomain protein Chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19**:**2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66**:**117-172. [DOI] [PubMed] [Google Scholar]
- 46.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12**:**343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wind, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22**:**327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wittschieben, B., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4**:**123-128. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97**:**41-51. [DOI] [PubMed] [Google Scholar]