Identification of a Receptor-Binding Domain of the Spike Glycoprotein of Human Coronavirus HCoV-229E (original) (raw)
Abstract
Human coronavirus HCoV-229E uses human aminopeptidase N (hAPN) as its receptor (C. L. Yeager et al., Nature **357:**420-422, 1992). To identify the receptor-binding domain of the viral spike glycoprotein (S), we expressed soluble truncated histidine-tagged S glycoproteins by using baculovirus expression vectors. Truncated S proteins purified by nickel affinity chromatography were shown to be glycosylated and to react with polyclonal anti-HCoV-229E antibodies and monoclonal antibodies to the viral S protein. A truncated protein (S547) that contains the N-terminal 547 amino acids bound to 3T3 mouse cells that express hAPN but not to mouse 3T3 cells transfected with empty vector. Binding of S547 to hAPN was blocked by an anti-hAPN monoclonal antibody that inhibits binding of virus to hAPN and blocks virus infection of human cells and was also blocked by polyclonal anti-HCoV-229E antibody. S proteins that contain the N-terminal 268 or 417 amino acids did not bind to hAPN-3T3 cells. Antibody to the region from amino acid 417 to the C terminus of S blocked binding of S547 to hAPN-3T3 cells. Thus, the data suggest that the domain of the spike protein between amino acids 417 and 547 is required for the binding of HCoV-229E to its hAPN receptor.
Human coronavirus HCoV-229E is an enveloped positive-stranded RNA virus in group 1 that causes common colds (18). Human aminopeptidase N (hAPN), a cell surface metalloprotease found on apical membranes of intestinal cells, lung and kidney epithelial cells, on macrophages, and at synaptic junctions, serves as receptor for HCoV-229E (57). The deduced amino acid sequence of the gene that encodes the 200-kDa spike glycoprotein (S) of HCoV-229E suggests that the S protein is composed of a 15-amino acid (aa) signal sequence, an N-terminal S1 domain (aa 16 to 560), an S2 domain (aa 561 to 1173) containing several heptad repeat regions, a transmembrane domain (aa 1117 to 1138), and a short cytoplasmic tail (aa 1139 to 1173) with a carboxy-terminal cysteine cluster (31). The S protein has 30 potential sites for N-glycosylation. It is likely that the spikes on the HCoV-229E viral envelope are trimers of the S glycoprotein, as shown for the spikes of a closely related coronavirus of pigs, transmissible gastroenteritis virus (TGEV) (9). This paper reports studies to identify the receptor-binding domain of the viral spike glycoprotein.
HCoV-229E only causes disease in humans, so all studies on the pathogenesis of this virus have been done with human volunteers (45). At present there are very few isolates of HCoV-229E that can be propagated in human cell cultures, including the strain isolated by Hamre and Procknow in the United States in 1966 (15) and the LP strain isolated by Tyrrell et al. in the United Kingdom in 1968 (46). In this paper we have used the Hamre strain of HCoV-229E.
To initiate infection, the spikes of HCoV-229E virions bind to hAPN on the plasma membrane of human cell lines or on the apical membranes of respiratory epithelial cells (47, 57). HCoV-229E virions are neutralized by incubation at 37°C with soluble hAPN (4a). Fusion of the TGEV envelope with host cell membranes occurs in endosomes (16). The molecular events that follow binding of a virion to its APN receptor have not been defined for group 1 coronaviruses. However, for mouse hepatitis virus in serogroup 2, binding at 37°C of the S protein to its receptor, murine CEACAM1a (CD66a), leads to conformational changes in the carboxyl-terminal domain (S2) that may facilitate fusion of the viral envelope with the host cell plasma membrane (44, 58).
Three coronaviruses in serogroup 1, HCoV-229E, TGEV, and feline coronavirus, and probably also canine coronavirus (CCoV), utilize the APN glycoprotein of their normal host species as a virus receptor (3, 7, 17, 22, 24, 43, 57). Although HCoV-229E can utilize either hAPN or feline APN (fAPN) as a receptor (43, 57), it cannot utilize porcine APN (pAPN) (7, 23). Kolb and collaborators identified a region of hAPN from aa 288 to 295 as essential for HCoV-229E infection. A different region, aa 717 to 813, on pAPN and the corresponding sites on fAPN and canine APN are essential for infection by TGEV, feline coronavirus, and CCoV, respectively (6, 17). Recent studies showed that the HCoV-229E receptor activity of hAPN can be abrogated by the addition of a single N-linked glycosylation site at amino acid 291 of hAPN, corresponding to a naturally occurring N-glycosylation site on pAPN (48). Thus, it is possible that the four group 1 coronaviruses studied may bind to a common site found on APN proteins of all of these host species and that the species specificity of APN recognition by the viruses depends on other domains of the APN proteins that differ from one host species to another and impede the approach of S proteins of group 1 coronaviruses from other hosts to the putative common conserved binding site (48). It is not yet known what domains of the S glycoproteins of human and porcine coronaviruses are responsible for their selective utilization of hAPN or pAPN, respectively, or what domain of S binds to the putative common receptor site on APN proteins of human and pigs.
This report describes the expression of purified soluble, truncated S glycoproteins of HCoV-229E by using a recombinant baculovirus expression system, and it analyzes the interactions of these truncated S proteins with polyclonal anti-HCoV-229E antiserum, anti-S monoclonal antibodies (MAbs), and hAPN on the surface of living cells. The region between aa 417 and 547 of the HCoV-229E S protein was found to be essential for binding to the hAPN receptor for HCoV-229E.
MATERIALS AND METHODS
Cells.
Spodoptera frugiperda (Sf9) cells (Invitrogen, Carlsbad, Calif.) were maintained at 27°C in TC-100 medium (JRH BioSciences, Lenexa, Kans.) with 10% fetal bovine serum (Gemini Bioproducts, Calabasas, Calif.) and 2% antibiotics (penicillin, streptomycin, and amphotericin B; Gibco/BRL, Gaithersburg, Md.). Murine 3T3 cells transfected with hAPN and 3T3 cells transfected with vector alone were obtained from T. Look (St. Judes, Memphis, Tenn.) and were cultured as previously described (57). The myeloma cell line P3X63-AG8.653 and porcine ST cells were obtained from the American Type Culture Collection (Rockville, Md.) and were propagated as recommended.
Virus.
HCoV-229E was obtained from the American Type Culture Collection (VR-740). The RW stock of HCoV-229E was propagated at 34°C in MRC5 cells as previously described (A. Bonavia and K. V. Holmes, submitted for publication). Plaque assays were done with MRC5 cells at 34°C.
Antibodies.
A polyclonal goat antiserum (G1) directed against sucrose density gradient-purified, NP-40-disrupted HCoV-229E virions, and preimmune goat serum were prepared in collaboration with L. Sturman, New York State Department of Health, Albany, N.Y.
Anti-HCoV-229E MAbs were prepared by immunizing BALB/c mice with 100 μg of semipurified HCoV-229E emulsified in complete Freund's adjuvant (Difco Laboratories, Detroit, Mich.). At 3-week intervals two boosters of 100 μg of semipurified HCoV-229E in incomplete Freund's adjuvant (Difco Laboratories) were administered, and mice were sacrificed 4 days after the last booster inoculation. Spleen cells from immunized mice were fused with P3X63-AG8.653 myeloma cells. Positive hybridoma clones were selected by immunofluorescence on L-132 cells infected with HCoV-229E.
WM15, a MAb directed against human aminopeptidase N (hAPN CD13), was obtained from Biodesign International (Kennebunk, Maine). Anti-hAPN MAb-BB1 was prepared by injecting Swiss Webster mice with membrane preparations of 3T3 cells expressing hAPN. Treatment of mouse cell lines or rodent cell lines expressing hAPN with anti-hAPN MAb-BB1 blocked HCoV-229E infection in these cells. A control mouse MAb of the same immunoglobulin G1 (IgG1) isotype against cholera toxin B subunit was kindly provided by R. Holmes (University of Colorado Health Sciences Center, Denver, Colo.). IgG from polyclonal goat anti-HCoV-229E antiserum G1 was purified on a protein A column (Pierce, Rockford, Ill.) and then was adsorbed with either S547 (G1/S547), S417 (G1/S417), or S268 (G1/S268) on Ni-nitrilotriacetic acid superflow columns (Qiagen, Valencia, Calif.).
Virus neutralization.
Five thousand PFU of HCoV-229E was incubated at 4°C with serial 10-fold dilutions of polyclonal goat anti-HCoV-229E antiserum in MRC5 medium (pH 7) or control goat preimmune serum in a total volume of 200 μl. After 2 h of incubation, 300 μl of medium was added and plaque assays were performed with MRC5 cells at 34°C by using an overlay medium of modified Eagle's medium containing 0.45% Seakem LEagarose (FMC Bioproducts, Rockland, Maine), 8% heat-inactivated fetal bovine serum, and antibiotics.
Expression of histidine-tagged, soluble, truncated HCoV-229E spike glycoproteins.
The S gene from HCoV-229E was reverse transcribed by using primer AB25 (Table 1) with Superscript 2 (Gibco, Grand Island, N.Y.) in a 20-μl reaction mixture, as described in the manufacturer's instructions. The gene was amplified by PCR by using 1 μl of cDNA and primers S-23 and S-3600 (Table 1) and then was cloned into pBCSk+ (Stratagene, La Jolla, Calif.). The cDNA encoding the S gene was reamplified by using _Pfu_-cloned polymerase (Stratagene) with primers BZ21 and BZ22 (Table 1) and then was cloned into pCITE at the _Nco_I and _Xba_I sites (Novagen, Madison, Wis.). The latter clone was used to amplify the cDNA sequences encoding the leader and the various regions of S with _Pfu-_cloned polymerase (Stratagene). We used the baculovirus expression vector pAcMP2(TH) (PharMingen, San Diego, Calif.) that encodes a thrombin cleavage site and six-histidine tag added to the C-terminal part of the expressed proteins, which were previously described (59). The cDNA encoding the N-terminal 547 aa (S547) including the signal sequence was amplified with oligonucleotides BZ21 and BZ22; 417 aa (S417) amplified with BZ21 and AB84 and 268 aa (S268) amplified with BZ21 and AB91. _Nco_I and _Xba_I restriction sites were added to the 5′ and 3′ ends, respectively, to allow cloning into pAcMP2(TH). Each construct was sequenced at the University of Colorado Health Sciences Center Cancer Center Core Sequencing Facility with primers BZ10, BZ14, AB29, AB31, AB41, and AB42 (see Table 1). The baculovirus expression constructs were cotransfected with BaculoGold DNA (PharMingen) into Sf9 cells, and progeny viruses were plaque purified according to the manufacturer's instructions. The resulting constructs, called S547, S417, and S268, produced soluble six-histidine-tagged glycoproteins (Fig. 1).
TABLE 1.
Cloning and sequencing primers
| Gene product | Primer | Sequence (5′ to 3′) | Locationa |
|---|---|---|---|
| Vector | BZ10 | CGGATTTCCTTGAAGAGAGTG | 4443 (c) |
| Vector | BZ14 | CAGTCGTCCAATGCAAAGCGT | 4268 |
| S | BZ21 | CGCGACCATGGTTGTTTTGCTTGTTGCA | 39 _Nco_I |
| S | BZ22 | CGCGATCTAGAAACAGCGTCTGTGCAATT | 1680 (c) _Xba_I |
| S | S-23 | GTCTCAACTAAATAAAATGTTTGT | 23 |
| S | S-3600R | ACAGCAGACACAAGTTGCAAT | 3600 (c) |
| ORF4a/b | AB25 | AGAGCCATTACTGTATGTGG | 56 (c) |
| S | AB29 | TGTGTCACAAACCTCTATTGC | 767 |
| S | AB31 | ATGTTGGTAGGTGGAGTGC | 1105 |
| S | AB41 | TCAGCATAAGAAGCTAACGC | 754 (c) |
| S | AB42 | CTTTAGGTAATGTAGAAGCCG | 667 |
| S | AB84 | ACTAGTCTAGACAATGAGCCTATAGTATAG | 1284 (c) _Xba_I |
| S | AB91 | GGGAAGGTCTAGAGGACAACTGGTCACATCT | 842 (c) _Xba_I |
FIG. 1.
Diagram of the baculovirus expression constructs for expression of soluble, six-histidine-tagged (6xH) truncated HCoV-229E spike glycoprotein (S). The number next to the S indicates the amino acid at which the construct was truncated. Each construct contains the HCoV-229E signal sequence. The region corresponding to the TGEV receptor-binding domain was identified by Godet et al. (14).
Even though there is no protease cleavage site in the HCoV-229E S protein as there is in the spike protein of mouse virus (MHV) (28), we used the S1/S2 nomenclature to refer to S1 as the amino-terminal half of the protein and S2 as the carboxy-terminal half.
Purification of soluble, histidine-tagged, truncated HCoV-229E S glycoproteins.
Large-scale preparations of supernatant medium containing the soluble truncated proteins were purified with HiTrap nickel affinity columns (Pharmacia, Piscataway, N. J.) as previously described (59). After analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with silver (data not shown), the fractions containing the truncated S proteins were pooled and stored at −80°C.
Immunoblots.
The soluble receptor glycoproteins and prestained molecular size standards (Bio-Rad, Hercules, Calif.) were separated by SDS-12.5% PAGE, transferred to Immobilon-P membranes (Millipore, Bedford, Mass.), and blocked overnight at 4°C in TBST (25 mM Tris [pH 7.6], 150 mM NaCl, 0.1% Tween 20) supplemented with 5% powdered milk. All subsequent steps were done in TBST buffer with 0.5% powdered milk. Blots were incubated for 1 h with a 1:3,000 dilution of polyclonal goat anti-HCoV-229E antiserum preabsorbed to mouse 3T3 cells. After being washed, the blots were incubated for 1 h with a 1:3,000 dilution of rabbit anti-goat IgG conjugated to horseradish peroxidase (HRP) (Cappel, Aurora, Ohio). Bound HRP-conjugated antibody complexes were detected with Renaissance Chemiluminescence Reagent (DuPont/NEN, Boston, Mass.), and images were exposed to autoradiography film (Kodak, Rochester, N.Y.).
PNGase treatments of truncated HCoV-229E S proteins.
Fifteen micrograms of the truncated HCoV-229E spike glycoprotein was incubated in Denaturing Buffer (New England Biolabs, Beverly, Mass.) at 100°C for 10 min. Upon cooling, G7 Buffer (New England Biolabs) and NP-40 (New England Biolabs) buffer were added to a 1× final concentration by following the manufacturer's directions. Fifteen hundred units of PNGase F (New England Biolabs) was added to each sample, and the samples were incubated for 1 h at 37°C. The deglycosylated proteins were separated by SDS-12.5% PAGE, transferred to Immobilon-P membrane (Millipore) paper, and immunoblotted by using polyclonal goat anti-HCoV-229E antiserum.
Enzyme-linked immunosorbent assay (ELISA).
Ninety-six-well Immulon plates (Dynex Technologies, Inc., Franklin, Mass.) were coated with (350 ng per well) the purified truncated HCoV-229E spike proteins in 0.05 M carbonate buffer (pH 9.6) (Sigma, St. Louis, Mo.). The plates were blocked overnight at 4°C in 5% bovine serum albumin (BSA)-B3 buffer (0.15 M NaCl, 0.05 M Tris-HCl, 0.8 mM EDTA, 0.05% Tween 20, 0.1% BSA, pH 7.4). Plates were incubated with anti-S MAbs (1:10) or polyclonal anti-HCoV-229E antiserum (1:30) for 1 h at room temperature. Bound antibodies were detected by using HRP-conjugated goat anti-mouse IgG or HRP-conjugated rabbit anti-goat IgG (Cappel) developed with _o_-phenylenediaminedihydrochloride peroxidase substrate (Sigma) by following the manufacturer's directions. Washes after every step and antibody dilution were performed in B3 buffer.
Receptor-binding assays.
Flow cytometry analysis was used to assay binding of truncated HCoV-229E S proteins to hAPN on live-cell membranes. Mouse 3T3 cells stably transfected with empty vector (3T3) or 3T3 cells expressing recombinant hAPN (hAPN/3T3) were incubated for 1 h at 4°C with different amounts of nickel affinity-purified truncated S proteins. After three washes, bound truncated HCoV-229E S proteins were detected by flow cytometry analysis by using various anti-S protein MAbs or polyclonal anti-HCoV-229E antiserum and their respective isotypic or normal serum controls. All incubations and washes were performed at 4°C in F buffer (0.1% BSA in phosphate-buffered saline, pH 7.4).
RESULTS
Characterization of truncated, soluble, histidine-tagged HCoV-229E spike proteins expressed by baculovirus.
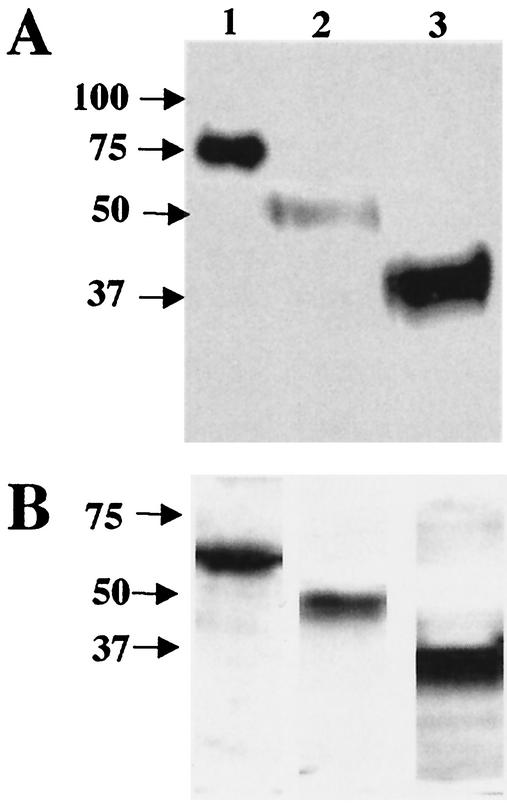
We constructed three baculovirus expression vectors that encoded soluble truncated, histidine-tagged HCoV-229E spike glycoproteins: S547, S417, and S268 (Fig. 1). The secreted proteins were purified by nickel affinity chromatography from the supernatant medium from baculovirus-infected Sf9 cell cultures. The Ni affinity-purified truncated HCoV-229E S proteins migrated in SDS-12.5% PAGE as broad bands (Fig. 2A). The molecular sizes of the glycoproteins were larger than expected on the basis of their predicted amino acid composition (Table 2). The larger apparent molecular sizes were largely due to N-linked glycosylation. That was shown by treating the proteins with PNGase F, which removes N-linked glycans (Fig. 2B). The three truncated S glycoproteins were recognized in immunoblots by polyclonal goat anti-HCoV-229E antiserum (Fig. 2A). These data suggest that the truncated S proteins produced by baculovirus were folded and processed like wild-type HCoV-229E spike glycoprotein. Blots probed with preimmune goat serum showed no bands (data not shown). In general, the sizes and the antigenicity of the soluble nickel affinity-purified, truncated S glycoproteins expressed by baculovirus were as expected.
FIG. 2.
Characterization of the purified, soluble, truncated HCoV-229E spike proteins. (A) Immunoblot of 1.2 μg of purified S547 (lane 1), S417 (lane 2), and S268 (lane 3) with polyclonal goat anti-HCoV-229E antiserum. (B) Immunoblot as for that in panel A of proteins treated with PNGase F.
TABLE 2.
Molecular sizes of truncated HCoV-229E nickel affinity-purified spike proteinsa
| Truncation | Predicted size (kDa) | Size (kDa) with possible glycosylation | Size (kDa) of baculovirus-expressed proteins | Size (kDa) of baculovirus-expressed proteins after PNGase treatment |
|---|---|---|---|---|
| S547 | 59.6 | 89.6 | 75-88 | 53-59 |
| S417 | 45.6 | 65.6 | 48-60 | 41-47 |
| S268 | 29.4 | 45 | 34-44 | 25-37 |
Purified, truncated HCoV-229E spike proteins share epitopes with wild-type HCoV-229E spike protein.
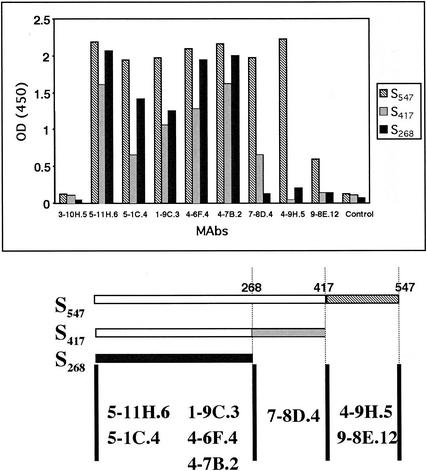
A panel of anti-HCoV-229E MAbs recognized the three truncated HCoV-229E spike glycoproteins in ELISA assays (Fig. 3). Anti-HCoV-229E MAbs 5-11H.6, 5-1C.4, 1-9C.3, 4-6F.4, and 4-7B.2 detected each of the three truncated S proteins. MAb 7-8D.4 detected S547 and S417 but not S268. MAbs 4-9H.5 and 9-8E.12 detected S547 but not S417 or S268. Figure 3 summarizes the likely sites of the epitopes on S that were recognized by each of the anti-S MAbs. MAb 3-10H.5 did not react to any of the truncated S proteins. This MAb recognized the N protein of HCoV-229E in an immunoblot assay (data not shown). The control MAb directed against an irrelevant protein did not react to any of the HCoV-229E S proteins. Therefore, eight MAbs directed against HCoV-229E virions reacted with Ni affinity-purified S547, S417, and/or S268. All antibodies were positive in immunofluorescence assays of susceptible infected cells, and none showed neutralizing activity (data not shown).
FIG. 3.
Binding of anti-HCoV-229E MAbs to the three truncated HCoV-229E spike glycoproteins. ELISA detection of 350 ng of S547, S417, and S268 by using anti-HCoV-229E MAbs followed by HRP-conjugated anti-mouse Igs. The diagram below shows the probable locations of MAb-binding epitopes in the linear map of the N-terminal domain of the HCoV-229E spike protein. OD (450), optical density at 450 nm.
S547 binds to the virus receptor hAPN on cell membranes.
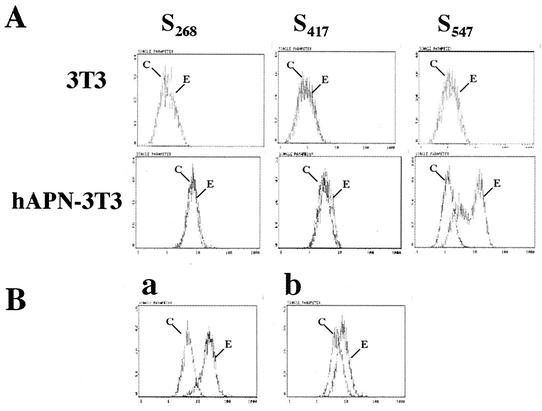
Since the identification of the receptor for HCoV-229E (57), it has been logically assumed that the S glycoprotein of HCoV-229E interacts with hAPN. We directly tested the hypothesis that the S glycoprotein binds to hAPN and that the receptor-binding site lies in the S1 region. Flow cytometry showed that S547, but not S417 or S268, bound to mouse 3T3 cells that express the receptor hAPN (hAPN/3T3) and that none of the three proteins bound to 3T3 cells that did not express the human coronavirus receptor hAPN (Fig. 4A). The amount of S547 that bound to hAPN-3T3 cells was concentration dependent, reaching saturation levels at approximately 1 μg of protein/106 cells (data not shown). Addition of up to 5 μg of S417 or S268 in this experiment showed no binding. These data suggest that the domain of the HCoV-229E spike that binds to the viral receptor lies between aa 417 and 547.
FIG. 4.
Binding of truncated HCoV-229E S glycoproteins to hAPN on cell membranes. (A) Detection of S547, S417, and S268 binding to 3T3 cells or hAPN-3T3 cells by using normal goat serum (C) or polyclonal goat anti-HCoV-229E antiserum (E). (B) Detection of S547 binding to hAPN-3T3 cells with preimmune goat serum (C) or polyclonal goat anti-HCoV-229E antiserum (E) after the cells were preincubated with isotype control (graph a) or anti-hAPN MAb-BB1 that blocks HCoV-229E infection of human cells or hAPN-expressing cells (graph b). The y axis indicates cell numbers, and the x axis indicates fluorescence intensity.
To investigate whether S417 bound to hAPN with low affinity and was therefore undetectable in the flow cytometry binding assay, we incubated cells expressing hAPN or no hAPN with 500 ng of S547 with increasing amounts of S417 (up to 40 μg) in a competition binding assay. The amount of S547 used in this experiment was below saturation levels. Binding of S proteins in this experiment was detected with MAb 4-9H.5 (which detected only S547 [Fig. 3]) or with MAb 5-11H.6 (which detected both S547 and S417 [Fig. 3]). Incubation of 500 ng of S547 with up to 40 μg of S417 caused no reduction in binding of S547 (data not shown). These data show that S417 did not compete with the binding of S547 to hAPN on cell membranes.
Neutralizing polyclonal goat anti-HCoV-229E antiserum and anti-hAPN MAb block binding of S547 to hAPN.
The specificity of the interaction between S547 and hAPN was further investigated by showing that binding of S547 to hAPN/3T3 cells was disrupted by pretreating the hAPN/3T3 cells with hAPN-specific anti-hAPN MAb BB1 that blocks HCoV-229E infection of human cells (Fig. 4B).
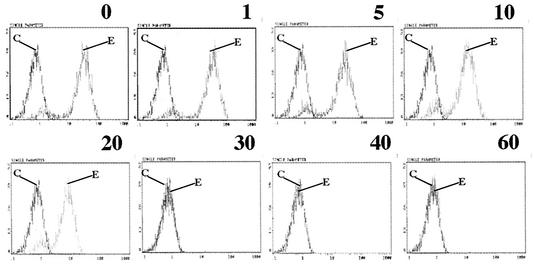
McIntosh and coworkers (30) showed that guinea pig anti-HCoV-229E antiserum neutralized the infectivity of HCoV-229E virus. We found that polyclonal goat anti-HCoV-229E antiserum neutralized 5,000 PFU of HCoV-229E at a dilution of 1:1,000 (data not shown). To determine whether neutralizing antiserum could block the binding of S547 to hAPN, we incubated S547 with different amounts of polyclonal goat anti-HCoV-229E antiserum preadsorbed to 3T3 cells before assaying for binding to hAPN. S547 binding was detected with MAb 5-11H.6. The epitope that 5-11H.6 recognizes in S547 was not masked by the polyclonal antiserum. Flow cytometry assays for binding of S547 to hAPN-3T3 showed that preincubation of S547 with the polyclonal goat anti-HCoV-229E antiserum blocked the interaction of S547 with hAPN in a concentration-dependent manner (Fig. 5).
FIG. 5.
Binding of S547 to hAPN-3T3 cells is blocked by preincubation of S547 with neutralizing polyclonal goat anti-HCoV-229E antiserum. Detection of S547 by flow cytometry with anti-HCoV-229E-S MAb5-11.H6 (E) and isotype control (C). The amount (in micrograms) of polyclonal goat anti-HCoV-229E antiserum used in the preincubation step is shown above each panel. The y axis indicates cell numbers, and the x axis indicates fluorescence intensity.
Antibody to the region of S between aa 417 and 547 blocks binding of S547 to hAPN/3T3 cells.
The experiments described above suggested that the region of HCoV-229E S between aa 417 and 547 was required for binding of S547 to hAPN. To prepare an antibody to this region that might directly block binding of S547 to hAPN-3T3 cells, we prepared region-specific anti-S polyclonal antibodies by purifying the polyclonal goat anti-HCoV-229E antiserum (G1) Igs by protein A (designated G1Ig) and then preabsorbing G1Ig with S547 (G1/S547), S417 (G1/S417), or S268 (G1/S268). An ELISA showed that G1 and G1Ig detected all three truncated HCoV-229E spike glycoproteins. G1/S547 did not detect any of the three truncated S glycoproteins (Fig. 6A). As expected, G1/S417 detected S547 but not S417 or S268. G1/S268 detected S547 and S417 but not S268 (Fig. 6A).
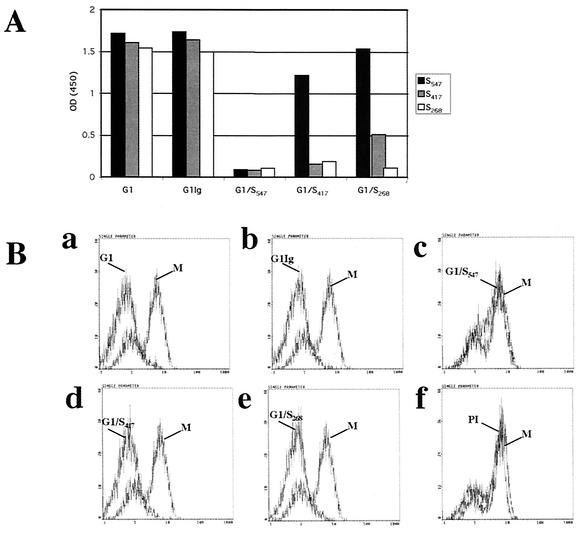
FIG. 6.
(A) ELISA of binding to S547, S417, and S268 of polyclonal anti-HCoV-229E (G1), purified polyclonal (G1Ig), and anti-HCoV-229E G1 antiserum immunoadsorbed against S547 (G1/S547), S417 (G1/S417), and S268 (G1/S268). (B) Binding of 1.5 μg of S547 to 106 hAPN/3T3 cells after incubation of S547 with various amounts of G1 (a), G1Ig (b), G1/S547(c), G1/S417 (d), and G1/S268 (e) preimmune goat serum (PI) (f) detected with anti-HCoV-229E S MAb 5-11H.6 or isotype control (M). The y axis indicates cell numbers, and the x axis indicates fluorescence intensity.
Flow cytometry assays with the region-specific polyclonal antibodies were done to detect binding of S547 to hAPN/3T3 cells preincubated with S547. Figure 6B shows that the anti-S antibodies G1, G1Ig, G1/S417, and G1/268 but not G1/S547 or preimmune goat serum were able to block the interaction of S547 with hAPN. Because these region-specific antibodies were derived from a polyclonal goat antiserum raised against gradient-purified, detergent-disrupted HCoV-229E virions, they also contained antibodies against the S2 portion of the spike glycoprotein and other viral structural proteins. The observation that adsorption of the antiviral antibody with S417 and S268 did not remove the antibodies that blocked binding of S547 to hAPN/3T3 cells strongly suggests that the region of S between aa 417 and 547 was essential for binding of S547 to hAPN.
DISCUSSION
The binding of viral attachment proteins (VAP) to their cognate cellular receptors and postbinding conformational changes in VAPs that lead to virus entry and uncoating have been studied intensively during the last 10 years. The receptor-binding domains of the VAPs of many different types of viruses have been mapped. In general, the amino acids that bind to the receptor are well conserved among related viruses that utilize the same receptor. Often the VAP binds to an exposed region on the surface of a receptor protein, frequently a site containing one or more hydrophobic residues (42). For example, a conserved RGD domain in the surface-exposed GH loop of the VP1 VAP of foot-and-mouth disease virus, a picornavirus, binds to integrin αvβ3 (21). The amino acid immediately following the RGD motif influences binding to another integrin, αvβ1 (20). Mutations in this amino acid of the VAPs of different foot-and-mouth disease virus strains may determine the ability of the virus to use different integrins as receptors in vivo. The hemagglutinin (HA) glycoprotein of influenza A virus is the VAP that binds to sialic acid receptors (52). HA forms a trimeric spike on the viral envelope. The crystal structures of HA proteins of many influenza A virus strains have been determined previously (54). Protease cleavage of HA by furin during virus maturation or by extracellular trypsin-like enzymes occurs at a site between the HA1 and HA2 proteins. The N-terminal domain, HA1, consists of a ball of predominantly beta sheets and contains the major antigenic determinants of the protein. The receptor-binding site is a shallow pocket at the tip of HA1 that contains six residues that are conserved among all influenza A and B viruses (53). The C-terminal HA2 domain contains a hydrophobic sequence called the fusion peptide at its amino terminus, followed by a coiled-coil domain that undergoes a profound conformational change upon shifting the pH to 5.5 (36, 50, 51), a transmembrane domain, and a short cytoplasmic domain. The structure and functions of HA provide a model for class I virus fusion glycoproteins of many other enveloped viruses, including the paramyxovirus simian virus 5, measles, Ebola, vesicular stomatitis virus, and human immunodeficiency virus type 1 (1, 11, 29, 56). In human immunodeficiency virus, the gp160 spike glycoprotein is cleaved to yield gp120 and gp41 (19). The N-terminal gp120 binds to CD4 on the cell surface, triggering a conformational change in the gp120 protein that exposes the binding site on gp120 for the chemokine coreceptor (33, 56). Interaction of the spike with the coreceptor triggers a conformational change that activates gp41 to position the fusion peptide in close proximity to the cell membrane and allow insertion of the fusion peptide into the membrane of the target cell (51). The N-terminal Ig-like domain of human CD4 binds to a recessed pocket in gp120, and the critical contact amino acids have been mapped (55).
Analyses of the deduced amino acid sequences of the 180- to 200-kDa coronavirus spike glycoproteins suggest that these proteins have the same general structure as the prototypic class 1 viral fusion protein, the HA protein of influenza A, although they are much larger than HA. The Chow and Fassman algorithms predict that the N-terminal domain of coronavirus S proteins consists primarily of beta sheets, and the C-terminal domain includes a coiled-coil domain, transmembrane domain, and short cytoplasmic domain. For some coronaviruses in group 2, such as MHV and bovine coronavirus, there is a protease cleavage site near the center of the S protein that defines the N-terminal S1 and the C-terminal S2 domains. Protease cleavage at that site can facilitate virus-induced cell fusion and viral infectivity (10, 39), although cleavage between S1 and S2 is not essential for viral infectivity (4). Cleavage has not been observed in S proteins of group 1 coronaviruses.
The spike glycoprotein (S) of murine coronavirus MHV is the most extensively studied coronavirus VAP. The principal receptors for MHV are isoforms of murine CEACAM1a (CD66a; formerly called Bgp, mmCGM, and MHVR) (2). Mutational analysis and X-ray crystallographic studies show that the protruding CC′ loop on the surface of the N-terminal Ig domain of murine CEACAM1a serves as the viral binding site (32, 42, 49). A hydrophobic amino acid, Ile41, that projects from this loop has been postulated to insert into a possible hydrophobic pocket in the viral S protein. The N-terminal 330 aa of the S1 protein of MHV is sufficient for binding to the murine CEACAM1a receptor protein (41). On MHV virions, binding of soluble murine CEACAM1a to the receptor-binding domain of S1 at 37°C induces a conformational change in the S2 protein (44, 58) and facilitates detachment of the S1 protein of MHV-JHM (12). There is no hydrophobic fusion peptide at the N terminus of the S2 protein, but several possible fusion peptides have been identified within the S2 protein (26, 27).
To date there are no structural data and only limited functional information about the spike glycoproteins of coronaviruses in serogroup 1, which includes HCoV-229E, TGEV, FCoV, and CCoV. Since each of these viruses uses APN as a receptor to enter cells (7, 43, 57), similar or conserved regions in the spike glycoproteins could be candidates for the binding site for viral S proteins. The amino acid identity between the S glycoproteins of serogroup 1 coronaviruses ranges from 41 to 45% (by using MacVector; International Biotechnologies, Inc., New Haven, Conn.). The best-studied S protein of the group 1 viruses is that of TGEV. In addition to binding to pAPN, TGEV virions treated with neuraminidase can bind via the S protein to N-acetyl neuraminic acid or N-glycolyl neuraminic acid moieties on the cell surface (25, 37). Two amino acids in the S1 domain play a role in sialic acid binding. A domain of more than 220 aa in the S1 domain of the TGEV S protein is not required for binding to pAPN, because deletions of this segment of S1 occur spontaneously, generating the porcine respiratory coronavirus that cause epizootics of mild respiratory disease in pigs (34, 35, 38). Like the TGEV viruses from which they are derived, porcine respiratory coronavirus isolates can also utilize pAPN as receptors (8). Epitopes for MAbs have been mapped to the S protein of TGEV (5). Because all strains of TGEV are neutralized by a MAb to epitope A, this region of S is a candidate for the receptor-binding domain (13, 40). There is a single report about the interaction of truncated TGEV S protein with pAPN (14). When a peptide from aa 506 to 655 of TGEV S protein was coexpressed in insect cells with pAPN, the two proteins could be coimmunoprecipitated. Therefore, this region of S was implicated as a possible receptor-binding domain. This aa 506 to 655 region of TGEV S protein corresponds by sequence alignment to aa 279 to 417 of the S protein of HCoV-229E, but there is only 28% identity of these peptides at the amino acid level. These coexpression experiments do not represent physiological binding of a viral spike to its receptor on a host cell. For example, coimmunoprecipitation could occur if the insect cells had not been thoroughly disrupted prior to antibody addition.
To identify the receptor-binding domain of the HCoV-229E spike protein, we expressed three C-terminal-truncated S proteins from the N-terminal domain of S1. S547 included the entire S1 domain. S417 was truncated at the C terminus corresponding to the putative receptor-binding domain of TGEV S protein, and S268 was the smallest N-terminal domain tested. The proteins were purified, and binding of MAbs demonstrated that they retained the antigenic determinants of the intact S protein. Epitopes recognized by eight nonneutralizing anti-S MAbs were mapped to these three peptides. We showed by flow cytometry that the soluble N-terminal domain of HCoV-229E (S547) could bind directly to hAPN on the membranes of hAPN/3T3 cells, while no binding was detected on 3T3 cells transfected with vector alone. Binding of the peptide to hAPN was specific and could be blocked either by pretreatment of the cells with anti-hAPN MAb-BB1 or by incubation of the S547 peptide with goat polyclonal anti-HCoV-229E antiserum. Neither S417 nor S268 was found to bind to hAPN/3T3 cells by this assay. This observation implicates the region between aa 417 and 547 of HCoV-229E S protein as an important determinant for binding to hAPN. Further evidence in support of this conclusion is the observation that region-specific polyclonal antibody directed against the region from aa 417 to the C terminus of S could block binding of S547 to hAPN on hAPN/3T3 cells. The putative receptor-binding domain of aa 417 to 547 of HCoV-229E protein is not homologous to the putative aa 506 to 655 receptor-binding domain of TGEV S protein (which corresponds to aa 279 to 417 of HCoV-229E S protein). The reason for this difference between TGEV and HCoV-229E is not clear. Possibly, although the N-terminal 417 aa of HCoV-229E S protein were not sufficient for binding to hAPN, they may contribute to the species specificity and/or the affinity of receptor binding.
Further mutational analysis and crystallographic studies of the S protein of HCoV-229E are needed to identify the specific amino acid residues on S that are required for binding to hAPN as well as residues important for possible induction of receptor-induced conformational changes in S. These studies will also elucidate the molecular determinants of species-specific recognition of APN proteins by S proteins of group 1 coronaviruses. Finally, understanding the interactions of HCoV-229E spike glycoprotein with its hAPN receptor may lead to the development of novel antiviral drugs to prevent or treat coronavirus-induced colds.
Acknowledgments
We are grateful to Dianna Blau and Jamie Breslin for thoughtful discussions and to Bonnie Bullis and Francine Lambert for technical assistance.
This work was supported by NIH grant RO1 26075 and an operating grant from the Canadian Institutes of Health Research (MT-9203). We also thank the University of Colorado Health Sciences Center Cancer Center Core (supported by NIH/NCI grant CA46934) for assistance with flow cytometry, DNA sequencing, and baculovirus expression systems.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3**:**309-319. [DOI] [PubMed] [Google Scholar]
- 2.Beauchemin, N., P. Draber, G. Dveksler, P. Gold, S. Gray-Owen, F. Grunert, S. Hammarstrom, K. V. Holmes, A. Karlsson, M. Kuroki, S. H. Lin, L. Lucka, S. M. Najjar, M. Neumaier, B. Obrink, J. E. Shively, K. M. Skubitz, C. P. Stanners, P. Thomas, J. A. Thompson, M. Virji, S. von Kleist, C. Wagener, S. Watt, and W. Zimmermann. 1999. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252**:**243-249. [DOI] [PubMed] [Google Scholar]
- 3.Benbacer, L., E. Kut, L. Besnardeau, H. Laude, and B. Delmas. 1997. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 71**:**734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos, E. C., W. Luytjes, and W. J. Spaan. 1997. The function of the spike protein of mouse hepatitis virus strain A59 can be studied on virus-like particles: cleavage is not required for infectivity. J. Virol. 71**:**9427-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Breslin, J. J., I. Mørk, M. K. Smith, L. K. Vogel, E. M. Hemmila, A. Bonavia, P. J. Talbot, H. Sjöström, O. Norén, and K. V. Holmes. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37°C. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 5.Correa, I., F. Gebauer, M. J. Bullido, C. Sune, M. F. Baay, K. A. Zwaagstra, W. P. Posthumus, J. A. Lenstra, and L. Enjuanes. 1990. Localization of antigenic sites of the E2 glycoprotein of transmissible gastroenteritis coronavirus. J. Gen. Virol. 71(Pt 2)**:**271-279. [DOI] [PubMed] [Google Scholar]
- 6.Delmas, B., J. Gelfi, E. Kut, H. Sjostrom, O. Noren, and H. Laude. 1994. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J. Virol. 68**:**5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmas, B., J. Gelfi, R. L'Haridon, L. K. Vogel, H. Sjostrom, O. Noren, and H. Laude. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357**:**417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delmas, B., J. Gelfi, H. Sjostrom, O. Noren, and H. Laude. 1993. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342**:**293-298. [DOI] [PubMed] [Google Scholar]
- 9.Delmas, B., and H. Laude. 1990. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 64**:**5367-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56**:**912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredericksen, B. L., and M. A. Whitt. 1995. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J. Virol. 69**:**1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher, T. M. 1997. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J. Virol. 71**:**3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebauer, F., W. P. Posthumus, I. Correa, C. Sune, C. Smerdou, C. M. Sanchez, J. A. Lenstra, R. H. Meloen, and L. Enjuanes. 1991. Residues involved in the antigenic sites of transmissible gastroenteritis coronavirus S glycoprotein. Virology 183**:**225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godet, M., J. Grosclaude, B. Delmas, and H. Laude. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68**:**8008-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamre, D., and J. J. Procknow. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121**:**190-193. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, G. H., B. Delmas, L. Besnardeau, L. K. Vogel, H. Laude, H. Sjostrom, and O. Noren. 1998. The coronavirus transmissible gastroenteritis virus causes infection after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment. J. Virol. 72**:**527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegyi, A., and A. F. Kolb. 1998. Characterization of determinants involved in the feline infectious peritonitis virus receptor function of feline aminopeptidase N. J. Gen. Virol. 79**:**1387-1391. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, K. V. 2001. Coronaviruses, p. 1187-1203. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 4th ed. Lippincott-Raven Publishers, New York, N.Y.
- 19.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157**:**187-253. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, T., W. Blakemore, J. W. Newman, N. J. Knowles, A. P. Mould, M. J. Humphries, and A. M. King. 2000. Foot-and-mouth disease virus is a ligand for the high-affinity binding conformation of integrin alpha5β1: influence of the leucine residue within the RGDL motif on selectivity of integrin binding. J. Gen. Virol. 81**:**1383-1391. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, T., A. Sharma, R. A. Ghazaleh, W. E. Blakemore, F. M. Ellard, D. L. Simmons, J. W. Newman, D. I. Stuart, and A. M. King. 1997. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin αvβ3 in vitro. J. Virol. 71**:**8357-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolb, A. F., A. Hegyi, J. Maile, A. Heister, M. Hagemann, and S. G. Siddell. 1998. Molecular analysis of the coronavirus-receptor function of aminopeptidase N. Adv. Exp. Med. Biol. 440**:**61-67. [DOI] [PubMed]
- 23.Kolb, A. F., A. Hegyi, and S. G. Siddell. 1997. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J. Gen. Virol. 78(Pt 11)**:**2795-2802. [DOI] [PubMed] [Google Scholar]
- 24.Kolb, A. F., J. Maile, A. Heister, and S. G. Siddell. 1996. Characterization of functional domains in the human coronavirus HCV 229E receptor. J. Gen. Virol. 77(Pt 10)**:**2515-2521. [DOI] [PubMed] [Google Scholar]
- 25.Krempl, C., M. L. Ballesteros, G. Zimmer, L. Enjuanes, H. D. Klenk, and G. Herrler. 2000. Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J. Gen. Virol. 81**:**489-496. [DOI] [PubMed] [Google Scholar]
- 26.Krueger, D. K., S. M. Kelly, D. N. Lewicki, R. Ruffolo, and T. M. Gallagher. 2001. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 75**:**2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, Z. L., and S. R. Weiss. 1998. Mutational analysis of fusion peptide-like regions in the mouse hepatitis virus strain A59 spike protein. Adv. Exp. Med. Biol. 440**:**17-23. [DOI] [PubMed] [Google Scholar]
- 28.Luytjes, W., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. van der Zeijst, M. C. Horzinek, and W. J. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161**:**479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malashkevich, V. N., B. J. Schneider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Natl. Acad. Sci. USA 96**:**2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIntosh, K., A. Z. Kapikian, K. A. Hardison, J. W. Hartley, and R. M. Chanock. 1969. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J. Immunol. 102**:**1109-1118. [PubMed] [Google Scholar]
- 31.Raabe, T., B. Schelle-Prinz, and S. G. Siddell. 1990. Nucleotide sequence of the gene encoding the spike glycoprotein of human coronavirus HCV 229E. J. Gen. Virol. 71(Pt 5)**:**1065-1073. [DOI] [PubMed] [Google Scholar]
- 32.Rao, P. V., S. Kumari, and T. M. Gallagher. 1997. Identification of a contiguous 6-residue determinant in the MHV receptor that controls the level of virion binding to cells. Virology 229**:**336-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280**:**1949-1953. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez, C. M., F. Gebauer, C. Sune, A. Mendez, J. Dopazo, and L. Enjuanes. 1992. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 190**:**92-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73**:**7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, S. B., K. Kawasaki, and S. Ohnishi. 1983. Hemolytic activity of influenza virus hemagglutinin glycoproteins activated in mildly acidic environments. Proc. Natl. Acad. Sci. USA 80**:**3153-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultze, B., L. Enjuanes, and G. Herrler. 1995. Analysis of the sialic acid-binding activity of the transmissible gastroenteritis virus. Adv. Exp. Med. Biol. 380**:**367-370. [DOI] [PubMed] [Google Scholar]
- 38.Sestak, K., I. Lanza, S. K. Park, P. A. Weilnau, and L. J. Saif. 1996. Contribution of passive immunity to porcine respiratory coronavirus to protection against transmissible gastroenteritis virus challenge exposure in suckling pigs. Am. J. Vet. Res. 57**:**664-671. [PubMed] [Google Scholar]
- 39.Storz, J., R. Rott, and G. Kaluza. 1981. Enhancement of plaque formation and cell fusion of an enteropathogenic coronavirus by trypsin treatment. Infect. Immun. 31**:**1214-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sune, C., C. Smerdou, I. M. Anton, P. Abril, J. Plana, and L. Enjuanes. 1991. A conserved coronavirus epitope, critical in virus neutralization, mimicked by internal-image monoclonal anti-idiotypic antibodies. J. Virol. 65**:**6979-6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, H., and F. Taguchi. 1996. Analysis of the receptor-binding site of murine coronavirus spike protein. J. Virol. 70**:**2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan, K., B. D. Zelus, R. Meijers, J. H. Liu, J. M. Bergelson, N. Duke, R. Zhang, A. Joachimiak, K. V. Holmes, and J. H. Wang. 2002.. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 21**:**2076-2086. [DOI] [PMC free article] [PubMed]
- 43.Tresnan, D. B., and K. V. Holmes. 1998. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 440**:**69-75. [DOI] [PubMed] [Google Scholar]
- 44.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003.. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77**:**841-850. [DOI] [PMC free article] [PubMed]
- 45.Tyrrell, D. A. 1979. Studies of rhinoviruses and coronaviruses at the Common Cold Unit, Salisbury, Wiltshire. Postgrad. Med. J. 55**:**117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyrrell, D. A., M. L. Bynoe, and B. Hoorn. 1968. Cultivation of “difficult” viruses from patients with common colds. Br. Med. J. 1**:**606-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, G., C. Deering, M. Macke, J. Shao, R. Burns, D. M. Blau, K. V. Holmes, B. L. Davidson, S. Perlman, and P. B. McCray, Jr. 2000. Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol. 74**:**9234-9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wentworth, D. E., and K. V. Holmes. 2001. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 75**:**9741-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wessner, D. R., P. C. Shick, J. H. Lu, C. B. Cardellichio, S. E. Gagneten, N. Beauchemin, K. V. Holmes, and G. S. Dveksler. 1998. Mutational analysis of the virus and monoclonal antibody binding sites in MHVR, the cellular receptor of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 72**:**1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89**:**674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, J. M. 1992. Membrane fusion. Science 258**:**917-924. [DOI] [PubMed] [Google Scholar]
- 52.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56**:**365-394. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, I. A., and N. J. Cox. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8**:**737-771. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289**:**366-373. [DOI] [PubMed] [Google Scholar]
- 55.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393**:**705-711. [DOI] [PubMed] [Google Scholar]
- 56.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280**:**1884-1888. [DOI] [PubMed] [Google Scholar]
- 57.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357**:**420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zelus, B. D., J. N. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2002.. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77**:**830-840. [DOI] [PMC free article] [PubMed]
- 59.Zelus, B. D., D. R. Wessner, R. K. Williams, M. N. Pensiero, F. T. Phibbs, M. deSouza, G. S. Dveksler, and K. V. Holmes. 1998. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1(b), and Bgp2 murine coronavirus receptors differ in mouse hepatitis virus binding and neutralizing activities. J. Virol. 72**:**7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]