Characterization of Mos1-Mediated Mutagenesis in Caenorhabditis elegans: A Method for the Rapid Identification of Mutated Genes (original) (raw)
Abstract
Insertional mutagenesis with a heterologous transposon provides a method to rapidly determine the molecular identity of mutated genes. The Drosophila transposon Mos1 can be mobilized to cause mutations in Caenorhabditis elegans (Bessereau et al. 2001); however, the mutagenic rate was initially too low for use in most forward genetic screens. To increase the effectiveness of _Mos1_-mediated mutagenesis we examined the conditions influencing Mos1 transposition. First, optimal transposition occurs 24 hr after expression of the transposase and is unlikely to occur in differentiated sperm or oocytes. Second, transposition is limited to germ-cell nuclei that contain donor elements, but the transposase enzyme can diffuse throughout the gonad syncytium. Third, silencing of transposition is caused by changes in the donor array that occur over time. Finally, multiple transposition events occur in individual germ cells. By using screening techniques based on these results, Mos1 mutagenicity was increased to within an order of magnitude of chemical mutagens.
IN the nematode Caenorhabditis elegans, screening for mutations causing visible phenotypes can assign a function to a gene. Yet, only <20% of the ∼20,000 predicted genes have been identified in genetic screens (C. elegans Sequencing Consortium 1998) because mutations in many genes produce wild-type or subtle mutant phenotypes (Park and Horvitz 1986). Screens for subtle phenotypes, such as changes in population behaviors, or the use of sensitized genetic backgrounds can be used to isolate new mutant strains (Jorgensen and Mango 2002). Identification of the mutated genes requires positional cloning; however, genetic mapping is especially laborious and time consuming when mapping synthetic phenotypes or when the mutant phenotype is very subtle. Single-nucleotide-polymorphism mapping techniques provide a significant improvement in the speed of positional cloning (Wicks et al. 2001) but they are still difficult to perform on certain phenotypes since polymorphic strains have exhibited phenotypic differences relative to the Bristol laboratory strain (Hodgkin and Doniach 1997).
Insertional mutagenesis with a transposon circumvents the need for genetic mapping: the transposon can be used as a sequence tag to rapidly identify the mutated gene. Endogenous transposable elements of the Tc1/mariner superfamily, especially Tc1 and Tc3, have been widely used for insertional mutagenesis in C. elegans (Moerman et al. 1986; Plasterk and van Luenen 1997). However, using Tc elements as mutagens has two major drawbacks. First, all known isolates of C. elegans contain multiple copies of Tc1 and Tc3, which makes it difficult to identify the relevant mutagenic insertion. Second, germline mobilization of Tc transposons cannot be controlled in mutator strains in which these elements are active. We have circumvented these two limitations by mobilizing the transposon Mos1 in the germline of C. elegans (Bessereau et al. 2001). Mos1 is a member of the Tc1/mariner family and was isolated from Drosophila mauritiana (Jacobson et al. 1986). The Mos1 element is absent from the C. elegans genome and controlled mobilization of Mos1 is achieved by conditional expression of the Mos1 transposase. We previously demonstrated that Mos1 mobilization is mutagenic; however, Mos1 insertion alleles were initially recovered at a frequency ∼100 times lower than that observed with chemical mutagens, a rate too low to be useful for difficult forward screens. Therefore, to optimize transposition and isolation of mutants, we characterized factors that affect Mos1 mobilization.
Mos1 mobilization requires two components:
Like all Tc1/mariner elements, Mos1 transposes by a conserved cut and paste mechanism (van Luenen et al. 1994). It contains a single gene encoding a transposase that is flanked by short terminal inverted repeats. The transposase enzyme binds to the inverted repeats and catalyzes the excision of an element from the genome and subsequent insertion at a new genomic location. In C. elegans the mobilization of Mos1 relies on two extrachromosomal arrays; one expresses the transposase enzyme and the other carries the substrate transposon. The enzyme array {oxEx166[Phsp:Mos1Transposase; Punc-122:GFP; lin-15(+)]} contains the coding region of the Mos1 transposase under the control of a heat-shock promoter. The substrate array {oxEx229[Mos1; Pmyo-2:GFP]} contains multiple copies of the Mos1 transposon. These two arrays were generated independently (Bessereau et al. 2001) and are maintained in separate strains. To mobilize Mos1, males carrying the enzyme array were crossed to hermaphrodites with the substrate array to generate double transgenic hermaphrodites that contain both arrays. Double transgenic hermaphrodites were subjected to heat shock to activate the heat-shock promoter and thereby express the transposase enzyme. In turn, the transposase enzyme catalyzes the transposition of a Mos1 element from the substrate array into a chromosomal location.
Time course of transposition:
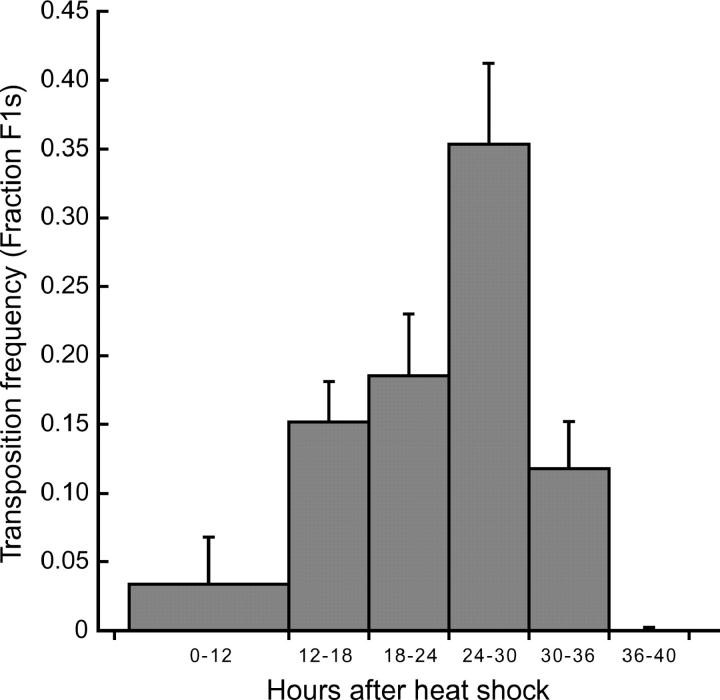
Initial experiments indicated that Mos1 mutagenesis was very inefficient (Bessereau et al. 2001). One possible source of this inefficiency is that we were scoring samples of animals in which transposition never took place. As a first step toward identifying the temporal and spatial conditions under which transposition occurs, we analyzed the time course of Mos1 transposition (Figure 1). Transposition was induced in young adults because heat-shocking L4 larvae caused most animals to die at the L4-adult molt and many of the surviving P0 animals were sterile. The progeny of heat-shocked animals were collected for 6-hr intervals. Heat shock caused the P0 animals to be paralyzed and they laid very few progeny in the first 12 hr after heat-shock treatment. It is likely that somatic transposition has an adverse effect on P0 animals since worms without the arrays were less affected by heat shock than were animals with the arrays. Within each time interval, the transposition frequency was measured by determining the percentage of F1 animals that contain at least one Mos1 insertion using PCR with _Mos1_-specific primers. Transposition was detected in F1 animals laid 12–18 hr after heat shock and peak transposition frequency was observed in animals laid 24–30 hr after heat shock. Collecting F1 animals that display the highest transposition frequency should result in obtaining a sample of F1's with the highest percentage of Mos1 insertions. Therefore, in all following experiments, F1 animals were collected 20–34 hr after heat shock.
Figure 1.—
Transposition time course. F1 animals were collected during the indicated intervals after heat shock (1 hr at 33°, 1 hr at 20°, and 1 hr at 33°). Within each interval, the transposition frequency was determined by assaying the percentage of F1 animals that contain at least one Mos1 insertion (minimum 20 F1's assayed). For F1 progeny that have lost the substrate array, PCR was conducted directly on individual F1 animals with _Mos1_-specific primers as described before (Bessereau et al. 2001). Transposition cannot be assayed directly in F1 animals that carry the substrate array because copies of the transposon remain in the array. Mos1 insertions in these animals were detected by performing PCR on pools of at least 5 F2 animals that did not carry the substrate array. Values are the average transposition frequency from three independent P0s and error bars represent standard error of the mean.
The increase in transposition observed over the first 24 hr could reflect increased transposase translation with time. However, the differentiation state of the germ cells also seems to play a profound role. First, Mos1 transposition does not occur in mature sperm. Heat shock was performed in young adult animals after spermatogenesis was complete (L'Hernault 1997). If transposition occurred in these mature sperm, then a basal rate of insertions should have been observed, including the early and late collection periods. But transposition was not observed 36 hr after heat shock (Figure 1), indicating that transposition does not occur in mature sperm cells. Second, transposition in oocytes seems to be limited to early meiotic nuclei. The gonad of adult hermaphrodites contains oocytes at different developmental stages: in the proximal arm of the gonad in an adult hermaphrodite, oocytes are arrested in meiosis at diakinesis of meiotic prophase I; above the reflex of the gonad in the distal arm nuclei are arrested in pachytene of meiosis I; and nuclei in the distal regions of the gonad are still dividing mitotically (Schedl 1997). Almost no insertions were detected in F1 animals laid during the first 12 hr after heat shock. The F1's laid within this time interval were probably fertilized zygotes or oocytes in diakinesis at the time of heat shock. Peak transposition rates were observed in animals laid 24–30 hr after heat shock, when nuclei were likely in pachytene arrest or late stages of mitosis.
Transposition is limited to those germ-cell nuclei that carry the substrate array:
Extrachromosomal arrays are unstable during meiosis (Mello et al. 1991). Therefore, the components required for transposition of Mos1 are not present in all germ-cell nuclei. To determine the genetic requirements for Mos1 transposition in a single germ-cell nucleus, F1 worms were sorted depending on whether they carried the substrate array, the enzyme array, both arrays, or neither array. Then the transposition frequency within each class was determined by assaying the percentage of F1 animals with at least one Mos1 insertion (Table 1). The transposition frequency in animals that lack the enzyme array is identical to the frequency in animals that contain the enzyme array (29 vs. 31%, P > 0.50). Since germ-cell nuclei share a common cytoplasm, these data suggest that the transposase enzyme can diffuse throughout the gonad syncytium and catalyze transposition in nuclei that do not contain the enzyme array. By contrast, the observed transposition frequency was lower in F1 animals without the substrate array when compared to F1's that carry the substrate array (45 vs. 19%, P < 0.001). These data indicate that the presence of the substrate array is required in a nucleus for transposition and suggest that after Mos1 elements have been excised from the array, the transposons do not diffuse out of the nuclei.
TABLE 1.
Transposition frequency within different classes of F1 animals
| Substrate array (+) | Substrate array (−) | Total | |
|---|---|---|---|
| Enzyme array (+) | 70/137 (0.51) | 44/229 (0.19) | 114/366 (0.31) |
| Enzyme array (−) | 59/152 (0.39) | 26/141 (0.18) | 85/293 (0.29) |
| Total | 129/286 (0.45) | 70/370 (0.19) | 199/659 (0.30) |
We detected some Mos1 inserts in F1 animals that did not carry the substrate array. Because the transposon is unable to diffuse into cells that lack the substrate array, these events must have occurred prior to loss of the substrate array. In addition, the transposition reaction itself may have a destabilizing effect on the meiotic segregation of the substrate array. To test this, we assayed array stability by determining the percentage of F1 animals that carried the substrate or enzyme extrachromosomal arrays after heat shock or in the absence of heat shock. We observed that heat-shock treatment of double transgenic animals decreased the stability of the substrate array [heat shock (+) 138/282 (0.49) vs. heat shock (−) 377/526 (0.72), P < 0.001]. Importantly, heat shock did not affect the stability of the enzyme array [heat shock (+) 149/282 (0.53) vs. heat shock (−) 282/526 (0.54)]. Together these results indicate that transposition affects the meiotic segregation of the substrate array, but does not affect the enzyme array. This suggests that double-stranded DNA breaks due to excision of individual elements have a destabilizing affect on the segregation of the substrate array.
The substrate array can be silenced:
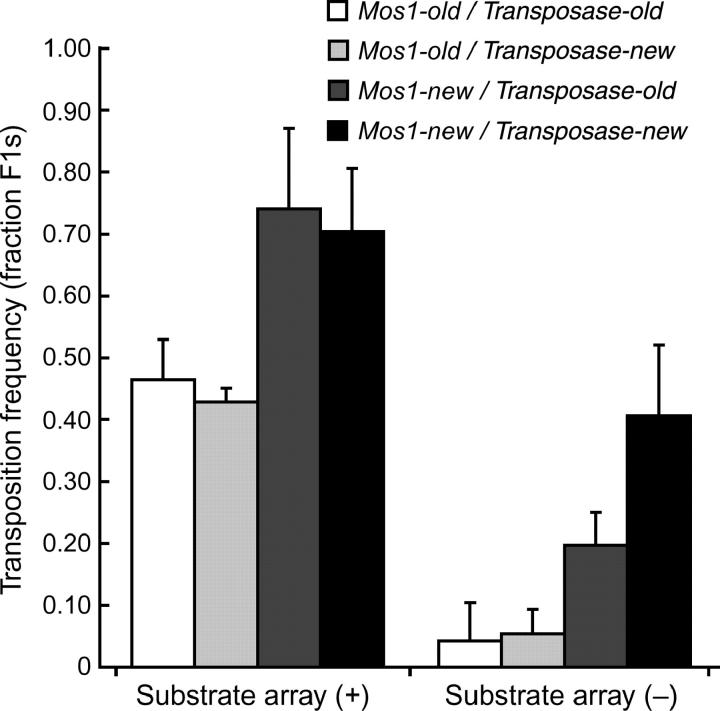
We observed a decline in the transposition frequency over a time scale of a year while using the same transgenic strains. Silencing of transposition occurred in the absence of active transposition because the substrate and enzyme arrays were propagated in separate strains. We examined whether transposition silencing was due to modification of either array (Figure 2). The transposition frequency was determined from transgenic strains that were cultivated for more than a year (Transposase-old and Mos1-old) or strains freshly thawed from liquid nitrogen storage (Transposase-new and Mos1-new). The transposition frequency was identical regardless of whether an old or new enzyme array was used. By contrast, the transposition frequency was significantly higher when Mos1-new was used compared to transposition from Mos1-old. Together, these results indicate that the substrate array becomes inactive over time. Silenced transgenes were shown to adopt a heterochromatic structure (Kelly et al. 2002), which in our experiments might prevent access of the transposase to the Mos1 copies in the substrate array. Alternatively, dsRNA from the transposase open reading frame contained in the Mos1 transposon could accumulate in the substrate strain and these dsRNA molecules could block transposase expression by RNA interference when these arrays are crossed together (Sijen and Plasterk 2003).
Figure 2.—
The substrate array can be silenced. Double transgenic animals were generated either from arrays that had been propagated in strains maintained at 20° for ∼1 year (-old) or from arrays that were from freshly thawed strains (-new). Young adults were heat-shocked to activate transposition. Transposition frequencies were determined in F1 animals carrying the substrate array (left) or in F1 animals in which the substrate array had been lost (right). Results are presented as the average of four independent experiments, except for the Transposase-new/Mos1-old category, which was measured in only two experiments. Error bars represent the standard error of the mean. Transposition frequencies are significantly higher using Mos1-new compared to Mos1-old (P = 0.0002, two-way ANOVA), while comparing Transposase-new and Transposase-old arrays did not reveal any significant difference (P = 0.13).
Multiple insertions occur in an individual germ cell:
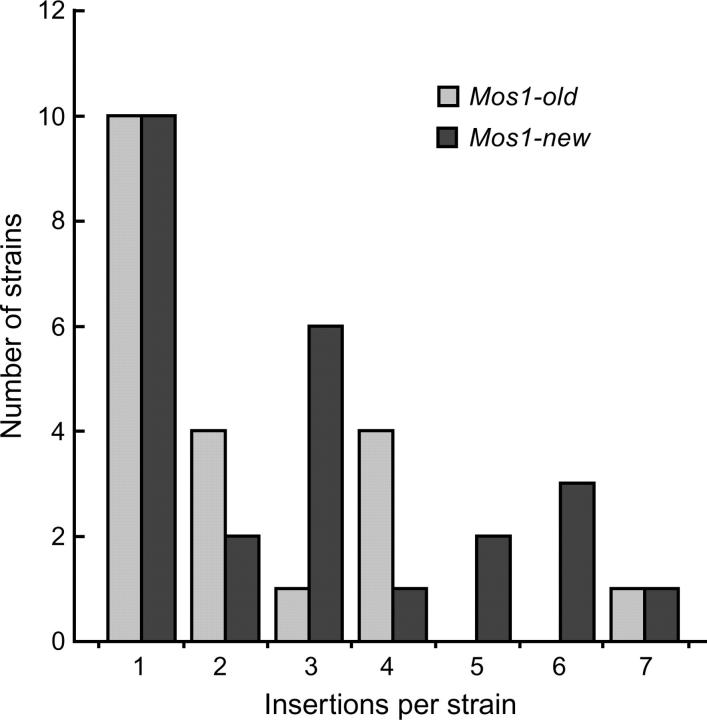
To determine the insertion frequency per F1 we performed Southern blot analysis using a Mos1 probe on strains in which transposition had occurred (Figure 3). The average number of insertions per strain was 2.5 (n = 45 strains). A majority of the strains examined contained one insertion and the highest number of insertions that occurred in a single germ cell was seven. The relative proportion of strains containing more than one insertion did not follow a Poisson distribution (P < 0.001). These data indicate that some nuclei possess a permissive environment for transposition. In addition, silencing of the substrate array was not a factor influencing the number of insertions per strain since the average insertion frequency was similar between Mos1-new (2.84, n = 25 lines), and Mos1-old (2.22, n = 20 lines), indicating that silencing is an “all or none” phenomenon that decreases the ability of the entire substrate array to contribute donor elements.
Figure 3.—
Multiple insertions occur in individual germ cells. Southern blot analysis was performed on lines containing Mos1 inserts obtained from substrate arrays propagated in strains maintained for 1 year at 20° (Mos1-old) or from substrate arrays derived from freshly thawed strains (Mos1-new). The number of strains that contain the indicated number of insertions is shown for each array. The distribution of the strains among the different classes does not fit a Poisson distribution (best fit to a Poisson distribution was calculated with Excel Solver after exclusion of the 0 class; by considering the 5 classes containing one to more than four insertions, Poisson distribution was rejected with P < 0.001).
Mutagenicity of Mos1:
To measure the effectiveness of Mos1, we compared the mutagenicity of Mos1 to the chemical mutagen EMS. We selected for mutants that were resistant to the anthelmintic drug levamisole. Six loci can be mutated to confer strong levamisole resistance and uncoordinated behavior and, in three other loci, mutations generate weakly resistant worms (Lewis et al. 1980a,b). A total of 6 levamisole-resistant mutants were isolated from 13,940 _Mos1_-mutagenized F1 animals (Table 2). In a parallel screen, 8 mutants were identified from the progeny of 1710 EMS-mutagenized F1's. Therefore, the mutagenic frequency of EMS was 2.34 × 10−3 mutations per haploid genome, which is consistent with the known mutagenic rate of EMS and the target size for levamisole resistance (Brenner 1974; Lewis et al. 1980a). Assuming that Mos1 hops occur only in the maternal germline (see above), then the derived mutagenicity of Mos1 is 4.30 × 10−4 mutations per haploid genome. Altogether, these results indicate that Mos1 is 5-fold less mutagenic than EMS; however, at a practical level this means that 10-fold more genomes must be screened compared to chemical mutagenesis.
TABLE 2.
Mos1 insertion alleles
| Screen | Transpositionfrequency | No. of F1's | Mutants | Insertion site |
|---|---|---|---|---|
| 1 | 18/31 (0.58) | 2,300 | lev-1(kr6) | Exon 3 |
| unc-63(kr13) | Splice donor | |||
| 2 | 16/23 (0.70) | 5,700 | lev-1(kr20) | Exon 3 |
| unc-63(kr19) | Exon 7 | |||
| 3 | 11/30 (0.37) | 5,900 | lev-1(kr25) | Promoter |
| lev-10(kr26) | Exon 7 | |||
| Total | 45/84 (0.53) | 13,900 | Six mutants (frequency = 4.32 × 10−4) | |
| EMS | 1,710 | Eight mutants (frequency = 4.68 × 10−3) |
The mutagenicity of the Mos1 system is in the same range as the mutagenicity obtained by mobilizing Tc elements in mutator strains (for review, see Anderson 1995), although a precise comparison is difficult since Tc mutagenesis can be highly variable. For example, mutations in the unc-22 gene were recovered in a mut-2 background at about the same frequency as with EMS (Collins et al. 1987). However, in a mut-7 strain, isolation of unc-93(e1500) suppressors or unc-22 mutants was ∼10 or 100 times less efficient than using EMS, respectively (R. Ketting and R. Plasterk, personal communication). The advantage of the Mos1 system as compared to Tc elements is the absence of endogenous Mos1 elements in the C. elegans genome, which greatly facilitates the identification of mutagenic insertions. For example, we were able to rapidly clone all the mutated genes in the levamisole screen. Although multiple insertions were present in _Mos1_-mutagenized levamisole-resistant mutants (data not shown), a single relevant insertion was easily identified after rough mapping to a chromosome or serial outcrossing. From the levamisole-resistance screens, we isolated three alleles of lev-1 and two alleles of unc-63, which encode acetylcholine receptor subunits (Fleming et al. 1997; Culetto et al. 2004), and one allele of lev-10, which codes for a transmembrane protein required for clustering acetylcholine receptors at neuromuscular junctions (Gally et al. 2004).
Mos1 can generate non-null mutations:
To determine the phenotypic consequences of Mos1 insertions, we examined the genetic nature of Mos1 insertion alleles. Most of the Mos1 insertion alleles are recessive loss-of-function mutations and are likely to represent the null phenotype. For example, unc-63(kr13) is a strong allele generated by an insertion in the splice donor site in the third intron of unc-63. lev-1(kr6) and lev-1(kr20) are strong loss-of-function mutations in which Mos1 is inserted in exon 3. These three mutations are phenotypically identical to null alleles. However, hypomorphic alleles were also isolated. lev-1(kr25) is a weak mutation caused by an insertion in the promoter region of the gene. This insertion is likely to disrupt regulatory regions of lev-1, resulting in lower levels of protein rather than a complete elimination of lev-1 expression. unc-63(kr19) is a semidominant allele caused by an insertion in exon 7 between transmembrane domains 3 and 4. It is likely that unc-63(kr19) causes an aberrant protein product that eliminates function of the levamisole-sensitive receptor even in the heterozygote.
Mobilization of transposons can generate mutations that are not caused by insertion of the transposon (Collins et al. 1987; Bessereau et al. 2001). These mutations are thought to arise by the chromosomal insertion and subsequent imprecise excision of an element and thus have been called “hit-and-run” mutations. All of the mutations in our levamisole-resistance screen still contained the Mos1 element. Thus, to examine the frequency of hit-and-run events, we determined whether all mutants with visible phenotypes isolated in our screens were caused by an insertion. Among 20 mutants isolated (data not shown), 3 did not have a Mos1 insertion linked to the mutant phenotype, corresponding to an apparent 15% rate of hit-and-run mutations. We identified the mutated genes by positional mapping and genetic complementation and we determined the molecular lesion of these three mutations. Two are single-base-pair mutations: bli-1(ox283) contains a G-to-A transition (tatttcagG→ATTTCCGTGC; lowercase indicates intron sequence) resulting in a glycine to aspartic acid residue change, and unc-13(e2914) contains a G-to-T transversion (caattttag→tGCCATGACT) that disrupts an intron splice acceptor site. These mutations are unlikely to be caused by a Mos1 hit-and-run event since neither mutation contains a Mos1 reexcision footprint. The third allele is a complex rearrangement of pha-4 that consists of an ∼5-kb duplication and 150-bp deletion (D. Updike and S. Mango, personal communication). This mutation could have been generated by DNA double-strand-break repair caused by Mos1 reexcision. Alternatively, the Mos transposase might be able to introduce nonspecific chromosomal breaks at low frequency. All six of the mutations isolated from the levamisole-resistance screen were due to a Mos1 insertion, while other screens have largely produced hit-and-run alleles (S. Mango, personal communication). This suggests that the screening methodology or the identity of the target genes can influence the likelihood of isolating insertion mutations.
Practical aspects of _Mos1-_mediated mutagenesis:
We have characterized transposition of Mos1 in the germline of C. elegans and identified methods to obtain F1 animals likely to contain insertions. First, peak transposition frequencies are seen in F1 animals laid 24–30 hr after heat shock. Therefore, F1 animals used in forward genetic screens should be collected during this time interval. Second, we observed silencing of the transposon array during maintenance of the strain. Thus, the transposition frequency should be assayed prior to screening to ensure adequate transposition frequencies. A rapid assay can be performed using PCR with _Mos1_-specific primers on a small sample of the progeny of heat-shocked animals. Finally, the highest transposition frequency was observed in F1 animals that carry the substrate array. Since this array is tagged with a pharyngeal GFP transgene, a fluorescent worm sorter could be used to obtain a large population of F1 animals with the highest probability of having a Mos1 insert. Ideally, using a Mos1 element carrying a reporter gene for enhancer and gene trapping would enable visual detection of transposition events close to or within genes. Unfortunately, we previously demonstrated that disruption of the Mos1 sequence with large insertions dramatically reduced the transposition frequency (Bessereau et al. 2001). This restriction is consistent with observations in Drosophila in which composite Mos1 elements tagged with reporter transgenes transposed very inefficiently (Lohe and Hartl 2002; Lozovsky et al. 2002).
Although the efficiency of Mos1 is lower than that of chemical mutagens, the unique sequence tag of Mos1 insertion alleles allows for rapid identification of the mutated gene. Some mutant strains contained multiple copies of Mos1 that interfered with identification of the relevant insertion by inverse PCR. In these cases, it is necessary to show linkage between a specific insertion and the mutant phenotype. A genomic insertion of Mos1 represents a polymorphism that is easy to identify and follow by single-worm PCR using primers that flank the insertion site. In this manner, even in strains that contain multiple insertions, the relevant insertion can be identified after a single outcross. Therefore, _Mos1_-mediated mutagenesis alleviates the need for traditional genetic mapping and provides a valuable tool for C. elegans genetics.
Acknowledgments
We thank S. Mango and D. Updike for communicating unpublished results, I. Nuez and H. Gendrot for technical support, M. W. Davis for isolation of bli-1(ox283), and K. Yook for isolation of unc-13(e2914). This work was supported by the Institut National de la Santé et de la Recherche Médicale (J-L.B.; ITM 2000-2002) and the National Science Foundation (E.J.). A-F.R. was supported by a fellowship from the Ministère de la Recherche. D.C.W. was supported by a fellowship from the National Institutes of Health.
References
- Anderson, P., 1995. Mutagenesis. Methods Cell Biol. 48**:** 31–58. [PubMed] [Google Scholar]
- Bessereau, J. L., A. Wright, D. C. Williams, K. Schuske, M. W. Davis et al., 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413**:** 70–74. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77**:** 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282**:** 2012–2018. [DOI] [PubMed] [Google Scholar]
- Collins, J., B. Saari and P. Anderson, 1987. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature 328**:** 726–728. [DOI] [PubMed] [Google Scholar]
- Culetto, E., H. A. Baylis, J. E. Richmond, A. K. Jones, J. T. Fleming et al., 2004. The caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor alpha subunit. J. Biol. Chem. 279**:** 42476–42483. [DOI] [PubMed] [Google Scholar]
- Fleming, J. T., M. D. Squire, T. M. Barnes, C. Tornoe, K. Matsuda et al., 1997. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. 17**:** 5843–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally, C., S. Eimer, J. E. Richmond and J. L. Bessereau, 2004. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature 431**:** 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., and T. Doniach, 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146**:** 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, J. W., M. M. Medhora and D. L. Hartl, 1986. Molecular structure of a somatically unstable transposable element in Drosophila. Proc. Natl. Acad. Sci. USA 83**:** 8684–8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, E. M., and S. E. Mango, 2002. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3**:** 356–369. [DOI] [PubMed] [Google Scholar]
- Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim et al., 2002. X-chromosome silencing in the germline of C. elegans. Development 129**:** 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault, S. W., 1997 Spermatogenesis, pp. 271–294 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Lewis, J. A., C. H. Wu, H. Berg and J. H. Levine, 1980. a The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95**:** 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J. A., C. H. Wu, J. H. Levine and H. Berg, 1980. b Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience 5**:** 967–989. [DOI] [PubMed] [Google Scholar]
- Lohe, A. R., and D. L. Hartl, 2002. Efficient mobilization of mariner in vivo requires multiple internal sequences. Genetics 160**:** 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovsky, E. R., D. Nurminsky, E. A. Wimmer and D. L. Hartl, 2002. Unexpected stability of mariner transgenes in Drosophila. Genetics 160**:** 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10**:** 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman, D. G., G. M. Benian and R. H. Waterston, 1986. Molecular cloning of the muscle gene unc-22 in Caenorhabditis elegans by Tc1 transposon tagging. Proc. Natl. Acad. Sci. USA 83**:** 2579–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E. C., and H. R. Horvitz, 1986. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics 113**:** 821–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R. H., and H. G. van Luenen, 1997 Transposons, pp. 97–116 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Schedl, T., 1997 Developmental genetics of the germ line, pp. 241–269 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Sijen, T., and R. H. Plasterk, 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426**:** 310–314. [DOI] [PubMed] [Google Scholar]
- van Luenen, H. G., S. D. Colloms and R. H. Plasterk, 1994. The mechanism of transposition of Tc3 in C. elegans. Cell 79**:** 293–301. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28**:** 160–164. [DOI] [PubMed] [Google Scholar]