Gene expression changes in foam cells and the role of chemokine receptor CCR7 during atherosclerosis regression in ApoE-deficient mice (original) (raw)
Abstract
Atherosclerosis regression is an important clinical goal. In previous studies of regression in mice, the rapid loss of plaque foam cells was explained by emigration to lymph nodes, a process reminiscent of dendritic cells. In the present study, plaque-containing arterial segments from apoE−/− mice were transplanted into WT recipient normolipidemic mice or apoE−/− mice. Three days after transplant, in the WT regression environment, plaque size decreased by ≈40%, and foam cell content by ≈75%. In contrast, both parameters increased in apoE−/− recipients. Foam cells were isolated by laser capture microdissection. In WT recipients, there were 3- to 6-fold increases in foam cells of mRNA for liver X receptor α and cholesterol efflux factors ABCA1 and SR-BI. Although liver X receptor α was induced, there was no detectable expression of its putative activator, peroxisome proliferator-activated receptor γ. Expression levels of VCAM or MCP-1 were reduced to 25% of levels in pretransplant or apoE−/− recipient samples, but there was induction at the mRNA and protein levels of chemokine receptor CCR7, an essential factor for dendritic cell migration. Remarkably, when CCR7 function was abrogated in vivo by treatment of WT recipients with antibodies to CCR7 ligands CCL19 and CCL21, lesion size and foam cell content were substantially preserved. In summary, in foam cells during atherosclerosis regression, there is induction of CCR7 and a requirement for its function. Taken with the other gene expression data, these results in vivo point to complex relationships among the immune system, nuclear hormone receptors, and inflammation during regression.
Keywords: cholesterol efflux, dendritic cell, macrophage, monocyte, nuclear hormone receptor
Mouse models of atherosclerosis regression are relatively few. However, similarities between atherosclerosis in humans and in mice deficient in apolipoprotein E (apoE−/−) or the LDL receptor (e.g., see ref. 1) suggest that molecular mechanisms underlying regression in mouse models could be relevant to the reduction of the large plaque burden in the middle-aged and older human population.
We have previously described a mouse model of regression. First, plaques were allowed to develop in apoE−/− mice, then a segment of either thoracic aortic (2) or aortic arch (3, 4) was transplanted into the abdominal aorta of a WT recipient, quickly changing the plasma lipid environment from hyper to normolipidemia. As a control, an aortic segment was transplanted into an apoE−/− mouse. By 1 month, in the regression environment (WT recipient), essentially all of the foam cells disappeared from plaques (4), with many no longer visible after 3 days (5). In contrast, in the progression environment (apoE−/− recipient), plaque size and foam cell content increased over time.
In a recent study, we showed that the rapid depletion of foam cells in the regression environment was correlated with a substantial number of these cells emigrating to lymph nodes (5). Interestingly, the emigrating cells expressed markers of dendritic cells (DCs), which, like macrophages, can derive from monocytes (6). DCs acquire and process antigens in peripheral tissues and then migrate to draining lymph nodes (7). This migration depends on the chemokine receptor CCR7 (8–10), which becomes up-regulated as the cells mature in response to activation stimuli encountered in the periphery (11–13). Because monocytes differentiate into either macrophages that remain resident in tissues or lymph node-homing DCs after transmigration from the circulation (6, 14), our recent study (5) raised the intriguing possibility that, during regression, macrophage foam cells acquire DC characteristics that permit them to emigrate to lymph nodes. Thus, the expression and functional activity of CCR7 in foam cells would be plausible in plaques undergoing regression.
Besides seeking evidence to support this possibility, we were also interested in other molecular changes in the foam cells in the regression environment, in which conditions would be expected to favor cholesterol efflux, in that high non-HDL (high-density lipoprotein), low HDL plasma levels are changed to low non-HDL, high HDL levels. By current understanding (e.g., refs. 15–18), cholesterol efflux pathways involve a variety of factors, including the nuclear orphan receptors peroxisome proliferator-activated receptor γ (PPARγ) and liver X receptors (LXRs), as well as the proteins ABCA1 and SR-BI, so we examined the expression of their corresponding mRNAs under regression and progression conditions. Because cholesterol loading of macrophages is thought to provoke inflammatory changes (e.g., refs. 18–20), we also reasoned that in a regression environment, targets of the NF-κB pathway (such as VCAM-1 and MCP-1) may become down-regulated.
For our studies, we applied a number of approaches, including (i) laser capture microdissection (LCM) to isolate foam cells from plaques in the different environments; (ii) real-time (quantitative)-PCR analyses of gene expression in the LCM-selected cells; and, (iii) blockade of CCR7 function in vivo by injection of WT recipients with antibodies to its two critical ligands, CCL19 and CCL21. Overall, the data indicate that in the regression environment, in foam cells there is the stimulation of cholesterol efflux-related genes independent of PPARγ and the suppression of the inflammatory state. Remarkably, the data also revealed that in plaques transplanted into WT recipients, CCR7 is induced in foam cells and is functionally required for regression.
Results
Changes in Plasma Lipid Levels After Transplantation.
ApoE−/− donor mice were fed Western-type diet (WD) for 20 weeks. Transplantation into a WT recipient mouse on a chow diet dramatically changed the lipid environment to which the plaques were exposed (Table 1), with a reduction by a factor of 10 in plasma TC (P < 0.0001), and a 2-fold increase in plasma HDL-C (P < 0.0001). The apoE−/− recipient mice on chow diet remained dyslipidemic. Although Table 1 shows recipient data 3 days after transplant, there were no significant changes throughout the study (data not shown).
Table 1.
Mouse HDL and plasma cholesterol levels
| HDL-C, mg/dl | Plasma total cholesterol, mg/dl | |
|---|---|---|
| ApoE−/− donors (n = 43) | 28 ± 2* | 1,109 ± 186 |
| ApoE−/− recipients (n = 11) | 26.3 ± 4.2* | 543 ± 36 |
| Wild-type recipients (n = 10) | 63.8 ± 11.1 | 102 ± 10 |
Correction of Dyslipidemia Decreases Lesion Size and the Content of Foam Cells.
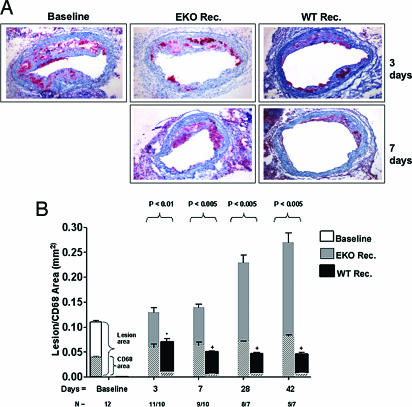
To determine the effects of a sustained correction of dyslipidemia, recipient animals were killed at 3, 7, 28, and 42 days after transplantation. Immunostaining for CD68+ cells (presumably macrophage foam cells) was performed on serial sections from the graft (Methods). Representative data and a graphical summary for all time points are shown in Fig. 1.
Fig. 1.
Reversal of dyslipidemia decreases plaque size and foam cell content. (A) Representative sections of aortic transplant segments from baseline (apoE−/−), and WT and apoE−/− recipients at 3 and 7 days after transplantation. Sections were immunostained with anti-CD68 antibody (red). (B) Quantification of intimal (outset bars) and foam cell areas (inset bars). N represents the number of animals in each group.
At 3 days, consistent with our previous study (5), the lesion (i.e., intimal) area of animals in the WT recipient group (0.07 ± 0.006 mm_2_) was already significantly decreased compared to the baseline (pretransplant) group (0.11 ± 0.01 mm_2_, P < 0.05). This change was largely attributable to a decrease in the plaque content of foam cells (area of CD68+ staining: 0.009 ± 0.001 in WT recipients vs. 0.04 ± 0.005 mm_2_ at baseline, P < 0.001). After 3 days, further decreases in lesion size were observed at 7, 28, and 42 days, although at a slower rate. In contrast to WT recipients, in apoE−/− recipients plaques continued to progress, with notable size increases observed at 28 and 42 days.
The Expression of the Chemokine Receptor CCR7 Is Up-Regulated in Foam Cells After Correction of Dyslipidemia.
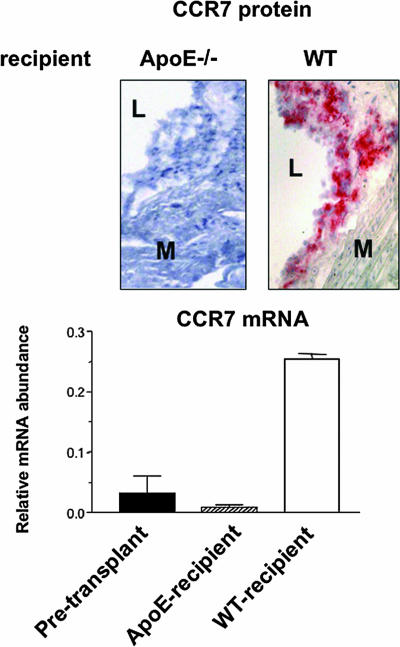
We previously demonstrated that depletion of CD68+ foam cells from the plaques 3 days after transplantation into the WT recipient was associated with the emigration of monocyte-derived cells from the grafts to either regional lymph nodes or the systemic circulation (5). Because this type of migratory behavior is typical for mature DCs and requires the chemokine receptor CCR7 (8), we measured, in laser-captured CD68+ cells, the expression of CCR7 at the mRNA and, in tissue, the protein levels before and 3 days after transplantation.
As shown in Fig. 2, in the cells selected from plaques from either apoE−/− donor or recipient mice, there was a low level of CCR7 mRNA expression and no detectable protein (although in the corresponding spleens, both CCR7 mRNA and protein were readily detectable; data not shown). In contrast, there was a ≈5-fold relative increase in CCR7 mRNA abundance in cells from plaques transferred to WT mice. Notably, this increase at the mRNA level was accompanied by strong staining for CCR7 protein (Fig. 2). Taken with our prior findings that emigrating cells had properties of both macrophages and immature dendritic cells (5), the CCR7 results raise the possibility that the emigration that rapidly follows the normalization of the plasma lipoprotein profile represents either the full maturation of immature lipid-laden dendritic cells or the acquisition of mature dendritic-like properties by macrophage–foam cells.
Fig. 2.
Up-regulation of the chemokine receptor CCR7 in foam cells in the regression environment. Sections were prepared from the aortic arches of baseline apoE−/− mice or from the grafts 3 days after transplantation into WT or apoE−/− recipients. (Upper) Staining results using the CCL19-Fc chimeric protein are shown. “L” and “M” indicate lumen and media. In an adjacent section, the CCR7+ area corresponded to staining for CD68 (data not shown). Results from baseline plaques were similar to those in the apoE−/− recipient (data not shown). (Lower) CCR7 and cyclophilin A mRNA levels were measured by QRT-PCR in laser captured CD68+ cells. The levels were then converted to final values based on standard curves for these RNAs in total RNA isolated from spleen. Shown are the mean ± SEM of the CCR7/cyclophilin A ratios from two pools of RNA, each pool consisting of three independent mice.
Regression Is Inhibited by Treatment with Antibodies to the CCR7 Ligands CCL19 and -21.
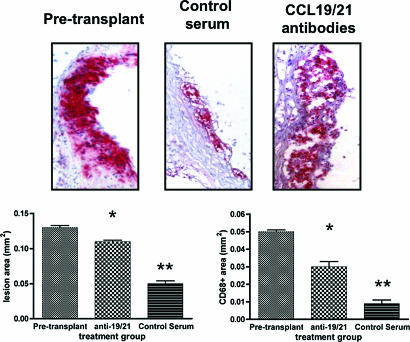
Although the above results suggest a functional role for CCR7 in foam cell depletion, we were interested in directly demonstrating this. Thus, based on a similar approach to study DC migration in skin (21), CD45.1+ WT recipients were treated with either antibodies to ligands for CCR7 (chemokines CCL19 and CCL21), or preimmune serum, shortly before and after transplantation of arches from CD45.2+ apoE−/− mice.
As shown in Fig. 3 Upper, in representative histologic sections from the three groups, there are abundant CD68+ cells in the pretransplant sample. The CD68+ cell content is obviously less in the control serum treatment group, consistent with Fig. 1 and ref. 5, whereas CD68+ cell content after treatment with the CCR7 ligand antibodies is not noticeably different from that at baseline. The group averages for lesion size and the contents of CD68+ cells (Fig. 3 Lower) support the visual impressions, namely that the dramatic reductions in lesion and CD68+ areas in the control serum group (to 38% and 18%, respectively, of pretransplant values) were substantially blunted (to 85% and 60%, respectively) by treatment with the anti-CCL19 and CCL21 antibodies.
Fig. 3.
Functional role of CCR7 in atherosclerosis regression. Donor apoE−/− mice were placed on the WD and, at 20 weeks of age, the aortic arches were taken either for baseline data (labeled as “Pretransplant”; n = 5 mice) or transplantation into WT recipients. Half of the recipient mice were injected ≈2 h before surgery and shortly after with goat anti-CCL-19 and anti-CCL-21 (labeled as “CCL19/21 antibodies” in Upper and “anti-19/21” in the graphs, n = 11), the other half with preimmune (“Control serum,” n = 7) goat IgG. At 3 days after transplant, grafts were harvested and sectioned for morphometric and immunostaining analyses. (Upper) Representative CD68-immunostained sections from each group. (Lower) Graphical summaries for the groups (means ± SEM). Statistical analysis was by ANOVA and the Bonferroni test of multiple comparisons. ∗, P < 0.001 vs. pretransplant; ∗∗, P < 0.001 vs. anti-19/21.
In a subset of mice (n = 3–5 for each group), lymph node analyses of the content of CD45.2 (donor) cells doubly positive for CD68 and either the DC marker CD11c or MHC-II showed results consistent with the lesional data. In the anti-CCL19/21 group, in which foam cells were retained in plaques, there tended to be fewer (≈50%; P > 0.05) such cells in either the iliac lymph node (representing local drainage) or hepatic lymph node (representing cells that entered the blood) than in the control serum group.
Overall, the results confirm that plaque foam cell content is inversely related to the emigration of foam cells (5), but, significantly, also reveal the functional importance of CCR7 in the regression process.
Effects of the Correction of Dyslipidemia on Cholesterol Efflux and Inflammatory Pathways.
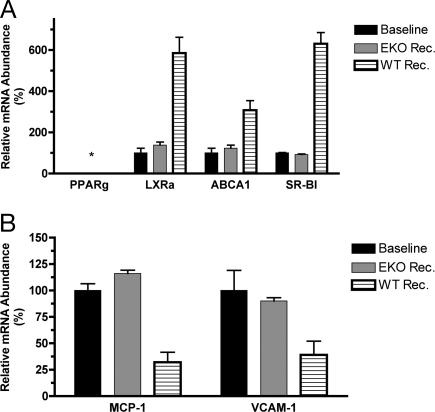
In the WT recipient, plaques become rapidly exposed to a plasma environment containing a relative increase in HDL and decrease in non-HDL cholesterol, changes which are expected to promote cholesterol efflux from foam cells and thereby reverse inflammatory changes associated with the lipid loading of macrophages (e.g., refs. 18–20). Thus, CD68+ cells were selected from the plaques of pretransplant and recipient mice, and the expression of genes related to cholesterol efflux (PPARγ, LXRα, ABCA1, and SR-BI) and inflammation (VCAM, MCP-1) was measured (Fig. 4).
Fig. 4.
Regulation of the expression of genes associated with cholesterol efflux and inflammation in foam cells after the reversal of dyslipidemia. Foam cells were laser captured from sections of aortic arches of baseline apoE−/− mice or of grafts 3 days after transplantation into WT or apoE−/− recipients. Total RNA was isolated, and the indicated mRNA levels were measured by QRT-PCR as described in Fig. 2, except that the final ratios for the baseline samples were set to 100%. Shown are the results for genes associated with cholesterol efflux (A) and inflammation (B). Data are based on two pools of RNA, each pool consisting of three independent mice, and are expressed as fold change over baseline (mean ± SEM).
Surprisingly, PPARγ was not detectable at either the mRNA or protein level in any of the samples, despite its ready detection in either the spleen or thioglycolate-elicited macrophages from the corresponding mice (data not shown). Previous studies (reviewed in refs. 17 and 18) have suggested a PPARγ–LXRα–ABCA1 regulatory cascade for cholesterol efflux. However, as also shown in Fig. 4A, despite the deficiency of PPARγ, the expression of LXRα, ABCA1, or SR-BI was increased in cells from the plaques placed in the WT recipient. In these same cells, there was decreased expression of the NF-κB-target inflammatory genes MCP-1 and VCAM-1 (Fig. 4B).
In Vitro Studies of the Role of PPARγ in Cholesterol Efflux.
PPARγ is thought to promote cholesterol efflux through LXR-dependent and -independent pathways (22). The lack of PPARγ expression raised the question of its requirement for cholesterol efflux from foam cells. We investigated this question in elicited peritoneal macrophages, in which PPARγ is expressed at the mRNA and protein levels. We used two strategies. In the first, cholesterol-loaded peritoneal macrophages from apoE−/− mice were treated with the selective PPARγ antagonist GW9662 (23); in the second, we studied cholesterol-loaded peritoneal macrophages from PPARγ−/− mice. Cholesterol acceptors used were HDL (preferred by SR-BI or ABCG1) or lipid-poor apoA-I (preferred by ABCA1) (24–26).
In the first study, inhibition of PPARγ by co-treatment with GW9662 did not affect efflux to either apoA-I (≈5% at 18 h in either group) or HDL (≈12% at 18 h in either group; all data for the efflux studies are available in Fig. 5, which is published as supporting information on the PNAS web site). This could not be explained by differences in LXRα, ABCA1, or SR-BI gene expression between the control and treated cells; in addition, the net efflux of cellular cholesterol as measured by isotopic methods was confirmed by measurement of the unesterified and esterified cholesterol mass (data not shown).
These results suggested that PPARγ was not required for efflux from cholesterol-loaded macrophages via HDL or apoA-I-mediated pathways, consistent with a recent report (27). Further testing in cholesterol-loaded peritoneal macrophages elicited from PPARγ+/+ and PPARγ−/− mice confirmed that deficiency of PPARγ did not affect the efflux of cholesterol to either apoA-I (≈4% at 18 h in either group) or HDL (≈10% at 18 h in either group).
PPARγ Agonist Pioglitazone Increases Foam Cell Gene Expression of PPARγ, LXRα, and ABCA1 in Vivo.
The apparent lack of requirement of PPARγ for stimulation of cholesterol efflux-related genes in vivo or for cholesterol efflux in vitro is in contrast to the anti-atherosclerosis effects in mice of PPARγ agonists (e.g., refs. 17 and 18). Besides non-PPARγ-dependent actions of the agonists (28, 29), another possible explanation is that they induce PPARγ expression and increase the activity of the LXR-ABCA1 pathway. To test this, apoE−/− mice fed a WD for 20 weeks were then continued on either a WD, or a WD diet containing the PPARγ agonist pioglitazone (Pio), for an additional 2 weeks (a treatment duration shown to stimulate PPARγ gene expression in mouse tissues; ref. 30). Foam cells from the aortic root of the mice were laser captured and the RNA was analyzed. Again, there was no detectable PPARγ mRNA expression in the control group foam cells, but in mice fed WD and Pio, expression was now comparable to that in elicited macrophages (Fig. 6, which is published as supporting information on the PNAS web site). In addition, compared to results in the controls, pioglitazone treatment significantly (P < 0.005) increased in foam cells the levels of LXRα and ABCA1 mRNAs by 7- and 2.5-fold, respectively (Fig. 6).
Discussion
We previously demonstrated that rapid correction of the dyslipidemia of the apoE−/− mouse leads quickly to plaque regression (2, 4, 5), which could be largely explained by the emigration of CD68+ cells to regional lymph nodes and the systemic circulation (5). In the current report, the major findings are: (i) in CD68+ cells placed into the regression environment, the chemokine receptor required for DC migration, CCR7 mRNA, and protein were up-regulated; (ii) CCR7 function was required for the decreased foam cell content in regressing plaques; (iii) there was no expression in vivo of PPARγ in foam cells under progressive or regressive conditions; (iv) the deficiency of PPARγ did not impair cholesterol efflux in cholesterol-loaded peritoneal macrophages; and, (v) under conditions expected to promote reverse cholesterol transport, the efflux pathway-related factors LXRα, ABCA1, and SR-B1 were up-regulated, and inflammation-related factors VCAM-1 and MCP-1 were decreased at the mRNA levels in vivo.
Before obtaining our emigration data (5), we would not have expected the induction of CCR7 because of the prevailing view that bone marrow-derived monocytes entering the arterial wall terminally differentiate into macrophages (and subsequently become foam cells). Because emigration is a classic feature of DCs and absolutely requires CCR7 (8), after our previous report (5), we hypothesized that, despite having the conventional features of macrophages in pretransplant plaques, foam cells acquired properties of mature DCs in the regression environment. The increases in CCR7 mRNA and protein in the CD68+ cells only in the regression environment (Fig. 2) certainly confirm this hypothesis. The CCL19/21 antibody blocking experiments (Fig. 3) importantly extend these observations by demonstrating a major functional role for CCR7 in the regression process.
Obviously, more studies are needed to determine in the regression environment how many features of the mature DC state the foam cells assume and the specific factors that stimulate CCR7 expression. A number of inflammatory agents, microbial products, growth factors, and cytokines promote DC maturation and CCR7 induction (7), but the molecules that directly regulate CCR7 expression are unknown. One candidate is LXR based on its increased expression in regression (Fig. 4), and on preliminary studies in vitro that show LXR agonists induce CCR7 mRNA and protein (J.E.F., Y. Ma, I. Torra, M. Garabedian, E.A.F., unpublished data).
In addition to inducing factors, stimulation of migration could also result from derepression. In particular, PAF and PAF-like lipids inhibit DC migration (5, 31). With the relative increase in the regression environment in HDL (and PAF-AH; ref. 32), there could either be removal of inhibitory lipids by HDL or their enzymatic detoxification. The latter possibility is consistent with our recent report that skin DC migration was increased in apoE−/− mice by infusion of HDL-associated PAF-AH (31).
Turning to PPARγ, recent studies have reported that it promotes cellular cholesterol efflux in vitro and exerts antiatherosclerotic effects in vivo (reviewed in refs. 17 and 18). Thus, we were surprised not to find foam cell expression of PPARγ in plaques from any condition (baseline, progression, or regression).
Although we had assumed that PPARγ would be expressed in apoE−/− plaques, to our knowledge, PPARγ expression has been shown only in early lesions of LDL-R−/− mice (33) and in human plaques (34). In studies of PPARγ in vitro, its expression is highly variable and depends a number of factors, such as the cell line, or for peritoneal macrophages, whether they were elicited (e.g., refs. 34 and 35). Overall, the published data taken with the present results suggest that in apoE−/− mice, PPARγ expression may be transient during plaque progression, but is not required for regression. The increases in vivo of PPARγ, LXRα, and ABCA1 gene expression in apoE−/− foam cells from pioglitazone treatment, therefore, may contribute to the decreased atherosclerosis progression in PPARγ agonist-treated apoE−/− mice (17, 18), in that such treatment may sustain PPARγ expression and stimulate cholesterol efflux. Although its promoter is not known to have a peroxisome proliferator response element, the induction of PPARγ we observed is consistent with its increased expression in the muscle and liver in rats and mice treated with pioglitazone (30, 36), and may be due to indirect effects on PPARγ transcription or mRNA stability.
Other recent studies in vitro have suggested that PPARγ is a primary inducer of LXRα, which then activates expression of ABCA1, thereby enhancing cholesterol efflux (e.g., ref. 37). However, our findings of the lack of PPARγ expression and increased levels of LXRα, ABCA1, and SR-B1, suggest that, at least in foam cells of apoE−/− mice after 20 weeks of WD feeding, PPARγ was not required for the activation of reverse cholesterol pathways. This suggestion was supported by our studies in cholesterol-loaded peritoneal macrophages, and is consistent with a recent report (27), but in contrast to another study in which efflux to HDL was impaired from elicited PPARγ−/− mouse peritoneal macrophages (38). However, those cells were not cholesterol-loaded, making it difficult to judge the relevance to the foam cell state.
Although PPARγ may not be required for cholesterol efflux, PPARγ agonists have been shown to promote it (reviewed in ref. 17), consistent with the results noted above that pioglitazone induced LXRα, and ABCA1 expression in foam cells in vivo.
We were also surprised by the induction of LXRα and ABCA1 expression in the regression environment. Not only was PPARγ not required for this induction, but it also occurred under conditions expected to promote cholesterol efflux. By current understanding (reviewed in ref. 18), depletion of cholesterol, and by extension, potent oxysterol LXRα ligands, would be expected to decrease the expression of LXRα and its target, ABCA1. Under regression conditions, SR-BI mRNA also rose, but this would be consistent with its inverse regulation by macrophage sterol content, a phenomenon that does not appear to include LXRα or -β (39).
There are a number of possible explanations for the increase in LXRα and ABCA1 gene expression in vivo. The foam cells may have accumulated such a high level of sterols that toxic effects impaired the activity of the LXRα promoter, and efflux relieved the toxicity. Another possibility is that the foam cells, in the process of becoming DC-like, became more phagocytic, consistent with the known properties of DCs (7). This may have resulted in the engulfment of oxysterols and other LXRα activator ligands.
Alternatively, the induction of LXRα may have been independent of the changes in the intra- or extracellular lipid milleu, for example, via a mechanism related to the inverse relationship between LXR/ABCA1 expression and the inflammatory state (18, 40) (although, currently, those data pertain particularly to microbial-derived ligands of the Toll-like receptors; ref. 20). Interestingly, LXR activation has been shown to decrease the responsiveness of genes that are targets of NF-κB (41), consistent with the inverse association in the present data between the mRNAs for LXRα and either VCAM-1 or MCP-1 (Fig. 4). Obviously, future studies will be needed to discern among these possibilities.
In conclusion, we have studied atherosclerosis regression and the molecular events in foam cells in vivo. In addition to gaining direct insights not obtainable with in vitro studies, the application of LCM to our mouse model has allowed analysis on a per-cell basis, a considerable advantage over the homogenization of aortic tissue. This approach has led to a number of provocative findings, notably, the demonstration of the induction and functional requirement of the chemokine receptor CCR7 in foam cells during regression, as well as the complexity in vivo of the interrelationships among members of the nuclear hormone receptor family.
Methods
Animals and Aortic Transplantation.
All procedures were approved by the Animal Care and Use Committee. The aortic arch transplantation model has been described (3–5). Basically, a donor arch is interpositioned with the abdominal aorta in the recipient mouse and blood flow is directed through the graft.
ApoE−/− (C57BL/6) mice were weaned at 1 month onto a 21% (wt/wt) fat, 0.15% cholesterol WD (Research Diets catalog no. D01022601), which was continued for 20 weeks. Mice were then divided into one group (pretransplant, n = 12) for baseline analyses and another group (n = 67) to be donors of aortic arch segments. The recipients were either apoE−/− (n = 33) or WT (n = 34) mice, maintained on standard chow diet, and killed at 3, 7, 28, or 42 days after transplantation.
To test the effects of a PPARγ agonist on foam cell gene expression in vivo, apoE−/− mice were treated as above until 20 weeks of age, at which time half were fed WD plus pioglitazone (20 mg per kg of mouse per day; the drug was milled into the food) for 2 weeks and the other half continued on WD.
The test of the functional requirement for CCR7 in regression was based on a similar approach to study DC migration in skin (21). Donor apoE−/− mice expressing CD45.2 were placed on the WD at weaning. At 20 weeks of age, the aortic arches were taken either for baseline data or for transplantation into WT recipients expressing CD45.1. Half of the recipients were injected (≈2 h before surgery and just after regaining consciousness) with goat anti-CCL19 and anti-CCL21 (50 μg each), and the other half were injected with 100 μg of preimmune (control) goat IgG (all from R & D Systems). In pilot testing, this protocol inhibited skin DC migration by ≈50%. As before (5), at 3 days after transplant, the graft and the lymph nodes (iliac and hepatic) were taken to measure plaque foam cell content (assessed by CD68 staining) and the appearance of donor monocyte-derived cells in the lymph nodes (assessed by flow cytometry of single cell suspensions using biotinylated antibodies to CD45.2 and other cell surface markers). It should be noted that interpretable lymph node data are available from a subset of the mice; the presence of a false-positive signal confounded the data from the remaining animals.
Lesion Assessment by Histology and Morphometry.
The pretransplant and grafted arches were removed, embedded in OCT, and frozen. Serial sections (6 μm thick) were obtained. For immunohistochemistry, sections were stained for CD68 (rat anti-mouse macrophages; Serotec; 2 μg/ml), as described (42). For ABCA1 (rabbit anti-human ABCA1; Novus Biochemicals; 10 μg/ml) or PPARγ (rabbit anti-mouse PPARγ; Calbiochem; 1:100 dilution), sections were stained for 1 h at room temperature with the primary antibody, followed by incubation with biotinylated goat anti-rabbit Ig for 1 h, reaction with streptavidin-linked alkaline phosphatase, color development with substrate, and haematoxylin counterstaining. Negative controls were performed with an irrelevant primary antibody.
CCR7 was detected by a similar approach, except that a chimeric protein was used, in which the N-terminal end is the binding domain of the CCR7 ligand CCL19, and the C-terminal end is the Fc Ig fragment. This reagent was provided by Timothy Springer (Harvard Medical School, Boston) and used as described (43).
Morphometric measurements were performed on digitized images of CD68-stained serial sections of each arch by using imagepro plus software (4). At least five sections per vessel were analyzed and the mean value used as the summary parameter.
For assessment of cholesteryl ester content, sections were stained with Oil red-O (44).
LCM.
To isolate foam cells from plaques, LCM was performed with the PixCell IIe (Arcturus Bioscience, Mountain View, CA) as we have reported (42) with some modifications. Briefly, 6-μm frozen sections were dehydrated in ethanol, placed twice in xylene, and air dried. At 100-μm intervals, sections were immunostained for CD68 and used as templates for the next five serial sections. RNA was isolated by the Qiagen RNeasy MicroIsolation kit and treated with DNase. The concentration of RNA was determined by the Ribogreen RNA Quantitation kit (Molecular Probes), and the RNA quality verified with the Agilent 2100 Bioanalyzer. Each RNA sample from laser captured foam cells represents a pool of three mice.
PPARγ-Conditional Null Mice.
Mice hemizygous for the MX-Cre transgene and homozygous for the PPARγ-floxed allele have been reported (38). A breeder pair was kindly provided by Christopher K. Glass (University of California, San Diego) and Frank Gonzalez (National Institutes of Health, Bethesda). Mice hemizygous for the MX-Cre transgene but having WT PPARγ alleles were used as littermate controls. Mice were genotyped by PCR of tail-tip DNA. Deletion of exon 2 was induced by i.p. injections of polyinosinic-polycytidylic RNA (pIpC; 2.5 mg/ml in PBS) (38) on days 1, 4, and 7. On day 10, independent of genotype, mice were injected ip with 3% thioglycollate and at day 14 elicited macrophages were isolated by peritoneal lavage.
Cholesterol Efflux Assays.
Human apo A-I was prepared as described (45). LDL and HDL3 were isolated by sequential ultracentrifugation of human plasma. Acetylated LDL (AcLDL) was prepared according to ref. 46. Cell culture reagents were purchased from Invitrogen. Compound F-1394, a potent inhibitor of acyl-coA/cholesterol acyl transferase (ACAT) activity, was a kind gift of Fujirebio (Tokyo). [1,2-_3_H(N)]cholesterol (40–60 Ci/mmol) was purchased from PerkinElmer.
Primary peritoneal murine macrophages were incubated with 3 μCi/ml [_3_H]cholesterol-labeled AcLDL (100 μg/ml) for 24 h in the presence of F-1394 (1 μM) in 0.2% BSA-DMEM. After an equilibration period of 18 h in DMEM-BSA and F-1394 compound, efflux was initiated by either the addition of apoA-I (50 μg/ml) or HDL3 (50 μg/ml) to the culture medium. To test the effect of PPARγ inhibition on efflux, cells were also cultured in the presence of GW-9662 (1.5 μM; Cayman Chemical), a selective PPARγ antagonist (23).
Aliquots of media were collected at 6 and 18 h, and tritium cpm was measured. Free and esterified cholesterol cellular mass were measured (with kits from Roche Applied Science) before and after the efflux period after extracting lipids with isopropanol. After lipid extraction, cells were lysed with 0.2 M NaOH, and protein was determined by Lowry assay. Cholesterol efflux was calculated as the percentage of total intracellular [_3_H]cholesterol recovered from the medium; i.e., 100 × ([_3_H]cholesterol in medium in presence of acceptor − [_3_H]cholesterol in medium in absence of acceptor)/[_3_H]cholesterol in cellular lysate before efflux).
Quantitative Real-Time RT-PCR (QRT-PCR).
RNA abundances were determined by QRT-PCR (42) using 100 pg of total RNA. The primer and probe sequences for PPARγ, LXRα, SR-BI, and CCR7 are shown in Table 2, which is published as supporting information on the PNAS web site, whereas those for CD68, ABCA1, MCP-1, VCAM-1, and cyclophilin A (control) are the same as described in refs. 42 and 47. QRT-PCR standard curves (for each mRNA species) were constructed by using serial dilutions of murine total RNA isolated from thioglycollate-elicited macrophages (PPARγ, LXRα, ABCA1, VCAM-1), TNF-α-treated smooth muscle cells (MCP-1), liver (SR-BI), or spleen (CCR7). All data were normalized to cyclophilin A and expressed as fold change over the controls. For the laser-captured cell data, the results are from two independent samples, each one representing a pool of foam cell RNA from three animals. For the in vitro data, the results represent two replicate experiments with three wells per treatment.
Other Measurements.
Plasma lipid levels were measured by standard enzymatic assays (48).
Statistical Analysis.
Data are expressed as mean ± SEM and were analyzed typically by two-tailed Student's t testing. P < 0.05 was considered significant. For the CCR7 functional experiment, the data for the three groups (baseline, control serum, anti-CCL19/anti-CCL21) were analyzed by one-way ANOVA. All tests were done with prism software.
Supplementary Material
Supporting Information
Acknowledgments
We thank Ms. Ilda Bander and Mr. Jeffrey Mayne for technical assistance and Drs. Igor Chereshnev and Jianhua Liu for performing the surgeries. We also thank Dr. James Rong for helpful discussions. These studies were supported by National Institutes of Health Grants HL61814/70524 (to E.A.F.), HL69446 (to G.J.R.), and HL63768/HL22633 (to G.H.R.). E.T. was partially supported by National Institutes of Health Training Grant HL70524.
Abbreviations
DC
dendritic cell
PPARγ
peroxisome proliferator-activated receptor γ
LXR
liver X receptor
HDL
high-density lipoprotein
LCM
laser capture microdissection
WD
Western-type diet.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Breslow J. L. Science. 1996;272:685–688. doi: 10.1126/science.272.5262.685. [DOI] [PubMed] [Google Scholar]
- 2.Reis E. D., Li J., Fayad Z. A., Rong J. X., Hansoty D., Aguinaldo J. G., Fallon J. T., Fisher E. A. J. Vasc. Surg. 2001;34:541–547. doi: 10.1067/mva.2001.115963. [DOI] [PubMed] [Google Scholar]
- 3.Chereshnev I., Trogan E., Omerhodzic S., Itskovich V., Aguinaldo J. G., Fayad Z. A., Fisher E. A., Reis E. D. J. Surg. Res. 2003;111:171–176. doi: 10.1016/s0022-4804(03)00039-8. [DOI] [PubMed] [Google Scholar]
- 4.Trogan E., Fayad Z. A., Itskovich V. V., Aguinaldo J. G., Mani V., Fallon J. T., Chereshnev I., Fisher E. A. Arterioscler. Thromb. Vasc. Biol. 2004;24:1714–1719. doi: 10.1161/01.ATV.0000139313.69015.1c. [DOI] [PubMed] [Google Scholar]
- 5.Llodra J., Angeli V., Liu J., Trogan E., Fisher E. A., Randolph G. J. Proc. Natl. Acad. Sci. USA. 2004;101:11779–11784. doi: 10.1073/pnas.0403259101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randolph G. J., Inaba K., Robbiani D. F., Steinman R. M., Muller W. A. Immunity. 1999;11:753–761. doi: 10.1016/s1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau J., Steinman R. M. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 8.Forster R., Schubel A., Breitfeld D., Kremmer E., Renner-Muller I., Wolf E., Lipp M. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Fontecha A., Sebastiani S., Hopken U. E., Uguccioni M., Lipp M., Lanzavecchia A., Sallusto F. J. Exp. Med. 2003;198:615–621. doi: 10.1084/jem.20030448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohl L., Mohaupt M., Czeloth N., Hintzen G., Kiafard Z., Zwirner J., Blankenstein T., Henning G., Forster R. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Sozzani S., Allavena P., D’Amico G., Luini W., Bianchi G., Kataura M., Imai T., Yoshie O., Bonecchi R., Mantovani A. J. Immunol. 1998;161:1083–1086. [PubMed] [Google Scholar]
- 12.Sallusto F., Schaerli P., Loetscher P., Schaniel C., Lenig D., Mackay C. R., Qin S., Lanzavecchia A. Eur. J. Immunol. 1998;28:2760–2769. doi: 10.1002/(SICI)1521-4141(199809)28:09<2760::AID-IMMU2760>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 13.Dieu M. C., Vanbervliet B., Vicari A., Bridon J. M., Oldham E., Ait-Yahia S., Briere F., Zlotnik A., Lebecque S., Caux C. J. Exp. Med. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randolph G. J., Beaulieu S., Lebecque S., Steinman R. M., Muller W. A. Science. 1998;282:480–483. [PubMed] [Google Scholar]
- 15.Tall A. R., Costet P., Wang N. J. Clin. Invest. 2002;110:899–904. doi: 10.1172/JCI16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yancey P. G., Bortnick A. E., Kellner-Weibel G., de la Llera-Moya M., Phillips M. C., Rothblat G. H. Arterioscler. Thromb. Vasc. Biol. 2003;23:712–719. doi: 10.1161/01.ATV.0000057572.97137.DD. [DOI] [PubMed] [Google Scholar]
- 17.Li A. C., Glass C. K. J. Lipid Res. 2004;45:2161–2173. doi: 10.1194/jlr.R400010-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Castrillo A., Tontonoz P. Annu. Rev. Cell Dev. Biol. 2004;20:455–480. doi: 10.1146/annurev.cellbio.20.012103.134432. [DOI] [PubMed] [Google Scholar]
- 19.Linton M. F., Fazio S. Int. J. Obes. Relat. Metab. Disord. 2003;27(Suppl. 3):S35–S40. doi: 10.1038/sj.ijo.0802498. [DOI] [PubMed] [Google Scholar]
- 20.Tobias P., Curtiss L. K. J. Lipid Res. 2005;46:404–411. doi: 10.1194/jlr.R400015-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Robbiani D. F., Finch R. A., Jager D., Muller W. A., Sartorelli A. C., Randolph G. J. Cell. 2000;103:757–768. doi: 10.1016/s0092-8674(00)00179-3. [DOI] [PubMed] [Google Scholar]
- 22.Li A. C., Binder C. J., Gutierrez A., Brown K. K., Plotkin C. R., Pattison J. W., Valledor A. F., Davis R. A., Willson T. M., Witztum J. L., et al. J. Clin. Invest. 2004;114:1564–1576. doi: 10.1172/JCI18730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leesnitzer L. M., Parks D. J., Bledsoe R. K., Cobb J. E., Collins J. L., Consler T. G., Davis R. G., Hull-Ryde E. A., Lenhard J. M., Patel L., et al. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- 24.Thuahnai S. T., Lund-Katz S., Dhanasekaran P., de la Llera-Moya M., Connelly M. A., Williams D. L., Rothblat G. H., Phillips M. C. J. Biol. Chem. 2004;279:12448–12455. doi: 10.1074/jbc.M311718200. [DOI] [PubMed] [Google Scholar]
- 25.Wang N., Lan D., Chen W., Matsuura F., Tall A. R. Proc. Natl. Acad. Sci. USA. 2004;101:9774–9779. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rye K. A., Barter P. J. Arterioscler. Thromb. Vasc. Biol. 2004;24:421–428. doi: 10.1161/01.ATV.0000104029.74961.f5. [DOI] [PubMed] [Google Scholar]
- 27.Babaev V. R., Yancey P. G., Ryzhov S. V., Kon V., Breyer M. D., Magnuson M. A., Fazio S., Linton M. F. Arterioscler. Thromb. Vasc. Biol. 2005;25:1647–1653. doi: 10.1161/01.ATV.0000173413.31789.1a. [DOI] [PubMed] [Google Scholar]
- 28.Chawla A., Barak Y., Nagy L., Liao D., Tontonoz P., Evans R. M. Nat. Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 29.Welch J. S., Ricote M., Akiyama T. E., Gonzalez F. J., Glass C. K. Proc. Natl. Acad. Sci. USA. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki A., Yasuno T., Kojo H., Hirosumi J., Mutoh S., Notsu Y. Jpn. J. Pharmacol. 2000;84:113–123. doi: 10.1254/jjp.84.113. [DOI] [PubMed] [Google Scholar]
- 31.Angeli V., Llodra J., Rong J. X., Satoh K., Ishii S., Shimizu T., Fisher E. A., Randolph G. J. Immunity. 2004;21:561–574. doi: 10.1016/j.immuni.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Forte T. M., Subbanagounder G., Berliner J. A., Blanche P. J., Clermont A. O., Jia Z., Oda M. N., Krauss R. M., Bielicki J. K. J. Lipid Res. 2002;43:477–485. [PubMed] [Google Scholar]
- 33.Tontonoz P., Nagy L., Alvarez J. G., Thomazy V. A., Evans R. M. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 34.Ricote M., Huang J., Fajas L., Li A., Welch J., Najib J., Witztum J. L., Auwerx J., Palinski W., Glass C. K. Proc. Natl. Acad. Sci. USA. 1998;95:7614–7619. doi: 10.1073/pnas.95.13.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricote M., Li A. C., Willson T. M., Kelly C. J., Glass C. K. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 36.Pelzer T., Jazbutyte V., Arias-Loza P. A., Segerer S., Lichtenwald M., Law M. P., Schafers M., Ertl G., Neyses L. Biochem. Biophys. Res. Commun. 2005;329:726–732. doi: 10.1016/j.bbrc.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 37.Chawla A., Boisvert W. A., Lee C. H., Laffitte B. A., Barak Y., Joseph S. B., Liao D., Nagy L., Edwards P. A., Curtiss L. K., et al. Mol. Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama T. E., Sakai S., Lambert G., Nicol C. J., Matsusue K., Pimprale S., Lee Y. H., Ricote M., Glass C. K., Brewer H. B., Jr., Gonzalez F. J. Mol. Cell. Biol. 2002;22:2607–2619. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu L., Cao G., Repa J., Stangl H. J. Lipid Res. 2004;45:889–899. doi: 10.1194/jlr.M300461-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Castrillo A., Joseph S. B., Vaidya S. A., Haberland M., Fogelman A. M., Cheng G., Tontonoz P. Mol. Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- 41.Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. Nat. Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 42.Trogan E., Choudhury R. P., Dansky H. M., Rong J. X., Breslow J. L., Fisher E. A. Proc. Natl. Acad. Sci. USA. 2002;99:2234–2239. doi: 10.1073/pnas.042683999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manjunath N., Shankar P., Wan J., Weninger W., Crowley M. A., Hieshima K., Springer T. A., Fan X., Shen H., Lieberman J., von Andrian U. H. J. Clin. Invest. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunnari J. J., Zand T., Joris I., Majno G. Exp. Mol. Pathol. 1989;51:1–8. doi: 10.1016/0014-4800(89)90002-6. [DOI] [PubMed] [Google Scholar]
- 45.Weisweiler P. Clin. Chim. Acta. 1987;169:249–254. doi: 10.1016/0009-8981(87)90325-1. [DOI] [PubMed] [Google Scholar]
- 46.Basu S. K., Goldstein J. L., Anderson G. W., Brown M. S. Proc. Natl. Acad. Sci. USA. 1976;73:3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong J. X., Shapiro M., Trogan E., Fisher E. A. Proc. Natl. Acad. Sci. USA. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choudhury R. P., Rong J. X., Trogan E., Elmalem V. I., Dansky H. M., Breslow J. L., Witztum J. L., Fallon J. T., Fisher E. A. Arterioscler. Thromb. Vasc. Biol. 2004;24:1904–1909. doi: 10.1161/01.ATV.0000142808.34602.25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information