Processes of copy-number change in human DNA: The dynamics of α-globin gene deletion (original) (raw)
Abstract
Ectopic recombination between locally repeated DNA sequences is of fundamental importance in the evolution of gene families, generating copy-number variation in human DNA and often leading to pathological rearrangements. Despite its importance, little is known about the dynamics and processes of these unequal crossovers and the degree to which meiotic recombination plays a role in instability. We address this issue by using as a highly informative system the duplicated α-globin genes in which ectopic recombination can lead to gene deletions, often very prevalent in populations affected by malaria, as well as reduplications. Here we show that spontaneous deletions can be accessed directly in genomic DNA by using single-DNA-molecule methods. These deletions proved to be remarkably common in both blood and sperm. Somatic deletions arise by a strictly intrachromosomal pathway of homologous exchange that also operates in the germ line and can generate mutational mosaicism, whereas sperm deletions frequently involve recombinational interactions between homologous chromosomes that most likely occur at meiosis. Ectopic recombination frequencies show surprisingly little requirement for long, identical homology blocks shared by paralogous sequences, and exchanges can occur even between short regions of sequence identity. Finally, direct knowledge of germ-line deletion rates can give insights into the fitness of individuals with these α-globin gene deletions, providing a new approach to investigating historical levels of selection operating in human populations.
Keywords: ectopic, recombination, selection, mutation, mosaicism
Ectopic recombination is initiated by the misalignment of nonallelic but homologous DNA sequences and can result in rearrangements such as deletions, duplications, inversions, and translocations (1, 2). Ectopic recombination contributes to the extensive copy-number variation seen in the human genome (3–5) and can lead to changes in gene number in gene families as well as segmental duplications and deletions, often with pathological consequences (6, 7). The dynamics of ectopic recombination have been extensively characterized in model organisms such as yeast (8). In contrast, relatively little is known about these processes in humans, with most analyses to date having focused on descriptions of pathological DNA changes seen in patients (9, 10).
Analysis of familial and de novo DNA rearrangements in genomic disorders, both in patients and by accessing mutations in sperm, has established the importance of ectopic recombination (nonallelic homologous recombination) between widely separated repeated DNA elements in driving large-scale duplications and deletions in the human genome (1, 11). For example, ectopic recombination between a pair of 24-kb-long repeat elements on chromosome 17 can lead to duplication or deletion of the intervening 1.4-megabase segment of DNA, resulting in the inherited disorders Charcot–Marie–Tooth type 1A (CMT1A) and hereditary neuropathy with liability to pressure palsies (HNPP), respectively (12–14). Ectopic exchange points within the repeat elements cluster within a 0.7-kb hot spot and often show patchy gene conversion near the site of crossover (14–16). Similar hot spots in widely separated repeat elements have been seen in other genomic disorders (1, 11, 17) and are reminiscent of hot spots for allelic recombination at meiosis (18), although whether they also function as allelic crossover hot spots remains unknown. There are distinct sex-dependent mechanisms of CMT1A/HNPP ectopic recombination, with duplications of paternal origin generated by unequal meiotic recombination between homologous chromosomes, whereas maternal duplications and deletions are produced by ectopic exchanges both within and between homologous chromosomes (19).
In contrast, little is known about the dynamics of ectopic recombination between locally repeated DNA sequences that can lead to copy-number changes in human gene clusters. Classic examples are provided by the α- and β-globin gene families in which homologous regions (homology blocks) shared by duplicated genes only kilobases apart can promote ectopic exchanges, leading to duplications, deletions, and the creation of fusion genes as in Hb Lepore (20, 21). Extensive work on these rearrangements in patients and populations has revealed basic information on patterns of local exchanges between these homology blocks (21, 22). However, little is known about the dynamics and processes of these ectopic exchanges given that they rarely arise de novo. For example, recent studies have shown that Hb Lepore rearrangements between δ- and β-globin genes occur with extremely low frequency in sperm, preventing detailed analysis of deletion processes (23).
The α-globin gene family provides a potentially very informative system for studying the dynamics of copy-number change in gene clusters. The α1- and α2-globin genes are associated with homologous regions that can be subdivided into several homology blocks with different degrees of sequence identity (24). Ectopic exchanges between these homology blocks can lead to various types of chromosome with a single α-globin gene (−α chromosomes) depending on the sites of unequal exchange (21, 25, 26). Chromosomes bearing these deletions can be extremely common in malarial regions and are most likely maintained by malaria selection (27, 28). Such chromosomes represent the most common pathological DNA rearrangement in the world, although in themselves they are not severe; −α/−α homozygotes show only a mild and sometimes asymptomatic form of α+-thalassemia (α-thalassemia trait with microcytosis), whereas αα/−α carriers are generally normal (29, 30). These α-globin gene deletions occur on diverse haplotypic backgrounds (21, 27, 31) and, therefore, must be recurrent in human populations. However, selection on this region of DNA makes it impossible to estimate rates of germ-line deletion from population frequencies, and the dynamics of deletion remain unknown.
Here we show that it is possible to detect de novo α-globin gene-deletion molecules directly in genomic DNA. We have used these deletions to study basic features of ectopic exchange between locally repeated DNA sequences, including the germ-line specificity of deletion, the role of meiotic recombination in the deletion process, and the dynamics of ectopic exchange.
Results and Discussion
Physical Enrichment of α-Globin-Deletion Molecules.
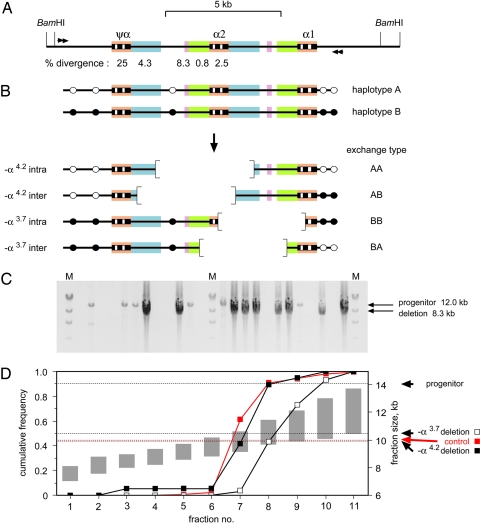
Studies of patients with α thalassemia have revealed two classes of deletion that can arise by ectopic exchange between homology blocks in the α-globin gene region (Fig. 1A): −α4.2 deletions that extend leftward from the α2-globin gene and lose 4.2 kb of DNA and smaller −α3.7 deletions that extend rightward (25) (Fig. 1B). We used physical enrichment (23) to recover simultaneously both classes of deletion directly from genomic DNA by digestion with BamHI to cleave outside the α-globin gene region (Fig. 1A) followed by gel electrophoretic fractionation to recover size fractions heavily depleted in progenitor molecules and enriched for mutant DNA molecules. Nested PCR analysis revealed deletions in these fractions plus some remaining progenitor molecules (Fig. 1C). These mutants were distributed correctly across the size fractions. Thus, the −α4.2 deletion molecules showed the same distribution as a control 10.0-kb genomic DNA fragment matched in size to this deletion. Likewise, the −α3.7 deletions showed a similar distribution but shifted to somewhat larger DNA fractions, as expected because these deletion molecules are 0.5 kb larger than −α4.2 mutants (Fig. 1D). This size validation (23) established the authenticity of >99% of these mutants and showed that they were not PCR artifacts arising from intact or broken progenitor DNA molecules.
Fig. 1.
Detection of de novo deletions in the α-globin gene region. (A) The region analyzed, showing the α-globin genes and pseudogene, plus BamHI cleavage sites used for size fractionation of genomic DNA and nested PCR primers (arrows) used to amplify deletion molecules. Similar sequences are colored to highlight homologies, with levels of sequence divergence between each homology block and its paralog immediately to the right shown below. (B) Leftward and rightward deletions that can arise by ectopic exchange. SNP heterozygosities [white (haplotype A) and black (haplotype B) circles] make it possible to distinguish interchromosomal recombinants showing exchange of flanking markers (AB and BA) from intrachromosomal deletions (AA and BB). (C) Examples of detection of deletion molecules. Aliquots of one of the fractions of blood DNA (10.2- to 11.4-kb BamHI DNA fragments), each derived from 1.1 × 105 amplifiable haploid genomes and containing ≈0.5 remaining 14.1-kb progenitor DNA molecules, were amplified by nested PCR, and products were analyzed by agarose gel electrophoresis. M, λ DNA × HindIII. Some reactions show PCR products derived from deletion mutants, and some others show progenitor molecules. Differential amplification results in mutant molecules being substantially overamplified relative to the longer progenitor. (D) Cumulative frequencies of deletion molecules across size fractions of BamHI-digested sperm DNA, determined from a total of 90 −α3.7 and 19 −α4.2 deletion mutants recovered from 6.7 × 106 progenitor molecules. Size ranges covered by fractions are shown as gray bars. The control is a 10.0-kb genomic BamHI fragment matched in size to the −α4.2 deletion mutant.
Deletion Rates.
These deletions were analyzed in blood and sperm DNA from two men, one of north European descent (man 1) and one of mixed north European/south Asian ancestry (man 2). Neither man carried a constitutional α-globin gene deletion or reduplication. Substantial instability was seen in all samples. Blood-deletion frequencies were very similar in both men, at 6.7 × 10−6 and 6.8 × 10−6 per haploid genome in man 1 and man 2, respectively. Sperm-deletion frequencies were significantly higher than in blood in both men (P < 0.001), with man 1 showing a 4-fold higher frequency than man 2 (68 × 10−6 vs. 16 × 10−6 per sperm, P < 0.001).
Structures of Deletions.
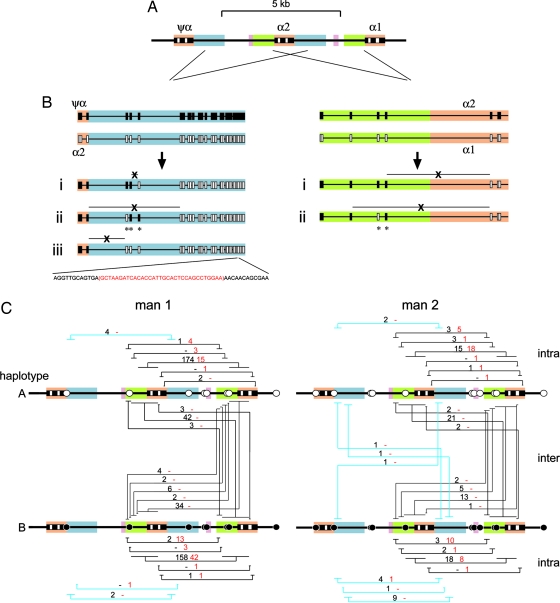
All deletion mutants recovered from blood and sperm were characterized for sequence differences between homology blocks [paralogous sequence variants (PSVs)] to locate ectopic exchange points. These exchange points were confirmed by sequence analysis of 131 assorted deletions representing 20% of all mutants recovered. Deletion molecules were also analyzed for single-nucleotide polymorphism (SNP) heterozygosities to determine whether exchanges were intrachromosomal (AA or BB type) or had occurred between homologous chromosomes (interchromosomal, AB or BA type) (Fig. 1B). Some of these SNPs mapped within homology blocks and created haplotype-specific PSVs that allowed us to investigate the effects of sequence mismatches on ectopic recombination.
Almost all deletions had arisen by simple exchange between homologous DNA sequences within intervals of sequence identity. Only three exceptional types were seen (Fig. 2B). First, nine −α4.2 deletions seen in sperm from man 2 that mapped to the same interval all showed the same switching of PSVs near the site of exchange, indicating patchy gene conversion accompanying ectopic exchange. A similar type of conversion was seen in five −α3.7 deletions. Finally, one −α4.2 deletion showed a 33-bp microdeletion located ≈1 kb away from the ectopic exchange point. The rarity of patchy conversions seen in these α-globin gene deletions is comparable to their incidence during allelic crossover at recombination hot spots (18, 32) but contrasts with the frequent conversions that accompany CMT1A/HNPP ectopic exchanges between distant repeat elements (14–16).
Fig. 2.
α-globin gene deletions detected in sperm and blood DNA. (A) The region analyzed, as described for Fig. 1. (B) Structure of −α4.2 (Left) and −α3.7 (Right) deletions, with PSVs marked as rectangles. (i) Examples of typical deletions mapping to a single interval between PSVs. A total of 539 and 128 such exchanges were seen in sperm and blood DNA, respectively. (ii) Complex mutants with PSV switching (∗) near the site of ectopic exchange. Nine such −α4.2 mutants were identified in the sperm of man 2. The complex −α3.7 deletion was seen in three blood mutants from man 1 and two sperm mutants from man 2. (iii) A simple exchange accompanied by a microdeletion in a distal Alu repeat (red sequence lost) seen in one blood mutant. (C) Distribution of ectopic exchange points. Progenitor haplotypes have heterozygous SNPs marked as black and white circles. Each interval of sequence identity containing exchange breakpoints is marked by a tied horizontal line, in blue for −α4.2 deletions, with the number of exchanges seen in sperm and blood DNA indicated in black and red, respectively, above the tie (−, no mutants). Intra- and interchromosomal deletions are shown separately. Some SNPs are located within homology blocks and, depending on the allele, can break long regions of identity into smaller regions, creating differences between haplotypes in the intervals within which ectopic exchanges can be mapped. These mutants were recovered from 6.5 × 106 and 12.9 × 106 progenitor molecules from sperm and blood DNA, respectively, from man 1 and 6.7 × 106 and 7.1 × 106 molecules from man 2.
Most deletions in blood and sperm were of the −α3.7 class, with only 4% showing the leftward −α4.2 deletion (Fig. 2C). All of the 132 blood mutants were intrachromosomal, with none showing exchange of flanking SNP markers. These mutants are most likely products of unequal mitotic recombination, although a possible contribution from replication slippage cannot be excluded. In contrast, 26% of sperm mutants (145 of 550) were interchromosomal and showed exchange of flanking markers, which indicates a clear difference in somatic and germ-line ectopic recombination pathways and suggests a significant role for homologue pairing and meiotic recombination in the generation of sperm deletions. The pattern of germ-line deletion via a predominantly intrachromosomal pathway again contrasts with CMT1A, in which paternal rearrangements arise almost exclusively by exchange between homologous chromosomes (19).
Mutational Mosaicism in Blood and Sperm.
Intrachromosomal deletions in both blood and sperm occurred with similar frequencies on each haplotype, although there were some significant distortions. For example, the most common type of −α3.7 deletion in blood from man 1 showed an excess on haplotype B (42 mutants vs. 18 on haplotype A, P = 0.002). These fluctuations most likely arise from mutational mosaicism. They were also seen in sperm in man 2 for the most common class of −α4.2 deletion, all nine of which were derived by intrachromosomal exchange on haplotype B and shared the same unusual conversion event (Fig. 2B). This germ-line mosaicism strongly suggests a premeiotic intrachromosomal component to sperm deletion, with in this case one sperm in 700,000 sharing the same premeiotic mutation that must have arisen in a single progenitor cell. Similar levels of sperm mosaicism have been seen for minisatellite deletion mutants (33) and can distort estimates of mutation rate. In contrast, there were no significant differences between the numbers and distributions of interchromosomal AB- and BA-type sperm deletions in either man (P > 0.6), consistent with each being a unique product of meiotic recombination.
Homology Block Lengths and Mutation Frequencies.
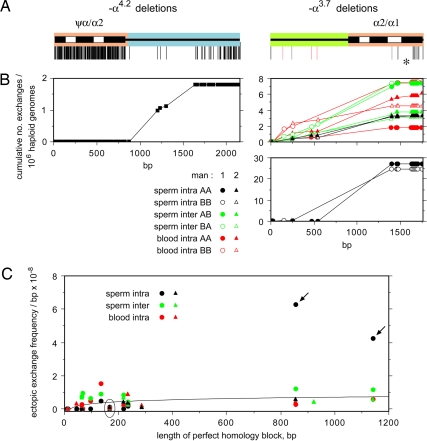
The bulk of ectopic exchanges mapped as expected to the longest regions of identical sequence shared between homology blocks (Fig. 2C). However, exchange distributions showed that ectopic exchange points, although avoiding regions of high divergence, were distributed fairly randomly across homology blocks (Fig. 3A and B). The only exceptions were −α3.7 intrachromosomal deletions in sperm from man 1, which were unusually frequent on both haplotypes in the longest region of sequence identity and largely responsible for the high overall sperm-deletion frequency seen in this man compared with man 2. There were no DNA sequence differences between the men in this region that could account for this frequency difference, and the only explanation we have is that premeiotic mosaicism has inflated these frequencies in man 1 and, by chance, to the same extent on both haplotypes. If correct, this inflation indicates that mutational mosaicism can cause substantial variation between men in deletion frequencies.
Fig. 3.
Distribution of ectopic exchange points within homology blocks. (A) The homologous regions analyzed, colored as described for Fig. 1 and with PSVs marked by lines below. SNPs in one haplotype that disrupt a region of perfect sequence identity in the other haplotype are marked in red. (B) Cumulative number of ectopic exchanges, per 106 haploid genomes, across homology blocks for all −α4.2 exchanges combined and for each type of −α3.7 exchange, with intrachromosomal sperm exchanges in man 1 shown separately. (C) Exchange frequencies per base pair estimated for each interval of sequence identity, with the best-fit logarithmic curve after excluding the outlying points (arrows). The circled points with a low rate correspond to the interval marked by an asterisk in A.
Ectopic exchange frequencies per base pair in each interval of sequence identity showed only a weak dependence on uninterrupted homology length (Fig. 3C), with exchanges occurring in intervals as short as 34 bp and with a frequency of ≈8 × 10−9 per base pair for the longest intervals. Therefore, there is no evidence that ectopic exchanges are inhibited by modest sequence divergence between homology blocks. This lack of inhibition is supported by SNPs that can act as PSVs and alter homology block lengths. For example, man 1 shows a 1,143-bp interval of sequence identity for AB-type exchanges, whereas two SNPs divide this interval into three for BA-type exchanges (Fig. 2C). Despite this disruption, the same numbers of exchanges mapped to this region in AB and BA recombinants (42 in each case), with BA exchanges distributed across the three intervals in proportion to their length (6, 2, and 34 exchanges in intervals 220, 66, and 855 bp long, respectively; P = 0.5).
Ectopic recombination frequencies, however, are not governed solely by lengths of high sequence similarity shared between homology blocks. Thus, a 166-bp interval at the 3′ end of the α2/α1 homology block consistently showed a low ectopic exchange frequency (Fig. 3). Likewise, the ψα/α2 homology block contains an 800-bp-long region of strong sequence similarity, compared with 1,600 bp for the α2/α1 homology block. If ectopic recombination frequencies were determined solely by homology length, then 33% of ectopic exchanges should generate −α4.2 deletions, compared with 4% observed (P < 0.001). It is possible that recombination-initiation events are, in fact, preferentially targeted to the longest regions of sequence identity, particularly within the α2/α1 homology block, and that subsequent migration leads to substantial diffusion of exchange-resolution points. This diffusion, however, would have to be greater than that seen at hot spots for allelic recombination, given their narrow width (18). Instead, it seems likely that other factors, such as features of nuclear architecture (34, 35), play an important role in controlling the frequency of recombinational interactions between different paralogous sequences.
Deletion Rates and Population Fitness.
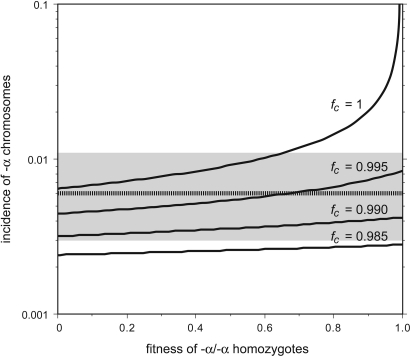
Chromosomes bearing these α-globin gene deletions are very common, particularly in Asia and Africa, where they can attain population frequencies as high as 90% (28, 36, 37), and are most likely maintained by malaria selection in favor of −α chromosomes (27, 28). In contrast, nonmalarial populations show a much lower incidence of deletion chromosomes (≈0.6% in northern Europeans; see Materials and Methods). However, the frequency of ectopic exchanges in sperm, primarily generating −α3.7 deletions, is high at 42 × 10−6 per sperm averaged over the two men tested. Given this sperm instability and assuming an equal deletion rate in the female germ line, then the low incidence of −α chromosomes in northern Europe can only have been maintained for a fully recessive deletion by significant selection against −α/−α homozygotes (fitness, <0.66) despite their showing only a mild and sometimes asymptomatic form of α+-thalassemia (Fig. 4). If their fitness were higher, then the population incidence of −α chromosomes would exceed 1.1%, the upper 95% confidence limit of the northern European frequency. Even if there were no instability in the female germ line, there would still have to be significant selection against homozygotes (fitness, <0.81; data not shown), and higher females rates would require even stronger selection against homozygotes. The nature of the selective forces operating against these largely normal individuals remains unknown.
Fig. 4.
The effect of fitness on the incidence of −α deletion chromosomes in nonmalarial populations. The expected population incidence of deletions at mutation/selection equilibrium was estimated from the observed germ-line deletion rate of 42 × 10−6 per sperm, averaged over the two men analyzed and assumed to be the same in the female germ line. Frequencies were estimated at various fitness levels of −α/−α homozygotes and with different fitness levels _f_c of αα/−α carriers. The dashed line provides the estimated incidence of −α chromosomes in north Europeans, with 95% confidence intervals indicated in gray. See Materials and Methods for details.
If there is a drop of fitness of αα/−α carriers, who can also show subtle hematological changes (29, 30), then the strength of selection operating against homozygotes could be weaker (Fig. 4). However, selection against carriers must be weak (fitness, >0.985), irrespective of the fitness of homozygotes. In malarial populations, the observed high incidence of chromosomes carrying a single α-globin gene can be maintained by modest balancing selection against normal αα/αα individuals; for example, a drop of fitness of only 1/10th relative to −α/−α homozygotes would be sufficient to maintain the incidence of deletion chromosomes in a population at 10%, with an equilibrium frequency that is largely independent of deletion rate.
Conclusions
This work has shown that it is possible to use single-DNA-molecule analysis to gain direct insights into the dynamics and processes of ectopic recombination that operate within human gene families and provide a major mechanism driving copy-number change within the human genome (1, 10, 14). These studies have revealed distinct mitotic and meiotic recombination pathways, with mosaicism of the former significantly influencing the frequency of deletion even in sperm, and have shown distinct differences between processes of ectopic exchange between locally repeated DNA sequences in the α-globin gene cluster and recombination between distant repeat elements as in CMT1A/HNPP ectopic exchanges. These studies also challenge the notion of minimum pairing segments of uninterrupted homology required for such exchanges (38–40). Finally, direct comparisons of germ-line mutation rates and population frequencies can provide insights into historical levels of selection operating in human populations.
Materials and Methods
DNA Samples.
Blood and semen samples were collected with approval from the Leicestershire Health Authority Research Ethics Committee and with informed consent. Sperm and blood DNAs were extracted and subsequently manipulated as described under conditions designed to minimize the risk of contamination (see ref. 41).
PCR Amplification.
The α-globin gene region is very GC-rich and difficult to amplify (29, 42). However, extensive variation of long PCR conditions and the use of additives (43) enabled us to develop a protocol to amplify this locus efficiently. PCRs were performed in 0.2-ml PCR tubes or 96-well plates in a PTC-225 Tetrad DNA engine (MJ Research, Cambridge, MA). These optimized PCRs used 0.2 μM PCR primers, 0.03 units/μl Taq polymerase (ABGene, Surrey, U.K.), and 0.003 units/μl Pfu polymerase (Stratagene) in 1.4 M betaine, 32 mM Tris·HCl (pH 8.8), 12.5 mM Tris base, 7.7 mM ammonium sulfate, 3.2 mM MgCl2, 4.7 mM 2-mercaptoethanol, 3.1 μM EDTA, 0.7 mM dNTPs, and 79 μg/ml BSA. Cycling was at 96° for 1 min, followed by 34 cycles of 96° for 20 sec, 63° for 30 sec, and 68° for 16 min. Primary PCRs used primers A13.6F (5′-TTT CCG GTG AGG CCT TTC CG-3′) and A25.7R (5′-CGC TGC ACT CCA ACC AGC C-3′), and secondary nested PCRs were with A13.6F2 (5′-CGG GGC AAT GGT GCA GCG-3′) and A25.6R2 (5′-GCT GCG GGA AGG ACA TCA C-3′). This PCR system allowed the efficient amplification of the entire 12.1-kb progenitor region at the single-DNA-molecule level with, on average, one amplifiable molecule detected per 4.2 pg of genomic DNA and giving a single-molecule PCR efficiency of 71%, typical of the efficiency of amplification of other long but not GC-rich DNA targets (32).
DNA Fractionation to Recover Deletion Mutants.
Genomic DNA (130–300 μg) was digested with BamHI (New England BioLabs), ethanol-precipitated, and dissolved in 400 μl of 5 mM Tris·HCl (pH 7.5). The digested DNA was mixed with 40 μl of loading dye (44 mM Tris-borate, pH 8.3/1 mM EDTA/30% vol/vol glycerol and bromophenol blue) plus 375 μg/ml ethidium bromide, loaded into a 5- × 0.3-cm slot in a 40-cm-long, 1.4-cm-deep 0.8% SeaKem HGT agarose gel (Cambrex Bio Science Rockland, Rockland, ME) and electrophoresed until a 6.6-kb λ DNA × HindIII marker had migrated 35 cm. Size fractions were collected ranging in length from 7.3 to 13.2 kb to include α-globin gene deletions (9.9 and 10.4 kb long for −α4.2 and −α3.7 deletions, respectively) and to exclude 14.1-kb progenitor DNA molecules. DNA in each fraction was recovered by electroelution onto dialysis membrane and then ethanol-precipitated and dissolved in 5 mM Tris·HCl, pH 7.5. Additional details of fractionation are provided in ref. 23.
Size Validation and Estimation of DNA Recovery.
The distribution across the fractions of DNA molecules very similar in size to −α4.2 deletion mutants was established by PCR amplification of an 8.0-kb interval from a control 10.0-kb BamHI DNA fragment from the MHC by using PCR primers R111.4F (5′-GAG GCT CCC TTT GGG ACT GC-3′) and R119.4R (5′-CCG GTG AGA GAA TAT AGC C-3′). This control BamHI fragment also allowed us to estimate overall recovery of DNA in these fractions; yields varied from 20% to 50%. Parallel PCR analysis of the 14.1-kb α-globin DNA fragment showed that >99.997% of progenitor molecules had been excluded from the fractions.
Recovery of Deletion Mutants.
Deletion molecules in size-fractionated genomic DNA were recovered by PCR amplification (as described above) of multiple aliquots of each fraction, with each aliquot containing at most 1.2 deletion molecules and no more than 0.9 remaining progenitor molecules as established from pilot experiments performed on each fraction to gain an initial estimate of deletion frequency. Secondary PCR products were analyzed by agarose gel electrophoresis and visualized by staining with ethidium bromide. The full screening of blood and sperm DNA in both men involved the analysis of 3,125 nested PCRs.
Characterizing Progenitor Haplotypes.
The progenitor α-globin gene region was amplified from each of the two men and sequenced by using BigDye Terminators v1.1 or v3.1 (Applied Biosystems) to confirm all PSVs and identify any sites heterozygous for an SNP. Single progenitor molecules were amplified from extreme dilutions of genomic DNA from each man, and the linkage phase of SNPs was established by typing these progenitor molecules by hybridization with allele-specific oligonucleotides as described in ref. 32.
Mapping Deletion Mutants.
The ectopic exchange-point location in each mutant was roughly localized by digesting mutant- and progenitor-amplified DNA with PvuII or RsaI and comparing electrophoretic profiles of restriction fragments. Exchange points were refined further by hybridization with oligonucleotides specific to PSVs (32). SNPs were likewise typed by hybridization of allele-specific oligonucleotides to these PCR products (32). In total, 131 assorted deletions also were sequenced to confirm exchange regions.
In some cases (10–14% of positive reactions), two different mutants were seen in the same PCR as shown by mixed PSV and/or SNP sites. In cases of a single mixed site, the constituent mutants could be deduced unambiguously. In other cases, we used sequence-specific PCR directed to a mixed PSV or SNP site to separate mutants before mutant characterization. Only 8 of 459 positive PCRs gave complex mixtures that could not be resolved; these mutant pools were excluded from further analyses. To correct for instances of more than one molecule of a given type being present in a PCR, a full inventory of PCRs that were positive and negative for each type of mutant in each size fraction was established and used to estimate Poisson-corrected numbers of molecules (32). These Poisson corrections were insignificant for most mutant classes and only 1.7-fold for the most abundant type of mutant.
Estimation of Deletion Incidence and Selective Fitness.
The population incidence of α-globin gene deletions was analyzed in 132 men in the United Kingdom self-reported as being of northern European descent, plus 97 CEPH (population named for the Centre d’Etude du Polymorphisme Humain) grandparents or parents of Utah, Amish, and French descent and an additional 182 unrelated men in the United Kingdom identified by grandparental place of birth as northern European in origin. These surveys yielded zero, three, and six αα/−α carriers, respectively. Previous studies have reported one carrier in a survey of 155 north Europeans (44) and no carriers in 140 individuals from the United Kingdom and 110 Icelandics (45). There is no evidence for significant heterogeneity in carrier frequency across these samples (P = 0.15), and therefore all were combined to obtain a mean incidence of −α chromosomes of 0.006 (95% confidence interval, 0.003–0.011). This estimate of the incidence of deletion chromosomes in nonmalarial areas is maximal and could be lower if any of the surveyed individuals have partial ancestry from regions with a high frequency of −α chromosomes.
This incidence of deletion chromosomes was compared with population frequencies expected for a given germ-line deletion rate, μ. Consider a population with −α chromosomes at frequency q and with normal αα/αα individuals showing a fitness _f_n of 1, αα/−α carriers a fitness of _f_c, and −α/−α homozygotes a fitness of _f_t. At equilibrium, the gain of new −α deletions from normals and carriers [(1 − q)2μ + f_c_q(1 − q)μ] will be balanced by the loss of deletions from carriers and homozygotes [(1 − _f_c)q(1 − q) + (1 − _f_t)_q_2]. If selection only operates on homozygotes (_f_c = 1, _f_t < 1), then this equilibrium frequency at q ≪ 1 simplifies to the standard form q = √[μ/(1 − _f_t)]. More complex models incorporating triplicated ααα chromosomes that could regenerate αα chromosomes by interchromosomal recombination in ααα/−α heterozygotes proved unnecessary, given the low incidence (0.2–0.6%) of ααα chromosomes in north Europe (28). The incidence of deletion chromosomes in populations under balancing selection by malaria (_f_n < 1, _f_c = 1, _f_t < 1) was likewise investigated by identifying stable equilibrium frequencies of the deletion for varying values of _f_n and _f_t.
Acknowledgments
We thank the two volunteers for providing blood and semen samples, M. Jobling and T. King for population DNA samples, and colleagues for helpful discussions. This work was supported by grants from the Medical Research Council, the Royal Society, and the Louis-Jeantet Foundation (to A.J.J.).
Abbreviations
CMT1A
Charcot–Marie–Tooth type 1A
HNPP
hereditary neuropathy with liability to pressure palsies
PSV
paralogous sequence variant.
Conflict of interest statement: No conflicts declared.
See accompanying Profile on page 8921.
References
- 1.Shaw C. J., Lupski J. R. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 2.Feuk L., Carson A. R., Scherer S. W. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 3.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 4.Sharp A. J., Locke D. P., McGrath S. D., Cheng Z., Bailey J. A., Vallente R. U., Pertz L. M., Clark R. A., Schwartz S., Segraves R., et al. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuzun E., Sharp A. J., Bailey J. A., Kaul R., Morrison V. A., Pertz L. M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 6.Bailey J. A., Gu Z., Clark R. A., Reinert K., Samonte R. V., Schwartz S., Adams M. D., Myers E. W., Li P. W., Eichler E. E. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 7.Stankiewicz P., Lupski J. R. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 8.Petes T. D., Hill C. W. Annu. Rev. Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- 9.Baumer A., Dutly F., Balmer D., Riegel M., Tukel T., Krajewska-Walasek M., Schinzel A. A. Hum. Mol. Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- 10.Potocki L., Chen K., Park S., Osterholm D. E., Withers M. A., Kimonis V., Summers A. M., Meschino W. S., Anyane-Yeboa K., Kashork C. D., et al. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 11.Emanuel B. S., Shaikh T. H. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 12.Pentao L., Wise C. A., Chinault A. C., Patel P. I., Lupski J. R. Nat. Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- 13.Reiter L. T., Murakami T., Koeuth T., Pentao L., Muzny D. M., Gibbs R. A., Lupski J. R. Nat. Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- 14.Lopes J., Tardieu S., Silander K., Blair I., Vandenberghe A., Palau F., Ruberg M., Brice A., LeGuern E. Hum. Mol. Genet. 1999;8:2285–2292. doi: 10.1093/hmg/8.12.2285. [DOI] [PubMed] [Google Scholar]
- 15.Reiter L. T., Hastings P. J., Nelis E., De Jonghe P., Van Broeckhoven C., Lupski J. R. Am. J. Hum. Genet. 1998;62:1023–1033. doi: 10.1086/301827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han L. L., Keller M. P., Navidi W., Chance P. F., Arnheim N. Hum. Mol. Genet. 2000;9:1881–1889. doi: 10.1093/hmg/9.12.1881. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Correa C., Dorschner M., Brems H., Lazaro C., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J. L., et al. Hum. Mol. Genet. 2001;10:1387–1392. doi: 10.1093/hmg/10.13.1387. [DOI] [PubMed] [Google Scholar]
- 18.Jeffreys A. J., Kauppi L., Neumann R. Nat. Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 19.Lopes J., Ravise N., Vandenberghe A., Palau F., Ionasescu V., Mayer M., Levy N., Wood N., Tachi N., Bouche P., et al. Hum. Mol. Genet. 1998;7:141–148. doi: 10.1093/hmg/7.1.141. [DOI] [PubMed] [Google Scholar]
- 20.Efremov G. D. Hemoglobin. 1978;2:197–233. doi: 10.3109/03630267809007068. [DOI] [PubMed] [Google Scholar]
- 21.Higgs D. R., Vickers M. A., Wilkie A. O. M., Pretorius I.-M., Jarman A. P., Weatherall D. J. Blood. 1989;73:1081–1104. [PubMed] [Google Scholar]
- 22.Metzenberg A. B., Wurzer G., Huisman T. H. J., Smithies O. Genetics. 1991;128:143–161. doi: 10.1093/genetics/128.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holloway K., Lawson V. E., Jeffreys A. J. Hum. Mol. Genet. 2006;15:1099–1111. doi: 10.1093/hmg/ddl025. [DOI] [PubMed] [Google Scholar]
- 24.Higgs D. R., Hill A. V., Bowden D. K., Weatherall D. J., Clegg J. B. Nucleic Acids Res. 1984;12:6965–6977. doi: 10.1093/nar/12.18.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Embury S. H., Miller J. A., Dozy A. M., Kan Y. W., Chan V., Todd D. J. Clin. Invest. 1980;66:1319–1325. doi: 10.1172/JCI109984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden D. K., Hill A. V. S., Higgs D. R., Oppenheimer S. J., Weatherall D. J., Clegg J. B. J. Clin. Invest. 1987;79:39–43. doi: 10.1172/JCI112804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flint J., Hill A. V. S., Bowden D. K., Oppenheimer S. J., Sill P. R., Serjeantson S. W., Bana-Koiri J., Bhatia K., Alpers M. P., Boyce A. J., et al. Nature. 1986;321:744–750. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 28.Hill A. V. S. Baillieres Clin. Haematol. 1992;5:209–238. doi: 10.1016/s0950-3536(11)80042-9. [DOI] [PubMed] [Google Scholar]
- 29.Higgs D. R. Baillieres Clin. Haematol. 1993;6:117–150. doi: 10.1016/s0950-3536(05)80068-x. [DOI] [PubMed] [Google Scholar]
- 30.Wang C., Beganyi L., Fernandes B. J. Lab. Haematol. 2000;6:163–166. [Google Scholar]
- 31.Flint J., Harding R. M., Boyce A. J., Clegg J. B. Baillieres Clin. Haematol. 1998;11:1–51. doi: 10.1016/s0950-3536(98)80069-3. [DOI] [PubMed] [Google Scholar]
- 32.Jeffreys A. J., Ritchie A., Neumann R. Hum. Mol. Genet. 2000;9:725–733. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 33.Jeffreys A. J., Neumann R., Wilson V. Cell. 1990;60:473–485. doi: 10.1016/0092-8674(90)90598-9. [DOI] [PubMed] [Google Scholar]
- 34.Vanin E. F., Henthorn P. S., Kioussis D., Grosveld F., Smithies O. Cell. 1983;35:701–709. doi: 10.1016/0092-8674(83)90103-4. [DOI] [PubMed] [Google Scholar]
- 35.Nicholls R. D., Fischel-Ghodsian N., Higgs D. R. Cell. 1987;49:369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- 36.Dozy A. M., Kan Y. W., Embury S. H., Mentzer W. C., Wang W. C., Lubin B., Davis J. R., Jr., Koenig H. M. Nature. 1979;280:605–607. doi: 10.1038/280605a0. [DOI] [PubMed] [Google Scholar]
- 37.Embury S. H., Lebo R. V., Dozy A. M., Kan Y. W. J. Clin. Invest. 1979;63:1307–1310. doi: 10.1172/JCI109426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonda D. K., Radding C. M. Cell. 1983;34:647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- 39.Rubnitz J., Subramani S. Mol. Cell. Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waldman A. S., Liskay R. M. Mol. Cell. Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeffreys A. J., Tamaki K., MacLeod A., Monckton D. G., Neil D. L., Armour J. A. L. Nat. Genet. 1994;6:136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- 42.Chong S. S., Boehm C. D., Higgs D. R., Cutting G. R. Blood. 2000;95:360–362. [PubMed] [Google Scholar]
- 43.Henke W., Herdel K., Jung K., Schnorr D., Loening S. A. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beutler E., West C. Blood. 2005;106:740–745. doi: 10.1182/blood-2005-02-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flint J., Hill A. V., Weatherall D. J., Clegg J. B., Higgs D. R. Br. J. Haematol. 1986;63:796–797. doi: 10.1111/j.1365-2141.1986.tb07564.x. [DOI] [PubMed] [Google Scholar]