A Family of ADP-Ribosylation Factor Effectors That Can Alter Membrane Transport through the trans-Golgi (original) (raw)
Abstract
A family of three structurally related proteins were cloned from human cDNA libraries by their ability to interact preferentially with the activated form of human ADP-ribosylation factor 3 (ARF3) in two-hybrid assays. The specific and GTP-dependent binding was later confirmed through direct protein binding of recombinant proteins. The three proteins share large (≈300 residues) domains at their N termini that are 60–70% identical to each other and a shorter (73 residues) domain at their C termini with 70% homology to the C-terminal “ear” domain of γ-adaptin. Although GGA1 is found predominantly as a soluble protein by cell fractionation, all three proteins were found to localize to the _trans_-Golgi network (TGN) by indirect immunofluorescence. The binding of GGAs to TGN was sensitive to brefeldin A, consistent with this being an ARF-dependent event. Thus, these proteins have been named Golgi-localizing, γ-adaptin ear homology domain, ARF-binding proteins, or GGAs. The finding that overexpression of GGAs was sufficient to alter the distribution of markers of the TGN (TGN38 and mannose 6-phosphate receptors) led us to propose that GGAs are effectors for ARFs that function in the regulation of membrane traffic through the TGN.
INTRODUCTION
ADP-ribosylation factors (ARFs) are 21-kDa GTP-binding proteins that constitute a subfamily of the Ras superfamily (Donaldson and Klausner, 1994; for reviews, see Boman and Kahn, 1995; Moss and Vaughan, 1998). Results from several laboratories have pointed to a central role for ARFs in regulating some aspect of membrane traffic, particularly to and from the Golgi, and more recently from the _trans_-Golgi network (TGN) (Stamnes and Rothman, 1993; Traub et al., 1993; Ooi et al., 1998; Hirst et al., 1999). ARFs were originally found to localize to the Golgi (Stearns et al., 1990b). Assays of endoplasmic reticulum (ER)-Golgi (Balch et al., 1992), intra-Golgi (Taylor et al., 1992), endosome fusion (Lenhard et al., 1992), and nuclear vesicle fusion (Boman et al., 1992) were used to define a role for ARF in each assay, and presumably in those transport processes. However, in several of these cases, ARF was found not to be required for the transport/fusion activity but only for inhibition of the assay by the nonhydrolyzable GTP analogue GTPγS (Gant and Wilson, 1997; Happe and Weidman, 1998). Thus, an obligate role for ARF in any of these steps is uncertain. However, a model that has emerged is that ARFs recruit multimeric coat complexes to the sites of vesicle buds and play a regulatory role in vesicle formation.
Six currently defined coat protein complexes are thought to be recruited to membranes by ARF or an ARF family member. Two of these coat complexes are heptameric (COP-I and COP-II) and four are tetramers (adaptin complexes AP-1, AP-2, AP-3, and AP-4). COP-I–coated vesicles function in the transport of vesicles between the Golgi and the intermediate compartment (Cosson et al., 1996; Gaynor and Emr, 1997). COP-II, whose recruitment to membranes involves activation of the ARF homologue SAR1, mediates export from the ER (Barlowe et al., 1994). The four adaptin complexes share structural relatedness between homologous subunits and are recruited to membranes in concert with ARFs. AP-1 is found associated with sites of protein export from the TGN and on TGN-derived vesicles (Stamnes and Rothman, 1993; Traub et al., 1993; Hirst and Robinson, 1998). AP-2 associates with plasma membranes in an ARF-sensitive manner (West et al., 1997) and is found on endosomes derived from the plasma membrane. AP-3 (Dell'Angelica et al., 1997; Simpson et al., 1997; Ooi et al., 1998) and AP-4 (Dell'Angelica et al., 1999; Hirst et al., 1999) are involved in transport within the endosomal-lysosomal system and in the transport of non-clathrin–coated vesicles from the TGN, respectively.
Much of the early work on the role of ARF in coat protein recruitment was fueled by results with the macrolide antibiotic brefeldin A (BFA) because of its remarkable ability in live cells to cause protein coats (e.g., COP-I [Donaldson et al., 1990] and AP-1 [Traub et al., 1993]) on the Golgi to rapidly fall off and the uncoated Golgi membranes to fuse with the ER (Lippincott-Schwartz et al., 1989). BFA was shown to inhibit an ARF-specific guanine nucleotide exchange factor in crude membrane fractions (Helms and Rothman, 1992; Randazzo et al., 1993) and later directly on purified ARF guanine nucleotide exchange factors (Peyroche et al., 1996, 1999; Mansour et al., 1999; Togawa et al., 1999). Thus, until another site of BFA action is defined, it is reasonable to conclude that a protein whose localization is rapidly (within seconds to a few minutes) and dramatically altered by BFA addition to live cells lies downstream of an ARF-dependent step.
It is evident from both genetic and biochemical studies in yeast and mammalian cells that, in addition to their effects on membrane traffic, ARFs regulate other cellular processes and must do so through regulated interactions with a variety of effectors, distinct from those involved in membrane traffic. In addition, it is not clear if the effects of ARF on vesicular traffic are direct or only mediated by interactions with coat proteins. To identify novel effectors for activated ARFs, we initiated a two-hybrid screen of human cDNA libraries with the use of a dominant activated mutant of ARF3 ([Q71L]ARF3) as bait. Among the effectors identified were a group of three proteins, termed GGA1, GGA2, and GGA3 (for Golgi-localizing, γ-adaptin ear homology domain, ARF-binding proteins), that share extensive sequence identity and that include a region at the C terminus with marked homology to an adaptin subunit. We describe the direct and GTP-dependent binding of ARF3 to GGA1 and provide data from in vivo studies of cells overexpressing GGA proteins that support a model in which GGAs act as ARF-sensitive components in protein transport from the TGN.
MATERIALS AND METHODS
Materials
Unless specified otherwise, chemicals and reagents were purchased from Sigma Chemical (St. Louis, MO). Radiolabeled nucleotides were purchased from Amersham (Arlington Heights, IL). Yeast and bacterial media reagents were purchased from Difco (Detroit, MI).
Yeast Two-Hybrid Assay Components
Yeast strains Y190 (_MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,-112 +URA3::GAL→lacZ, LYS2::GAL(UAS)→HIS3 cyh_r) and Y187 (_MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,-112 met_− URA3::GAL→lacZ), plasmids pAS2 and pACT2, and a B-cell library in pACT were the generous gift of Steve Elledge (Baylor University, Houston, TX). The yeast strains are constructed with two readouts for an interaction of histidine auxotrophy and β-galactosidase expression with the use of the Gal4 binding domain (BD) and activation domain system (Durfee et al., 1993). Plasmid pBG4D was obtained from Dr. Rob Brazas (University of California, San Francisco) to make C-terminal constructs of ARF with the BD. The ORF of human ARF3 was cloned into pAS2 with the use of _Nde_I and _Bam_HI sites and into pBG4D at the _Bam_HI site. In addition to wild-type ARF3, we also subcloned two mutant alleles of ARF, [Q71L]ARF3, a dominant activating mutant, and [N126I]ARF3, a dominant negative mutant. Any DNA amplified by PCR was sequenced to confirm that no errors were introduced by the polymerase.
The expression of each ARF-BD fusion protein was confirmed by immunoblotting. Proteins in yeast lysates were resolved by electrophoresis on SDS-PAGE gels, transferred to nitrocellulose, and probed with either mAb 12CA5 against the hemagglutinin (HA) epitope incorporated at the C terminus of the BD or ARF3-specific polyclonal antibody R-1023 (Cavenagh et al., 1996).
Two-Hybrid Screen
Transformation of Y190 with a plasmid encoding the activated ARF mutant construct [Q71L]ARF3-BD yielded strain YAB324. This strain was transformed with each of two human cDNA libraries, a B-cell library in pACT (8 × 105 transformants) and a kidney library in pGAD10 (9 × 105 transformants; Stratagene, La Jolla, CA), and transformants were selected for histidine auxotrophy on minimal medium lacking histidine and containing 25 mM 3-aminotriazole. After reselection on 3-aminotriazole, colonies were assayed for β-galactosidase activity, as described previously (Breeden and Nasmyth, 1985). Filters were incubated at 30°C for 30 min to 3 h, with color scoring at regular intervals. Obvious blue color development within 3 h was considered positive. The [Q71L]ARF3-BD plasmid, carrying a gene conferring sensitivity to cycloheximide, was next lost by growth of potential positive strains on plates containing cycloheximide. Only those strains that tested negative in the β-galactosidase assay after losing the ARF plasmid were investigated further. These were then tested in two-hybrid assays in Y190 strains carrying either lamin or cdk2 as partners to confirm the specificity of the interaction with [Q71L]ARF3. As a final selection and to enrich for potential ARF effectors or GTPase-activating proteins (GAPs), each library plasmid was also transformed into Y190 carrying either the wild-type ARF3-BD construct, pAB155, or the negative dominant mutant [N126I]ARF3 construct, pABH3-3. Only those library inserts encoding proteins that interacted preferentially with the activated mutant of ARF3 were pursued.
Library plasmids were rescued from each positive strain and confirmed to be of library origin by restriction digestion analysis. Each library plasmid was then transformed into Y190 or YAB324 cells to test for two-hybrid activity alone or to confirm the positive reactivity with [Q71L]ARF3-BD, respectively. Library inserts in those plasmids encoding proteins that interacted less well with ARF3 than with the activated mutant were then sequenced in both directions by automated DNA sequencing. Initial sequence analyses were performed with the use of BLAST (Altschul et al., 1990) and FASTA (Pearson and Lipman, 1988) against the GenBank and dbEST databases. Alignments of complete sequences were obtained with the use of Multalign (Corpet, 1988).
Yeast Methods
All yeast transformations were performed with the use of the LiOAc method of Ito et al. (1983) with denatured herring sperm DNA as a carrier (Schiestl and Gietz, 1989). Plasmids were rescued from the yeast as described by Hoffman and Winston (1987), followed by plasmid purification on a Qiagen (Chatsworth, CA) miniprep column.
Northern Blotting
Northern blotting was performed with the use of the Multiple Human Tissue I blot (Clontech, Palo Alto, CA), as described in the manufacturer's protocol. Each probe was ∼500 base pairs (bp) in length and was labeled by incorporating [α-32P]dCTP by random priming with the use of the kit from Life Technologies (Grand Island, NY). Blots were exposed to Kodak (Rochester, NY) XAR-5 film for 5–46 h. The same filter was reprobed and used to develop Northern blots for each of the GGAs. Complete stripping was confirmed by exposure to film before reprobing and by the differences in message size, without residual bands appearing. An actin probe was used as a control for RNA loading after all GGA probes were completed.
5′ Rapid Amplification of cDNA Ends
The 5′ end of the GGA1 cDNA was amplified by PCR from a human brain Marathon cDNA library (Clontech) with the use of two gene-specific primers and adaptor primer AP1. The PCR product was subcloned into pCR2.1 by TA cloning (Invitrogen, Carlsbad, CA). Colonies were selected by colony hybridization with a third gene-specific primer, confirmed by restriction digestion, and sequenced by automated sequencing. No identical sequences were found in either GenBank or dbEST.
Antibody Production
Antibodies to GGA1 were raised in two rabbits by HTI Bioproducts (Ramona, CA) with the use of GST-GGA1(145–639) as the antigen. Subcutaneous injections in complete Freund's adjuvant were followed by three boosts in incomplete Freund's adjuvant. All data shown were obtained with the use of serum from rabbit R-79709.
Immunoblotting
Immunoblotting was performed as described previously (Cavenagh et al., 1996; Boman et al., 1999). Antibodies to GGA1 (R-79709) or ARF3 (R-1023) were used at a 1:2000 dilution. Bound antibodies were detected with HRP-linked secondary antibodies (Amersham) and ECL detection reagents (Amersham).
Recombinant Proteins
ARF proteins were expressed in bacteria and purified as described previously (Randazzo et al., 1995). The cDNA encoding GGA1(145–639) was subcloned into pGEX5X-2 (Pharmacia, Piscataway, NJ) after PCR amplification to add appropriate restriction sites. This construct directs the expression of the N-terminal fusion protein GST-GGA1(145–639). Expression of the fusion protein was induced with isopropylthio-β-galactoside in BL21 cells and detected on Coomassie brilliant blue–stained SDS-PAGE gels or by immunoblotting with an anti-GST antibody (Stratagene). Fusion proteins were purified by lysing the bacteria with a French press and removing insoluble material by centrifugation at 100,000 × g (35,000 rpm, Ti45 rotor, 30 min, 4°C). Glutathione-agarose beads were added to the supernatant (S100) and incubated for 1 h at 4°C with constant mixing; then they were washed three times with 10 volumes of TND (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT). GST-GGA1 was eluted from the beads with the use of TNG (50 mM Tris, pH 8.0, 100 mM NaCl, 10 mM reduced glutathione) in three batches of 2 volumes each at room temperature. Proteins were concentrated as needed with a Microcon 10 ultrafiltration unit (Amicon, Beverly, MA). All protein concentrations were determined by the amido black method (Schaffner and Weissmann, 1973).
Glutathione-Agarose Affinity Chromatography
The S100 from bacterial lysates containing GST alone or GST-GGA1 was incubated with glutathione-agarose beads for 30 min at 4°C. The beads were washed twice with TND and once with binding buffer (25 mM HEPES, pH 7.0, 2 mM EDTA, 1 mM MgCl2, 100 mM NaCl, 1 mM DTT, 100 μg/ml BSA, 0.1% Triton X-100) and then incubated with ARF3 or [Q71L]ARF3 (10 μM) that had been preincubated with either GTPγS or GDP (100 μM) for 20 min at room temperature. The beads were then washed three times with binding buffer containing 10 μM nucleotide. Bound proteins were eluted from the beads with an equal volume of 2× Laemmli's sample buffer and run on a 12% SDS-PAGE gel. Duplicate gels were either stained with Coomassie brilliant blue or developed by immunoblotting with the use of a polyclonal rabbit antiserum (R-1023; Cavenagh et al., 1996) to detect human ARF3.
Nucleotide-binding Assays
Quantitative nucleotide-binding assays were performed by filter trapping as described (Kahn and Gilman, 1986; Randazzo et al., 1995) with the use of [35S]GTPγS as ligand.
Microinjection, Transfections, and Immunofluorescence
Normal rat kidney (NRK) fibroblasts were grown in RPMI medium with l-glutamine (Life Technologies) plus 10% FBS at 37°C and 5% CO2. Purified GST-GGA1(145–639) was exchanged into 100 mM KPO4, pH 7.4, and concentrated to 4 mg/ml. Protein was mixed with 0.6 mg/ml lysine-fixable FITC-dextran (Molecular Probes, Eugene, OR) to allow detection of injected cells and injected into the cytosol of NRK cells with an Eppendorf Scientific (Westbury, NY) micromanipulator and transjector.
GGA1, GGA1(145–639), GGA2, and GGA3 were subcloned into pcDNA3.1 (Invitrogen) containing an HA epitope tag at the N terminus (a gift from Drs. Lixin Zhang and Haian Fu, Emory University). Transient transfections of NRK cells with the pcDNA3-derived plasmids were performed with the use of the FuGene6 transfection reagent (Boehringer Mannheim, Indianapolis, IN) in the presence of serum. Control transfections contained pcDNA3 with no insert. Cells were fixed 18–24 h after transfection.
Cells were fixed with 2% formaldehyde in PBS for 10 min at room temperature, followed by permeabilization with 0.2% saponin. Cells were stained for indirect immunofluorescence with antibodies to GGA1 (rabbit polyclonal R-79709), ARF (mouse monoclonal 1D9; Cavenagh et al., 1996), mannosidase II (a gift from K. Moremen [University of Georgia, Athens, GA] and M. Farquhar [University of California, San Diego, CA]; Moremen et al., 1991), β-COP (EAGE peptide rabbit polyclonal; Pepperkok et al., 1993), mannose 6-phosphate receptor (M6PR; a gift from K. von Figura, Georg-August Universität, Göttingen, Germany), TGN38 (kindly provided by G. Banting, University of Bristol, Bristol, United Kingdom), AP-1 (A36120, mouse monoclonal to γ-adaptin; Transduction Laboratories, Lexington, KY), or AP.6 (raised against the α-subunit of AP-2; a gift from Dr. Linton Traub, Washington University, St. Louis, MO). Secondary antibodies against mouse or rabbit immunoglobulin G were labeled with Texas Red or FITC, respectively (Cappel Laboratories, Durham, NC). Cells were observed and photographed with the use of a Zeiss (Thornwood, NY) Axiophot or Olympus (Tokyo, Japan) BX-60 microscope equipped with a 100× objective. Controls were performed to ensure against bleed through between different filters when double staining was performed.
During the characterization of the GGA1 antiserum, it was noted that when cells were permeabilized with 0.1% Triton X-100, we observed mitochondrial staining, as determined by colocalization with Mitotracker (Molecular Probes, Eugene, OR). This is likely the result of cross-reactivity with a 72-kDa band seen on immunoblots, because it was shown to localize to mitochondria by cell fractionation (see RESULTS) and the mitochondrial staining pattern was not affected by overexpression of GGA1. In addition, no evidence of a mitochondrial distribution was detected in cells expressing the HA-tagged version of GGA1 after staining with the HA antibody.
RESULTS
Two-Hybrid Screening
To identify proteins that interact with the active (GTP-bound) conformation of human ARF3, we initiated a two-hybrid screen in yeast with the use of [Q71L]ARF3, fused to the Gal4 binding domain at the C terminus, as bait. ARF3 was used because it and ARF1 are the most abundant isoforms expressed in most cells and are >97% identical. This mutant was used because it is resistant to GTP hydrolysis and is more likely to exist in the GTP-bound form in cells (Kahn et al., 1995). Both human kidney and B-cell libraries were screened, as described in MATERIALS AND METHODS, and 23 independently isolated positive clones were obtained. Eleven of these positive clones fell into one group and have been described in a separate publication (Boman et al., 1999). A single clone was identified as POR1/Arfaptin2 (Van Aelst et al., 1996; Kanoh et al., 1997). Of the remaining positive clones, four encode the first two members of a new protein family, which are described in more detail below. Three plasmids from the kidney library contained identical 2.2-kilobase (kb) inserts encoding 495 amino acids of a novel protein that is not found in GenBank or Swiss-Prot. A fourth clone contained a plasmid with a 1.6-kb insert from the B-cell library that encoded 500 amino acids of another novel protein. The two ORFs had extensive sequence similarities, as described in more detail below. Database (GenBank, Swiss-Prot, and dbEST) searching with each of these sequences revealed significant homology throughout the lengths of each ORF with a third ORF, which is present as an unknown but fully sequenced human cDNA in GenBank (accession number KIAA0154). The KIAA0154 cDNA was obtained, subcloned into pACT2, and found to interact strongly with [Q71L]ARF3. Because of the high degree of sequence relatedness between these three sequences, and for reasons explained in more detail below, we named the proteins Golgi-localizing, γ-adaptin ear homology domain, ARF-binding proteins: GGA1 (kidney clones), GGA2 (B-cell clone), and GGA3 (KIAA0154). The cDNA sequences of each of the GGAs have been deposited in GenBank. The accession numbers are AF190862, AF190863, and AF190864 for GGA1, GGA2, and GGA3, respectively.
Based on sequence homology with GGA3 and typical for large cDNAs pulled from two-hybrid libraries, the clones encoding GGA1 or GGA2 were truncated at their 5′ ends. Alignment of the N-terminal regions revealed 60–75% identity, but GGA3 had an extension of 100–150 amino acids. The sequence of the 5′ end of GGA2 was determined on the computer (and later confirmed by direct DNA sequencing of a full-length clone), whereas 5′ rapid amplification of cDNA ends was performed to determine the 5′ end of the ORF of GGA1.
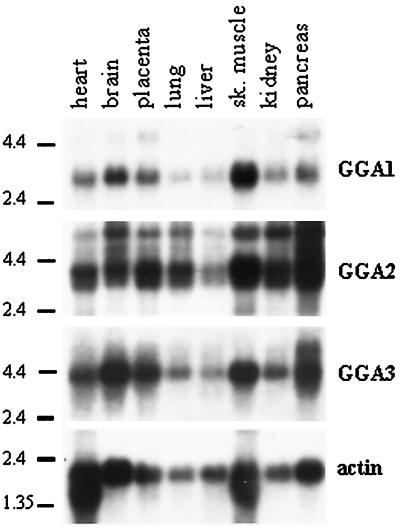
GGA3 Message
Northern blot analysis with the use of full-length cDNA for GGA3 revealed a single message of ∼3.9 kb in all eight tissues tested (Figure 1), with pancreas, brain, skeletal muscle, and placenta the most abundant sources. The GenBank sequence KIAA0154 is 3.8 kb, consistent with this being a full-length cDNA. In pancreas, a second message of 5.2 kb is apparent. The 5′ and 3′ untranslated regions (UTRs) of the GGA3 clone are 6 and 1600 bp, respectively. Searches of the dbEST database identified many sequences identical to KIAA0154 at the 3′ end, consistent with the ubiquity of the message seen in Northern blots, but none that extend the length of the 5′ UTR.
Figure 1.
All three GGAs are ubiquitously expressed in adult human tissues. Northern blotting was performed sequentially, as described in MATERIALS AND METHODS, with the use of probes derived from GGA1, GGA2, or GGA3 and a multiple-tissue blot that contained mRNA from eight tissues. The locations of RNA size markers are indicated on the left of each blot. Actin was used as a control for RNA integrity and was the last probe used.
GGA2 Message
Northern blot analysis of GGA2 revealed a doublet at 3.4/3.1 kb in all tissues, a less prominent third band at 5.5 kb, and an even weaker signal seen at ∼7.5 kb (Figure 1). The 3.4-kb band was the most abundant. A cDNA sequence of 3.3 kb for GGA2 was pieced together by sequential BLAST analyses of the dbEST database. Multiple expressed sequence tags (ESTs) overlapped with the 5′ region of our known GGA2 sequence. Clone zs05e07 was the longest among several clones that appeared to start within five bases of one another and contained a putative initiating methionine in frame with our GGA2 sequence. Clone zs05e07 was obtained, sequenced to confirm the ORF, and used for subcloning of full-length GGA2. GGA2 has a very short (23-base) 5′ UTR. The dbEST database contains clones from many libraries that are identical to our GGA2 sequence, suggesting that this is the correct sequence of GGA2.1 Altogether, the GGA2 contig spanned 3.3 kb, consistent with the major band on Northern blots.
GGA1 Message
Northern blot analysis indicated a single GGA1 message, 3.0 kb in length, present in all eight tissues (Figure 1). The 2.2-kb inserts from the library plasmids encoding GGA1 contained a 3′ UTR of 860 bp. Based on alignment with many EST clones, this appears to be the entire 3′ UTR. Hence, we predicted that up to 800 bases at the 5′ end of GGA1 were absent from our library plasmid. No sequences in the dbEST database overlapped with the 5′ end of GGA1, so 5′ rapid amplification of cDNA ends was performed with the use of a human brain cDNA library, as described in MATERIALS AND METHODS. The longest of the sequenced clones (∼650 bp) contained a putative start methionine in frame with the GGA1 ORF and 16 bases of 5′ UTR. The size of the predicted full-length message [2.8 kb without the poly(A) tail] is consistent with the size of the message determined by Northern blotting. The combination of alignment with the other two GGAs, short 5′ UTRs, and the fact that they accounted for nearly all the message seen in Northern blots led us to conclude that we had the full-length ORFs for all three cDNAs.
GGA Proteins
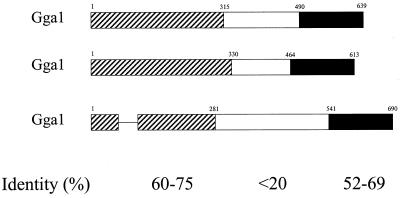
The ORFs of the GGA1, GGA2, and GGA3 cDNAs encode proteins with predicted molecular masses of 70.4, 67.2, and 74.5 kDa, respectively. Alignment of these predicted proteins (Figure 2A) led us to organize the sequences into three regions, as shown in Figure 2B. The N-terminal domains of ∼300 amino acids are 60–75% identical to one another (pairwise) and contain stretches of up to 15 residues of absolute conservation. One gap of 33 amino acids was inserted into the GGA3 sequence for this alignment (indicated by a dash in Figure 2B). Both the COILS (Lupas et al., 1991) and PSORTII (Nakai and Kanehisa, 1992) programs predict the presence of a coiled-coil domain in all three GGA proteins that align in Figure 2A (residues 187–228, 205–243, and 155–199 in GGA1, GGA2, and GGA3, respectively). Domain searching revealed the presence in the N termini of all three GGA proteins of homology with the “HRS domain” or “VHS domain” present in VPS-27, HRS, and STAM proteins. The middle domains, ranging in size from 150 to 280 amino acids, are divergent from one another, with little sequence identity to each other or to other known proteins. The C-terminal 150 amino acids are 54–68% identical to each other. This C-terminal domain also has homology to the “ear domain” of γ-adaptin, a subunit of the clathrin adaptor complex AP-1, with 44% amino acid identity and 70% homology over 73 residues at the C termini (Figure 2A). BLAST searches identified a rat homologue of GGA1 that is nearly identical in protein sequence and several mouse ESTs and two potential yeast (Saccharomyces cerevisiae) homologues that are each >20% identical.
Figure 2.
Alignment of the protein sequences of GGA1, GGA2, and GGA3 showing regions of homology to γ-adaptin and predicted coiled-coil domains. (A) The amino acid sequences for GGA1, GGA2, and GGA3 were aligned with the use of CLUSTAL. Identical residues are indicated by asterisks (*) in the line below, and conservative substitutions are indicated by colons (:). Residues predicted to be in a coiled-coil configuration are italicized (begins at residue 188 for GGA1). The alignment with the C terminus of γ-adaptin (-Ad) is also shown, with the line below that showing identities in all four sequences (uppercase) or in three of the four sequences (lowercase). (B) The three domains with different levels of sequence conservation between GGAs are shown schematically. Bars representing the protein domains are scaled to reflect the relative size of each domain. The percent pairwise identity within each domain is indicated, as determined by Bestfit (Genetics Computer Group, Madison, WI).
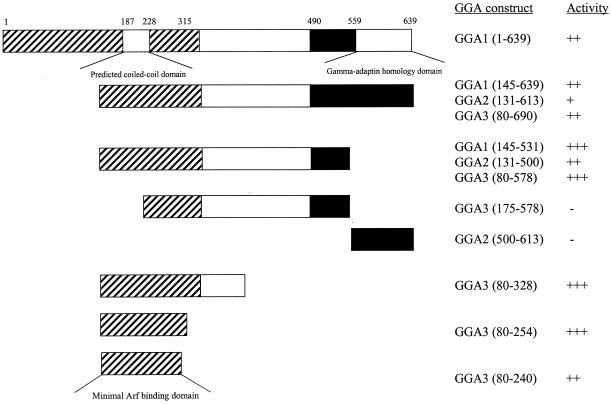
To determine which domain(s) was required for binding to ARF3, full-length GGA1 and a series of deletion constructs of all three GGAs were subcloned into pACT2 and tested for β-galactosidase activity in two-hybrid assays with [Q71L]ARF3 (Figure 3). Expression of each of these constructs was confirmed by immunoblotting of yeast extracts with the HA antibody. The C-terminal domains were not involved in the interaction with [Q71L]ARF3, because this domain alone had no activity in this assay and deletion of this region from any of the GGAs was not detrimental to interaction (Figure 3). Rather, removal of this domain increased the strength of the two-hybrid signal between [Q71L]ARF3 and each of the GGAs. Further truncations from the C termini to include the middle, divergent domain neither enhanced nor reduced the interaction with [Q71L]ARF3. In contrast, deletion of the first half of the N-terminal conserved domains eliminated the interaction with [Q71L]ARF3 (Figure 3). We conclude that the N-terminal regions of GGA proteins contain the ARF-binding domains. Based on the results of truncations of the N-terminal domain of GGA3, and if we assume that the ARF-binding domain is homologous in all three GGA proteins, we can further limit the site of interaction to residues 114–274, 130–290, and 80–240 of GGA1, GGA2, and GGA3, respectively. Homology searches with this portion of each GGA did not identify any proteins known to bind ARFs or of likely biological significance. Note that this region overlaps the predicted coiled-coil domain described above (residues 187–228 of GGA1).
Figure 3.
ARF binds to the N-terminal, most highly conserved region of GGAs. A variety of deletion constructs of all three GGA ORFs were tested in two-hybrid assays when paired with [Q71L]ARF3, as described in MATERIALS AND METHODS. The three domains of GGAs, shown in Figure 2B, are indicated, along with the specific GGA fragments that were cloned into the two-hybrid expression vector and the results of β-galactosidase assays. The minimal predicted fragment that includes the ARF-binding domain, determined by consensus from all three GGAs, is shown at the bottom.
GGA1 Binds Directly to ARF3 in a GTP-dependent Manner
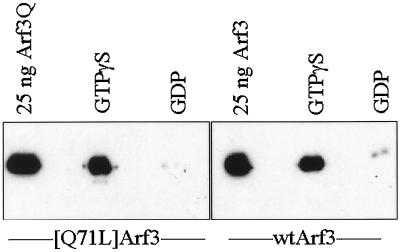
To confirm that the interactions between ARF3 and GGA proteins are 1) direct, 2) dependent on GTP binding to ARF, and 3) independent of the [Q71L] mutation, an in vitro GST-affinity binding assay was developed. GST fusions of each GGA protein were expressed in bacteria. All three fusion proteins were expressed at high levels in bacteria. However, GST fusions of GGA2 and GGA3 were insoluble, whereas GST-GGA1(145–639) was soluble in bacterial lysates. The reasons for this difference in solubility are unknown. Efforts then focused on the GGA1 fusion protein for biochemical analyses.
We first tested whether ARF3 could be retained in a GTP-specific manner by GST-GGA1(145–639) on glutathione-agarose beads. Purified ARF3 (wild-type or the [Q71L] mutant) was preloaded with either GTPγS or GDP and then incubated with GST-GGA1 on glutathione-agarose beads. ARF3·GTPγS was retained by GST-GGA1(145–639) (Figure 4) but not by GST alone. In contrast, ARF3·GDP was not retained by either GST-GGA1(145–639) (Figure 4) or GST. Identical results were obtained for both wild-type and [Q71L]ARF3. These results reveal that ARF3 binds directly to GGA1 in a GTP-dependent manner and that the interaction does not require mutation of glutamine 71. We also assayed GST-GGA1(145–639) for ARF exchange factor and ARF GAP activity and found it devoid of each.
Figure 4.
GST-GGA1(145–639) binds to ARF3 in a GTP-dependent manner. Purified recombinant ARF3 or [Q71L]ARF3 was preincubated with GTPγS or GDP and then added to glutathione-agarose beads containing bound GST-GGA1(145–639), as described in MATERIALS AND METHODS. After extensive washing of the beads, the proteins that remained bound were analyzed on immunoblots with the use of antisera specific to ARFs. Note that both ARF3 and [Q71L]ARF3 bound to GST-GGA1(145–639) only when in the activated, GTPγS-bound state, and not when bound to GDP. Purified ARF3 proteins were used as a positive control for the Western blots and are shown on the left in each case.
Localization of GGA1
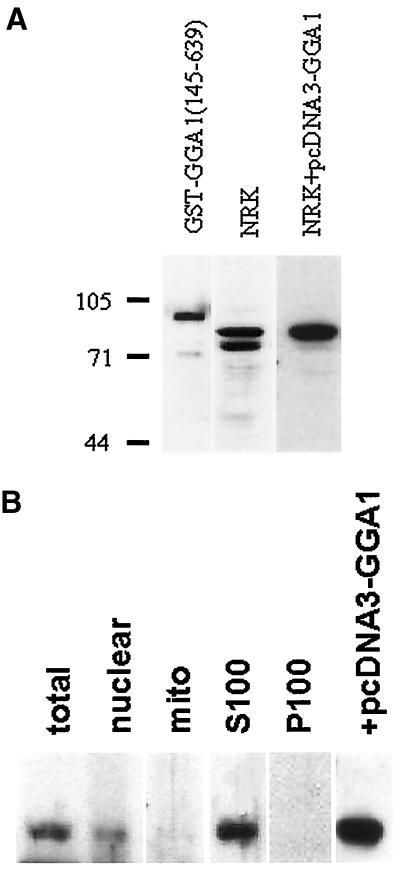
Antibodies to GGA1 were raised in rabbits with the use of purified GST-GGA1(145–639) as the antigen. These antisera were characterized with respect to specificity and sensitivity by immunoblotting. A 1:2000 dilution of serum (R-79709) was able to detect 1 ng of GST-GGA1(145–639) as a major band at the expected, apparent molecular mass of 90 kDa (Figure 5A, left lane). Western blots of NRK lysates identified two prominent bands of 72 and 78 kDa (Figure 5A, center lane) that were blocked by preincubation of the antibody with antigen. Overexpression of full-length GGA1 in NRK cells caused a clear increase in immunoreactivity of the 78-kDa band, with no change in the lower immunoreactive band (Figure 5A, right lane). Note that the development times for the immunoblots of NRK lysates in Figure 5A (center and right lanes) were quite different; otherwise, the signal from the overexpressed GGA1 would completely obscure the cross-reactive 72-kDa band. In other experiments, GGA2 and GGA3 were also overexpressed in NRK cells, and no change in immunoreactivity was detected with R-79709. We conclude that the band migrating as a 78-kDa species is full-length GGA1 and that the GGA1 antibodies do not strongly cross-react with the other members of this family.
Figure 5.
GGA1 antibodies detect the soluble endogenous or overexpressed GGA1 proteins on immunoblots. (A) Rabbit polyclonal antiserum R-79709 was used as the primary antibody at a dilution of 1:2000 to detect GGA1 in immunoblots of purified GST-GGA1(145–639) antigen (1 ng; left lane), NRK cell lysate (20 μg; center lane), or an NRK cell lysate from cells overexpressing GGA1 (20 μg; right lane). The GGA1 was identified as the upper band in the doublet seen in NRK cells. Note that the immunoreactivity of the lower band is the same in the two right lanes but that the right lane was exposed to film for a shorter time to allow clear visualization of the upper band. (B) NRK cells were fractionated, as described in MATERIALS AND METHODS, and fractions were probed with the GGA1 antiserum to reveal a predominantly soluble location.
NRK cells were fractionated by differential centrifugation to yield crude nuclear, mitochondrial, other membranous, and cytosol protein fractions (Figure 5B). Note that the nuclear fraction is simply a low-speed spin that also contains unbroken cells. The 78-kDa band, corresponding to GGA1, is predominantly soluble because it partitions into the S100 fraction, with some also present in the nuclear fraction. In contrast, the 72-kDa band was found only in the nuclear and mitochondrial fractions. The clear differences in fractionation are consistent with the 72-kDa band being a cross-reacting protein.
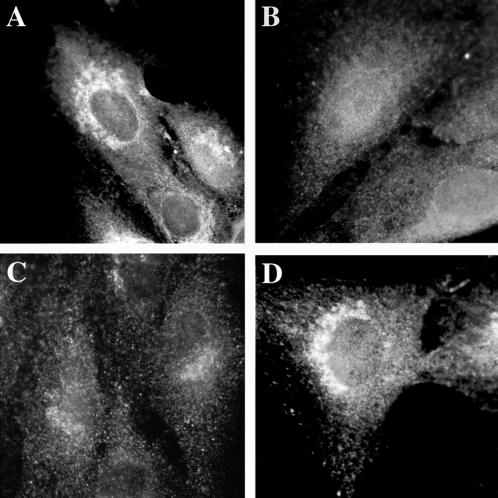
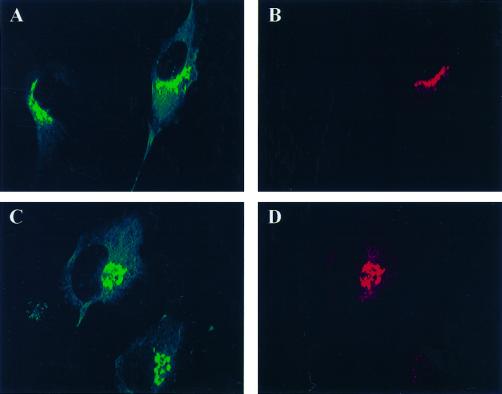
The localization of both endogenous and overexpressed GGA1 was studied in cultured mammalian cells (Figure 6). Weak perinuclear staining of endogenous GGA1 was observed when NRK, CHO, or HeLa cells were stained by indirect immunofluorescence (Figure 6A). Perinuclear staining of endogenous or overexpressed GGA1 was lost by preincubating the antiserum with recombinant GST-GGA1(145–639) (Figure 6B). The perinuclear staining was increased markedly, but otherwise was very similar to endogenous staining, after cells were either microinjected (Figure 6D) or transfected (see Figures 8–10) with vectors directing expression of full-length GGA1. When the overexpressed protein was GGA1 carrying the HA epitope at the N terminus, we obtained images with the GGA1 antibody and the HA antibody that were indistinguishable (Figure 7, compare A and G with D). Together with the evidence that GGA1 is predominantly a soluble protein, these data support the conclusion that GGA1 is peripherally associated with membranes that reside in the perinuclear region.
Figure 6.
Endogenous and overexpressed GGA1 proteins localize to the perinuclear region in NRK cells. NRK cells were cultured, fixed, and permeabilized as described in MATERIALS AND METHODS. (A) The rabbit polyclonal GGA1 antiserum (R-79709) was used as the primary antibody at a dilution of 1:500 to increase the signal from endogenous protein. (B) Specific staining was prevented by previous incubation of the primary antibody with antigen. (C) Exposure of the cells to BFA (30 μM) for 1 min. (D) Staining very similar to that of endogenous protein was seen after the GGA1-GST fusion protein was introduced into cells by microinjection.
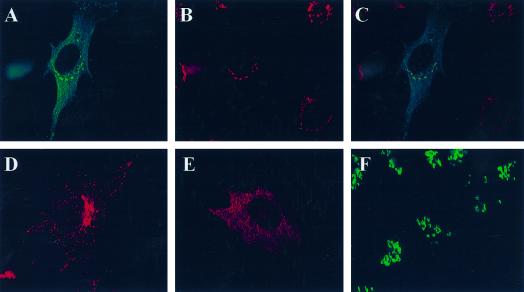
Figure 8.
Like GGA1, GGA2 and GGA3 each localize to the _trans_-Golgi region and are sensitive to BFA. HA-GGA2 (A–C) or HA-GGA3 (D–F) was transiently transfected into NRK cells, and indirect immunofluorescence was performed as described in MATERIALS AND METHODS. The top panels show the colocalization of HA-GGA2 (A) with TGN38 (B), and the overlaid images are shown in C. The bottom panels show the staining of NRK cells expressing HA-GGA3. The typical pattern of staining for GGA3 is shown in D. The doubly stained images in E and F reveal the BFA sensitivity of the perinuclear localized GGA3. Cells treated with 10 μM BFA for 2 min show much more diffuse staining of HA-GGA3 (E), whereas the staining of mannosidase II in the same cells is unchanged (F) at this early time point.
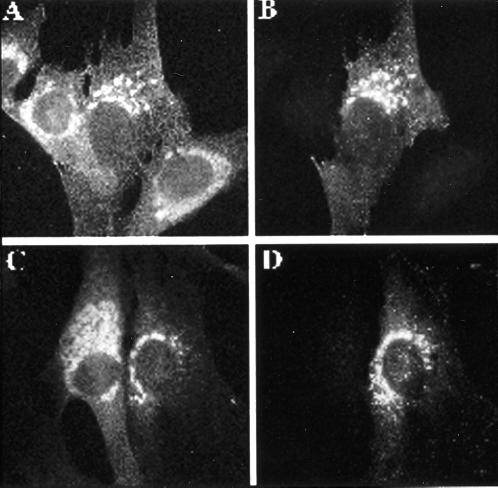
Figure 10.
Overexpression of GGA1 prevents [Q71L]ARF1-induced expansion of the Golgi. A/B and C/D show two different examples of doubly stained images revealing that GGA1 expression blocks the Golgi vesiculation resulting from [Q71L]ARF1 expression. GGA1 was transiently transfected into NRK cells stably transfected with inducible human [Q71L]ARF1. Three hours after transfection with the GGA1 plasmid, cells were induced with interferon to express human [Q71L]ARF1. After 21 h of induction, cells were labeled with antibodies to ARF (1D9; A and C) or GGA1 (R-79709; B and D).
Figure 7.
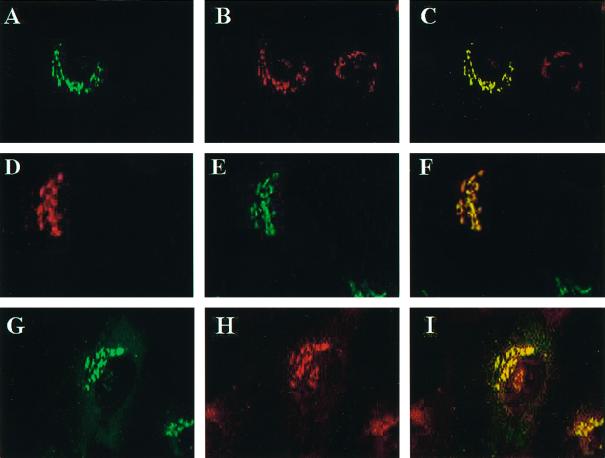
GGA1 localizes to the TGN and colocalizes with ARFs. Full-length wild-type or HA-tagged GGA1 was expressed in NRK cells after transient transfection, and cells were fixed and permeabilized as described in MATERIALS AND METHODS. Staining of GGA1 is shown in A, D, and G; A and G show GGA1 staining with rabbit polyclonal antiserum (R-79709), and D shows the staining profile of GGA1-HA revealed with the mAb to the HA epitope (12CA5). Cells in B were stained with the TGN38 mAb, and those in E were stained with the polyclonal mannosidase II antiserum. The primary antibody in H was the pan-ARF antibody 1D9. The images in the left and middle panels were merged and are shown on the right in each case. Note the near colocalization of GGA1 with the marker of the medial-Golgi (mannosidase II) and the improved colocalization of GGA1 with TGN38 and ARFs.
A number of other antibodies were then used for indirect immunofluorescence to further define the location of GGA1 in cultured cells. NRK cells overexpressing either wild-type or HA-tagged GGA1 were used because their specific signals were much stronger. Double staining with GGA1 and mannosidase II antibodies revealed very similar patterns. However, when the images were overlaid, it became clear that these two antisera stained overlapping but distinct compartments (Figure 7, D–F). Because mannosidase II is a lumenal marker for the medial-Golgi stacks and the GGA1 staining was so similar, it was likely that GGA1 was simply localizing to distinct stacks in the Golgi.
When cells were costained with GGA1 and TGN38 antisera, we saw patterns that were superimposable when the images were overlaid (Figure 7, A–C). Antibodies directed against two other markers of the TGN, M6PR and AP-1, were also found to colocalize with GGA1. Thus, we conclude that GGA1 is localized primarily to the TGN.
The localization of GGA1 to the TGN was extended to include TGN-derived vesicles by costaining of NRK cells with GGA1 and AP-1 antibodies. Both antibodies stained the TGN. In addition, the punctate, vesicular staining in the cytosol, described above for the GGA1 antibody, overlaid extensively with the AP-1 staining. In contrast, antibodies directed against the α-adaptin subunit of AP-2 did not colocalize with GGA1.
We also tested for GGA1 colocalization with ARFs. ARFs were originally shown to localize to the _cis_- and medial-Golgi (Stearns et al., 1990b), and more recent evidence supports the presence of ARFs on the TGN (Stamnes and Rothman, 1993; Traub et al., 1993; Hirst and Robinson, 1998). When cells were stained with the monoclonal anti-ARF (1D9) antibody and the GGA1 antiserum, there was clear overlap in the high-density staining of each around the nucleus, consistent with partial colocalization of GGA1 and ARFs (Figure 7, G–I). However, the punctate staining throughout the cytosol, detected with 1D9 and seen with overexposure of anti-GGA1 images, did not colocalize.
The sensitivity of Golgi-localized proteins to the fungal metabolite BFA has been correlated with in vivo ARF interaction, because BFA can inhibit some ARF exchange factor activities (Peyroche et al., 1996, 1999; Togawa et al., 1999). NRK cells overexpressing GGA1 were treated with BFA and then fixed and stained with GGA1 and mannosidase II, β-COP, or ARF antibodies. The mannosidase II distribution is sensitive to this concentration of BFA only after >5 min. β-COP and ARF staining have been shown previously to be peripherally associated with Golgi membranes and rapidly (0.5–5 min) dissociate upon exposure of NRK cells to BFA (Donaldson et al., 1992; Zhang et al., 1994). Within 1 min of BFA treatment, endogenous and overexpressed GGA1 (Figure 6C), ARF, and β-COP staining in the perinuclear region all become more diffuse. Within 5 min, nearly all of the GGA1 and β-COP staining was diffuse, whereas the mannosidase II staining of the Golgi stacks was intact. These data are consistent with a model in which the localization of GGA1 to the Golgi compartment is ARF dependent.
Localization of GGA2 and GGA3
Because we still lack antisera specific for GGA2 or GGA3, we determined the location of transiently expressed, N-terminal, HA epitope–tagged versions of the two proteins. In each case, the densest staining was seen near the nucleus, similar to staining seen with the GGA1 antiserum (Figure 8). This perinuclear staining was also revealed to be BFA sensitive in each case, because it was quickly (<5 min) dispersed by exposure to 10 μM BFA (Figure 8, E and F).
Furthermore, the GGA2 and GGA3 staining were coincident with the staining of TGN38 (Figure 8), M6PR, and AP-1 and overlapped with mannosidase II and β-COP staining. Similar results were obtained in NRK, CHO, and HeLa cells. That all three GGAs localize to the same membranes was confirmed when NRK cells were transfected with plasmids directing expression of both GGA1 and GGA2-HA or GGA1 and GGA3-HA and indirect immunofluorescence was performed with both GGA1 and HA antibodies (Figure 9).
Figure 9.
GGA1, GGA2, and GGA3 localize to the same membranes. NRK cells were transiently transfected with plasmids directing the overexpression of GGA1 and HA-GGA2 (A and B) or GGA1 and HA-GGA3 (C and D), as described in MATERIALS AND METHODS. Cells in A and C were stained with the GGA1 antiserum R-79709, and cells in B and D were stained with the HA antibody 12CA5.
Although the densest staining for all three GGAs was in the TGN, there were also distinct features of the staining of each GGA. GGA1 exhibited very little punctate staining in the cytosol. GGA3 displayed distinct punctate staining in cytosol that did not colocalize with markers of mitochondria, ER, or _cis_-Golgi. In contrast, GGA2 was predominantly seen as punctate spots that were more diffusely arranged, i.e., less concentrated around the nucleus. Diffuse staining throughout the cell was also evident, although it is difficult to see in the figures. In all three cases, the BFA sensitivity was observed only for the perinuclear TGN staining and not for the punctate staining in the cytosol.
Relationship between ARF Activity and GGAs at the Golgi
The induced expression of the dominant activating mutant of ARF1 (Zhang et al., 1994), ARF3, or ARF4 (C. Zhang and R.A. Kahn, unpublished observations) in mammalian cells causes dramatic expansion of the ER, causes engorgement and vesiculation of the Golgi stacks, inhibits protein secretion, and blocks protein exit from the ER (Dascher and Balch, 1994; Zhang et al., 1994). These effects have been proposed to result from the ARF-induced recruitment of the COP-I coats, which lack the ability to uncoat, as a result of the lack of GTP hydrolysis by these mutated ARFs. We reasoned that the increased presence of GGAs at the same membranes should either exacerbate (if GGAs and COP-I both function in a common pathway) or oppose (if GGAs and COP-I function in distinct pathways) these effects of the activating mutant on vesicular traffic and organelle morphology.
NRK cell lines that had been stably transfected with a plasmid that allowed the interferon-inducible expression of [Q71L]ARF1 were used (Zhang et al., 1994). Increased expression of GGA1 in these cells was achieved by either microinjection of GST-GGA1(145–639) or transient transfection of the pcDNA3.1-based vector, with similar results, and was visualized with either FITC-dextran or staining with R-79709. In untransfected or mock-transfected cells, [Q71L]ARF1-expressing cells have dramatically swollen and expanded Golgi, when visualized with ARF antibodies, as described previously (Zhang et al., 1994) and shown in Figure 10. In contrast, the Golgi in cells that overexpress GGA1 do not expand or vesiculate and look very much like control NRK cells when viewed by light microscopy (Figure 10). Thus, instead of enhancing the effects on Golgi morphology that are associated with the expression of [Q71L]ARF1, GGA1 overexpression prevented these changes in Golgi structure. These data support the conclusions that GGA1 is active at Golgi membranes and can act antagonistically to the ARF effector(s) that promotes Golgi expansion/vesiculation.
Role of GGA1 in Membrane Traffic
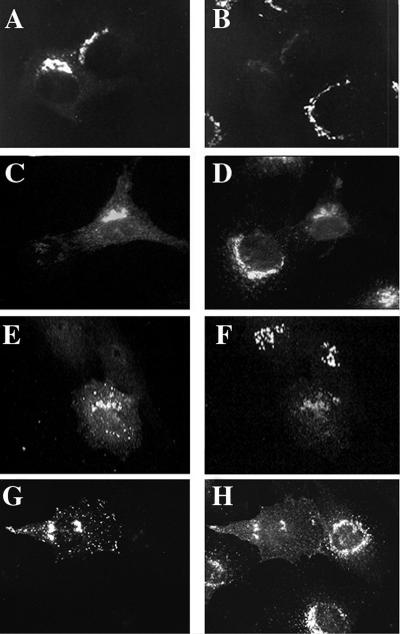
Both M6PR and TGN38 are transmembrane proteins that cycle between the TGN and the cell surface, with the majority of staining normally evident at the TGN of NRK cells. The overexpression of GGA1 caused a reduction of TGN38 staining at the TGN that approached a complete loss of staining in some cells (Figure 11, A and B). This effect was similarly observed with the overexpression of GGA3 (Figure 11, E and F). Although decreased staining in the perinuclear region was clear and observed consistently, increased TGN38 staining at the plasma membrane was not always evident. We conclude that this is the result of more diffuse distribution over the entire cell surface. When cells were stained for M6PR, a variety of effects were apparent, correlating with the amount of GGA1 expressed in each cell. The predominant effect was a redistribution of M6PR from the TGN to a primarily plasma membrane location in GGA1-overexpressing cells (Figure 11, C and D). This experiment was also performed with HA-GGA3, and an even more dramatic redistribution of M6PR receptors was observed (Figure 11, G and H); in this instance, the increased staining at the plasma membrane is evident.
Figure 11.
Markers of the TGN compartment, TGN38 and M6PR, are redistributed in cells overexpressing GGA1 or HA-GGA3. GGA1 (A and B), HA-GGA1 (C and D), or HA-GGA3 (E–H) were transiently transfected into NRK cells, as described in MATERIALS AND METHODS. Doubly labeled images are shown of cells stained for GGA1 (A) and TGN38 (B); HA-GGA1 (C) and M6PR (D); HA-GGA3 (E) and TGN38 (F); and HA-GGA3 (G) and M6PR. Note the loss of signal from TGN38 and M6PR in cells overexpressing GGAs compared with untransfected cells in the surrounding area.
Both β-COP, a marker for COP-I, and p58, a marker for the intermediate compartment, cycle between the intermediate compartment and the _cis_-Golgi and were monitored for changes in localization upon GGA1 overexpression. The distribution and intensity of p58 staining were unaffected by GGA1 overexpression, whereas the distribution and intensity of β-COP were only moderately affected by excess GGA1. Specifically, the β-COP signal at the Golgi was reduced slightly, but the punctate, more diffuse cytosolic staining was unaffected. At the highest levels of GGA1 expression, the morphology of the Golgi, as seen with mannosidase II staining, was altered, i.e., it became smaller, more diffuse, and, in a few instances, lost all discernible Golgi stacks.
DISCUSSION
We describe the isolation and initial characterization of three members of a new family of ubiquitously expressed ARF effectors in mammalian cells, termed GGAs. The conclusion that these are direct effectors of ARF signaling in cells is based on several criteria: 1) GGAs bind directly to ARFs in a GTP-dependent manner in both the yeast two-hybrid assay and on an affinity matrix; 2) localization in intact cells and sensitivity to BFA are consistent with interaction with ARFs; 3) increased expression leads to changes in membrane traffic, particularly in the region of the TGN; and 4) they share a region with sequence homology to γ-adaptin, a component of the AP-1 tetrameric coat complex and a putative effector for ARF action in cells. We propose that GGAs are involved in the regulation by ARFs of vesicular traffic exiting the TGN and discuss potential overlap between GGAs and other putative ARF effectors.
The full-length ORFs of all three GGA proteins were cloned from human tissue sources and encode proteins that are similar in length, roughly 33% identical when all three are aligned and 37–47% identical in pairwise alignments. They also have N- and C-terminal regions of 300 and 150 residues, respectively, that are 55–75% identical. ARFs bind to the conserved N-terminal domain. Both Northern blot data from a variety of human tissues and the presence of multiple EST sequences from libraries, derived from a variety of tissues, are consistent with all three GGAs being expressed in a wide variety of human tissues. Other than the similarities in sequence among the GGAs, there was little functional information gleaned from the sequences themselves.
One interesting exception to this are the C termini of GGAs, which possess significant (44% identity and 70% homology over 73 residues) homology with the ear domain of γ-adaptin, a subunit of the AP-1 complex. This region of γ-adaptin is divergent from the other large subunits of the AP complexes. Some TGN-targeting information resides in the ear domain, although it appears to be secondary to targeting signals in the N-terminal domains of γ- and α-adaptins (Page and Robinson, 1995). An intriguing possibility for the GGAs is that the C-terminal domain is involved in interactions with other (unknown) proteins to specify their localization at the TGN. We are currently testing this hypothesis. Other possible functions of the γ-adaptin ear involve proper assembly with the other adaptin subunits, although not through direct protein–protein interactions, and the assembly of adaptin multimers (Page and Robinson, 1995). Preliminary efforts to identify protein complexes containing GGA1, including immunoprecipitation and gel filtration chromatography, have not proven productive. Should conditions be found to stabilize such a (potential) protein complex, it will be possible to determine the presence of ARFs under different conditions. Alternatively, it is possible that the biologically important interactions of GGAs require membrane association, making their identification by copurification more problematic. The colocalization of GGA1 and AP-1 in vesicles argues against the GGAs serving as adaptors for a distinct group of vesicles leaving the TGN and provides a useful source for further dissection of GGA-containing protein complexes.
The binding of ARF to the GGA proteins was found to be highly specific and almost certainly regulated in cells. In addition to the controls built into the two-hybrid screen, we also found that all ARF isoforms interact well with GGAs but that the structurally related ARF-like (Arl) proteins (Arl1 and Arl2; 40–50% identical to ARFs) do not (Boman et al., 1999). The facts that all three GGAs preferentially interacted with the activated mutant of ARF3 over the wild-type protein and that GST-GGA1(145–639) bound ARF3 in a GTP-dependent manner on an affinity matrix are consistent with the proposed role of GGAs as immediate downstream effectors of ARF proteins.
Three lines of experimental evidence support the conclusion that ARFs and GGAs colocalize in cells. First, double labeling of fixed cells with antibodies to ARFs and GGAs revealed evidence of overlap in their locations in cells. Compared with other markers of organelles, it is clear that the highest concentration of GGA1 is at the TGN, and ARFs have been physically and functionally localized to this compartment (Stearns et al., 1990a). Epitope tagging of GGA1 was shown to have no detectable effect on the localization of this protein; by extension, we tentatively conclude that the same will hold true for GGA2 and GGA3. When the N-terminal–tagged GGA proteins were transiently expressed in mammalian cells, they colocalized in large part with TGN markers, but each protein also revealed additional staining: faint punctate cytosolic staining with HA-GGA1, which is more pronounced with HA-GGA3, and more scattered punctate staining around the TGN and more diffuse staining throughout the cell with HA-GGA2. Thus, although all three GGAs are present on TGN membranes and partially colocalize with ARFs, each may also migrate to a distinct location likely to be functionally related to the TGN.
Second, the localization of GGAs to the TGN is rapidly and reversibly blocked upon exposure to BFA. Because the only clearly identified targets for BFA are ARF exchange factors (Peyroche et al., 1999), which are inhibited upon binding of BFA, it is currently thought that the actions of BFA on cells follow from a deficiency of active ARF. Thus, we conclude that the localization of GGAs at the TGN is dependent on their binding to activated ARF. Although the most intensively studied aspect of BFA action is the ARF-dependent recruitment of COP-I to Golgi membranes, it is also known that BFA induces alterations in the localization of markers of the TGN, specifically M6PR, resulting in increased staining at the plasma membrane. The recruitment of other coat complexes, specifically AP-1 (Stamnes and Rothman, 1993; Traub et al., 1993), AP-3 (Simpson et al., 1997; Faundez et al., 1998; Ooi et al., 1998), and AP-4 (Dell'Angelica et al., 1999; Hirst et al., 1999), has also been shown to be BFA sensitive and, thus, predicted to require active ARF.
Third, and somewhat paradoxically, we found that when GGA1 was transiently overexpressed in NRK cells, there was a clear enhancement in the staining of ARF at the TGN (Zhu et al., 2000). The simplest interpretation of this observation is that the effectors (GGAs) bind and stabilize the active conformation of ARFs, leading to the accumulation of the activated form of ARF, but an alternative explanation has also been proposed (Zhu et al., 2000). Thus, we conclude that the GGAs have all the characteristics of immediate downstream effectors that mediate the actions of ARF at TGN membranes.
The identification of the three GGA proteins binding to the TGN brings to at least seven the number of different proteins, or protein complexes, that are regulated by interactions with ARFs at the Golgi: COP-I, AP-1, AP-4, PLD1, and the three GGAs. This high density of ARF effectors begs the question of how specificity in signal propagation is achieved in intact cells. To begin to address this question, we first asked if GGAs were mediators of one or more of the activities of ARF at the Golgi, all of which were ascribed previously to recruitment of coat proteins, e.g., COP-I or AP-1, or activation of phospholipase D (PLD1). The inducible expression of the dominant activated ARF mutant [Q71L]ARF1 in mammalian cells has been shown to cause enlargement of the ER and the Golgi lumen, vesiculation of the Golgi into heterogeneously sized vesicles and later cell death (Zhang et al., 1994), and inhibition of protein export from the ER (Dascher and Balch, 1994). If GGAs are mediators of one or more of these phenotypes, we predicted an exacerbation of these morphological changes upon overexpression. In contrast, overexpression of any of the GGAs prevented this dramatic Golgi expansion induced by [Q71L]ARF1 expression (Figure 10). This effect is further evidence of a functional interaction between GGAs and ARFs in cells, but rather than being mediators of the Golgi expansion, GGAs mediate a distinct pathway that can compete with the mediator(s) of Golgi vesiculation, perhaps by direct competition for the binding of the activated ARF. Thus, in contrast to COP-I and PLD1, which may each work in a single ARF-regulated pathway, the GGAs are likely to mediate a distinct process from these two other ARF effectors. In a related study (Kuai et al., 2000), a series of mutations in the switch I and II regions of ARF3 were characterized with regard to differential effector interactions. These mutants allowed the functional separation of defects in binding to GGAs, recruitment of COP-I, and activation of PLD1, supporting the conclusion that none of these five effectors (PLD1, COP-I, or the three GGAs) can be the sole mediator of the Golgi vesiculation caused by an excess of ARF activity.
By monitoring the effects of GGA overexpression on a variety of markers of the secretory pathway, we conclude that the primary site and role of GGA action is most likely the regulation of protein export from the TGN. The redistribution of both TGN38 and M6PR, both transmembrane proteins, away from the TGN is among the strongest evidence to support this model. This effect is most evident in cells that overexpress GGA3. This conclusion is further supported by the observation that GGA1 colocalizes with AP-1 but not with AP-2. With high levels of GGA expression, we also observed changes in the markers of the _cis_- and medial-Golgi: β-COP and mannosidase II, respectively. We take this as further evidence that the functions of GGAs in cells include the regulation of vesicular transport. We have not detected any changes in the staining of PDI (a marker for the ER lumen) or p58 (a marker for the intermediate compartment) with overexpression of any GGA protein.
In several, but not all, experiments, the increased expression of any of the three GGA proteins caused a decrease in the staining of transferrin or transferrin receptors (A.L. Boman, C.-j. Zhang, and R.A. Kahn, unpublished observation). This may suggest the ability of GGA proteins to modify the recycling endosome pathway, but it is apparently at odds with the observation that GGA1 does not colocalize with AP-2. Although we conclude that the compartments that are affected first and most dramatically by increased GGA expression are those in the TGN-endosome secretory pathway, we clearly cannot exclude actions of GGAs at other membrane sites. Indeed, the findings that GGA1 is soluble and likely recruited to membranes along with ARFs and that ARFs are active at virtually all membranes in cells make it likely that GGAs will be found to have roles in vesicular transport at other sites as well.
Another aspect of the complexity of ARF signaling at Golgi/TGN membranes is that we anticipate either that some level of ARF isoform specificity must exist or, more likely, that recruitment involves additional factors that may be differentially localized or regulated. Candidates for the latter possibility include ARF GAPs, because evidence has recently been presented (Goldberg, 1999) that supports the simultaneous binding of ARF GAP and COP-I to activated ARF. The apparent redundancy in the GGA family offers another possible explanation for or source of its specificity, and the differences in staining between the different GGAs offer preliminary evidence for specific differences in their actions.
Clearly, one of the challenges in the near future is to determine the relationship between the ARF-dependent recruitment of GGAs and coat complexes to Golgi membranes. Our choice of M6PR as marker for the TGN was not arbitrary, because M6PRs have been implicated as regulators of AP-1 recruitment. With the discovery of a role for ARF in AP-1 recruitment, it was proposed that ARFs activate M6PRs to promote coat formation at the TGN (Le Borgne et al., 1993; Le Borgne and Hoflack, 1997). The use of M6PR-deficient cell lines to assess the requirement for M6PR in AP-1 recruitment has led to conflicting conclusions (Le Borgne and Hoflack, 1997; Zhu et al., 1999). One model proposes that activated ARF initiates the formation of a specific binding site for AP-1, which in turn binds clathrin to promote the maturation of the budding vesicle. With the discovery of three new, direct binders of ARF·GTP at the TGN (the GGAs), we speculate that the recruitment of GGAs to the TGN may be a prerequisite for adaptin complex binding or that the binding of GGAs and adaptins could occur simultaneously. We are currently testing this and related models in an effort to provide more direct evidence for the roles of ARFs and GGAs in membrane traffic at the TGN.
ACKNOWLEDGMENTS
We thank the following people for their gifts of reagents: S. Elledge (two-hybrid plasmids and yeast strains), R. Brazas (plasmid pBG4D), K. Moremen and M. Farquhar (mannosidase II antisera), L. Traub (AP.6 antibody), K. von Figura (M6PR antibodies), G. Banting (p58 antibody), and L. Zhang and H. Fu (plasmid). We also thank the other members of the Kahn laboratory for helpful discussions and R. Ahmed for the use of his microscope during the early phases of this work. It was discovered during the preparation of this article that Margaret Robinson and colleagues have similar data on the characterization of GGA proteins, and we acknowledge and appreciate the collegial sharing of information that resulted.
Abbreviations used:
ARF
ADP-ribosylation factor
BFA
brefeldin A
bp
base pairs
EST
expressed sequence tag
GAP
GTPase-activating protein
GGA
Golgi-localizing, γ-adaptin ear homology domain, ARF-binding protein
GST-GGA1(145–639)
fusion protein consisting of GST and amino acid residues 145–639 of GGA1
kb
kilobase
M6PR
mannose 6-phosphate receptor
NRK
normal rat kidney
TGN
_trans_-Golgi network
UTR
untranslated region
Footnotes
1
During the preparation of this article, it was noted that a new, apparently full-length protein sequence corresponding to GGA2 had appeared in GenBank (accession number AAC05813) as a result of sequencing of genomic human DNA from chromosome 16p12. The encoded protein is the same length as GGA2 and is nearly identical to our GGA2 sequence. Only two differences were noted: our residue tryptophan 281 is a proline and our proline 425 is an alanine in AAC05813. The sources of these differences are unknown, but these data otherwise agree with the sequence we obtained.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balch WE, Kahn RA, Schwaninger R. ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J Biol Chem. 1992;267:13053–13061. [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Boman AL, Kahn RA. ARF proteins: the membrane traffic police? Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- Boman AL, Taylor TC, Melancon P, Wilson KL. A role for ADP-ribosylation factor in nuclear vesicle dynamics. Nature. 1992;358:512–514. doi: 10.1038/358512a0. [DOI] [PubMed] [Google Scholar]
- Boman AL, Zhu X, Kuriyama R, Kahn RA. ARF interacts with mitotic kinesin-like protein-1 (MKLP1) in a GTP-dependent fashion. Cell Motil Cytoskeleton. 1999;44:119–132. doi: 10.1002/(SICI)1097-0169(199910)44:2<119::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Cavenagh MM, Whitney JA, Carroll K, Zhang C, Boman AL, Rosenwald AG, Mellman I, Kahn RA. Intracellular distribution of ARF proteins in mammalian cells: ARF6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Demolliere C, Hennecke S, Duden R, Letourneur F. Delta- and zeta-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Balch WE. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- Dell'Angelica EC, Mullins C, Bonifacino JS. AP-4, a novel protein complex related to clathrin adaptors. J Biol Chem. 1999;274:7278–7285. doi: 10.1074/jbc.274.11.7278. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Cassel D, Kahn RA, Klausner RD. ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc Natl Acad Sci USA. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Klausner RD. ARF: a key regulatory switch in membrane traffic and organelle structure. Curr Opin Cell Biol. 1994;6:527–532. doi: 10.1016/0955-0674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Lippincott-Schwartz J, Bloom GS, Kreis TE, Klausner RD. Dissociation of a 110-kD peripheral membrane protein from the Golgi apparatus is an early event in brefeldin A action. J Cell Biol. 1990;111:2295–2306. doi: 10.1083/jcb.111.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Faundez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell. 1998;93:423–432. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Gant TM, Wilson KL. ARF is not required for nuclear vesicle fusion or mitotic membrane disassembly in vitro: evidence for a non-ARF GTPase in fusion. Eur J Cell Biol. 1997;74:10–19. [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell. 1999;96:893–902. doi: 10.1016/s0092-8674(00)80598-x. [DOI] [PubMed] [Google Scholar]
- Happe S, Weidman P. Cell-free transport to distinct Golgi cisternae is compartment specific and ARF independent. J Cell Biol. 1998;140:511–523. doi: 10.1083/jcb.140.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyzes exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Hirst J, Bright NA, Rous B, Robinson MS. Characterization of a fourth adaptor-related protein complex. Mol Biol Cell. 1999;10:2787–2802. doi: 10.1091/mbc.10.8.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Robinson MS. Clathrin and adaptors. Biochim Biophys Acta. 1998;1404:173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Clark J, Rulka C, Stearns T, Zhang CJ, Randazzo PA, Terui T, Cavenagh M. Mutational analysis of Saccharomyces cerevisiae ARF1. J Biol Chem. 1995;270:143–150. doi: 10.1074/jbc.270.1.143. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J Biol Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- Kanoh H, Williger BT, Exton JH. ARFaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J Biol Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- Kuai J, Boman AL, Arnold R, Zhu X, Kahn RA. Effects of activated ARF on Golgi morphology require neither activation of PLD1 nor recruitment of COPI. J Biol Chem. 2000;275:4022–4032. doi: 10.1074/jbc.275.6.4022. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Hoflack B. Mannose 6-phosphate receptors regulate the formation of clathrin-coated vesicles in the TGN. J Cell Biol. 1997;137:335–345. doi: 10.1083/jcb.137.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Schmidt A, Mauxion F, Griffiths G, Hoflack B. Binding of AP-1 Golgi adaptors to membranes requires phosphorylated cytoplasmic domains of the mannose 6-phosphate/insulin-like growth factor II receptor. J Biol Chem. 1993;268:22552–22556. [PubMed] [Google Scholar]
- Lenhard JM, Kahn RA, Stahl PD. Evidence for ADP-ribosylation factor (ARF) as a regulator of in vitro endosome-endosome fusion. J Biol Chem. 1992;267:13047–13052. [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mansour SJ, Skaug J, Zhao XH, Giordano J, Scherer SW, Melancon P. p200 ARF-GEP1: a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Touster O, Robbins PW. Novel purification of the catalytic domain of Golgi alpha-mannosidase II: characterization and comparison with the intact enzyme. J Biol Chem. 1991;266:16876–16885. [PubMed] [Google Scholar]
- Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LJ, Robinson MS. Targeting signals and subunit interactions in coated vesicle adaptor complexes. J Cell Biol. 1995;131:619–630. doi: 10.1083/jcb.131.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Scheel J, Horstmann H, Hauri HP, Griffiths G, Kreis TE. Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Weiss O, Kahn RA. Preparation of recombinant ADP-ribosylation factor. Methods Enzymol. 1995;257:128–135. doi: 10.1016/s0076-6879(95)57018-7. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Yang YC, Rulka C, Kahn RA. Activation of ADP-ribosylation factor by Golgi membranes: evidence for a brefeldin A- and protease-sensitive activating factor on Golgi membranes. J Biol Chem. 1993;268:9555–9563. [PubMed] [Google Scholar]
- Schaffner W, Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990a;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Willingham MC, Botstein D, Kahn RA. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci USA. 1990b;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TC, Kahn RA, Melancon P. Two distinct members of the ADP-ribosylation factor family of GTP-binding proteins regulate cell-free intra-Golgi transport. Cell. 1992;70:69–79. doi: 10.1016/0092-8674(92)90534-j. [DOI] [PubMed] [Google Scholar]
- Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- Traub LM, Ostrom JA, Kornfeld S. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- West MA, Bright NA, Robinson MS. The role of ADP-ribosylation factor and phospholipase D in adaptor recruitment. J Cell Biol. 1997;138:1239–1254. doi: 10.1083/jcb.138.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Rosenwald AG, Willingham MC, Skuntz S, Clark J, Kahn RA. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Boman, A.L, Amor, J.C., Cieplak, W., Jr., and Kahn, R.A. (2000). Effectors increase the affinity of ARF for GTP and lead to increased binding. J. Biol. Chem. (in press). [DOI] [PubMed]
- Zhu Y, Traub LM, Kornfeld S. High-affinity binding of the AP-1 adaptor complex to trans-Golgi network membranes devoid of mannose 6-phosphate receptors. Mol Biol Cell. 1999;10:537–549. doi: 10.1091/mbc.10.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]