Pause Sites Promote Transcriptional Termination of Mammalian RNA Polymerase II (original) (raw)
Abstract
Polymerase II (Pol II) transcriptional termination depends on two independent genetic elements: poly(A) signals and downstream terminator sequences. The latter may either promote cotranscriptional RNA cleavage or pause elongating Pol II. We demonstrate that the previously characterized MAZ4 pause element promotes Pol II termination downstream of a poly(A) signal, dependent on both the proximity of the pause site and poly(A) signal and the strength of the poly(A) signal. The 5′→3′ exonuclease Xrn2 facilitates this pause-dependent termination by degrading the 3′ product of poly(A) site cleavage. The human β-actin gene also possesses poly(A) site proximal pause sequences, which like MAZ4 are G rich and promote transcriptional termination. Xrn2 depletion causes an increase in both steady-state RNA and Pol II levels downstream of the β-actin poly(A) site. Taken together, we provide new insights into the mechanism of pause site-mediated termination and establish a general role for the 5′→3′ exonuclease Xrn2 in Pol II termination.
Transcriptional termination can be defined as cessation of RNA synthesis followed by polymerase-DNA dissociation. Correct termination serves to maintain an active cellular pool of RNA polymerase II (Pol II) and to insulate downstream promoters from elongating Pol II (11). Despite the fundamental importance of this process, its mechanism remains enigmatic. Transcripts of all protein-encoding genes, with the exception of histone genes, contain a poly(A) signal at their 3′ end composed of an AAUAAA sequence positioned 10 to 50 nucleotides upstream of a GU-rich downstream sequence element (see references 20 and 38 for reviews). The 5′ product of endonucleolytic cleavage between these two sequences is polyadenylated and therefore stabilized. The 3′ product of the cleavage is unstable and rapidly degraded.
A substantial body of evidence argues that cleavage and polyadenylation of Pol II transcripts occur cotranscriptionally, effectively releasing mRNA from the transcription site. Thus, defects in mRNA 3′-end processing cause the accumulation of pre-mRNA at nuclear transcription foci both in yeast (13) and in mammals (6). Furthermore, numerous studies have shown that the C-terminal domain of the Pol II large subunit promotes efficient cleavage and polyadenylation (see references 14, 18, and 21 for reviews). It is also well documented that for mRNAs with multiple poly(A) signals, selection of a downstream poly(A) site results in mRNA with internal unprocessed poly(A) signals (7, 10). If 3′ processing occurred posttranscriptionally, then usage of upstream poly(A) signals would always predominate. Finally, the poly(A) site is necessary for efficient transcriptional termination. However, the site of Pol II release is often distinct from the gene's poly(A) site. Thus, Pol II terminates transcription at sites positioned between 100 bp and several kb downstream of the poly(A) site (2, 8, 10, 12, 29). These observations suggest a requirement for additional, distal sequences in the termination process.
A sequence (G5AG5) which binds the transcription factor MAZ in vitro is thought to pause Pol II and promote termination of the human C2 complement gene (3). Moreover, in vitro studies demonstrated that four MAZ binding sites positioned downstream of a synthetic poly(A) site (SPA) promoted more-efficient 3′ end formation than did mutated MAZ binding sites (34, 35). MAZ binding sites have also been shown to change patterns of alternative and _trans-_splicing through pausing of Pol II, delaying subsequent transcription of splicing signals (23, 24, 28). Despite much study, the mechanism of MAZ site-dependent Pol II transcriptional termination in vivo is unknown.
Another sequence, positioned 800 bp downstream of the poly(A) site, was previously identified as being essential for transcriptional termination of the human β-globin gene (9). RNA transcribed from this sequence encodes a cotranscriptional cleavage (CoTC) activity essential for termination. CoTC provides two naked RNA termini. It was revealed that the Pol II-associated terminus is a substrate for the 5′→3′ exoribonuclease Xrn2 (32). Cotranscriptional degradation of this RNA by Xrn2 was shown to lead to transcriptional termination. Interestingly, the yeast homologue Rat1 has also been shown to influence termination of Pol II-transcribed genes through degradation of the 3′ product of poly(A) site cleavage (16). It is thought that a 5′ phosphate group on the terminal residue of an RNA chain creates a preferred substrate for Xrn2/Rat1 (27). Significantly, experiments have indicated that CoTC-generated ends possess a 5′ phosphate (30), as does the 3′ product of poly(A) site cleavage (26). It is therefore possible that poly(A) site cleavage can act as a precursor to Pol II transcriptional termination in an Xrn2-dependent manner in humans.
A role for the poly(A) site in transcriptional termination is firmly established. However, a requirement for cleavage at the poly(A) site to elicit proper Pol II release has not been demonstrated. Electron micrographs of plasmids, injected into Xenopus oocytes, showed that when cleavage at the poly(A) site could be directly visualized, termination was more rapid. However, 80% of the time, cleavage at the poly(A) site was not detected and transcription proceeded further into the 3′ flanking region before Pol II release occurred. Whether cleavage at the poly(A) signal also occurred at the time of release could not be determined with this experimental system (18a). It seems plausible that termination of closely spaced genes or genes that are highly expressed may require rapid cotranscriptional poly(A) site cleavage to elicit more rapid termination.
Here, we show that either the MAZ4 pause sequence or a related G-rich region adjacent to the poly(A) signal of the human β-actin gene is able to promote efficient transcriptional termination in conjunction with a strong poly(A) signal. Our data indicate that when Pol II is paused close to the poly(A) site, exoribonucleolytic degradation by Xrn2 of the Pol II-associated product of poly(A) site cleavage promotes termination.
MATERIALS AND METHODS
Plasmid construction.
CoTC (or wild type), ΔCoTC, and ΔpA plasmids have been described in reference 32. Tat plasmid and the VA plasmid and probe have been described previously (9). The nuclear run-on (NRO) probes have been described previously (9, 32). The β4 selection probe has been described in references 1, 9, and 32. MAZ4 and ZAM4 constructs were made by blunt-end insertion of a MAZ4-containing fragment generated by KpnI/EcoRI digestion from MLPIII SPA MAZ4 (35) into a vector prepared by VF/VR (32) amplification of the CoTC plasmid. The mMAZ4 construct was made by blunt-end insertion of KpnI/EcoRI mMAZ4 fragment generated from MLPIII SPA mMAZ4 plasmid (34) into a vector prepared by VF/VR amplification of the CoTC plasmid as described previously (32). MAZ4 ΔpA, mMAZ4 ΔpA, and ΔCoTC ΔpA constructs were made by site-directed mutagenesis of the MAZ4, mMAZ4, and ΔCoTC constructs, respectively, using oligonucleotides described in reference 8, mutating AATAAA to GAATTC. MAZ4/U3 was prepared by blunt-end ligation of the KpnI/EcoRI MAZ4 fragment described above into ΔCoTC vector cut with AflIII. The β/SPA plasmid was made by insertion of the annealed oligonucleotide pair SPAF (GAT CCA ATA AAA GAT CTT TAT TTT CAT TAG ATC TGT GTG TTG GTT TTT TGT GTG GAT C) and SPAR (GAT CCA CAC AAA AAA CCA ACA CAC AGA TCT AAT GAA AAT AAA GAT CTT TTA TTG GAT C) into a vector prepared by VF/VR amplification of the CoTC plasmid (32). SPA/β was made by blunt-end ligation of the annealed oligonucleotide pair SPAF/SPAR into a vector prepared by PCR amplification of the CoTC plasmid with oligonucleotidess p(A)F (CCT TGG GAA AAT ACA CTA TAT C) and p(A)R (AAT CCA GAT GCT CAA GGC CC). Then, this generated plasmid was amplified by oligonucleotidess VF and VR to produce SPA vector. An insert containing β-globin p(A) was then prepared by PCR amplification of the CoTC plasmid with βp(A)F (GGG GAT ATT ATG AAG GGC CTT G) and βp(A)R (AAC TAG CTC TTC ATT TCT TTA TG). Blunt-end ligation of this insert into SPA vector formed SPA/β. Plasmids 30C and 30T were constructed by ligation of the annealed oligonucleotide pairs C30/G30 and T30/A30, respectively, into a vector prepared by PCR amplification of the CoTC plasmid with oligonucleotides VF/VR. The globin/actin fusion construct was made by ligation of the β-actin termination region amplified from human genomic DNA using oligonucleotides A (pr1) (F) (TTA CCC AGA GTG CAG GTG TG) and B (pA) (R) (TCC CAT AGG TGA AGG CAA AG) into β-globin backbone plasmid prepared by PCR amplification of the CoTC plasmid with oligonucleotides VF/VR. The globin/actin ΔpA mutant construct was made by site-directed mutagenesis of the globin/actin construct using oligonucleotides mutating AATAAA to GAATTC (8).
Nascent RNA analysis.
Transfection procedures, NRO, and hybrid selection NRO have been described previously (32). Termination efficiencies (TE) were calculated as follows. The average ratio of signals for DNA probes A/B3 and U3/B3 of the test construct was divided by the same value for the ΔCoTC construct and expressed as a percentage. TE values are based on average values from at least three independent experiments.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analysis on the endogenous β-actin gene was carried out as previously described in the Upstate Biotechnology (Lake Placid, NY) protocol and in references 15 and 32. The Pol II antibody (H-224) was obtained from Santa Cruz Biotechnology, Inc. The immunoprecipitated, nonprecipitated, and input DNAs were used as templates for real-time quantitative PCR performed using a Corbett Research Rotor-Gene GG-3000 machine. The PCR mixture contained QuantiTect SYBR green PCR master mix (QIAGEN), 2 to 3 μl of the template DNA, and the following primers: prom (F), GAG GGG AGA GGG GGT AAA A; prom (R), AGC CAT AAA AGG CAA CTT TCG; ex3 (F), CAT CCT CAC CCT GAA GTA CCC; ex3 (R), TAG AAG GTG TGG TGC CAG ATT; ex5 (F), GGA CAC CGC AAA GAC CTG TA; ex5 (R), CCA GGC AGT GAT CTC CTT CT; ex6 (F), CAA GAT GAG ATT GGC ATG GCT; ex6 (R), CTC CAA CCG ACT GCT GTC ACC; B(F), GGG ACT ATT TGG GGG TGT CT; B(R), TCC CAT AGG TGA AGG CAA AG; C(F), TGG GCC ACT TAA TCA TTC AAC; C(R), CCT CAC TTC CAG ACT GAC AGC; D(F), CAG TGG TGT GGT GTG ATC TTG; and D(R), GGC AAA ACC CTG TAT CTG TGA. Cycling parameters were 95°C for 15 min, followed by 45 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer.
Steady-state RNA analysis.
For S1 analysis, 10 μg of test construct and 1 μg of Tat plasmid were transfected into subconfluent HeLa cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's guidelines. Cytoplasmic RNA was isolated, and 5 to 10 μg of RNA was analyzed by S1 analysis as previously described (8). Probes were made from the β-globin gene constructs CoTC, β/SPA, and SPA/β (see Fig. 1C and 3B) by EcoRI digestion of each plasmid, followed by end labeling with [α-32P]dATP (Amersham) with Klenow DNA polymerase (New England Biolabs) using the manufacturer's guidelines. The VA probe was made as previously described (8).
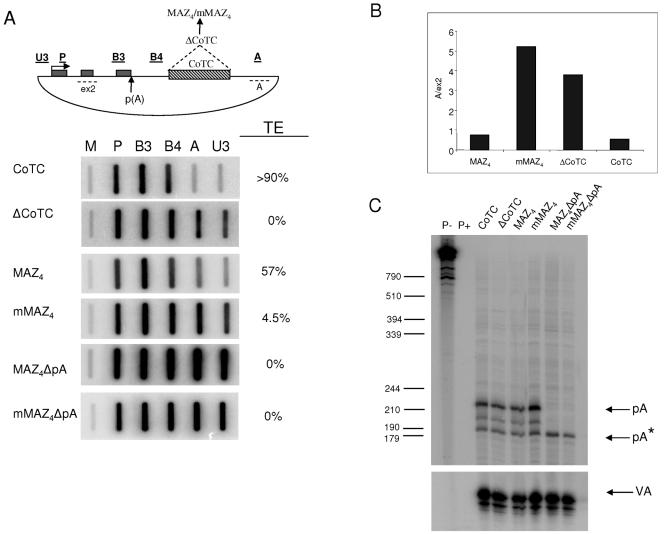
FIG. 1.
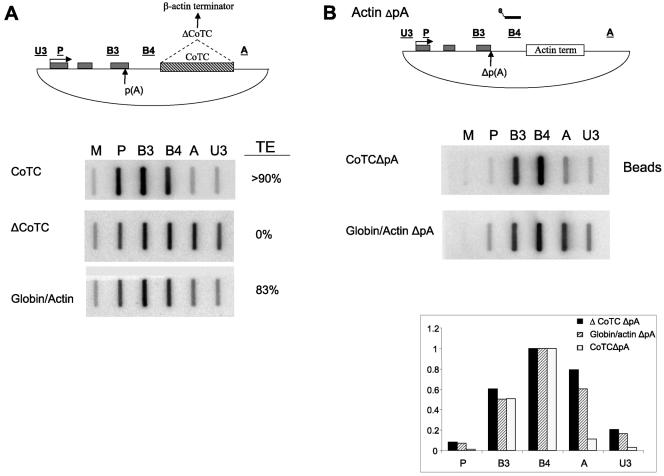
MAZ element promotes poly(A)-dependent transcriptional termination in vivo. (A) β-Globin construct with exons shown as gray boxes and CoTC element as hatched box. NRO probe positions are underlined, and RT-PCR probe positions are dotted lines. The panels below show NRO signals for CoTC, ΔCoTC, MAZ4, mMAZ4, MAZ4ΔpA, and mMAZ4ΔpA constructs. M (empty M13 vector) shows the background signal. The TE is shown against each NRO analysis. See Materials and Methods for details. (B) Quantitative real time RT-PCR analysis of transcription termination efficiency of CoTC, ΔCoTC, MAZ4, and mMAZ4 constructs. Termination efficiency is measured as the ratio of accumulation of steady-state RNA at positions A to ex2 (A/ex2). (C) S1 analysis of cytoplasmic, steady-state RNA of CoTC, ΔCoTC, MAZ4, mMAZ4, MAZ4ΔpA, and mMAZ4ΔpA transfections. Lanes P− and P+ are S1 probes (made with the CoTC plasmid) incubated without or with the S1 nuclease treatment. Note that no background bands were detected over the poly(A) site region in these lanes. Positions of the pA, pA* (cryptic), and VA (cotransfection control) bands detected in S1 analysis are indicated (based on the indicated positions of size markers). Note that read-through transcripts enhanced by mutation of the β-globin poly(A) signal were not detected in cytoplasmic RNA as they are retained in the nucleus, where they are degraded.
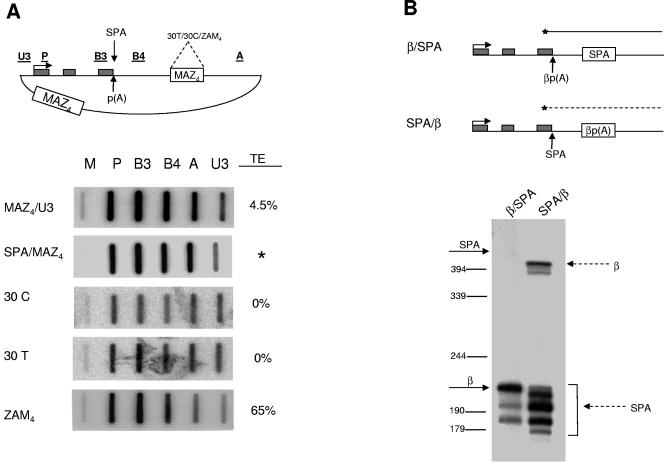
FIG. 3.
Proximity and strength of poly(A) site are important for pause site-mediated transcriptional termination. (A) Human β-globin gene, with exons shown as gray boxes. Positions of SPA and β poly(A) site insertions are indicated; various plasmid modifications (insertion of 30 T, 30 C, and ZAM4 elements) are also depicted on the diagram at the corresponding locations. The panels below show NRO signals for MAZ4/U3, SPA/MAZ4, 30T, 30C, and ZAM4 constructs. TE is indicated. The asterisk indicates that in the case of the SPA/MAZ4 construct, TE could not be determined. (B) Positions of the S1 probes over the β/SPA (solid line) and SPA/β (dashed line) constructs are indicated. The panel below shows S1 analysis of cytoplasmic, steady-state RNA of β/SPA and SPA/β transfections. Positions of the SPA- and β poly(A)-specific bands are indicated (based on size markers indicated).
For reverse transcription (RT)-PCR analysis, total RNA was isolated using TRIzol reagent (Invitrogen), treated with DNase I RNase free (Roche). One microgram of total RNA was reverse transcribed with RT-ex2 (GTG CAG CTT GTC ACA GTG C) and RT-MAZ (CAT CAT TGG AAA ACG TTC TTC) primers positioned immediately downstream of respective PCR primers ex2 and A for the transfected β-globin constructs (see Fig. 1A) or corresponding reverse primers for the β-actin gene (see Fig. 4A), using SuperScript III (Invitrogen) enzyme following the manufacturer's instructions. The real-time quantitative PCR was performed using a Corbett Research Rotor-Gene GG-3000 machine. The PCR mixture contained QuantiTect SYBR green PCR master mix (QIAGEN), 3 μl of template cDNA, primers ex2 (F) (TTG GAC CCA GAG GTT CTT TG), ex2 (R) (GAG CCA GGC CAT CAC TAA AG), A(F) (TTG CCT TCC TGT TTT TGC TC), and A(R) (CCG CTG TTG AGA TCC AGT TC) for the β-globin gene, and primers ex5 (F), ex5 (R), ex6 (F), ex6 (R), B(F), B(R), C(F), C(R), D(F), and D(R) for the actin gene, as used in the ChIP analysis. The β-actin A primers were A(F) (CAA CTT GAG ATG TAT GAA GGC) and A(R) (CCC CAA TAA GCA GGA ACA GA). Cycling parameters were 95°C for 15 min, followed by 45 cycles of 95°C for 15 s, 57.5°C for 30 s (β-globin)/60°C for 30 s (β-actin), and 72°C for 30 s. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer.
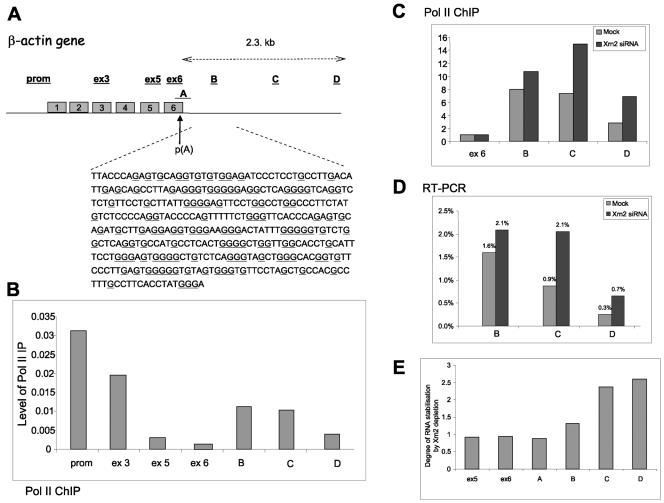
FIG. 4.
Pause-mediated transcriptional termination of endogenous β-actin gene. (A) Human β-actin gene, with exons shown as gray boxes. The positions of the probes for RT-PCR and ChIP are underlined. The sequence presented corresponds to a 380-bp pause element from the 3′ flanking region of the β-actin gene. G nucleotides are underlined. (B) Pol II ChIP analysis across β-actin gene using anti-Pol II antibody. (C) Pol II ChIP analysis across β-actin gene using primers over ex6 and regions B to D of the 3′ flanking sequence. Light-gray bars represent mock-depleted cells, and dark-gray bars are Xrn2-depleted cells. The level of Pol II density over the ex6 probe was taken as 1. (D) Real time RT-PCR analysis of the β-actin gene in mock-treated (light-gray bars) and Xrn2-depleted (dark-gray bars) cells. The level of the steady-state message was corrected relative to the level of the RNA over the ex5 probe (taken as 100%). (E) Ratios of total β-actin transcript levels detected with versus without Xrn2 depletion using real-time RT-PCR primers for exon 5, exon 6, uncleaved actin poly(A) (A), and 3′ flanking regions B, C, and D. All the quantitative ChIP and RT-PCR data were based on several independent experiments.
Xrn2 RNAi and Western blot analysis.
The RNA interference (RNAi) procedure and mRNA knockdown detection have been previously described (32). Western blotting was performed with an ECL kit (Amersham Biosciences) on 25 and 50 μg of total HeLa cell protein extract. Rabbit polyclonal anti-Xrn2 antibody was raised against an N-terminal fragment (amino acids 1 to 298) of Xrn2 expressed in bacteria (Eurogentec) and used at a 1:1,000 dilution for Western blot analysis. Actin antibody (Sigma) was used at a 1:2,000 dilution.
RESULTS
MAZ element promotes poly(A)-dependent transcriptional termination in vivo.
Previous work in this lab identified the existence of a sequence, G5AG5 (herein called a MAZ element), positioned downstream of the poly(A) site in the liver-specific C2 complement gene. This sequence is a potential binding site for the DNA-interacting protein MAZ and was shown to promote Pol II pausing and termination of the C2 complement gene in vivo (2, 3). In vitro evidence demonstrated that four copies of the MAZ site positioned downstream of a strong SPA promoted efficient poly(A) site cleavage and termination in HeLa nuclear extracts (35). It seemed likely that cleavage of the RNA transcript at the poly(A) site and Pol II pausing, induced by the MAZ site, were requirements for transcriptional termination. We have sought to further investigate the role of cleavage at the poly(A) site and Pol II pausing in mediating transcriptional termination in vivo. To do this, we took advantage of the in vivo system previously used to analyze the termination mechanism of the human β-globin gene (Fig. 1A) (8). Transcriptional termination of this gene requires the CoTC sequence, positioned ∼800 nucleotides from the poly(A) site, since its removal results in a loss of transcriptional termination. To test whether the MAZ sequence can promote Pol II pausing and transcriptional termination in vivo, we replaced the CoTC sequence of the β-globin gene with four copies of the MAZ (MAZ4 construct) or mutant MAZ sites (mMAZ4 construct), previously shown to have no pausing activity in vitro (34). As positive controls for transcriptional termination or transcriptional read-through, we used the previously described wild-type construct containing the 3′ flanking CoTC sequence (CoTC construct) or the ΔCoTC construct. Finally, to look at the requirement for poly(A) site cleavage in pause-dependent transcriptional termination, we mutated the poly(A) site to form MAZ4ΔpA and mMAZ4ΔpA constructs. These constructs were transfected into HeLa cells, and RNA polymerase II density was analyzed by NRO using probes located across the transfected plasmids (diagram in Fig. 1A). High Pol II density is indicated by strong signals over β-globin gene probes P, B3, and B4, while signal intensities over probes A (positioned downstream of CoTC or MAZ elements) and U3 (located just upstream of the promoter) give a measure of the termination efficiency of the given construct (32). To obtain a quantitative measure of termination, each NRO data set was given a TE value based on the amount of the averaged read-through signal over probes A and U3 (see Materials and Methods). As shown in Fig. 1A, transcriptional termination occurred very efficiently in the CoTC construct, with low signals observed over probes A and U3 compared to that observed with the upstream P, B3, and B4 probes. In contrast, termination efficiency was negligible in the ΔCoTC construct as indicated by the high signals over probes A and U3. Significantly, MAZ4 was able to promote transcriptional termination at 57% efficiency, somewhat less than that of the CoTC construct. Importantly, only a low level of termination activity was promoted only by mMAZ4 sites, giving a profile similar to that of the ΔCoTC construct. As expected, the poly(A) site mutation in the MAZ4 construct (MAZ4ΔpA) completely abolished transcriptional termination, showing that Pol II pausing is not sufficient for termination and that an efficient functional poly(A) site is also required. Note that a weak cryptic poly(A) site is still active in the MAZ4ΔpA construct (Fig. 1C) but insufficient to promote Pol II termination. Finally, the poly(A) site mutation in mMAZ4 (mMAZ4ΔpA) did not alter the NRO profile significantly, as termination remained impaired.
As an alternative approach, we employed real-time RT-PCR. Reverse transcription was primed from exon 2 and from region A, producing two cDNAs (Fig. 1A and see Materials and Methods). The amount of A- and ex2-specific steady-state RNA was measured by PCR. The ratio of accumulation of steady-state RNA at A relative to ex2 gives a measure of termination efficiency (Fig. 1B). As expected, a low ratio was observed with the CoTC construct and a high ratio for the ΔCoTC construct, indicating high and low efficiencies of transcriptional termination, respectively. The ratio of A to ex2 RNA from the MAZ4 transfection was similar to that from the CoTC construct, confirming that MAZ4 can promote efficient transcriptional termination. Also in agreement with the NRO data, the ratio of A to ex2 obtained for mMAZ4 was comparable to that obtained for the ΔCoTC construct. In general, these steady-state RT-PCR data gave quantitative values in agreement with the nascent NRO analysis (Fig. 1A).
We finally sought to confirm that the different constructs employed in the NRO analysis (Fig. 1A) gave the expected levels and size of polyadenylated mRNA. HeLa cells were transfected with each construct shown in Fig. 1A, and cytoplasmic RNA was S1 analyzed with an end-labeled DNA probe to detect 3′ processed β-globin mRNA. VA gene transcription from the cotransfected VA plasmid was also analyzed by S1 with a specific VA probe (Fig. 1C). Three β-globin-specific bands were detected. The top band (labeled pA) corresponds to the major poly(A) site and, consequently, is the strongest band. The middle band is an overdigestion product caused by the AT-rich nature of the RNA/DNA duplex close to the RNA 3′ end. The bottom band (labeled pA*) corresponds to a cryptic poly(A) site. As expected, the ratios of the pA band to the VA cotransfection control were the same for the CoTC, ΔCoTC, MAZ4, and mMAZ4 constructs. This shows that the various termination efficiencies of these constructs are not due to an increase in the kinetics of processing at the β-globin poly(A) site. The pA band was completely absent in cells transfected with MAZ4ΔpA and mMAZ4ΔpA constructs, confirming that the poly(A) site mutation inactivates the major β-globin poly(A) site. Interestingly, in its absence, the MAZ4 construct promoted a small (∼twofold) increase in the use of the upstream cryptic poly(A) site compared to the mMAZ4 construct.
In summary, we demonstrate that four wild-type MAZ but not four mutant MAZ elements can promote efficient transcriptional termination of the heterologous β-globin construct when transfected into HeLa cells. Since these sequences are of equal size, this precludes the possibility that transcriptional termination by MAZ4 is due to a simple distance effect. We also establish that the MAZ sites promote termination in a poly(A) site-dependent manner.
The 5′→3′ exonuclease Xrn2 is involved in pause-dependent transcriptional termination.
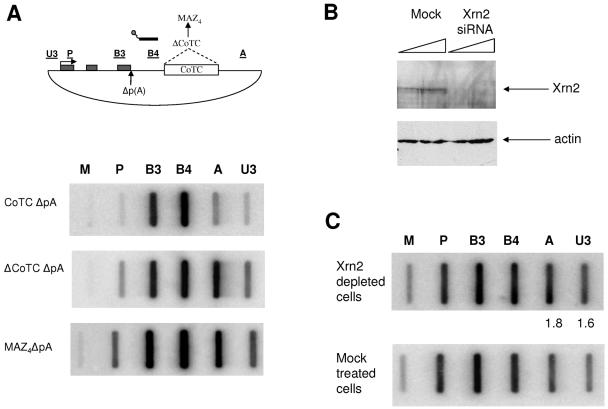
Previously, we have demonstrated that the 5′→3′ exonuclease Xrn2 plays a role in promoting human β-globin transcriptional termination from sites of CoTC. Similarly, it was shown that the Xrn2 homologue Rat1 induces Pol II termination in Saccharomyces cerevisiae, with the only difference being that Rat1 is thought to utilize the downstream product of poly(A) site cleavage as a substrate for degradation rather than a CoTC-generated end as in the case of the β-globin gene. We therefore sought to determine whether human Xrn2 can also use the 3′ product of poly(A) site cleavage as an exonuclease entry site (Fig. 2). It is conceivable that pause sequences, such as MAZ4, when placed close to the poly(A) site, may act to retain Pol II in close proximity with the poly(A) site. When cleavage takes place, Pol II may then be close enough to be affected by Xrn2 activity. An important assumption for this hypothesis is that MAZ sites do not encode CoTC activity, akin to that found in the human β-globin 3′ flanking region. To test this possibility, hybrid selection NRO analysis was employed (9). Since CoTC cleavage does not require a poly(A) site, MAZ4ΔpA, CoTCΔpA, and ΔCoTCΔpA constructs were analyzed to increase the detectable signals downstream of these inserts (Fig. 2A). Using hybrid selection NRO, we selected nascent transcripts continuous with region B4 (just upstream of the CoTC or MAZ4 element) through hybridization to a biotinylated antisense RNA probe spanning this region (see Materials and Methods). Selected transcripts were then hybridized to a filter containing the same NRO probes shown in Fig. 1A. Low levels of A and U3 signals were observed in the CoTCΔpA construct, demonstrating transcript cleavage within the CoTC sequence positioned between the B4 selection probe and probe A. In contrast, high A signals were observed in the B4-selected RNA fractions from the ΔCoTCΔpA and MAZ4ΔpA transfections. U3 signals were also elevated, though less so than with A signals, due to some nonspecific degradation occurring on these longer plasmid transcripts. This demonstrates that most transcripts extend from the selection probe B4 through the MAZ4 insert. The strong signal over B3 observed in all three constructs is due to polymerases from this region transcribing into the B4 selection region during the NRO transcription reaction. Signals immediately upstream of the selection probe are routinely observed in hybrid selection NRO analysis (8, 33). Overall, these results show that there is no detectable CoTC activity associated with transcripts from the MAZ4 insert. Therefore, the sole cleavage event in the primary transcript from the gene construct takes place at the poly(A) site.
FIG. 2.
The 5′→3′ exonuclease Xrn2 is involved in pause-dependent transcriptional termination. (A) β-Globin construct, with the position of the biotinylated β4 probe just downstream of the poly(A) site. The panels below show NRO transcripts selected by biotinylated β4 probes from CoTCΔpA, ΔCoTCΔpA, and MAZ4ΔpA transfections. (B) Western blot analysis of protein extracts obtained from HeLa cells treated with Xrn2 or mock siRNAs. Total protein (25 or 50 μg) was analyzed using Xrn2- or actin-specific antibodies, respectively. As indicated, Xrn2 but not actin was depleted to background levels. (C) RNAi-mediated depletion of human Xrn2 protein inhibited Pol II transcriptional termination mediated by the MAZ4 element. NRO analysis of MAZ4 construct transfected into mock-depleted and Xrn2-depleted cells is shown. Quantitative analysis shows a 1.8- and 1.6-fold increase in Pol II density over probes A and U3, respectively, comparing Xrn2-depleted versus mock-treated cells (based on two independent experiments).
To test whether Xrn2 is involved in the MAZ4-dependent termination mechanism, we transfected the MAZ4 construct into HeLa cells depleted of Xrn2 using RNAi. The efficiency of Xrn2 knockdown in these cells was judged to be around 10-fold based on RT-PCR analysis of Xrn2 mRNA levels (data not shown). This was confirmed by Western blot analysis (Fig. 2B) using a newly generated Xrn2 polyclonal antibody (see Materials and Methods). As shown in Fig. 2B, Xrn2 small interfering RNA (siRNA) treatment reduced the levels of Xrn2 protein to background compared to mock-treated cells. Actin levels were unchanged in the same protein samples. NRO analysis of the MAZ4 construct transfected into these siRNA-treated HeLa cells showed a reproducible near-twofold reduction in termination efficiency upon Xrn2 knockdown compared to the mock-treated cells. This suggests that Xrn2 can promote poly(A) site-dependent termination, possibly through degradation of the 3′ product of poly(A) site cleavage. However, this may occur only when Pol II is retained close to the poly(A) site through a pausing event. The observed effect is small but in line with previously described studies in which Xrn2/Rat1 depletion caused a two- to threefold decrease in Pol II transcriptional termination (16, 32). Since the MAZ4 element has a less-dramatic effect on transcriptional termination (TE, 57%) than the CoTC sequence (TE, >90%), we anticipate that Xrn2 will have a proportional effect: a 2- to 3-fold-lower TE in the case of CoTC-driven termination and a 1.5- to 2-fold-lower TE with MAZ4-driven termination.
The proximity of the MAZ4 element to the poly(A) site is important for transcriptional termination.
To further assess the function of MAZ sites in Pol II pausing and termination, we generated a further construct, MAZ4/U3, in which the MAZ4 insert is positioned ∼2 kb away from the poly(A) site, just upstream of the region spanned by NRO probe U3. This construct tested the effect of pausing of Pol II at a more distant position to the poly(A) site. NRO analysis of this construct (Fig. 3), compared to that of the previously tested CoTC, ΔCoTC, and MAZ4 constructs (Fig. 1) demonstrates that moving the MAZ4 sequence away from the poly(A) signal significantly reduces termination efficiency. In effect, MAZ4/U3 has a TE similar to that of the ΔCoTC construct. These results imply that the MAZ sites must be positioned close to the poly(A) site to exert their effects (as is the case with the MAZ4 construct). This indicates that MAZ sites serve to pause Pol II close to the poly(A) site, allowing termination to occur. Xrn2-mediated RNA degradation of the 3′ product of poly(A) site cleavage may well be an integral part of this mechanism.
Poly(A) site strength is important for pause site-mediated transcriptional termination.
To look at the effect of poly(A) site cleavage kinetics on MAZ4-mediated transcriptional termination, we replaced the β-globin poly(A) site in the MAZ4 construct with another poly(A) site (SPA), forming SPA/MAZ4. To compare the relative strengths of the βp(A) and SPA, we carried out a poly(A) site competition assay (Fig. 3B). Two constructs were generated, one with the SPA positioned ∼250 bp downstream of the βp(A) (β/SPA) and the other with the βp(A) positioned ∼250 bp downstream of the SPA (SPA/β). Cells were transfected with these constructs, and S1 nuclease analysis was carried out using an end-labeled probe to detect cleavage at either poly(A) site (see diagram in Fig. 3B). The ratio of cleavage at βp(A) to that at SPA gives a measure of the strength of each poly(A) site. In particular, 5′ poly(A) signals have a strong positional advantage, so that a 3′ poly(A) site can be utilized only if it is a much stronger mRNA processing signal (2). With βp(A) upstream of SPA, there is a strong βp(A)-specific band and no SPA-specific band, indicating that the downstream SPA is not able to out-compete the upstream βp(A). In contrast, when the βp(A) is positioned downstream of the SPA, it is still able to partially out-compete the SPA [note the clear βp(A)-specific bands in Fig. 3B]. Multiple bands corresponding to SPA represent S1 overdigestion products caused by AT richness of the SPA insert. Their additive signal defines the level of SPA usage. This was confirmed in earlier analysis of SPA activity (17). Overall, this experiment shows that the SPA is a significantly weaker poly(A) site than the βp(A). To assess the effect of the SPA upon MAZ4-dependent termination, the SPA/MAZ4 construct was transfected into HeLa cells and Pol II density was analyzed by NRO (Fig. 3A). Comparison between the NRO profiles of SPA/MAZ4 and MAZ4 shows that termination is significantly less efficient with SPA/MAZ4 than with MAZ4. Since we have demonstrated that SPA is a weaker poly(A) site, we predict that the kinetics of poly(A) site recognition and cleavage may be critical for efficient MAZ4-dependent termination. We note that in SPA/MAZ4, although the high A signal is indicative of no termination, the U3 signal is lower than expected. This may imply that some Pol II termination over the plasmid backbone still occurs. Perhaps the more rapidly formed cleavage product of the βp(A) would be recognized by Xrn2 more rapidly and therefore promote more-efficient Pol II termination. In agreement with this, we observed above that the β-globin weak, cryptic poly(A) site is not sufficient to promote transcriptional termination (Fig. 1A and C). These results show that intimate coordination of both poly(A) site and downstream terminator sequence strength is necessary to bring about efficient Pol II termination. Moreover, they support a model in which the degradation of the 3′ product of poly(A) site cleavage by Xrn2 contributes toward efficient termination.
Our results on MAZ4-dependent Pol II termination might indicate that any highly GC-rich sequence may be sufficient to promote Pol II termination. We therefore made additional constructs to test this possibility. In two constructs, the MAZ4 element was replaced by a stretch of either 30Ts (30T construct) or 30Cs (30C construct). We also inverted the MAZ4 insert (ZAM4). All three constructs were transfected into HeLa cells and analyzed by NRO (Fig. 3A). The Pol II density profiles obtained with both 30T and 30C were essentially identical to that obtained with the ΔCoTC construct, showing no detectable increase in termination efficiencies. In contrast, when inverted, MAZ sequences were still able to promote termination as efficiently as when in the correct orientation.
Overall, these experiments show that the MAZ sequences exert their effects on transcriptional termination in a sequence-specific manner that appears to be strand independent. Previous reports suggested that a DNA-binding protein may bind to MAZ sequences, although an actual role for DNA-binding proteins in MAZ-specific pausing in vivo has not yet been established (3). This earlier study also indicated that MAZ4 pausing showed a twofold-higher activity in the correct orientation. Possibly, this difference with our present data reflects the different assays employed.
Pause-mediated transcriptional termination of the endogenous β-actin gene.
If pause-dependent Pol II termination were a general feature of human genes, then G-rich sequences such as MAZ4 would be predicted to be a common feature of the human genome. We therefore searched for genes that contain G-rich sequences close to their poly(A) sites and identified the human β-actin as one such example (Fig. 4A). To determine whether this G-rich sequence promotes transcriptional pausing and consequent termination activity, we performed Pol II ChIP experiments on HeLa cell nuclei using primer pairs across the promoter (prom) and the open reading frame (ex3, ex5, and ex6) and within the 3′ flanking region of the β-actin gene (primers B to D). ChIP analysis provides information about the Pol II density across a gene, although unlike NRO analysis, actively transcribing Pol II and inactive Pol II are not distinguishable. As shown in Fig. 4B, Pol II density decreased along the open reading frame of the β-actin gene but then significantly increased past the poly(A) signal, finally gradually decreasing further into the 3′ flanking region of the endogenous β-actin gene. These data are suggestive of Pol II pausing immediately past the poly(A) signal, followed by a gradual release of the polymerase in the 3′ flanking region beyond. We did not observe complete disappearance of the Pol II signal even 2.3 kb downstream of the poly(A) signal (probe D). Possibly, termination of the β-actin gene is inefficient so that full Pol II release may occur even further downstream of the region probed in this experiment. Significantly, we also demonstrated that RNAi-mediated depletion of Xrn2 protein in HeLa cells causes a significant increase in Pol II ChIP signals over probes B to D (Fig. 4C). These data therefore indicate that Xrn2 affects polymerase release downstream of the β-actin poly(A) site.
We also used real-time RT-PCR to look at the termination profile of the endogenous β-actin gene. Reverse transcription was primed from exon 5 and regions B, C, and D, producing four cDNAs (see Fig. 4A for primer locations). The amount of steady-state RNA at each position was then deduced by PCR analysis (Fig. 4D). As expected, the amount of the steady-state RNA transcript decreased dramatically after the poly(A) site, as unprocessed transcripts are very unstable. However, detectable transcript levels were evident at the primer B position [300 nucleotides away from the poly(A) site] and then again gradually decreased along the gene (primers C and D), supporting the ChIP data (Fig. 4C). Upon depletion of Xrn2, 3′ flanking β-actin transcripts increased significantly (Fig. 4D), again indicating a role for this enzyme in degrading these transcripts. We finally carried out real time RT-PCR analysis to determine whether Xrn2 degradation specifically targets transcripts downstream of the actin poly(A) signal (Fig. 4E). As shown in Fig. 4E, Xrn2 depletion had little effect on levels of exon 5 and 6 transcripts or uncleaved transcripts over the poly(A) site [detected with primers across the poly(A) site]. In contrast, 3′ flanking transcripts B to D showed a significant and increasing level of stabilization, reflecting their sensitivities to Xrn2 degradation. These data strongly implicate Xrn2 in the degradation of 3′ flanking transcripts exposed by cleavage at the gene's poly(A) site.
In summary, results from the above-described quantitative ChIP and RT-PCR experiments suggest an Xrn2-dependent mode of termination for the β-actin gene. Due to the buildup of the Pol II signal observed downstream of the poly(A) site, we suggest that termination of the β-actin gene is mediated through a pausing mechanism similar to MAZ4. Our data also indicate that Xrn2 plays a critical role in recognizing cleavage at the poly(A) site by degrading the nascent transcript up to Pol II stalled at the G-rich sequences.
β-Actin pause element promotes poly(A)-dependent transcriptional termination of heterologous construct in vivo.
To further define the β-actin gene pause site, we tested its ability to mediate transcriptional termination in the heterologous β-globin gene construct. A 380-bp region of the β-actin gene [located 100 nucleotides downstream of the poly(A) site] was isolated from HeLa genomic DNA by PCR and cloned into the β-globin plasmid, replacing the CoTC element (Fig. 5A). This globin/actin fusion construct was transfected along with CoTC and ΔCoTC constructs into HeLa cells, and NRO experiments were then performed. As shown in Fig. 5A, we demonstrate that the 380-bp β-actin pause element promotes transcriptional termination of the β-globin construct, as judged from the reduced signals over probes A and U3. We have also demonstrated that the transcription termination effect mediated by the β-actin pause element is poly(A) site dependent, as mutation of the poly(A) sequence causes a loss of termination (data not shown). Compared to the above-described ChIP experiment detecting total Pol II density across the gene (Fig. 4A), NRO analysis detects only actively transcribing polymerases, which may account for differences between the two analyses. Thus, the NRO data imply efficient Pol II termination (TE, 83%), while the endogenous Pol II ChIP analysis indicates significant Pol II deposition beyond the immediate 3′ flanking region of the β-actin gene.
FIG. 5.
β-Actin gene pause element promotes poly(A)-dependent transcriptional termination of heterologous construct in vivo. (A) Diagram of the globin/actin fusion construct, with exons shown as gray boxes, and CoTC element, substituted by the β-actin pause element, shown as a hatched box. Panels below show NRO signals for CoTC, ΔCoTC, and globin/actin fusion constructs. TE is indicated. (B) Globin/actin fusion construct, with the position of the biotinylated β4 probe just downstream of poly(A) site indicated. The panels below show transcripts selected by biotinylated β4 probes from CoTCΔpA and globin/actin ΔpA transfections. Corrected hybridization signals (with B4 taken as 1) are shown in the graph below.
We finally investigated by hybrid selection NRO the possibility that the 380-bp β-actin pause element possesses CoTC activity similar to the β-globin CoTC activity (Fig. 5B). For this experiment, we employed the globin/actin ΔpA construct, as CoTC cleavage does not require a poly(A) site. As a positive control for CoTC cleavage, we used the CoTCΔpA construct. We observed high signals over probe A for the globin/actin ΔpA construct, indicating that most transcripts extend from the selection probe B4 through the β-actin termination element and that there is no significant CoTC activity associated with transcripts from the β-actin pause element. Therefore, the only cleavage event in the primary transcript is associated with the poly(A) site. These data support the view that Xrn2 recognizes the cleaved poly(A) site RNA when degrading the β-actin transcript.
Our results from both ChIP and NRO analyses demonstrate that β-actin gene termination is mediated through a pause-dependent mechanism and that Xrn2 has a major role in this process by recognizing RNA 5′ ends generated by cleavage at the poly(A) site. In effect, β-actin gene termination provides a clear physiological example of pause-dependent Pol II termination, as demonstrated for the MAZ4 element.
DISCUSSION
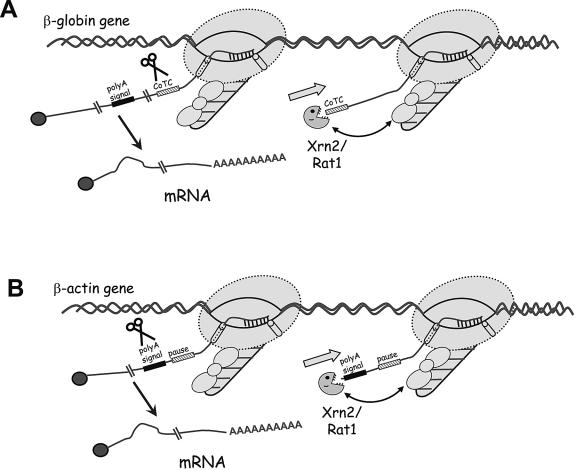
Two previous studies, one from our laboratory, demonstrated a role for a 5′→3′ exonuclease (Xrn2 in humans and Rat1 in yeast) in the transcriptional termination of mRNA-encoding genes (16, 32). Xrn2 was found to be involved in termination of the human β-globin gene (32). Degradation of the 3′ product of CoTC was implicated in this termination mechanism that also required prior transcription of a poly(A) signal. In contrast, Rat1 was implicated in the degradation of the 3′ product of poly(A) site cleavage, in line with the original torpedo model of transcriptional termination (5, 22). This discrepancy in substrate choice can be reconciled, since no examples of CoTC activity in yeast have been found. However, as described in these studies, replacement of the CoTC element with the MAZ4 sequence, containing no CoTC activity, reveals that Xrn2 can also recognize the 3′ product of poly(A) site cleavage, like yeast Rat1. We demonstrate that this pause sequence promotes Pol II termination in a poly(A) site-dependent manner affected by both poly(A) site strength and proximity to the poly(A) site. A G-rich sequence similar to that of MAZ4, found in the β-actin 3′ flanking region, also promotes termination. Moreover, upon Xrn2 depletion, the RNA downstream of the poly(A) site was significantly stabilized and there was a concomitant increase in Pol II density. The mechanism of action of these G-rich pause elements is as yet unknown. MAZ (_M_YC-_a_ssociated _z_inc finger protein) was originally identified as a transcription factor that interacts with the MYC promoter (4) and that may also interact with a MAZ site originally identified in the termination region of the human C2 complement gene (3, 4). However, RNAi-mediated depletion of MAZ had no significant effect on the termination activity of the MAZ4 construct used in these studies (data not shown). It is possible that MAZ sites are recognized by redundant factors or, alternatively, that the inherent structure of these G-rich sequences (either at the DNA or RNA level) may act to promote Pol II pausing. Our data support a model whereby pausing of Pol II at positions close to the poly(A) site promotes termination at least in part, due to exonucleolytic degradation of the 3′ product of poly(A) site cleavage (Fig. 6). Although this study, together with our previous data, provides a link between Xrn2 and transcriptional termination, as envisaged by the earlier torpedo model, we favor a synthesis of both the antitermination and torpedo models.
FIG. 6.
Model for two types of Pol II transcriptional termination in mammalian cells. (A) CoTC-dependent termination of β-globin gene. CoTC presents a free 5′ RNA end which is recognized by the 5′→3′ exonuclease Xrn2/Rat1p. Subsequent degradation of this downstream cleavage product by Xrn2 leads to Pol II transcription termination and release of free Pol II. (B) Pause-dependent termination of β-actin gene. Due to the Pol II pausing/slowing elongation rate at the pause sequence, cleavage at the poly(A) site presents a free 5′ RNA end which is recognized by the 5′→3′ exonuclease Xrn2/Rat1p. Subsequent degradation of this downstream cleavage product by Xrn2 leads to Pol II transcription termination and release of Pol II. The double-headed arrow indicates potential interaction between Xrn2 and CTD-bound proteins.
Links between transcriptional termination and the Pcf11 component of the poly(A) site cleavage machinery have recently been made. Unexpectedly, Pcf11 mutants that inhibited cleavage were not deficient in termination of a CYC1 reporter gene in yeast, suggesting that the two processes can be uncoupled under some circumstances (25). Recent transcription studies of both yeast and Drosophila spp. showed that the addition of Pcf11 to stalled Pol II elongation complexes (ECs) stimulated Pol II-DNA dissociation in an ATP-independent manner, possibly through the formation of a bridge between the C-terminal domain and the nascent transcript that would constrain movements conducive to elongation (36, 37). These studies suggest that the length of RNA between the Pol II exit channel and Pcf11 must be short so as not to dissipate the force exerted by Pcf11 on the EC. Given this model, stalled or slow polymerases may be more susceptible to this mode of termination, perhaps providing biological relevance to Pol II pause sites. Interestingly, shortening nascent EC-associated transcripts to <51 nucleotides was previously shown to increase the incidence of EC arrest in vitro (31). Perhaps degradation of Xrn2/Rat1 to within <50 nucleotides of the EC leads to stalling and subsequent EC disassembly through the actions of Pcf11 and other essential termination factors. CoTC may similarly cause EC arrest by attenuating the length of the Pol II-associated transcript.
Previously, it has been demonstrated that CoTC is likely to occur before the cleavage at the β-globin poly(A) site (9). It is interesting to speculate that the inability of the ΔCoTC construct to support termination may be due to delayed production of Xrn2 substrates. Previously, the presence of downstream MAZ sites was shown to enhance the efficiency of SPA cleavage and subsequent termination in vitro (34). In addition, it was shown that the downstream product of SPA cleavage remained attached to the ternary complex and was degraded, cotranscriptionally, while still attached (35). Therefore, pausing Pol II downstream of the β-globin poly(A) site may enhance termination by giving more time for cotranscriptional poly(A) site cleavage. Evidence presented here strongly suggests that poly(A) site cleavage degradation of the cotranscriptionally formed 3′ product of Xrn2 may be an integral part of this mechanism. Termination may also depend on steric changes in the elongation complex and the proximity of the polymerase to the poly(A) site upon this change (Fig. 6).
Termination elements in the 3′ flanking region of genes are likely to coordinate temporal and spatial interactions between the polymerase, the poly(A) site, the nascent transcript, and the DNA template. Therefore, the strengths of the promoter, the poly(A) site, and the termination element of a gene must be coordinated in order to bring about the most efficient termination. This was shown in a recent study in which identical termination elements were placed downstream of both “strong” and “weak” poly(A) sites, and only with the strong poly(A) site present did termination occur efficiently (19). The results presented here demonstrate that Xrn2 can play a role in transcriptional termination in the absence of 3′ flanking region CoTC and establish the importance of polymerase pausing in the process. Thus, Xrn2, like Rat1, can utilize the downstream product of poly(A) site cleavage as a substrate. This implies that Xrn2 may also play a global role in termination of mRNA genes in humans, as Rat1 does in yeast.
Acknowledgments
We are grateful to members of the N.J.P. lab for advice and encouragement. We thank Joan Monks for help with the preparation of recombinant Xrn2 protein for production of anti-Xrn2 antibody.
These studies were supported by a Programme grant from the Wellcome Trust to N.J.P. S.W. is supported by an EP Abraham Research studentship.
REFERENCES
- 1.Ashe, H. L., J. Monks, M. Wijgerde, P. Fraser, and N. J. Proudfoot. 1997. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11**:**2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashfield, R., P. Enriquez-Harris, and N. J. Proudfoot. 1991. Transcriptional termination between the closely linked human complement genes C2 and factor B: common termination factor for C2 and c-myc? EMBO J. 10**:**4197-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashfield, R., A. J. Patel, S. A. Bossone, H. Brown, R. D. Campbell, K. B. Marcu, and N. J. Proudfoot. 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 13**:**5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossone, S. A., C. Asselin, A. J. Patel, and K. B. Marcu. 1992. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA 89**:**7452-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connelly, S., and J. L. Manley. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 2**:**440-452. [DOI] [PubMed] [Google Scholar]
- 6.Custodio, N., M. Carmo-Fonseca, F. Geraghty, H. S. Pereira, F. Grosveld, and M. Antoniou. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18**:**2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denome, R. M., and C. N. Cole. 1988. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol. Cell. Biol. 8**:**4829-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dye, M. J., and N. J. Proudfoot. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105**:**669-681. [DOI] [PubMed] [Google Scholar]
- 9.Dye, M. J., and N. J. Proudfoot. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3**:**371-378. [DOI] [PubMed] [Google Scholar]
- 10.Enriquez-Harris, P., N. Levitt, D. Briggs, and N. J. Proudfoot. 1991. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 10**:**1833-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greger, I. H., and N. J. Proudfoot. 1998. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17**:**4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagenbuchle, O., P. K. Wellauer, D. L. Cribbs, and U. Schibler. 1984. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell 38**:**737-744. [DOI] [PubMed] [Google Scholar]
- 13.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413**:**538-542. [DOI] [PubMed] [Google Scholar]
- 14.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395**:**93-96. [DOI] [PubMed] [Google Scholar]
- 15.Kadener, S., J. P. Fededa, M. Rosbash, and A. R. Kornblihtt. 2002. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. USA 99**:**8185-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, M., N. J. Krogan, L. Vasiljeva, O. J. Rando, E. Nedea, J. F. Greenblatt, and S. Buratowski. 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432**:**517-522. [DOI] [PubMed] [Google Scholar]
- 17.Levitt, N., D. Briggs, A. Gil, and N. J. Proudfoot. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev. 3**:**1019-1025. [DOI] [PubMed] [Google Scholar]
- 18.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385**:**357-361. [DOI] [PubMed] [Google Scholar]
- 18a.Oshein, Y. N., N. J. Proudfoot, and A. L. Beyer. 1999. EM visualisation of transcription by RNA polymerase II: downstream termination required by poly(A) signal but not transcript cleavage. Mol. Cell 3**:**379-387. [DOI] [PubMed] [Google Scholar]
- 19.Plant, K. E., M. J. Dye, C. Lafaille, and N. J. Proudfoot. 2005. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Gγ-globin gene. Mol. Cell. Biol. 25**:**3276-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proudfoot, N. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25**:**290-293. [DOI] [PubMed] [Google Scholar]
- 21.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16**:**272-278. [DOI] [PubMed] [Google Scholar]
- 22.Proudfoot, N. J. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14**:**105-110. [DOI] [PubMed] [Google Scholar]
- 23.Roberts, G. C., C. Gooding, H. Y. Mak, N. J. Proudfoot, and C. W. Smith. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26**:**5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robson-Dixon, N. D., and M. A. Garcia-Blanco. 2004. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 279**:**29075-29084. [DOI] [PubMed] [Google Scholar]
- 25.Sadowski, M., B. Dichtl, W. Hubner, and W. Keller. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22**:**2167-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheets, M. D., P. Stephenson, and M. P. Wickens. 1987. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol. Cell. Biol. 7**:**1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens, A., and T. L. Poole. 1995. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem. 270**:**16063-16069. [DOI] [PubMed] [Google Scholar]
- 28.Takahara, T., B. Tasic, T. Maniatis, H. Akanuma, and S. Yanagisawa. 2005. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol. Cell 18**:**245-251. [DOI] [PubMed] [Google Scholar]
- 29.Tantravahi, J., M. Alvira, and E. Falck-Pedersen. 1993. Characterization of the mouse βmaj globin transcription termination region: a spacing sequence is required between the poly(A) signal sequence and multiple downstream termination elements. Mol. Cell. Biol. 13**:**578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira, A., A. Tahiri-Alaoui, S. West, B. Thomas, A. Ramadass, I. Martianov, M. Dye, W. James, N. J. Proudfoot, and A. Akoulitchev. 2004. Autocatalytic RNA cleavage in the human beta-globin pre-mRNA promotes transcription termination. Nature 432**:**526-530. [DOI] [PubMed] [Google Scholar]
- 31.Ujvari, A., M. Pal, and D. S. Luse. 2002. RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 277**:**32527-32537. [DOI] [PubMed] [Google Scholar]
- 32.West, S., N. Gromak, and N. J. Proudfoot. 2004. Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432**:**522-525. [DOI] [PubMed] [Google Scholar]
- 33.West, S., K. S. Zaret, and N. Proudfoot. Transcriptional termination sequences in the mouse serum albumin gene. RNA, in press. [DOI] [PMC free article] [PubMed]
- 34.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3**:**593-600. [DOI] [PubMed] [Google Scholar]
- 35.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19**:**3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Z., J. Fu, and D. S. Gilmour. 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 19**:**1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Z., and D. S. Gilmour. 2006. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol. Cell 21**:**65-74. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63**:**405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]