Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3 (original) (raw)
Abstract
Agrobacterium tumefaciens causes crown gall disease on a variety of plants. During the infection process Agrobacterium transfers a nucleoprotein complex, the VirD2 T-complex, and at least two Vir proteins, VirE2 and VirF, into the plant cell via the VirB/VirD4 type IV secretion system. Recently, we found that T-DNA could also be transferred from Agrobacterium to Saccharomyces cerevisiae. Here, we describe a novel method to also detect trans-kingdom Vir protein transfer from Agrobacterium to yeast, using the Cre/lox system. Protein fusions between Cre and VirE2 or VirF were expressed in Agrobacterium. Transfer of the Cre-Vir fusion proteins from Agrobacterium to yeast was monitored by a selectable excision event resulting from site-specific recombination mediated by Cre on a _lox_-flanked transgene in yeast. The VirE2 and VirF proteins were transported to yeast via the _virB_-encoded transfer system in the presence of coupling factor VirD4, analogous to translocation into plant cells. The yeast system therefore provides a suitable and fast model system to study basic aspects of trans-kingdom protein transport from Agrobacterium into host cells. Using this method we showed that VirE2 and VirF protein transfer was inhibited by the presence of the Osa protein. Besides, we found evidence for a novel third effector protein, VirE3, which has a similar C-terminal signature to VirE2 and VirF.
INTRODUCTION
The plant pathogen Agrobacterium tumefaciens has a tumour-inducing (Ti) plasmid of which part, the transfer (T)-region, is transferred to plant cells during the infection process. As a result, the infected plant cells are triggered to divide, leading to the formation of crown gall tumours (reviewed in 1–8). The tumorous growth of the infected plant cells is caused by expression of the oncogenes located on the T-DNA. The vir region, also present on the Ti plasmid, encodes the Vir proteins, which mediate the processing of the T-region and the transfer of a single-stranded (ss) DNA copy of this region, the T-strand, into the recipient cells. One of the Vir proteins, the nicking enzyme VirD2, remains covalently attached to the 5′ end of the T-strand. VirD2 pilots the T-strand to the plant cell nucleus by virtue of its nuclear localisation signal (NLS). Recently, we showed that, in addition to the nucleoprotein complex, A.tumefaciens translocates the Vir proteins VirE2 and VirF directly into plant cells (9). Transport occurs via the VirB/VirD4 type IV secretion channel (7). The ssDNA-binding protein VirE2 and the F-box protein VirF play a role in the process of plant transformation. The VirE2 protein protects the T-strand and is involved in its transport into the nucleus (10). VirF can interact through its F-box with plant homologues of the yeast Skp1 protein (11). Skp1 and F-box proteins are subunits of a class of E3 ubiquitin ligases, called SCF complexes. These SCF complexes target specific proteins through interaction with F-box proteins for ubiquitin-mediated proteolysis (12,13). However, the target of VirF remains unknown so far. The virF operon contains a single gene, but the adjacent virE operon embraces three genes, namely virE1, virE2 and virE3. The VirE1 protein plays an important role in the transport of VirE2 by acting as a chaperone of this protein and thus preventing at the same time premature binding to the T-strand and VirE2 aggregate formation (14,15). The function of the VirE3 protein has not been established, but virFvirE3 double mutants are more strongly attenuated in virulence than virF mutants (B.Schrammeijer, P.Zuiderwijk and P.J.J.Hooykaas, unpublished results).
Agrobacterium tumefaciens can also transfer T-DNA to Saccharomyces cerevisiae (16) and filamentous fungi (17). The VirE2 and VirF proteins are not necessary for this process. The question was therefore raised whether A.tumefaciens nevertheless still translocates the Vir proteins VirE2 and VirF into yeast in a process that resembles translocation into plant cells. To study this, we made use of the site-specific recombination system Cre/lox of bacteriophage P1 (reviewed in 18), analogous to what was used to show direct Vir protein translocation from Agrobacterium into plants (9). Delivery of a bacterially expressed functional Cre recombinase fused to the VirE2 or VirF proteins into yeast cells was detected by selection for loss of a lox flanked (f_lox_ed) URA3 marker gene due to Cre recombination activity (Fig. 1). Our results indicate that A.tumefaciens indeed translocates VirE2 and VirF proteins directly into S.cerevisiae and that this translocation occurs via the VirB/VirD4 transport system as in plants. The yeast system thus offers a suitable and fast model system for the study of trans-kingdom protein translocation from Agrobacterium. We applied the system to show that the presence of the IncW plasmid pSa Osa protein, which inhibits tumour formation by Agrobacterium (19–21) blocks VirE2 and VirF protein transfer. Moreover we found evidence for the translocation of a third effector protein, the VirE3 protein, into yeast.
Figure 1.
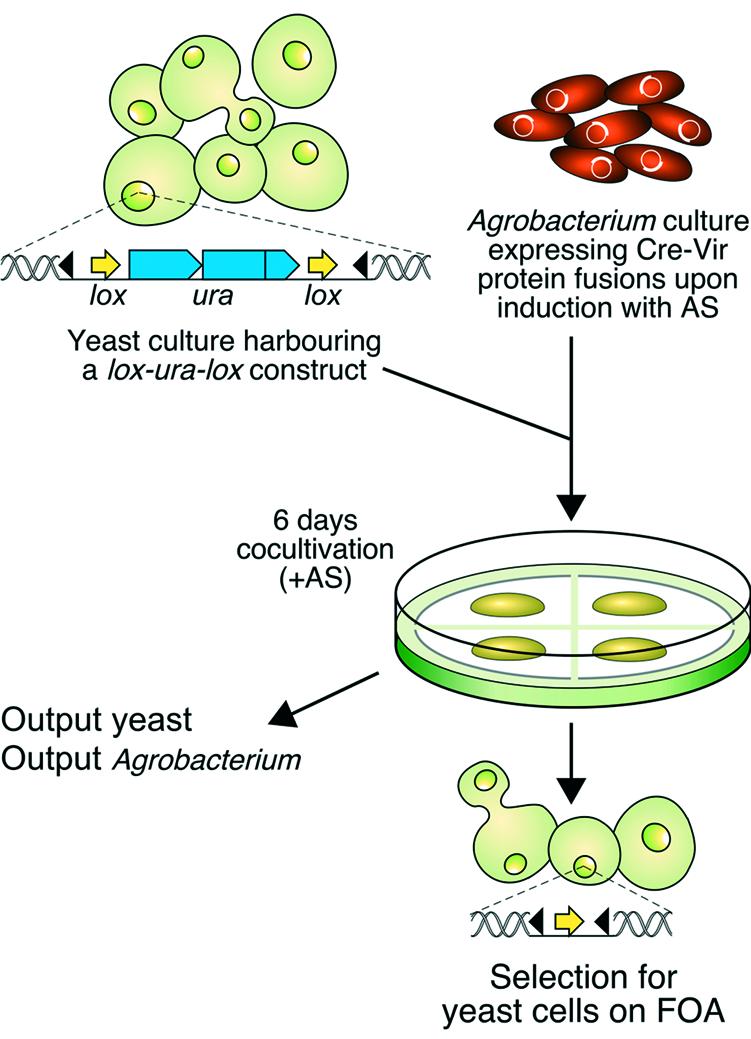
Experimental strategy to detect Vir protein translocation from Agrobacterium into yeast. The yeast cells contain a lox-URA3-lox marker integrated at the PDA locus. Upon co-cultivation with acetosyringone (AS)-induced Agrobacterium cells expressing Cre-Vir fusion protein (see Materials and Methods) for 6 days the mixture of cells is plated on medium supporting yeast growth and containing FOA. Delivery of a functional Cre enzyme from Agrobacterium into yeast is detected by Cre-mediated site-specific recombination on the lox sites resulting in loss of the URA3 gene allowing growth on medium containing FOA. Part of the mixture is used to determine the output of Agrobacterium and yeast cells (see Materials and Methods). Black triangles, Agrobacterium left and right border sequences.
MATERIALS AND METHODS
Construction of a S.cerevisiae strain containing a f_lox_ed URA3 gene
The URA3 gene was cloned as a _Hin_dIII fragment from pJJ244 (22) into the filled-in _Eco_RI site of pIC-2lox (23) resulting in pSDM3011. The resulting f_lox_ed URA3 gene was subcloned as a _Hin_dIII fragment into pUC4α10 (24), which contains part of the PDA1 locus (pSDM3012). Finally, the _PDA1_-lox-URA3-lox-PDA1 cassette was cloned into binary vector pBin19 (25) resulting in pSDM3013. Agrobacterium tumefaciens strain LBA1126 (26) was electroporated (27) with pSDM3013 and used in a co-cultivation experiment with S.cerevisiae strain RSY12 (MATa leu2-3, 112 his3-11, 15 ura3Δ::HIS3) (28). Transformants, prototrophic for uracil, were selected on minimal yeast (MY) medium (29) containing 200 µM cefotaxim (Duchefa, B.V.), leucine (30 mg/l) and histidine (20 mg/l) but lacking uracil. Transformants were further characterised with polymerase chain reaction (PCR) and Southern blot analysis. Strain LBY2, in which the loxURA3lox gene had integrated at the PDA1 locus on chromosome V by homologous recombination (gene replacement), was selected and used in all co-cultivation experiments.
Construction of cre control and cre fusion plasmids
Cre control. The cre recombinase gene was cloned as an _Sph_I/_Eco_RI fragment from pUC19Cre (30) into pUC21 (31) (pSDM3120). The ATG start codon was removed by replacing the _Bgl_II/_Nru_I fragment in pSDM3120 by a _Bgl_II/_Nru_I-digested PCR fragment, amplified with primers cre 1 (5′-ggcagatctgTCCAATTTACTG-3′) and cre 2 (5′-GATAA TCGCGAACATCTTCAGG-3′) (restriction sites underlined) using pSDM3120 as template (pSDM3121). A 2.2-kb _Sal_I fragment from pRAL3248 (32), containing virE promoter, virE1 and virE2(Δ_30 3_′ bp), was cloned into the _Xho_I site of pSDM3121 (pSDM3122). Digestion with _Bgl_II and _Bst_YI (partial) followed by ligation of the vector (pSDM3126) resulted in transcriptional control of cre from the virE promoter at the start codon position of virE2 (pvirE-virE1-cre). This cassette was cloned into the _Sma_I/_Xba_I sites of the broad host range, non-mobilisable plasmid pRL662 (9) (pSDM3147 or cre control).
virE2::cre fusion. VirE2 was fused translationally to the 5′ end of the cre gene in several steps. The pvirE-virE1-virE2(Δ_30 3_′ bp)::cre cassette from pSDM3122 was cloned into the _Sma_I/_Xba_I sites of pRL662 (pSDM3148). The 3′ end of the virE2 gene was restored as follows. The _Bgl_II/Nru_I fragment of pSDM3122 (cre1-205 Δ_ATG) was cloned into pIC19R (33) and named pSDM3151. A 34-bp _Sal_I/_Bgl_II linker (5′-TCGACCGCGTAGCCAAAGCGTCAACAGCTTTCGA-3′) representing the 3′ 30 bp of virE2 but lacking the stop codon was cloned into pSDM3151 (pSDM3152). The _Sal_I fragment of pRAL3248 [pvirE-virE1-virE2(Δ_30 3_′ bp)] was cloned into pSDM3152 resulting in a translational fusion of full length virE2 with cre (pSDM3157). The _Nru_I fragment in pSDM3148 was finally replaced by the _Nru_I fragment of pSDM3157 resulting in plasmid pSDM3166 carrying the pvirE-virE1-virE2::cre cassette.
cre::virE2 fusion. To remove the STOP codon of the cre gene primers cre 6 (5′-acgcgtcgactATCGCCATCTTC-CAGCAGGCGC-3′) and cre 7 (5′-cCATCGATTGATTA CGGCGCTAAGG-3′) were used in a PCR amplification with pSDM3126 as template DNA. After digestion with _Cla_I (underlined) and _Sal_I (underlined) the fragment was used to replace the corresponding fragment in pSDM3126 (pSDM3127 or creΔSTOP). An _Xho_I/_Sma_I fragment from pRAL3248 (32) was inserted into _Xho_I/_Eco_RV-digested pBluescriptIISK– (34) followed by insertion of an _Xho_I/_Stu_I linker (5′-tcgaGATCTTTCTGGCAATGAGAAATCAGG-3′) resulting in pBluevirE2_Δ_ATG. VirE2_Δ_ATG was then fused in frame to the 3′ end of cre as an _Xho_I/_Not_I fragment into the _Sal_I/_Not_I sites of pSDM3127 (pSDM3128). Finally, a _Stu_I/_Xba_I fragment was cloned into the _Sma_I/_Xba_I sites of pRL662 resulting in pSDM3129 harbouring the pvirE-virE1-cre::virE2 cassette.
NLS::virF::cre fusion. A _Sac_I/_Eco_RV 5′ virF flank from pTi15955, containing the ribosome binding site, was subcloned from pRAL7088 (35) into pBluescriptIISK– (pSDM3183). The NLS from simian virus 40 (SV40), containing an ATG start codon, was synthesised (Eurogentec, Belgium) with _Eco_RV and _Sal_I sites, and cloned into pSDM3183 (pSDM3184). VirF_Δ_ATG_Δ_495–609 was cloned downstream the SV40 NLS in pSDM3184 (_Xho_I) as a _Sal_I/_Xho_I fragment from pSDM3193 (11) and named pSDM3185. To remove the STOP codon in virF a 175-bp fragment of the 3′ coding region was amplified with primers PF170 (5′-ATCCCTAACTTGGTCTTCAAC-3′) and PF583 (5′-cttagatcTAGACCGCGCGTTGATCGAGG-3′) containing a _Bgl_II site at the 5′ end (underlined) using pRAL7088 (35) as template DNA. After subcloning the PCR fragment into the pGEM T-vector (Promega) VirF_Δ_STOP was cloned into _Xho_I/_Bgl_II-digested pSDM3121-L, which resulted after insertion of a _Stu_I/_Bgl_II linker (5′-CCTCGAGCCCGGGATA-3′) into pSDM3121 (pSDM3186). The 3′ virF::cre fusion was cloned into the _Xho_I site of pSDM3185 (pSDM3187 or NLS::virF::cre) and subsequently into _Sac_I/_Pst_I digested pUC28 (36). The final _pvirF_-NLS::virF::cre cassette was cloned into _Eco_RI digested pRL662 (pSDM3153).
NLS::cre::virF fusion. The 5′ virF flank_–SV40NLS_ cassette from pSDM3184 was subcloned into pIC19H (_Sac_I/_Sal_I) (33), followed by a _Hin_dIII/_Sal_I cloning into pIC19R and finally cloned as an _Xho_I/_Sal_I fragment into the _Xho_I site of pSDM3121 (pSDM3188). Exchange of the _Cla_I/_Sal_I 3′ cre fragment from pSDM3188 by the _Cla_I/_Sal_I fragment from pSDM3127 resulted in a pvirF-NLS::cre_Δ_STOP cassette (pSDM3179). VirF_Δ_ATG was cloned as a _Sal_I/_Eag_I fragment from pSDM3193 (11) into the _Sal_I/_Not_I sites of pSDM3179 (pSDM3189). The pvirF-NLS::cre::virF cassette was finally inserted into _Hin_dIII/_Xba_I digested pRL662 (pSDM3154).
NLS::cre::virF_Δ_1–126 fusion. Deletion of the 126 5′ base pairs of the virF gene (Δ1–126) results in an N-terminal truncated VirFΔ42N protein lacking the F-box. The _SaI_I/_Eag_I fragment from pSDM3194 (11) was cloned into the _Sal_I/_Not_I sites of pSDM3179, resulting in pSDM3190 with a pvirF-NLS::cre::virF_Δ_1–126 cassette. This cassette was transferred into pRL662 as a _Hin_dIII/_Xba_I fragment (pSDM3155).
NLS::cre::virE3. The virE3 gene was cloned as an _Eco_RV/_Sac_I fragment from pRAL3248 (32) into pIC20R (33) resulting in pSDM3006. First, the 3′ coding region was subcloned in _Sal_I/_Pst_I digested pBluescriptIIKS (pSDM3503). The ATG start codon was removed by PCR amplification with primers virE3-1 (5′-ACGCgtcgacagatctGCGTGAGCACTA CGAAGAAAAG) and virE3-2B (5′-AGCCTATTTCGC CACGAAACCC) with pSDM3006 as template DNA. _Sal_I-digested PCR fragment (underlined) was cloned into pSDM3503 (pSDM3504). The final virE3_Δ_ATG gene was ligated into _Sal_I/_Xba_I digested pSDM3197, which resulted from cloning a _Hin_dIII/_Xba_I fragment from pSDM3179 in pRL662, and removal of the second _Sal_I site in proximity to the virF promoter sequence. The final plasmid harbouring the pvirF-NLS::cre::virE3 cassette was named pSDM3507.
The in frame fusions of all plasmids as well as the accurate SV40NLS and 3′ virF fragment sequences were confirmed by sequence analysis.
Construction of the pOsa plasmid. The osa gene under control of the tac promoter was cloned from pUFR047::ptacosa into pRL662 (pOsa or pSDM3180).
Agrobacterium tumefaciens strains
The stably maintained non-mobilisable plasmids pSDM3166 (virE2::cre), pSDM3129 (cre::virE2), pSDM3153 (NLS::virF::cre), pSDM3154 (NLS::cre::virF), pSDM3155 (NLS::cre::virF_Δ_1–126) and pSDM3507 (cre::virE3) were electroporated in Agrobacterium strain LBA1100, harbouring a disarmed octopine pTi plasmid named pAL1100 (37), according to den Dulk-Ras and Hooykaas (27). Furthermore, pSDM3129 and pSDM3155 were introduced in LBA2561, a LBA1100 derivative deleted for the virF gene (35), as well as in a series of LBA1100 derived vir mutants (37) having a Tn3_hoho1_ transposon insertion in virA (LBA1142), virB4 (LBA1143), virB7 (LBA1144), virC2 (LBA1146), virD1 (LBA1150), virD2 (LBA1147), virD4 (LBA1148), virE2 (LBA1149) or virG (LBA1145). p_NLS::cre::virF_Δ_1–126_ was also introduced into LBA2565 (LBA1100Δ_virE3_). The strains LBA1147 and LBA1150 contained plasmid pSDM3191 (or pD3,D4) to complement for polar mutation of downstream located virD3 and virD4. As control pSDM3191 was introduced in LBA1100. pD3,D4 was made by cloning the virD3 and virD4 genes behind the virD promoter as a 4.4-kb _Bam_HI fragment from pMP3 (38) into pSDM3015. pSDM3015 is a derivative of pBin19 (25) in which the T-region was replaced by the pIC19R/H polylinker sequence (33).
Agrobacterium strain LBA1100 was electroporated with the integrative cre control plasmid pSDM3126 or cre::virE2 fusion plasmid pSDM3128, resulting in LBA1100::cre and LBA1100_::cre::virE2_, respectively. Selection for integration of the introduced plasmid via homologous recombination at the virE1 locus of pAL1100 was done on LC medium (10 g/l bacto-tryptone, 5 g/l bacto-yeast extract and 8 g/l NaCl, pH 7) containing carbenicillin (100 µg/ml) and integration was confirmed by Southern blot analysis. The same was performed for strain LBA2561 with plasmid pSDM3190, resulting in LBA2561::NLS::cre::virF_Δ_1–126 expressing the Cre::VirFΔ42N protein. Here, integration of the plasmid occurred at the 5′ virF flank of pAL1100Δ_virF_. pSDM3180 (pOsa) was introduced in LBA1100::cre::virE2 and LBA2561::NLS::cre::virF_Δ_1–126.
Protein transport experiments
Agrobacterium strains expressing the cre or _cre_-fusion gene were grown overnight at 29°C in 5 ml minimal medium (MM) (39) supplemented with spectinomycin (250 µg/ml) and, in case a pRL662-based plasmid was present, also with gentamycin (40 µg/ml). Cells were harvested and diluted at an A600 of 0.25 in 5 ml induction medium [IM: MM salts and 40 mM 2-(N-morpholino) ethanesulfonic acid, pH 5.3, 10 mM glucose, 0.5% (w/v) glycerol and 200 µM acetosyringone (AS)]. The cells were then grown for 5 h at 28°C.
Saccharomyces cerevisiae strain LBY2 was grown overnight at 30°C in 10 ml yeast-peptone-dextrose (YPD) medium (40). Cells were 1:10 diluted in fresh 100 ml YPD medium and grown for 5 h at 30°C. The cells were subsequently washed with and resuspended in 500 µl IM. Aliquots of 100 µl of the Agrobacterium and Saccharomyces cultures were mixed 1:1, the mixture was placed on 0.45 µm cellulose nitrate filters on IM containing 5 mM glucose and the amino acids leucine and uracil at a concentration of 30 mg/l. To prevent loss of pRL662 gentamycin was added to the medium. After 6 days of co-cultivation at 22°C, the mixture was resuspended in 2 ml NaCl solution (9 g/l) and 100 µl aliquots (undiluted and a 10–2 dilution) were plated out on solid MY medium supplemented with leucine, uracil, 200 µM cefotaxim and 0.1% fluoro-orotic acid (FOA). The number of colonies that had lost the URA3 gene as indicated by growth on FOA were calculated after 4 days growth at 30°C. The output number of Agrobacterium cells (n.b. of viable cells after co-cultivation) was determined on solid LC medium supplemented with spectinomycin or gentamycin by plating a 10–5 and a 10–6 dilution of the co-cultivation mix. The output number of Saccharomyces cells was determined on solid MY medium with cefotaxim and the amino acids leucine and uracil by plating a 10–4 and a 10–5 dilution of the co-cultivation mix. The efficiency of excision of the URA3 gene is indicated as the number of yeast colonies on FOA divided by the number of surviving yeast colonies (output yeast).
Results
Translocation of the Cre::VirE2 and NLS::Cre::VirF fusion proteins from A.tumefaciens into S.cerevisiae
Agrobacterium tumefaciens can introduce T-DNA not only into plants but also into yeast and other fungi. In plants transfer of the Vir proteins VirE2 and VirF, which function in the plant cell in the process of tumour formation, accompanies T-DNA transfer. These transferred proteins are not essential for T-DNA transfer to yeast (16). We studied whether translocation of these proteins from A.tumefaciens into yeast occurs nevertheless as this might offer a fast and simple system for the study of this process of trans-kingdom protein translocation. To this end, we employed a similar strategy (see Fig. 1) as used to demonstrate protein translocation from Agrobacterium into plants (9). This strategy is based on the Cre/lox system from bacteriophage P1. Translational fusions of the cre gene with the 3′ or 5′ end of the virE2 and virF gene were placed under control of the virE and virF promoter, respectively. The SV40 NLS was added to the Cre-VirF fusion proteins to ensure nuclear targeting. The virE2 fusion genes were coordinately expressed with the virE1 gene. The cre gene transcriptionally fused to the virE1 gene and under control of the virE promoter was used as a control. To analyse protein transfer in the absence of T-DNA, the donor strains expressing the Cre fusion proteins contained a disarmed octopine Ti plasmid. The recipient yeast strain LBY2 carried a f_lox_ed URA3 gene integrated at the PDA1 locus on chromosome V. Delivery of a functional Cre enzyme into this yeast was monitored by Cre-mediated loss of the f_lox_ed URA3 gene due to site-specific recombination on the lox sites. Positive selection for loss of the URA3 gene was performed on medium containing FOA. Expression and recombination activity of the fusion proteins in A.tumefaciens was reported earlier (9). Results of a representative co-cultivation experiment are shown in Table 1. Co-cultivation with donor strains expressing the Cre recombinase alone, the VirE2::Cre or the NLS::VirF::Cre protein resulted in only a few Ura– colonies, which was similar to the number of Ura– colonies obtained after co-cultivation with an Agrobacterium strain lacking Cre altogether [Table 1; LBA1100 (none)]. However, when Cre was fused to the N-terminus of VirE2 (Cre::VirE2) or VirF (NLS::Cre::VirF) a large increase in the frequency of Ura– colonies per output recipient yeast (see Materials and Methods and legend to Table 1 for calculation of the frequency) was obtained (Table 1). These results show that delivery of Cre from A.tumefaciens into S.cerevisiae only occurs efficiently when Cre is fused to the N-terminus of VirE2 or VirF as is the case with transfer into plants.
Table 1. Direct Vir protein translocation from Agrobacterium into yeasta.
| Agrobacterium strainb | No. yeast coloniesc | Output yeastd 10–4 | Output Agroe 10–6 | Excision efficiencyf | |
|---|---|---|---|---|---|
| Not diluted | 10–2 Dilution | ||||
| 1100 (none) | 1 | 0 | 41 | 23 | 2 × 10–6 |
| 1100 (Cre) | 3 | 0 | 95 | 14 (10–4) | 3 × 10–6 |
| 1100 (VirE2::Cre) | 2 | 0 | 78 | 23 | 3 × 10–6 |
| 1100 (Cre::VirE2) | 365 | 3 | 65 | 11 | 6 × 10–4 |
| 1100 (NLS::VirF::Cre) | 4 | 0 | 86 | 25 | 5 × 10–6 |
| 1100 (NLS::Cre::VirF) | Full | 151 | 124 | 19 | 1 × 10–2 |
| 1100 (NLS::Cre::VirFΔ42N) | Full | 145 | 121 | 20 | 1 × 10–2 |
Recently, we showed that VirF contains an F-box domain at its N-terminus by which it can bind to F-box interacting plant proteins (11). In co-cultivation experiments with plant cells, removal of the F-box from the NLS::Cre::VirF fusion protein (NLS::Cre::VirFΔ42N) led to a 5-fold increase in the frequency of a Cre-mediated excision event, possibly due to an increased stability of the protein or the inability of the protein to bind with F-box interacting proteins in the cytoplasm (9). In transport experiments with yeast as a recipient, the F-box deleted version did not result in an increase in the number of colonies that had lost the URA3 gene (Table 1) compared with NLS::Cre::VirF. Furthermore, an NLS::Cre::VirF fusion protein in which the two most conserved amino acids of the F-box, leucine 26 and proline 27, were replaced by alanine [NLS::Cre::VirF(LP→AA)] did not result in an increased detection of excision (data not shown). Nevertheless, to be able to compare the transport studies in yeast and plants we used NLS::Cre::VirFΔ42N for further transport studies to yeast.
Protein transport to yeast is dependent on the VirB/VirD4 secretion system
The results shown in the previous paragraph indicate that Agrobacterium translocates the VirE2 and VirF proteins into yeast. To analyse whether translocation from Agrobacterium into yeast has the same requirements as translocation into plants, the fusion proteins were expressed from a non-mobilisable broad host range plasmid in a set of vir mutants (donors) harbouring an octopine Ti plasmid lacking the T-region. Results of a representative co-cultivation experiment are given in Table 2A and B, respectively. Co-cultivation of yeast with donor strains carrying a mutation in virA or virG did not result in the generation of Ura– colonies at a frequency higher than the background level [LBA1100 (wild type vir) expressing only Cre]. This was expected since the VirA/VirG two component regulatory system is responsible for transcriptional activation of all the vir genes. Similar low numbers of Ura– colonies were detected after co-cultivation of yeast with donor strains having a (polar) mutation in virB4, virB7 or virD4 indicating that transport of VirE2 and VirF occurs via the VirB/VirD4 type IV secretion system (TFSS). Mutations in virD1 or virD2 led to a block and decrease, respectively, of protein transport, but as the transposon mutations in these genes were (partially) polar on the distal virD4 gene, this could be due to a defect in VirD4 expression. Therefore, we introduced a plasmid expressing VirD4 in these mutants and tested again for protein transport. This time the strains were proficient in Vir protein translocation into yeast, showing that neither virD1 nor virD2 are necessary for protein transfer. This was expected since VirD1 together with VirD2 is involved in efficient T-DNA processing and VirD2, by covalently binding the 5′ end of the T-strand, also in transfer of the nucleoprotein complex from Agrobacterium into the host cell nucleus (8). Cre::Vir protein transport from Agrobacterium to yeast did take place from donor strains with mutations in the virC2 gene, the virE2 or virF genes. Apparently, the VirC2 and the wild type VirE2 and VirF proteins are not necessary for the transport of the fusion proteins. Furthermore, NLS::Cre::VirFΔ42N transport is independent of VirE3 (Table 2B) and VirE1 (data not shown). These results show that the VirB and VirD4 proteins that are required for VirE2 and VirF protein translocation into plants are equally important for Vir protein translocation into yeast. The yeast system thus offers a good model system to study protein translocation from Agrobacterium. We used it in the next two paragraphs to study transport of a putative third effector protein and to study the effect of tumourigenesis-inhibitor Osa on protein translocation.
Table 2. The effect of vir gene mutations.
| Agrobacterium straina | No. yeast coloniesc | Output yeastd 10–4 | Output Agroe 10–6 | Excision efficiencyf | |
|---|---|---|---|---|---|
| Not diluted | 10–2 Dilution | ||||
| A. On Cre::VirE2 protein translocation from Agrobacterium into yeastb | |||||
| 1100 (Cre) | 6 | 0 | 25 | 5 | 2 × 10–5 |
| 1100 (Cre::VirE2) | Full | 11 | 24 | 73 | 5 × 10–3 |
| virA (Cre::VirE2) | 12 | 0 | 30 | 58 | 4 × 10–5 |
| virB4 (Cre::VirE2) | 26 | 0 | 37 | 7 (10–5) | 7 × 10–5 |
| virB7 (Cre::VirE2) | 5 | 0 | 17 | 66 | 3 × 10–5 |
| virG (Cre::VirE2) | 5 | 0 | 28 | 61 | 2 × 10–5 |
| virC2 (Cre::VirE2) | Full | 12 | 15 | 54 | 8 × 10–3 |
| virD1 (Cre::VirE2) | 5 | 0 | 16 | 98 | 3 × 10–5 |
| 3_′_virD2 (Cre::VirE2) | 43 | 0 | 26 | 34 | 2 × 10–4 |
| virD4 (Cre::VirE2) | 5 | 0 | 18 | 16 | 3 × 10–5 |
| virE2 (Cre::VirE2) | Full | 23 | 19 | 39 | 1 × 10–2 |
| virF (Cre::VirE2) | Full | 8 | 25 | 72 | 3 × 10–3 |
| virD1+pD3, D4 (Cre::VirE2) | Full | 13 | 46 | 72 | 3 × 10–3 |
| 3_′_virD2+pD3, D4 (Cre::VirE2) | Full | 11 | 30 | 40 | 4 × 10–3 |
| B. On NLS::Cre::VirFΔ42N protein translocation from Agrobacterium into yeastb | |||||
| 1100 (Cre) | 9 | 0 | 241 | 5 | 4 × 10–6 |
| 1100 (NLS::Cre::VirFΔ42N) | Full | 80 | 74 | 25 | 1 × 10–2 |
| virA (NLS::Cre::VirFΔ42N) | 1 | 0 | 43 | 15 | 2 × 10–6 |
| virB4 (NLS::Cre::VirFΔ42N) | 8 | 0 | 89 | 2 (10–5) | 9 × 10–6 |
| virB7 (NLS::Cre::VirFΔ42N) | 3 | 0 | 53 | 26 | 6 × 10–6 |
| virG (NLS::Cre::VirFΔ42N) | 27 | 0 | 387 | 74 | 7 × 10–6 |
| virC2 (NLS::Cre::VirFΔ42N) | Full | 51 | 74 | 12 | 7 × 10–3 |
| virD1 (NLS::Cre::VirFΔ42N) | 12 | 0 | 68 | 18 | 2 × 10–5 |
| 3_′_virD2 (NLS::Cre::VirFΔ42N) | 301 | 0 | 55 | 29 | 5 × 10–4 |
| virD4 (NLS::Cre::VirFΔ42N) | 1 | 0 | 104 | 10 | 1 × 10–6 |
| virE2 (NLS::Cre::VirFΔ42N) | Full | 26 | 56 | 8 (10–5) | 5 × 10–3 |
| virF (NLS::Cre::VirFΔ42N) | Full | 102 | 76 | 18 | 1 × 10–2 |
| virE3 (NLS::Cre::VirFΔ42N) | Full | 80 | 81 | 30 | 1 × 10–2 |
| 1100+pD3, D4 (NLS::Cre::VirFΔ42N) | Full | 106 | 104 | 24 | 1 × 10–2 |
| virD1+pD3, D4 (NLS::Cre::VirFΔ42N) | Full | 131 | 115 | 19 | 1 × 10–2 |
| 3_′_virD2+pD3, D4 (NLS::Cre::VirFΔ42N) | Full | 152 | 107 | 12 | 1 × 10–2 |
Identification of a third effector protein, VirE3
The translocated VirE2 and VirF proteins do not share an obvious transport domain. However, deletion analysis has shown that the transport signal is located in the C-terminal part of these proteins. A common Arg-Pro-Arg motif can be identified in this region and may form part of the transport signal (9). An Arg-Pro-Arg sequence is also present in the C-terminal region of the virulence protein VirE3. Although the function of this protein is still an enigma, it is worth noting that this protein has been conserved in all the different types of Ti and Ri plasmids. As the Arg-Pro-Arg motif indicated that VirE3 might be a transported effector protein of the Agrobacterium virulence system, we fused the N-terminus of VirE3 to NLS::Cre and assayed in the yeast system whether translocation of the fusion protein took place. The results of a representative experiment are summarised in Table 3. The data revealed that translocation into yeast cells did indeed occur with an efficiency intermediate between that of VirF and VirE2, thus identifying VirE3 as a third effector protein involved in the transformation of host cells by Agrobacterium. No transport of NLS::Cre::VirE3 was detected from a virD4 mutant, which is defective in type IV secretion (data not shown).
Table 3. Detection of VirE3 protein translocation from Agrobacterium into yeasta.
| Agrobacterium strainb | No. yeast coloniesc | Output yeastd 10–4 | Output Agroe 10–6 | Excision efficiencyf | |
|---|---|---|---|---|---|
| Not diluted | 10–2 Dilution | ||||
| 1100 (Cre) | 6 | 0 | 201 | 94 | 3 × 10–6 |
| 1100 (NLS::Cre::VirFΔ42N) | Full | 115 | 326 | 76 | 3 × 10–3 |
| 1100 (Cre::VirE2) | 192 | 1 | 250 | 31 | 8 × 10–5 |
| 1100 (NLS::Cre::VirE3) | 295 | 0 | 275 | 85 | 1 × 10–4 |
Osa inhibits Vir protein translocation from Agrobacterium into yeast
VirE2 or virF mutants (T-DNA donors) can be ‘extracellularly’ complemented for tumour formation on plants by co-infection with a helper strain lacking the T-region but containing the vir region including virE2 or virF, respectively (32,41–43). It has been found that the presence of the osa gene from IncW plasmid pSa in A.tumefaciens abolishes tumourigenicity on plants (44). Later, Lee et al. (45) showed that ‘extracellular’ complementation for tumour formation of a virE2 mutant on plants was inhibited by the presence of the osa gene in the helper strain but not by its presence in the T-DNA donor strain. These results indirectly indicate that Osa inhibits VirE2 transport rather than T-DNA transfer from Agrobacterium to plants. We have studied this possible inhibition of VirE2 and VirF transport by Osa in a direct assay. To this end, pOsa (pSDM3180) (containing the osa gene behind the tac promoter) was used in our protein transfer system to study the inhibitory effect of Osa on the translocation of Cre::VirE2 and NLS::Cre::VirFΔ42N from Agrobacterium into yeast. Results of a representative co-cultivation experiment are given in Table 4. Translocation of Cre::VirE2 or NLS::Cre::VirFΔ42N from Agrobacterium into LBY2 resulted in a large increase of the frequency of Ura– colonies per output recipient compared with the control strain that expresses only Cre. In the presence of pOsa a complete inhibition of Cre::VirE2 and NLS::Cre:VirFΔ42N translocation from Agrobacterium into yeast was observed, providing direct evidence for inhibition of Vir protein transport from Agrobacterium by Osa.
Table 4. The inhibitory effect of the Osa protein on Cre::Vir protein translocation from Agrobacterium into yeasta.
| Agrobacterium strainb | No. yeast coloniesc | Output yeastd 10–4 | Output Agroe 10–6 | Excision efficiencyf | |
|---|---|---|---|---|---|
| Not diluted | 10–2 Dilution | ||||
| 1100 (none) | 1 | 0 | 41 | 23 | 2 × 10–6 |
| 1100::cre | 1 | 0 | 39 | 41 | 2 × 10–6 |
| 1100::cre::virE2 | 416 | 4 | 52 | 29 | 8 × 10–4 |
| 1100::cre::virE2 + pOsa | 0 | 0 | 61 | 18 | <2 × 10–6 |
| 2561::NLS::cre::virF_Δ_1–126 | Full | 37 | 36 | 46 | 1 × 10–2 |
| 2561::NLS::cre::virF_Δ_1–126 +pOsa | 7 | 0 | 71 | 17 | 1 × 10–5 |
DISCUSSION
Here, we report a novel and fast method to detect A.tumefaciens Vir protein transport. We show that Agrobacterium not only transfers T-DNA to the yeast S.cerevisiae during co-cultivation (16) but also translocates the Vir proteins VirE2, VirE3 and VirF directly into yeast cells.
Virulence studies on plants have shown that an Agrobacterium strain mutated in the virE2 gene is avirulent on most plant species while a strain mutated in the virF gene is attenuated in virulence on certain host plants (43,46,47). The attenuated virulence of virF mutants is aggravated by a mutation in the virE3 gene (B.Schrammeijer, P.Zuiderwijk and P.J.J.Hooykaas, unpublished results). Both VirE2 and VirF play a role in the plant cell during tumourigenesis: plants expressing either VirE2 or VirF become hosts for virE2 and virF mutants, respectively (48,49). VirE2 attaches to the T-strand by cooperative binding thereby protecting the T-strand from degradation in the plant cell (50). Such coating is important for efficient entrance of the T-strand into the plant nucleus (8,10). VirF is an F-box protein that interacts with homologues of the yeast Skp1 protein in plants (11). F-box proteins and Skp1 form a part of SCF complexes, which are responsible for the ubiquitin-mediated proteolysis of specific plant proteins. VirF as part of such an SCF complex in plants, may be involved in the targeted proteolysis of specific host proteins during tumourigenesis by A.tumefaciens (11).
Recently, it was shown in our group that T-DNA transfer from virE2 or virF mutants to S.cerevisiae was not or only slightly affected compared with the wild type strain (16) (H. van Attikum, personal communication). Here, we show that although VirE2 and VirF are not essential for T-DNA transfer from Agrobacterium to yeast these Vir proteins are still translocated into yeast. Translocation of VirE2 and VirF fusion proteins from Agrobacterium into yeast occurs via the VirB secretion system in the presence of coupling factor VirD4 as was shown for plants (9). This indicates that both Vir proteins are actively translocated through the same pore/pilus like structure through which the T-strand is transferred into the yeast cell (16). In addition, we found now that VirE3 is also transported through this VirB/D4 channel.
The VirB/VirD4 transport system of A.tumefaciens is the prototype of the TFSS (reviewed in 7). Other members of this family of TFSSs are the conjugative DNA transport systems encoded by the (broad host range) plasmids IncW, IncN and IncP. In several animal pathogens, like Helicobacter pylori, Bordetella pertussis, Legionella pneumophila, Rickettsia prowazekii and Brucella suis, similar secretion systems are present that may not be involved in DNA transfer, but in protein export. Evidence for protein secretion has been found for the Bordetella and Helicobacter systems (reviewed in 51). It is entirely possible that the TFSSs involved in DNA translocation have evolved from a protein transport system. In this scenario the relaxase bound to the 5′ end of the DNA acts as a pilot protein that is recognised by the TFSS.
Translocation of VirE2 is independent of VirF and translocation of VirF is independent of VirE2 and VirE3. The VirE3 protein is encoded by a gene, which was identified downstream of the virE2 gene on Ti plasmids (52–54). Mutations in virE3 aggravate the attenuated virulence of virF mutants. We now show that the translocation of VirF into yeast is not affected by the absence of VirE3 from the bacterium, and that VirE3 itself is a translocated effector protein. Further study is necessary to reveal the role of VirE3 during tumourigenesis on plants.
For delivery of Cre into yeast cells mediated by VirE2 or VirF transport signals, the Cre protein must be fused to the N-terminus of either Vir protein, as was the case for delivery of Cre into plant cells (9). Interestingly, the 37 C-terminally located amino acids of VirF were sufficient to introduce Cre into yeast (data not shown), as in plants (9). These results show that the same transport signals located in the C-terminus of VirE2 and VirF are important to mediate translocation into plant and yeast cells. In agreement with a C-terminal transport signal, VirE3 was able to transport Cre into yeast as an N-terminal fusion. The common Arg-Pro-Arg motif that is present in the C-termini of VirE2, VirE3 and VirF may form part of the transport domain. Currently, we are analysing the C-terminal signal sequences in more detail.
To ensure nuclear targeting of the Cre::VirF fusion constructs the SV40 NLS sequence was fused translationally to the N-terminus. Guralnick et al. (55) suggested that the NLS sequences present in VirE2 are unable to mediate entrance into the nucleus of Drosophila embryos or Xenopus oocytes and that they are rather plant specific. Recently, Rhee et al. (56) showed in a genetic assay the inability of the VirE2 NLS to function in nuclear import in yeast cells. Here, we show that the Cre::VirE2 fusion protein is able to enter the yeast cell nucleus. Therefore, nuclear targeting is most probably mediated by the cryptic NLS sequence present in Cre (57).
Removal of the N-terminus from VirF, including the F-box (42 amino acids), resulted in a 5-fold higher efficiency of Cre-mediated deletion in Arabidopsis (9). The same high efficiency was obtained in Arabidopsis for the NLS::Cre::VirF(LP→AA) fusion protein (data not shown). Deletion or mutation of the F-box from VirF may stabilise the VirF fusion protein in plants by preventing VirF from binding to F-box interacting proteins (Skp1 orthologs) in the plant cytoplasm (11), ultimately leading to an increase in the numbers of NLS::Cre::VirFΔ42N or NLS::Cre:: VirF(LP→AA) molecules that reach the plant cell nucleus (9). However, the absence or mutation of this F-box from VirF did not lead to the detection of enhanced Cre activity in yeast. This difference might be the result of distinct binding affinities in yeast and plants between the full length VirF protein and the Skp1 orthologs, making the full length VirF protein more stable in yeast. Another explanation might be that a different factor, such as the number of yeast cells competent for transformation, is limited for translocation to yeast, thus concealing the effect of deletion of the F-box.
Previously, it was suggested that oncogenic suppression by Osa involves inhibition of VirE2 transport rather than T-DNA transfer from A.tumefaciens to plants (45). We have confirmed this in our direct protein transport assay. Moreover, we have shown that inhibition of protein transfer is not restricted to VirE2, but involved at least VirF also. The molecular mechanism by which Osa inhibits protein transport is unknown. In a previous article it was proposed that Osa may do this by interacting with VirE1, which is needed for VirE2 transport from the bacterium (45). However, as transport of VirF, which does not need VirE1 (data not shown), is inhibited by Osa we also think it is unlikely that Osa’s inhibitory effects are a result of its interaction with VirE1. Recently, we have done experiments to see whether Osa may compete with VirE2 and VirF export by being a favoured translocation protein itself. However, preliminary results indicate that Osa is not transported by the VirB/VirD4 secretion channel (data not shown). Therefore, at the moment we favour the idea that Osa, being a membrane protein, either inhibits the function of one of the proteins of the VirB/VirD4 type IV secretion channel or binds to the transported Vir proteins (VirE2, VirF) and thereby prevents these from being exported. Further research has to clarify the mechanism by which Osa inhibits Vir protein transport from Agrobacterium into the host cell.
Our results indicate that the A.tumefaciens system, which has so far been used as a tool to introduce genes of interest into plants, yeast and other fungi, also offers potential for delivering proteins of interest fused to VirE2, VirE3 or VirF or their transport domain(s) into plants, yeast and fungi.
Most of the techniques to mutate yeast genes are based on homologous recombination events during mitosis (58). Nutritional markers are commonly used as selection markers in yeast (59). However, the availability of different nutritional markers for selection is limited. An alternative is the kan R marker, which has been shown to be a good marker in yeast (60). Recently, mutagenesis methods were developed in which sequences homologous to the target site in the yeast genome (61) contain a selection marker flanked by specific target sites (e.g. _lox_-_kan_MX-lox) for recombinases (e.g. Cre recombinase). The selection marker is now removed by introducing a plasmid expressing the Cre protein in the yeast cell. Furthermore, this plasmid has to be removed again from the yeast strain. The method of protein transport (of a functional recombinase) described in this paper avoids the need to introduce and subsequently remove a Cre expressing plasmid. Therefore, with this Cre-Vir/lox system we provide a fast method to mediate site-specific recombination events in yeast. Besides, the trans-kingdom protein transfer to yeast offers a fast and reliable alternative for translocation studies of virulence effectors from bacteria carrying a TFSS.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor E. W. Nester (Seattle) for kindly providing plasmid pUFR047::ptacosa. Thanks are due to P. Bundock for critical reading of the manuscript and P. Hock for preparing Figure 1. This work was supported by the Netherlands Foundation for Chemical Research (SON) with financial aid from the Netherlands Organisation for Scientific Research (NWO).
REFERENCES
- 1.Christie P.J. (1997) Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol., 179, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelvin S.B. (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol., 51, 223–256. [DOI] [PubMed] [Google Scholar]
- 3.Hooykaas P.J.J. and Beijersbergen,A.G.M. (1994) The virulence system of Agrobacterium tumefaciens. Annu. Rev. Phytopathol., 32, 157–179. [Google Scholar]
- 4.Kado C.I. (1991) Molecular mechanisms of crown gall tumorigenesis. Crit. Rev. Plant Sci., 10, 1–32. [Google Scholar]
- 5.Winans S.C. (1992) Two-way chemical signaling in _Agrobacterium_–plant interactions. Microbiol. Rev., 56, 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zambryski P.C. (1992) Chronicles from the _Agrobacterium_-plant cell DNA transfer story. Annu. Rev. Plant Physiol. Plant Mol. Biol., 43, 465–490. [Google Scholar]
- 7.Zupan J., Muth,T.R., Draper,O. and Zambryski,P. (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J., 23, 11–28. [DOI] [PubMed] [Google Scholar]
- 8.Zupan J. and Zambryski,P. (1997) The Agrobacterium tumefaciens DNA transfer complex. Crit. Rev. Plant Sci., 16, 279–295. [Google Scholar]
- 9.Vergunst A.C., Schrammeijer,B., den Dulk-Ras,A., de Vlaam,C.M., Regensburg-Tuïnk,T.J. and Hooykaas,P.J. (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science, 290, 979–982. [DOI] [PubMed] [Google Scholar]
- 10.Ziemienowicz A., Merkle,T., Schoumacher,F., Hohn,B. and Rossi,L. (2001) Import of Agrobacterium T-DNA into plant nuclei. Two distinct functions of VirD2 and VirE2 proteins. Plant Cell, 13, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrammeijer B., Risseeuw,E., Pansegrau,W., Regensburg-Tuïnk,T.J., Crosby,W.L. and Hooykaas,P.J. (2001) Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol., 11, 258–262. [DOI] [PubMed] [Google Scholar]
- 12.Bai C., Sen,P., Hofmann,K., Ma,L., Goebl,M., Harper,J.W. and Elledge,S.J. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, 86, 263–274. [DOI] [PubMed] [Google Scholar]
- 13.Skowyra D., Craig,K.L., Tyers,M., Elledge,S.J. and Harper,J.W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell, 91, 209–219. [DOI] [PubMed] [Google Scholar]
- 14.Sundberg C.D. and Ream,W. (1999) The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J. Bacteriol., 181, 6850–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Z., Sagulenko,E., Ding,Z. and Christie,P.J. (2001) Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J. Bacteriol., 183, 3855–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bundock P., den Dulk-Ras,A., Beijersbergen,A. and Hooykaas,P.J. (1995) Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J., 14, 3206–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Groot M.J., Bundock,P., Hooykaas,P.J. and Beijersbergen,A.G. (1998) _Agrobacterium tumefaciens_-mediated transformation of filamentous fungi. Nat. Biotechnol., 16, 839–842. [DOI] [PubMed] [Google Scholar]
- 18.Ow D. and Medberry,S. (1995) Genome manipulation through site-specific recombination. Crit. Rev. Plant Sci., 14, 239–261. [Google Scholar]
- 19.Farrand S.K., Kado,C.I. and Ireland,C.R. (1981) Suppression of tumorigenicity by the IncW plasmid pSa in Agrobacterium tumefaciens. Mol. Gen. Genet., 181, 44–51. [Google Scholar]
- 20.Loper J.E. and Kado,C.I. (1979) Host range conferred by the virulence-specifying plasmid of Agrobacterium tumefaciens. J. Bacteriol., 139, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., Li,C.M. and Nester,E.W. (2000) Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc. Natl Acad. Sci. USA, 97, 7545–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J.S. and Prakash,L. (1990) Yeast Saccharomyces cerevisiae markers in pUC18 polylinkers. Yeast, 6, 363–366. [DOI] [PubMed] [Google Scholar]
- 23.Meijer A.H., Ouwerkerk,P.B. and Hoge,J.H. (1998) Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast, 14, 1407–1415. [DOI] [PubMed] [Google Scholar]
- 24.Steensma H.Y., Holterman,L., Dekker,I., van Sluis,C.A. and Wenzel,T.J. (1990) Molecular cloning of the gene for the E1 alpha subunit of the pyruvate dehydrogenase complex from Saccharomyces cerevisiae. Eur. J. Biochem., 191, 769–774. [DOI] [PubMed] [Google Scholar]
- 25.Bevan M. (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res., 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bundock P. and Hooykaas,P.J. (1996) Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl Acad. Sci. USA, 93, 15272–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.den Dulk-Ras A. and Hooykaas,P.J. (1995) Electroporation of Agrobacterium tumefaciens. Methods Mol. Biol. 55, 63–72. [DOI] [PubMed] [Google Scholar]
- 28.Schiestl R.H. and Petes,T.D. (1991) Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 88, 7585–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zonneveld B.J.M. (1986) Cheap and simple yeast media. J. Microbiol. Methods, 4, 287. [Google Scholar]
- 30.Mozo T. and Hooykaas,P.J. (1992) Design of a novel system for the construction of vectors for _Agrobacterium_-mediated plant transformation. Mol. Gen. Genet., 236, 1–7. [DOI] [PubMed] [Google Scholar]
- 31.Vieira J. and Messing,J. (1991) New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene, 100, 189–194. [DOI] [PubMed] [Google Scholar]
- 32.Melchers L.S., Maroney,M.J., den Dulk-Ras,A., Thompson,D.V., van Vuuren,H.A., Schilperoort,R.A. and Hooykaas,P.J. (1990) Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol. Biol., 14, 249–259. [DOI] [PubMed] [Google Scholar]
- 33.Alting-Mees M.A. and Short,J.M. (1989) pBluescript II: gene mapping vectors. Nucleic Acids Res., 17, 9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrammeijer B., Hemelaar,J. and Hooykaas,P.J. (1998) The presence and characterization of a virF gene on Agrobacterium vitis Ti plasmids. Mol. Plant Microbe Interact., 11, 429–433. [DOI] [PubMed] [Google Scholar]
- 35.Benes V., Hostomsky,Z.A. and Paces,V. (1993) M13 and pUC vectors with new unique restriction sites for cloning. Gene, 130, 151–152. [DOI] [PubMed] [Google Scholar]
- 36.Marsh J.L., Erfle,M. and Wykes,E.J. (1984) The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene, 32, 481–485. [DOI] [PubMed] [Google Scholar]
- 37.Beijersbergen A., Den Dulk-Ras,A., Schilperoort,R.A. and Hooykaas,P.J.J. (1992) Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science, 256, 1324–1327. [DOI] [PubMed] [Google Scholar]
- 38.Vogel A.M. and Das,A. (1992) The Agrobacterium tumefaciens virD3 gene is not essential for tumorigenicity on plants. J. Bacteriol., 174, 5161–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooykaas P.J.J., Roobol,C. and Schilperoort,R.A. (1979) Regulation of the transfer of Ti-plasmids of Agrobacterium tumefaciens. J. Gen. Microbiol., 110, 99–109. [Google Scholar]
- 40.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- 41.Gardner R.C. and Knauf,V.C. (1986) Transfer of Agrobacterium DNA to plants requires a T-DNA border but not the virE locus. Science, 231, 725–727. [DOI] [PubMed] [Google Scholar]
- 42.Otten L., Piotrowiak,G., Hooykaas,P., Dubois,M., Szegedi,E. and Schell,J. (1985) Identification of an Agrobacterium tumefaciens pTiB6S3 vir region fragment that enhances the virulence of pTiC58. Mol. Gen. Genet., 199, 189–193. [Google Scholar]
- 43.Otten L.H., De Greve,J., Leemans,R., Hain,P., Hooykaas,P. and Schell,J. (1984) Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet., 195, 159–163. [Google Scholar]
- 44.Close S.M. and Kado,C.I. (1991) The osa gene of pSa encodes a 21.1-kilodalton protein that suppresses Agrobacterium tumefaciens oncogenicity. J. Bacteriol., 173, 5449–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee L.Y., Gelvin,S.B. and Kado,C.I. (1999) pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export. J. Bacteriol., 181, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hooykaas P.J.J., Hofker,M., den Dulk-Ras,H. and Schilperoort,R.A. (1984) A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid, 11, 195–205. [DOI] [PubMed] [Google Scholar]
- 47.Iyer V.N., Klee,H.J. and Nester,E.W. (1982) Units of genetic expression in the virulence region of a plant tumor-inducing plasmid of Agrobacterium tumefaciens. Mol. Gen. Genet., 188, 418–424. [DOI] [PubMed] [Google Scholar]
- 48.Citovsky V., Zupan,J., Warnick,D. and Zambryski,P. (1992) Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science, 256, 1802–1805. [DOI] [PubMed] [Google Scholar]
- 49.Regensburg-Tuïnk A.J. and Hooykaas,P.J. (1993) Transgenic N.glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A.tumefaciens. Nature, 363, 69–71. [DOI] [PubMed] [Google Scholar]
- 50.Rossi L., Hohn,B. and Tinland,B. (1996) Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA, 93, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christie P.J. (2001) Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol., 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalogeraki V.S., Zhu,J., Stryker,J.L. and Winans,S.C. (2000) The right end of the vir region of an octopine-type Ti plasmid contains four new members of the vir regulon that are not essential for pathogenesis. J. Bacteriol., 182, 1774–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrammeijer B., Beijersbergen,A., Idler,K.B., Melchers,L.S., Thompson,D.V. and Hooykaas,P.J. (2000) Sequence analysis of the _vir_-region from Agrobacterium tumefaciens octopine Ti plasmid pTi15955. J. Exp. Bot., 51, 1167–1169. [DOI] [PubMed] [Google Scholar]
- 54.Zhu J., Oger,P.M., Schrammeijer,B., Hooykaas,P.J., Farrand,S.K. and Winans,S.C. (2000) The bases of crown gall tumorigenesis. J. Bacteriol., 182, 3885–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guralnick B., Thomsen,G. and Citovsky,V. (1996) Transport of DNA into the nuclei of xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell, 8, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee Y., Gurel,F., Gafni,Y., Dingwall,C. and Citovsky,V. (2000) A genetic system for detection of protein nuclear import and export. Nat. Biotechnol., 18, 433–437. [DOI] [PubMed] [Google Scholar]
- 57.Le Y., Gagneten,S., Tombaccini,D., Bethke,B. and Sauer,B. (1999) Nuclear targeting determinants of the phage P1 cre DNA recombinase. Nucleic Acids Res., 27, 4703–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baganz F., Hayes,A., Marren,D., Gardner,D.C. and Oliver,S.G. (1997) Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast, 13, 1563–1573. [DOI] [PubMed] [Google Scholar]
- 60.Delneri D., Gardner,D.C., Bruschi,C.V. and Oliver,S.G. (1999) Disruption of seven hypothetical aryl alcohol dehydrogenase genes from Saccharomyces cerevisiae and construction of a multiple knock-out strain. Yeast, 15, 1681–1689. [DOI] [PubMed] [Google Scholar]
- 61.Guldener U., Heck,S., Fielder,T., Beinhauer,J. and Hegemann,J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res., 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]