Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p (original) (raw)
Abstract
Tail-anchored proteins are distinct from other membrane proteins as they are thought to insert into the endoplasmic reticulum (ER) membrane independently of Sec61p translocation pores. These pores not only mediate import but are also assumed to catalyze export of proteins in a process called ER-associated protein degradation (ERAD). In order to examine the Sec61p dependence of the export of tail-anchored proteins, we analyzed the degradation pathway of a tail-anchored ER membrane protein, the ubiquitin-conjugating enzyme 6 (Ubc6p). In contrast to other ubiquitin conjugating enzymes (Ubcs), Ubc6p is naturally short-lived. Its proteolysis is mediated specifically by the unique Ubc6p tail region. Degradation further requires the activity of Cue1p-assembled Ubc7p, and its own catalytic site cysteine. However, it occurs independently of the other ERAD components Ubc1p, Hrd1p/Der3p, Hrd3p and Der1p. In contrast to other natural ERAD substrates, proteasomal mutants accumulate a membrane-bound degradation intermediate of Ubc6p. Most interestingly, mutations in SEC61 do not reduce the turnover of full-length Ubc6p nor cause a detectable accumulation of degradation intermediates. These data are in accordance with a model in which tail-anchored proteins can be extracted from membranes independently of Sec61p.
Keywords: proteasome/Sec61p/tail-anchored protein/Ubc6p/yeast
Introduction
Tail-anchored proteins are characterized by a single C-terminally located hydrophobic domain, which anchors them to membranes. Members of this protein class are located in various cellular organelles and include Ubc6p, soluble _N_-ethylmaleimide-sensitive factor attachment protein receptors (SNARES) and the Bcl-2 protein family (Kutay et al., 1993). Studies on synaptobrevin suggest that they can be imported first into the endoplasmic reticulum (ER) membrane in a post-translational manner and subsequently be distributed along the secretory pathway. Surprisingly, ER membrane insertion seems to occur independently of Sec61 translocation pores (Kutay et al., 1995). This distinguishes tail-anchored proteins from other membrane proteins, which are all thought to depend on this translocation machinery (Rapoport et al., 1996). Nevertheless, the insertion of tail-anchored proteins might not be totally independent of membrane-localized components. At least for synaptobrevin, membrane insertion still depends on an unidentified proteinaceous factor and ATP (Kutay et al., 1995).
The Sec61p pore per se seems to have no directionality as it is also involved in a process called ER- associated degradation (ERAD) (Sommer and Wolf, 1997; Bonifacino and Weissman, 1998). Here, retrograde transport of ER-resident proteins for polyubiquitylation and subsequent degradation by the cytosolic 26S proteasome have been shown to depend on Sec61p (Pilon et al., 1997; Plemper et al., 1997; Zhou and Schekman, 1999), the central subunit of the Sec61p translocons (Johnson and van Waes, 1999). Apart from Sec61p, ERAD involves a variety of other components, among which enzymes of the ubiquitin system are pivotal (Sommer and Wolf, 1997; Bonifacino and Weissman, 1998; Hershko and Ciechanover, 1998). In the yeast Saccharomyces cerevisiae, three ubiquitin-conjugating enzymes (E2s, Ubcs), namely Ubc1p, Ubc6p and Ubc7p, act on ERAD substrates (Biederer et al., 1996; Friedlander et al., 2000). The activity of two of them is located at the cytosolic side of the ER membrane. Ubc6p carries a tail region in addition to the so-called UBC domain (ubiquitin conjugation consensus sequence) that contains a transmembrane segment at the extreme C-terminus (Sommer and Jentsch, 1993; Yang et al., 1997). The soluble Ubc7p is recruited to the lipid bilayer via interaction with Cue1p, an integral ER membrane protein (Biederer et al., 1997). An additional ERAD component is the integral membrane protein Hrd1p/Der3p, which has been demonstrated to function as a ubiquitin ligase (E3) in ERAD together with Ubc1p and Ubc7p (Bays et al., 2001). Although E3–E2 complexes mediate most ubiquitylation, an E3 acting along with Ubc6p is unknown as yet. In at least some cases, ERAD also depends on Hrd3p and Der1p (Hampton et al, 1996; Knop et al. 1996). These are integral membrane proteins, identified in genetic screens. However, their function is less well understood.
The energy source of retrograde transport must be different from that of protein translocation. It has been demonstrated that polyubiquitylated ERAD substrates can be degraded directly at the ER membrane and that polyubiquitylation and retrograde transport are coupled processes (Biederer et al., 1997; Mayer et al., 1998). It has been proposed that export might be driven either by arresting the retrogradely transported protein on the cytosolic side by attachment of a polyubiquitin chain (Biederer et al., 1997) or by ATP hydrolysis at the 19S cap of the 26S proteasome (Mayer et al., 1998; Chillaron and Haas, 2000).
Here we show that the tail-anchored Ubc6p is not only an ERAD component but also a proteolytic substrate, as it is constitutively degraded by the cytosolic proteasome. In order to investigate whether degradation and concomitant export of tail-anchored proteins might differ from that of other membrane proteins, we analyzed the degradation pathway of the substrate Ubc6p. Interestingly, Ubc6p degradation depends on its own catalytic activity, on Cue1p and Ubc7p, but not on other ERAD components including the Ubc7p-associated E3, Hrd1p/Der3p. Since the proteolytic breakdown of Ubc6p occurred independently of Sec61p, its extraction from ER membranes might parallel its Sec61p-independent import into this membrane.
Results
Ubc6p is a naturally short-lived protein
We observed that the level of Ubc6p is elevated in strains deleted for UBC7 or the Ubc7p membrane receptor CUE1 (Figure 1A). To determine whether an impaired degradation process caused higher levels of Ubc6p, we measured the content of Ubc6p at different time points after translation was abrogated by treatment with cycloheximide. Figure 1B shows that the level of Ubc6p decreased over time and hence indicates that Ubc6p is a short-lived protein. We also analyzed the turnover of Ubc6p in cells deleted for UBC7 and CUE1. Ubc6p remained completely stable over the investigated time period of 3 h (Figure 1B). These results suggest that the elevated Ubc6p levels in the Δubc7 and Δcue1 strains are caused by an abrogation of its degradation. In contrast, a deletion of the third Ubc involved in ERAD, UBC1, did not stabilize Ubc6p (Figure 1C).
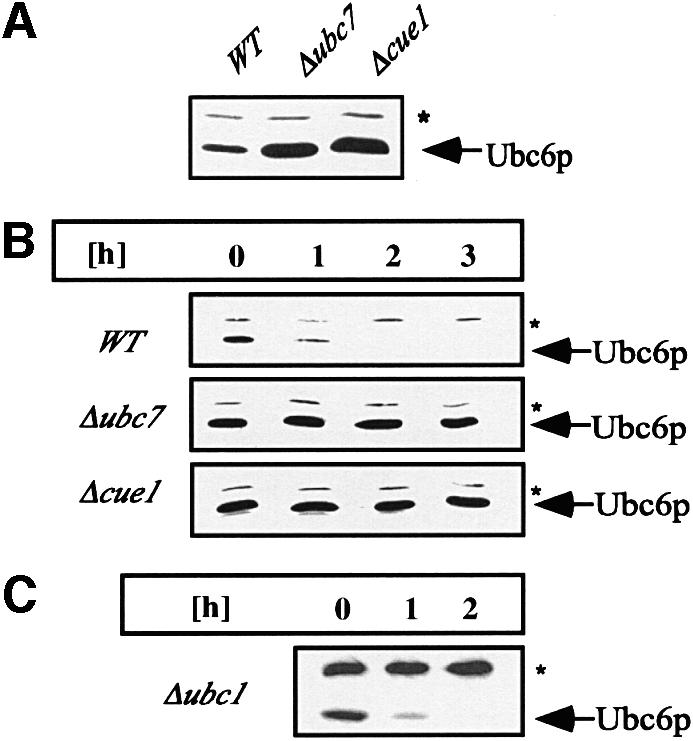
Fig. 1. Ubc6p degradation depends on Ubc7p and Cue1p, but noton Ubc1p. (A) Immunoblots loaded with equal amounts of isolated membranes from the strains indicated were probed with αUbc6p antibody. (B) The stability of Ubc6p was analyzed in cycloheximide decay experiments of the same strains: after abrogation of protein translation by the addition of cycloheximide, aliquots were taken at the indicated time points, and membranes were prepared and probed for the amount of Ubc6p. (C) Cycloheximide decay experiment in a strain depleted of UBC1. The asterisk marks Sec61p, which served as a loading control.
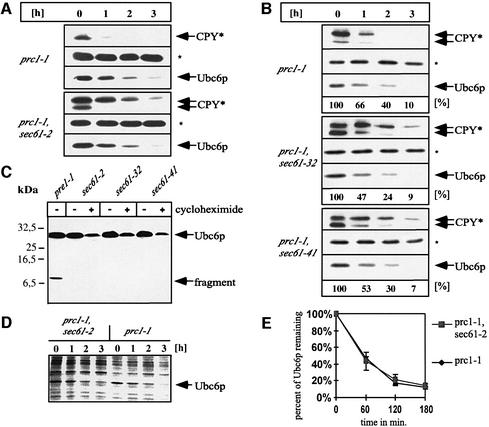
Radioactive pulse–chase experiments in the absence of translation inhibitors revealed similar degradation kinetics for Ubc6p in a wild-type strain (Figure 6D). Therefore, the degradation of Ubc6p is not a cycloheximide-induced phenomenon, but rather takes place constitutively. Quantification of the above data revealed a half-life for Ubc6p of ∼55 min in synthetic minimal medium (Figure 6E).
Fig. 6. Sec61p function in Ubc6p degradation. (A and B) Cycloheximide decay experiment conducted in sec61 mutant strains and the corresponding wild type. Membranes were immunoblotted and probed for the indicated proteins. The asterisk marks Sec61p, which served as a loading control. The experiments in (A) and (B) were conducted at 25°C. CPY* demonstrates the translocation defects of these strains. The lower band reacting with the αCPY antibody indicates the non-glycosylated precursor, which results from an impaired import. The amount of Ubc6p was determined using a luminescence imager. The relative intensity is given as a percentage of the value at the zero time point. (C) The indicated yeast strains treated with cycloheximide for 3 h at 25°C. Aliquots before and after cycloheximide treatment were taken and membranes immunoblotted for Ubc6p degradation intermediates. (D) Radioactive pulse–chase experiment of the same strains as in (A) after a 1 h pre-shift to 37°C. Total cell lysates were immuno precipitated for Ubc6p. (E) Quantification of experiments as shown in (D). Three independent experiments were averaged and the standard deviation was calculated and is indicated by error bars (values for prc1-1 are 100%; 45.1 ± 1.9%; 17.5 ± 3.6%; 12.0 ± 1.7%; and for prc1-1, sec61-2 are 100%; 43.4 ± 10.8%; 21.3 ± 6.1%; 14.5 ± 1.7%).
We wondered which part of the protein might render Ubc6p unstable. Since Ubc4p (Figure 2) and Ubc1p are stable proteins (J.Walter and T.Sommer, unpublished observation), we argued that the UBC domain does not contain a degradation signal. However, we found Ubc6p to be stable when the transmembrane anchor was deleted (Figure 2). As the membrane domain is part of the unique tail domain, we speculated that this region might signal Ubc6p degradation. We tested this notion further by fusing Ubc4p to the tail of Ubc6p, resulting in a C-terminally anchored Ubc4p (Ubc4ptail). This chimera behaved like an integral membrane protein (data not shown). A cycloheximide decay experiment revealed that it is short lived, demonstrating that the tail domain of Ubc6p is a transferable signal conferring a short half-life to an otherwise stable Ubc (Figure 2).
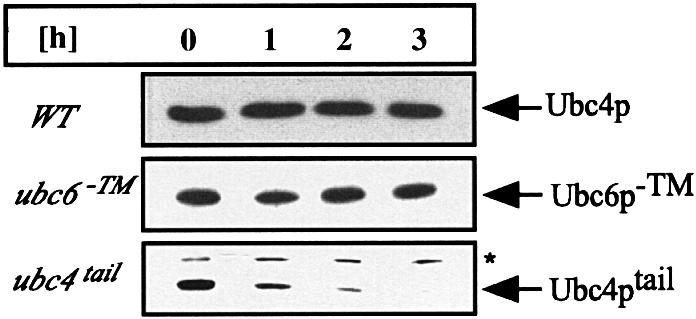
Fig. 2. Ubc6p degradation is mediated by its tail domain. The stability of Ubc4p, Ubc6p–TM and Ubc4ptail was tested in cycloheximide decay experiments and analyzed by immunoblotting. Experiments were conducted as in Figure 1B, except that in the upper two panels total cell lysates were probed. Ubc4p was tested in a wild-type strain. Ubc6p deleted for its transmembrane anchor (Ubc6p–TM) and the Ubc4p–Ubc6ptail fusion (Ubc4ptail) were expressed from a low-copy plasmid in a Δubc6 strain. The asterisk marks Sec61p, which served as a loading control.
Ubc6p is degraded by the ubiquitin–proteasome pathway depending on functional Ubc7p and its own catalytic activity
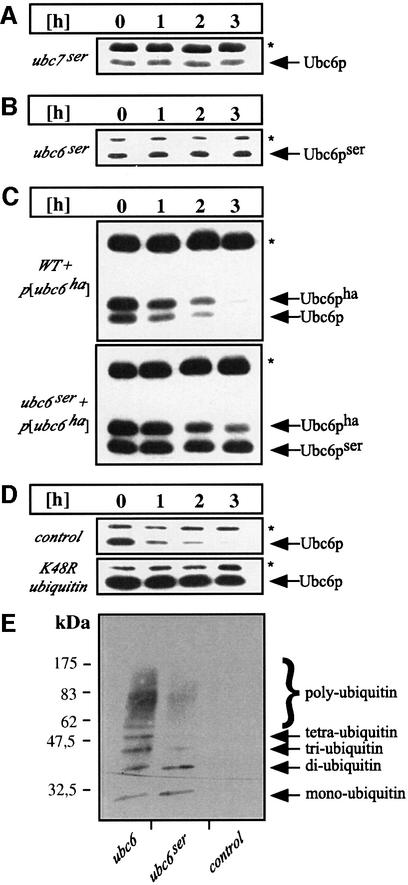
Ubc7p and Ubc6p have been proposed to form heterodimers (Chen et al., 1993) and to act in a concerted manner on a variety of ERAD and other substrates (Chen et al., 1993; Biederer et al., 1996; Hiller et al., 1996). We tested whether Ubc7p functions as a binding partner that targets Ubc6p for degradation or whether the catalytic activity of Ubc7p is required for Ubc6p proteolysis. Therefore, Ubc6p turnover was measured in a strain that was mutated in the active site cysteine of UBC7 (_ubc7_ser). Under these conditions, Ubc6p was fully stabilized (Figure 3A), indicating that the catalytic activity of Ubc7p is required for the degradation of Ubc6p. As the stabilization in this strain as well as in the Δubc7 and Δcue1 deletion strains was complete, we can exclude that redundant degradation pathways, e.g. transport to vacuolar proteases, act on Ubc6p. Furthermore, we have investigated whether Ubc6p levels are increased upon induction of the unfolded protein response (UPR; Mori, 2000). It has been described that components of the ERAD system are under the transcriptional control of the UPR transcription factor Hac1p (Friedlander et al., 2000, Travers et al., 2000). However, neither the induction of the UPR by overexpression of the active form of Hac1p (Hac1ip) nor the abrogation of the UPR by ire1 deletion influenced the expression of Ubc6p or altered its half-life (data not shown).
Fig. 3. Ubc6p degradation depends on polyubiquitylation, catalytically active Ubc7p and its own activity. Cycloheximide decay experiments conducted as in Figure 1B and analyzed by immunoblotting. (A and B) The genomic UBC7 and UBC6 genes were mutated in their active site cysteine resulting in _ubc7_ser and _ubc6_ser, which were tested for the stability of Ubc6p and Ubc6pser, respectively. (C) The HA-tagged Ubc6p (Ubc6pha) was expressed from a low-copy plasmid in a wild-type strain and in the _ubc6_ser strain. The asterisk marks Sec61p, which served as a loading control. (D) The dependence of Ubc6p on polyubiquitylation was tested by the co-expression of mutant ubiquitinK48R. (E) Δubc6 cells were transformed with high-copy plasmids coding for Ubc6p, Ubc6pser or an empty vector. Microsome preparations from equal amounts of cells were immunoprecipitated with anti-Ubc6p antibody, and western blots were analyzed with an anti-ubiquitin antibody. The Ubc6p immunoprecipitation was controlled by blotting aliquots for Ubc6p (data not shown).
In order to investigate whether Ubc6p activity might be required for its own degradation, we monitored the turnover of the active site mutant Ubc6pser in a strain deleted for wild-type UBC6. Figure 3B demonstrates that this mutant protein is stable, suggesting that Ubc6p activity is indeed required for its own degradation. In order to distinguish a scenario in which the catalytic activity has to be present on the molecule that is degraded and the alternative possibility that wild-type Ubc6p can function in trans, we performed the following experiment. A hemagglutinin (HA)-tagged Ubc6p (Ubc6pha) was co-expressed with Ubc6pser. Surprisingly, Ubc6pser was still stable, while Ubc6pha was degraded with kinetics comparable to wild-type Ubc6p (see below) (Figure 3C, lower panel). These experiments support a hypothesis in which the catalytic activity of Ubc6p is required for Ubc6p degradation and has to be present on the short-lived molecule itself. Alternatively, one might argue that the cysteine to serine mutation causes misfolding of Ubc6p and thereby prevents recognition by the ubiquitin– proteasome pathway. However, we consider this unlikely for several reasons. Ubc6pser exhibits a dominant-negative effect on ERAD (J.Walter and T.Sommer, unpublished observation). This effect is also seen in Figure 3C on the degradation of Ubc6pha. Ubc6pha is turned over more slowly in the Ubc6pser background (lower panel) compared with the wild-type situation (upper panel), indicating that catalytically inactive Ubc6pser still interacts with the ubiquitin–proteasome machinery. Moreover, Ubc6pser still receives its basic mono- and diubiquitylation and thus is recognized by the ubiquitin conjugation machinery (see below). Finally, misfolded proteins are usually efficient targets for ubiquitylation and subsequent degradation.
We next tested whether Ubc6p turnover depends on polyubiquitylation and whether Ubc6p is itself a target for ubiquitin conjugation. Since polyubiquitin chains linked via Lys48 of ubiquitin are a prerequisite for proteasomal degradation, the co-expression of the mutant ubiquitinK48R abrogates proteolysis (Finley et al., 1994). We tested the stability of Ubc6p under these conditions in a cycloheximide decay experiment and found Ubc6p to be stabilized markedly (Figure 3D). Additionally, we immunoprecipitated wild-type Ubc6p and Ubc6pser for analysis by immunoblotting with anti-ubiquitin antibodies. As shown in Figure 3E, immunoreactive species were detected in samples containing wild-type Ubc6p, but not in the control lacking Ubc6p. The pattern of these bands is typical for a polyubiquitin ladder, starting with monoubiquitylated forms and going up to a high molecular weight range. Interestingly, we observed that Ubc6pser is still conjugated to ubiquitin. However, mostly mono- and diubiquitylated species are visible while high molecular weight species are markedly reduced. Since it has been demonstrated that a polyubiquitin chain of at least four K48-linked molecules is necessary to initiate degradation by the proteasome (Thrower et al., 2000), our observation is consistent with the fact that Ubc6pser is a stable protein. Surprisingly, we also observed mono- and diubiquitylated species of Ubc6pser in cells lacking Ubc7p (data not shown).
Taken together, these data suggest that Ubc6p is targeted for degradation by covalent high molecular weight ubiquitylation and that a stable Ubc6p mutant version shows reduced polyubiquitylation. However, it remains to be seen at which position of Ubc6p ubiquitylation occurs.
In proteasomal mutant strains, a degradation fragment of Ubc6p accumulates
Most substrates of the ubiquitin system are degraded by the 26S proteasome (Hershko and Ciechanover, 1998). Hence, mutants affecting proteasomal function should stabilize Ubc6p. We investigated the degradation of Ubc6p in a strain hosting the pre1-1 allele. PRE1 encodes a subunit of the 20S core complex of the proteasome (Coux et al., 1996). The pre1-1 mutation results in a functionally impaired proteasome (Heinemeyer et al., 1991). Figure 4A shows that Ubc6p degradation was markedly reduced in this strain. A similar stabilization was observed in the active site double mutant pre2_-K108R/pre3_-T20A (Heinemeyer et al., 1997; data not shown).
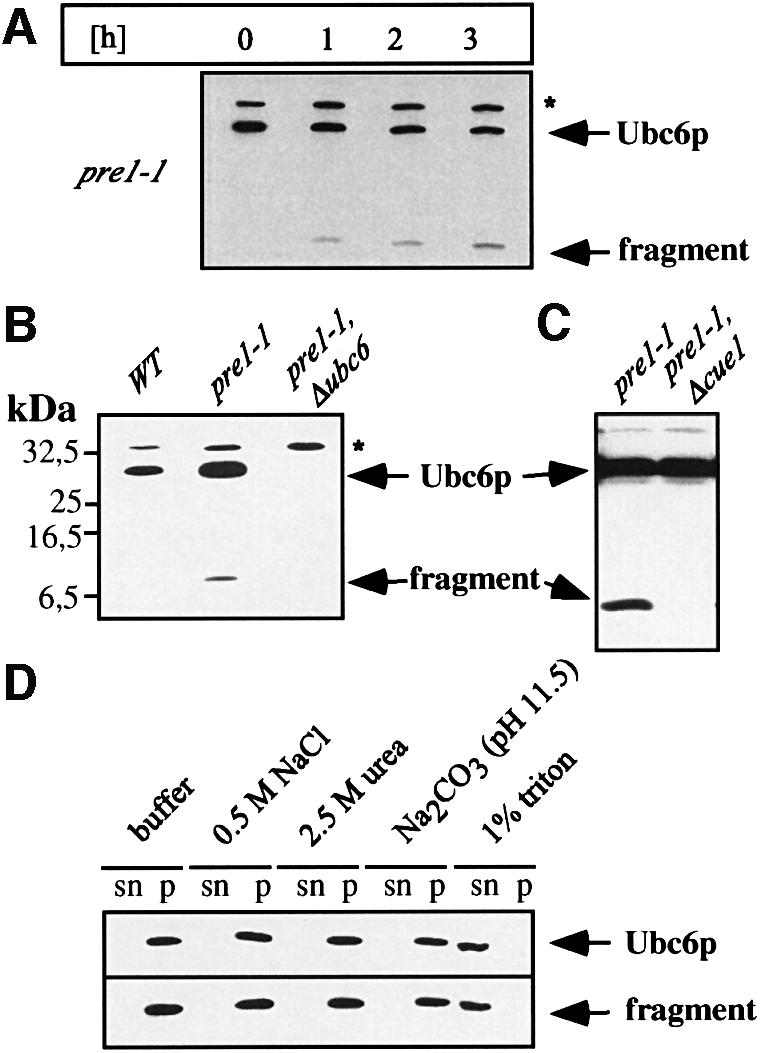
Fig. 4. Proteasomal mutants stabilize Ubc6p and cause the accumulation of a degradation intermediate. (A) Cycloheximide decay experiment of the proteasomal mutant strain pre1-1, conducted as in Figure 1B and analyzed by immunoblotting. A fragment accumulated, which reacted with the αUbc6p antibody. (B) Western blot of membrane preparations from the strains indicated. (C) Western blot of total cell lysates from the strains indicated. (D) Membranes from the pre1-1 strain were extracted with agents that strip peripheral proteins from the membranes, and with detergent, as indicated. Equal amounts of supernatant (sn) and pellet (p) were blotted for Ubc6p. The asterisk marks Sec61p, which served as a loading control.
Surprisingly, we detected a fragment migrating in SDS–PAGE at a position of ∼8 kDa, which reacted with the Ubc6p antibody. This fragment could also be detected in the ATPase mutant cim5-1 (Ghislain et al., 1993) and in pre2_-K108R/pre3_-T20A (data not shown), but not in strains hosting fully active proteasomes. It accumulated concomitantly with the degradation of Ubc6p upon inhibition of protein translation in the pre1-1 strain (Figure 4A). In a pre1-1 strain containing, in addition, an ubc6 deletion, we failed to observe this fragment, thus demonstrating that it truly originated from Ubc6p and not from a background protein (Figure 4B). Therefore, we deduced that the immunoreactive band represents a degradation intermediate of Ubc6p. It is important to note that other breakdown intermediates, except for the one described above, are not detectable in membrane preparations and total cell lysates (Figure 4C, and data not shown). Since it is possible that mutations in the 26S proteasome induce other unrelated proteases that might generate this intermediate, we tested whether its appearance is still dependent on Cue1p and thereby also on Ubc7p. As shown in Figure 4C, it is absent in pre1-1Δcue1 cells, and thus is most likely to be a breakdown intermediate generated by the aberrantly functioning proteasome. Next, we determined the topology of this fragment. Isolated membranes from pre1-1 cells were treated with urea, high salt or high pH and were sedimented by high-speed centrifugation. Subsequently, we monitored the supernatant (sn) and the pellet (p) for Ubc6p and the fragment (Figure 4D). Like wild-type Ubc6p, the fragment remained in the membrane fraction, but was solubilized with detergent. These procedures have been reported to wash off membrane-associated, but not membrane-spanning proteins (Coligan et al., 1997). This indicates that the degradation intermediate comprises the C-terminal Ubc6p membrane anchor.
The Ubc6p degradation does not depend on Hrd1p/Der3p, Hrd3p, Der1p or Sec61p function
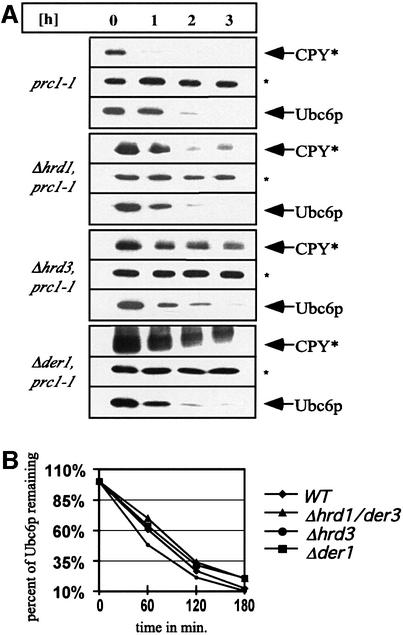
Hrd1p/Der3p, Hrd3p, Der1p and Sec61p are the classical ERAD components involved in the turnover of a variety of substrates (Sommer and Wolf, 1997). While the role of Der1p and Hrd3p has not been elucidated in detail, Hrd1p/Der3p is a RING-H2 finger protein and has been shown to function as an E3 in ERAD (Bays et al., 2001). We asked whether these proteins might also be required for the degradation of Ubc6p, and performed cycloheximide decay experiments in the Δhrd3, Δhrd1/der3 and Δder1 deletion strains. Figure 5 shows that Ubc6p is degraded in these strains with the same or only marginally changed kinetics compared with wild-type strains. Thus, Hrd1p/Der3p, Hrd3p and Der1p are not required for the degradation of Ubc6p.
Fig. 5. Ubc6p degradation does not depend on Hrd1p/Der3p, Hrd3p or Der1p. (A) The immunoblots show cycloheximide decay experiments, conducted as in Figure 1B, of Δhrd1/der3, Δhrd3 and Δder1 deletion strains and of the corresponding wild-type strains. All strains expressed the prc1-1 allele, which codes for the classical ERAD substrate CPY*. It served as a control for the stabilizing effects on ERAD. The asterisk (*) marks Sec61p, which served as a loading control. (B) Quantification of the luminescence of the Ubc6p signals in an experiment as in (A).
The extraction of proteins from the ER membrane during ERAD is thought to require the Sec61p translocon. SEC61 deletions are not viable, but different mutants exhibit transport defects and can be used to examine the function of this protein (Pilon et al., 1997; Plemper et al., 1997; Zhou and Schekman, 1999). Strains containing the mutant sec61-2, -32 and -41 alleles of SEC61 have been reported to stabilize ERAD substrates such as the mutant carboxypeptidase Y (CPY*) and mutant α-factor at the permissive temperature of 25°C (Pilon et al., 1997; Plemper et al., 1997). Therefore, we analyzed the turnover of Ubc6p in these cells at the same temperature. In contrast to CPY*, we observed no difference in the degradation kinetics of Ubc6p (Figure 6A and B). Furthermore, we investigated Ubc6p degradation under conditions where the translocon is severely impaired. It has been shown that Sec61-2p is short lived at the non-permissive temperature of 37°C. This results in a largely reduced number of transloci (Biederer et al., 1996). These conditions cannot be used for substrates that are imported via Sec61p such as CPY*, because of the strong accumulation of precursor molecules. However, they can be employed for substrates that are integrated into the ER membrane independently of Sec61p. Even under these conditions, Ubc6p turnover remained unchanged (Figure 6D and E). Thus, we concluded that the proteolysis of the full-length Ubc6p is not affected by mutations in SEC61, in contrast to the ERAD substrate CPY*. However, it might be possible that the full-length Ubc6p can still be attacked by the proteasome when the translocon is non-functional. In consequence, breakdown intermediates may accumulate that comprise the membrane anchor and a few additional amino acids. We looked for these fragments, which may be similar to that in the pre1-1 strain, but were unable to detect them in the mutant sec61 strains (Figure 6C).
Discussion
The data presented here demonstrate that Ubc6p is a naturally short-lived protein degraded by the ubiquitin– proteasome pathway. It is strongly stabilized in cells lacking Cue1p-assembled Ubc7p but also when the active site of Ubc7p is mutated. It is polyubiquitylated and, as a consequence, Ubc6p is stabilized by overexpression of ubiquitinK48R, a dominant-negative inhibitor of the formation of polyubiquitin chains.
Several features of its degradation distinguish Ubc6p from other membrane-bound substrates but also from other Ubcs. Constitutive degradation is not a general hallmark of ubiquitin-conjugating enzymes, since Ubc4p- (Figure 2), Ubc1p- and Cue1p-assembled Ubc7p (data not shown) are not subjected to proteolysis. Ubc6p turnover relies on its unique tail region, in particular on the membrane-spanning segment. This degradation signal can even be transferred to another Ubc domain as its fusion to the otherwise stable Ubc4p results in an unstable protein. However, the tail region is not the only prerequisite for degradation, since the turnover of Ubc6p strictly depends on the active site cysteine. This residue seems to be required for the efficient high molecular weight ubiquitylation of Ubc6p. Interestingly, the fact that only functional Ubc6p is degraded when it is co-expressed with non-functional Ubc6pser could be explained by a self-ubiquitylation in cis. However, in vitro ubiquitin conjugation experiments have to be performed to confirm this assumption.
In contrast to ERAD, Ubc6p degradation occurs independently of Ubc1p, Hrd1p/Der3p, Hrd3p and Der1p. While not much is known about the molecular function of Der1p and Hrd3p, the Hrd1p/Der3p independence is surprising, as Hrd1p/Der3p has been suggested to function as an E3 together with Ubc1p and Ubc7p (Bays et al., 2001). Our data show that Ubc7p can act independently of Hrd1p/Der3p at the ER membrane.
In proteasomal mutants, a specific membrane-bound degradation intermediate of Ubc6p accumulates. Other intermediates of this proteolysis have not been detected, either at the membrane or in the cytosol. We consider it unlikely that this fragment is generated by other unrelated proteases, since its appearance is strictly dependent on Cue1p-assembled Ubc7p. This observation is in agreement with the model that the 26S proteasome acts directly on membrane-bound substrates (Mayer et al., 1998). How ever, such a degradation intermediate is not normally observed for ERAD substrates and has only been reported for chimeric proteins consisting of a multispanning ER membrane protein and a cytosolic degradation signal, Deg1 (Mayer et al., 1998; Wilhovsky et al., 2000). One possible explanation for this difference is that the Deg1-containing fusion proteins and Ubc6p are degraded in a manner similar to cytosolic proteins rather than ER membrane proteins (see below).
The degradation of ERAD substrates investigated so far is thought to depend on the pore formed by Sec61p (Sommer and Wolf, 1997; Bonifacino and Weissman, 1998). In contrast, Ubc6p is not stabilized in any of the analyzed Sec61p mutants. In no case does Sec61p become rate limiting for the proteolysis of full-length Ubc6p, even under conditions where the translocon is severely affected. However, the degradation of Ubc6p might be a two-step process, in which the cytosolic domain is degraded first and in a second step Ubc6p is extracted from the membranes as indicated by the accumulation of a degradation intermediate in proteasomal mutants. In this model, a decrease in the amount of full-length Ubc6p independently of Sec61p would not be surprising. However, the absence of a degradation intermediate similar to that observed in the pre1-1 cells (at least in detectable quantities) in sec61 mutants suggests that the translocon does not play any role during the extraction of Ubc6p from the ER membrane. Taking together the independence of the translocon and the accumulation of a degradation intermediate in proteasomal mutants makes it feasible to speculate that Ubc6p might be extracted directly from the ER membrane by the 26S proteasome when a polyubiquitin chain of sufficient length is formed.
Ubc6p degradation differs markedly from that of other ERAD substrates. This is likely to be a consequence of Ubc6p’s particular topology as a tail-anchored protein. Thus, our observations lead to the speculation that tail-anchored proteins in general might be extracted from membranes independently of the normally used translocation pores. In a minimal model, export of tail-anchored proteins could depend just on the cytosolic ubiquitin– proteasome system and does not have to be restricted to the ER membrane. A second example for such a pathway is the proteasome-dependent degradation of Bcl-2, a protein of the mitochondrial outer membrane (Dimmeler et al., 1999). However, further work and particularly in vitro experiments will be necessary to determine fully the minimal requirements of these processes.
Materials and methods
Yeast and bacterial methods
Yeast rich and minimal media were prepared as described and standard genetic methods were employed (Ausubel et al., 1989). The strains used are listed in Table I. YTX236 and YTX272 were prepared by transformation of YUL22 and YW01 with the ubc6::LEU integration vector as described (Sommer and Jentsch, 1993). The strains RSY1283, RSY1284 and RSY1285 (Zhou and Schekmann, 1999) were altered in their PRC1 locus as described (Hiller et al., 1996), resulting in YTX283, YTX284 and YTX285, respectively. YJU32 was prepared by replacing the genomic DER1 sequence from _Eco_RV (–5) to _Cla_I (374) by the HIS3 gene in YTX140. YJU26, YJU35, YJU39, YUL22 and YJU41 were prepared by cloning the respective sequences of sec61-2, _ubc6_ser (Sommer and Jentsch, 1993), _ubc7_ser (Jungmann, 1993) and pre1-1 (Seufert and Jentsch, 1992) into pRS406 or pRS306 followed by genomic integration into YTX140, YW01 or YTX141 (Biederer et al., 1997) and selection for 5′-fluoro-orotic acid (FOA) resistance. Correct integration was verified using restriction sites that had been generated or deleted in the mutated sequence. YJU49 was constructed by replacing the PRC1 gene in YW01 from _Sph_I (3) to _Bgl_II (1296) with the LEU2 gene. A _Spe_I site (–3) was introduced into the HRD3 gene, followed by the insertion of the LEU2 marker, thereby removing the sequence from _Spe_I to _Sma_I (2465). This construct was integrated into a YW01-based strain, crossed with a YJU49-based strain. YJU72 was finally isolated by tetrad analysis.
Table I. Yeast strains used in this work.
| Strain | Geonotype | Reference |
|---|---|---|
| YW01 | MATα, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | Seufert et al. (1990) |
| YW05 | MATα, ubc1::HIS3, trp1-1(am), ura3-52, lys2-801, leu2-3,-112 | Seufert et al. (1990) |
| YTX106 | MATα, ubc7::LEU2, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | Biederer et al. (1996) |
| YTX115 | MATα, cue1::LEU2, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | Biederer et al. (1997) |
| YTX140 | MATa, prc1-1, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | Biederer et al. (1997) |
| YTX236 | MATα, pre1-1, ubc6::LEU2, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YTX272 | MATα, ubc6::LEU2, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YTX283 | MATα, sec61::HIS3, prc1-1, can-100, leu2,3,-112, his3-11,15, trp1-1, ura3-1, ade2-1; PDQ1 _(SEC61_-Nterm His tag, pLAC111) | this study |
| YTX284 | MATα, sec61::HIS3, prc1-1, can-100, leu2,3,-112, his3-11,15, trp1-1, ura3-1, ade2-1; psec61-32 (pLAC111) | this study |
| YTX285 | MATα, sec61::HIS3, prc1-1, can-100, leu2,3,-112, his3-11,15, trp1-1, ura3-1, ade2-1; psec61-41 (pLAC111) | this study |
| YJU26 | MATa, prc1-1, sec61-2, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YJU32 | MATa, prc1-1, der1::HIS, his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YJU35 | MATa, ubc6ser**, prc1-1**, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YJU37 | MATa, prc1-1, hrd1::TRP1, his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | Friedlander et al. (2000) |
| YJU39 | MATa, ubc7ser**, prc1-1**, trp1-1(am), his3- Δ 200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YJU41 | MATα, prc1::LEU2, pre1-1, cue1::HIS3, trp1-1(am), ura3-52, lys2-801, leu2-3,-112 | this study |
| YJU49 | MATα, prc1::LEU2, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| YJU72 | MAT?, prc1::LEU2, HRD3::LEU2, trp1, his3, ura3, lys2, leu2 | this study |
| YUL22 | MATα, pre1-1, trp1-1(am), his3-Δ200, ura3-52, lys2-801, leu2-3,-112 | this study |
| CMY765 | MATα, cim5-1, ura3-52, leu2Δ1, his3Δ-200 | Ghislain et al. (1993) |
| YWH26 | MATα, pre2-K108R, pre3-T20A, his3-11, 15 can_s, leu2-3,-112, ura3, Gal2_ | Heinemeyer et al. (1997) |
| YWCG4a | MATα, his3-11, 15 can_s, leu2-3,-112, ura3, Gal2_ | Heinemeyer et al. (1997) |
Ubc6p deleted for its transmembrane anchor (Ubc6p–TM) was as published (Sommer and Jentsch, 1993). _Ubc4_tail was prepared by cloning the tail region of Ubc6p via its _Xho_I site into the Ubc4p locus upstream of the stop codon. The construct was expressed from pRS416. The HA-tagged Ubc6p (Ubc6pha) (Sommer and Jentsch, 1993) was cloned into pRS416, via the _Hin_dIII restriction sites and expressed in YTX140 and YJU35. YJU72 carrying a prc1 deletion was transformed with a low-copy plasmid expressing prc1-1. As a wild-type control, YJU49 was tested for Ubc6p and CPY* stability. The mutant ubiquitinK48R was expressed from pUB203 (Finley et al., 1994) under the control of a Cu2+-inducible promoter in YW01. Expression was induced by adding 100 µM Cu2+ to the medium 5.5 h before the experiment. pRS415 in YW01 served as a control.
SDS–PAGE, western blotting and antibodies
Standard methods for SDS–PAGE, western blotting and immunodecoration were used (Ausubel et al., 1989). Quantification of the luminescence signal was conducted with a luminescence imager (Fuji). A Ubc6p standard was analyzed in parallel to ensure the linearity of the system.
An N-terminally His6-tagged version of Ubc6pha without the trans membrane domain was expressed in Escherichia coli and isolated by binding to an Ni-NTA–agarose column (Qiagen). Polyclonal antibodies were affinity purified from rabbit serum.
Polyclonal αCue1p antibodies were produced as published (Biederer et al., 1997). The αUbc4p antibody was kindly provided by S.Jentsch. The αCPY* antibody was obtained from Molecular Probes. Mouse α-ubiquitin antibody was obtained from Babco.
Protein chemistry
Membrane fractions were prepared as described (Biederer et al., 1996), except that microsomes were sedimented by ultracentrifugation (200 000 g, 15 min in a 100.3 Beckman rotor). Total cell lysates were either prepared by replacing this high-speed centrifugation with a trichloroacetic acid (TCA) precipitation or, alternatively, by disrupting cells in a buffer containing 1% SDS and clearing the lysates by high-speed centrifugation. The membrane localization of the fragmented Ubc6p in the pre1-1 strain was performed as described (Sommer and Jentsch, 1993). The radioactive pulse–chase experiments were conducted as described (Friedlander et al., 2000), except that the strains were shifted to 37°C 1 h prior to labeling and were kept at that temperature for the whole experiment including labeling.
In the cycloheximide decay experiments, 15 OD600 log-phase cells were harvested and resuspended in 4 ml of pre-warmed SD medium containing 100 µg/ml cycloheximide (‘Actidione’, obtained from Fluka). Aliquots were taken at the indicated times, degradation was stopped by the addition of NaN3, and membrane fractions were prepared and tested for the proteins of interest. For the soluble proteins, Ubc6p–TM and Ubc4p, total cell lysates were prepared. CPY*, although soluble, proved to be included reproducibly in membrane fractions and was analyzed in those. The same was true for the precursor of CPY in strains exhibiting import defects (Plemper et al., 1997; our own observations). All experiments were conducted at 30°C unless stated otherwise.
Ubiquitylated Ubc6p species were analyzed in 20 OD600 log-phase cells. These were incubated briefly in 10 mM azide, 20 mM _N_-ethylmaleimide (NEM) at 0°C. Membranes were prepared as described above, except that the lysis buffer contained in addition 20 mM NEM, and the proteasome inhibitor ALLN (5 µg/ml) was added to all buffers. Membranes were solubilized in 1% SDS, 50 mM Tris pH 7.5, 20 mM NEM, diluted with 9 vols of a buffer containing 1.1% Trition X-100, 165 mM NaCl, 5.5 mM EDTA and 50 mM Tris pH 7.5. Lysates were cleared by centrifugation and immunoprecipitated for Ubc6p.
Acknowledgments
Acknowledgements
The authors thank D.Finley, S.Jentsch, C.Mann, R.Schekmann, D.Wolf and M.Zhou for the generous gift of strains and plasmids, and U.Lenk for the kind gift of YUL22. We appreciate the helpful comments on the manuscript of the Sommer group and C.Taxis. This work was partially supported by a grant from the Deutsche Forschungs Gemeinschaft and the Deutsch–Israelische Projektkooperation (DIP) to T.S.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Stuhl,K. (1989) Current Protocols in Molecular Biology. John Wiley and Sons, New York, NY.
- Bays N.W., Gardener,R.G., Seelig,L.P., Joazeiro,C.A. and Hampton,R.Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol., 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1996) Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin–proteasome pathway. EMBO J., 15, 2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science, 278, 1806–1809. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. and Weissman,A.M. (1998). Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Johnson,P., Sommer,T., Jentsch,S. and Hochstrasser,M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell, 74, 357–369. [DOI] [PubMed] [Google Scholar]
- Chillaron J. and Haas,I.G. (2000) Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol. Biol. Cell, 11, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan J.E., Dunn,B.M., Ploegh,H.L., Speicher,D.W. and Wingfield,P.T. (1997) Current Protocols in Protein Science. John Wiley and Sons, New York, NY.
- Coux O., Tanaka,K. and Goldberg,A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem., 65, 801–847. [DOI] [PubMed] [Google Scholar]
- Dimmeler S., Breitschopf,K., Haendeler,J. and Zeiher,A.M. (1999) Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J. Exp. Med., 189, 1815–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Sadis,S., Monia,B.P., Boucher,P., Ecker,J.D., Croole,S. and Chau,V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination deficient yeast mutant. Mol. Cell. Biol., 14, 5501–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander R., Jarosch,E., Urban,J., Volkwein,C. and Sommer,T. (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nature Cell Biol., 2, 379–384. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy,A. and Mann,C. (1993) S.cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature, 366, 358–362. [DOI] [PubMed] [Google Scholar]
- Hampton R.Y., Gardener,R.G. and Rine,J. (1996) Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell, 7, 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Kleinschmidt,J.A., Saidowsky,J., Escher,C. and Wolf,D.H. (1991) Proteinase yscE, the yeast proteasome/multicatalytic– multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J., 10, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Fischer,M., Krimmer,T., Stachon,U. and Wolf,D.H. (1997) The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J. Biol. Chem., 272, 25200–25209. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hiller M.M., Finger,A., Schweiger,M. and Wolf,D.H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science, 273, 1725–1728. [DOI] [PubMed] [Google Scholar]
- Johnson A.E. and van Waes,M.A. (1999) The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol., 15, 799–842. [DOI] [PubMed] [Google Scholar]
- Jungmann J., Reins,H.-A., Schobert,C. and Jentsch,S. (1993) Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature, 361, 369–371. [DOI] [PubMed] [Google Scholar]
- Knop M., Finger,A., Braun,T., Hellmuth,K. and Wolf,D.H. (1996) Der1p, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J., 15, 753–763. [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hartmann,E. and Rapoport,T. (1993) A class of membrane proteins with a C-terminal anchor. Trends Cell Biol., 3, 72–75. [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger,G., Hartmann,E., Wiedenmann,B. and Rapoport,T.A. (1995) Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J., 14, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T.U., Braun,T. and Jentsch,S. (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J., 17, 3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. (2000) Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell, 101, 451–454. [DOI] [PubMed] [Google Scholar]
- Pilon M., Schekman,R. and Römisch,K. (1997) Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J., 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper K.R., Böhmler,S., Bordallo,J., Sommer,T. and Wolf,D.H. (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature, 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Rapoport T.A., Jungnickel,B. and Kutay,U. (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem., 65, 271–303. [DOI] [PubMed] [Google Scholar]
- Seufert W. and Jentsch,S. (1992) In vivo function of the proteasome in the ubiquitin pathway. EMBO J., 11, 3077–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., McGrath,J.P. and Jentsch,S. (1990) Ubc1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugation enzymes involved in protein degradation. EMBO J., 9, 4535–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T. and Jentsch,S. (1993) A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature, 365, 176–179. [DOI] [PubMed] [Google Scholar]
- Sommer T. and Wolf,D.H.(1997) Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J., 11, 1227–1233. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Peckart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K.J., Patil,C.K., Wodicka,L., Lockhart,D.J., Weissman,J.S. and Walter,P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell, 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Wilhovsky S., Gardener,R. and Hampton,R. (2000) HRD gene dependence of endoplasmic reticulum associated degradation. Mol. Biol. Cell, 11, 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Ellenber,J., Bonifacino,J.S. and Weissman,A.M. (1997) The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem., 272, 1970–1975. [DOI] [PubMed] [Google Scholar]
- Zhou M. and Schekman,R. (1999) The engagement of Sec61p in the ER dislocation process. Mol. Cell, 4, 925–934. [DOI] [PubMed] [Google Scholar]