De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination (original) (raw)
Abstract
Activation-induced cytidine deaminase (AID) is required for class-switch recombination (CSR), somatic hypermutation, and gene conversion of Ig genes. Although AID has sequence similarity to an RNA-editing enzyme Apobec-1, how AID functions in CSR and somatic hypermutation is unknown. Because involvement of RNA-editing but not DNA-editing in CSR requires de novo protein synthesis after AID expression, we examined whether protein synthesis inhibitors could block CSR in the presence of the AID activity. For this purpose we constructed AID fused with the hormone-binding domain of the estrogen receptor (AID-ER), which was introduced into AID-deficient spleen B cells. When such transfectants were treated with an estrogen analogue, 4-hydroxytamoxifen (OHT), CSR was induced within 1 h. Cycloheximide or puromycin drastically suppressed OHT-induced CSR in AID-ER expressing AID−/− B cells when added 1 h before OHT but not after OHT, suggesting that de novo protein synthesis is required for an event downstream to AID expression in CSR. The results lend the weight to RNA-editing hypothesis for the function of AID.
Naïve B lymphocytes that have completed successful VDJ recombination in the bone marrow express the IgM class of Ig and migrate to the secondary lymphoid organs such as spleen and lymph nodes. Antigen-stimulated mature B cells begin to proliferate vigorously in lymphoid follicles and to form germinal centers, in which Ig loci are further altered by class switch recombination (CSR) and somatic hypermutation (SHM). SHM introduces region-restricted point mutations into the variable (V) region sequence of the Ig genes, giving rise to a diversified repertoire of the V region that is subject to selection for high affinity. On the other hand, CSR changes the effector function of Ig by replacing the heavy-chain constant region (CH) genes without affecting the antigen specificity (1, 2).
CSR takes place between two S regions located 5′ to each CH gene, resulting in looped-out deletion of intervening DNA segments as circular DNA. The CSR reaction can be dissected into three steps: (i) chromatin opening of the target S region, (ii) recognition and cleavage of target DNA by recombinase, and (iii) repair and ligation of cleaved ends (2). The first step is mediated by so-called germ-line transcription from the cytokine-inducible I promoter located 5′ to each S region (3, 4). Several candidates are proposed as recognition target in the second step; (i) RNA/DNA heteroduplex formed between germ-line transcripts (GLT) and S region DNA and (ii) the stem-loop DNA structure induced by active transcription and palindromic nature of S sequences (5). An endonuclease that should be involved in the recognition and cleavage of the S region is not yet identified. It is proposed but not convincingly proven that the ligation step is mediated by nonhomologous end-joining proteins including Ku70 and Ku80 which also play an essential role in VDJ recombination (6–9).
Activation-induced cytidine deaminase (AID) has been shown to be induced specifically in activated B cells and essential to both CSR and SHM (10–12). In addition, AID expression alone was sufficient to induce SHM and CSR in artificial substrates in fibroblasts (13, 14). Therefore, AID is the only B cell-specific factor for CSR and SHM. Because AID deficiency did not affect either GLT formation or nonhomologous end-joining, AID is most likely to be involved in the cleavage step of CSR (15).
AID has a sequence similarity to an RNA-editing enzyme Apobec-1 (10). Apobec-1 is a catalytic subunit of the apolipoprotein B (apoB) mRNA-editing complex, which deaminates cytosine at a specific position in apoB 100 mRNA precursor, thus introducing a premature stop codon. The edited mRNA encodes the protein component of chylomicron, whereas the unedited mRNA encodes the carrier protein in low-density lipoprotein. Thus, AID might be an editing enzyme of a preexisting mRNA precursor and the edited mRNA may code for an enzyme involved in CSR and SHM (RNA-editing model). In this case, recombinase must be de novo synthesized after AID expression. Alternatively, AID itself might directly deaminate deoxycytidine in DNA as AID can deaminate deoxycytidine in vitro (DNA-editing model) (10). Deoxyuridine in DNA is removed by uracil-DNA glycosylase followed by apurinic/apyrimidinic endonuclease (base excision repair). In fact, B cells deficient in UNG uracil-DNA-glycosylase were shown to have reduced CSR activity (16). Because the process subsequent to deoxycytidine deamination should be catalyzed by a number of ubiquitous DNA repair enzymes, DNA-editing model predicts that CSR is not dependent on de novo protein synthesis after AID expression.
Here, we report establishment of a system that allows dissection of the events immediately after AID expression. When we used this system, we found that the AID function in CSR depends on de novo protein synthesis, in agreement with RNA-editing hypothesis.
Materials and Methods
Construction of Vector and Cell Line.
A PCR-amplified coding sequence of mouse AID with a C-terminal FLAG epitope was cloned in-frame into a _Bam_HI site located immediately upstream of coding sequence of hormone binding domain of estrogen receptor ER-T2 mutant (17) in pERT2 plasmid (18). AID-ER sequence is shown in Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org. The AID-ER sequence was introduced into a tetracycline-regulatable expression vector with internal ribosome entry site (IRES)-GFP (11) to generate pAID-ER-GTOP. After linearization, pAID-ER-GTOP was introduced to FTZ14, which is a transfectant of CH12F3–2 cells (10) harboring pTet-tTAk-zeo transgene, to establish AER lines. The AID-ER sequence was ligated with a constitutive expression vector pEF-BOSbsr, which is a vector obtained by modifying pEF-BOS (19) with introduction of the blasticidin S-resistance gene cassette from pSV2bsr (Kakenyaku, Japan), to generate pAID-ER-BOSbsr. NER7 transfectant was prepared by introduction of pAID-BOSbsr into HTTn202 cells, which are transfectants of NTZ1 cells with an artificial CSR substrate used in our previous study (13). NTZ1 cells are a NIH 3T3 transfectant line harboring pTet-tTAk-zeo, a variant of pTet-tTAk (20) with the zeocin resistance gene cassette. The AID-ER sequence was also ligated to a retroviral vector pFB (Stratagene) together with the IRES-GFP segment obtained from pIRES2-EGFP (CLONTECH), to generate pAID-ER-FBG.
Mice and Cell Culture.
AID−/− mice on (CBA × C57BL/6) × C57BL/6 background were maintained in our animal facility and used at 2–8 months of age. Red-blood-cell-depleted total splenocytes from AID−/− mice were cultured for 2 days in RPMI medium 1640 containing 10% FCS, 50 μM 2ME, 2 mM l-glutamine, 100 μ/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 50 μg/ml lipopolysaccharide (LPS, Sigma), and 15 ng/ml mouse IL-4 (PeproTech, Boston). Two days after culture, the percentage of B220+ cells was >80% by FACS analysis. NER7 cells were cultured in DMEM containing 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
PCR.
Total RNA was extracted from cultured cells by using TRIzol (Invitrogen) according to the manufacturer's instructions. Genomic DNA were extracted with SDS/proteinase K lysis, followed by phenol/chloroform extraction. Digestion circularization (DC)-PCR was done by using primers described previously (21) with modifications indicated below. Five micrograms of genomic DNA were digested with 75 units of _Eco_RI (Takara) overnight. Three-hundred nanograms of digested DNA were self-ligated with 1,750 units of T4 DNA ligase (Takara) in a 100-μl reaction volume at 16°C overnight. One-hundred and fifty nanograms and 15 ng DNA were used for Sμ-Sγ1 and nicotinic acetylcholine receptor β subunit gene DC-PCR, respectively. DC-PCR was initiated by a denaturing step of 95°C for 9 min, followed by 30 or 40 cycles of PCR (94°C for 30 s, 65°C for 3 min) with AmpliTaq Gold (Applied Biosystems). The PCR products were electrophoresed in 2% agarose gel, transferred to Hybond _N_+ membrane (Amersham Pharmacia), and probed with 32P-labeled products of IgG1 DC-PCR by using Sμ-Sγ1 primers. For RT-PCR, cDNA was synthesized with Superscript II reverse transcriptase (Invitrogen) by using 2 μg of total RNA and 0.1 μg of poly[d(T)12–18] (Amersham Pharmacia) in 20-μl reaction volume, 0.1 μl of which was used for PCR.
Amplification of α circle transcripts was done as described (22). Amplification of γ1 GLT was done by initial denaturing step of 94°C for 5 min, followed by 22 cycles of PCR (94°C for 30 s, 58°C for 30 s, 72°C for 1min) by using recombinant Taq polymerase (Takara) with a primer pair of Iγ1F and Cγ1R (11). Amplification of hypoxanthine-guanine phosphoribosyl transferase was initiated by a denaturing step of 94°C for 5 min, followed by 22 cycles of PCR (94°C for 30 s, 50°C for 30 s, 72°C for 1 min) by using recombinant Taq polymerase (Takara) with described primers (23).
Antibodies and Reagents.
Biotinylated anti-mouse IgG1, biotinylated anti-mouse IgG3, and allophycocyanin-conjugated anti-mouse B220 antibodies were purchased from PharMingen. Anti-human ER, anti-PI3K p85, and anti-RNA polymerase II antibodies were purchased from Santa Cruz Biotechnology. Anti-α-tubulin antibody was purchased from Oncogen. Cycloheximide and puromycin were purchased from Sigma.
Cell Fractionation and Western Blot.
Total cell lysates and subcellular fractions were obtained by lysing cells in an hypotonic solution and homogenizing with a Dounce homogenizer, followed by sucrose layer sedimentation (24). After washing with a lysis buffer, the eluates were subjected to 5–15% gradient SDS/PAGE gels (Bio-Rad) and electroblotted to nitrocellulose membranes, which were then incubated in a blocking buffer of 5% skim milk in TBS with 0.1% Tween-20. Primary antibody incubations were done overnight at 4°C in the blocking buffer. After washing, secondary antibody incubations were done at the room temperature for 40 min in the blocking buffer. Blots were developed with enhanced chemiluminescence (Amersham Pharmacia).
Retrovirus Infection.
Recombinant retrovirus preparation using pAID-ER-FBG and Plat-E cells (25) and its infection procedure were described before (26). Spleen cells were preactivated for 2 days before infection. Twenty-four hours after infection, cells were used for assay.
Results
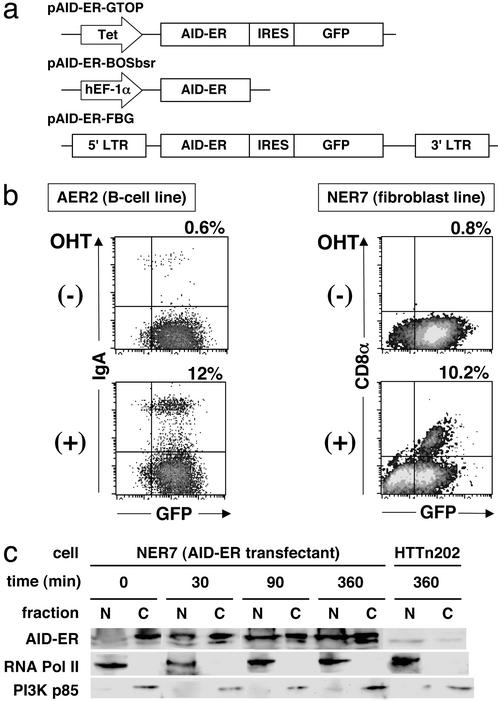
To examine whether de novo protein synthesis is required for the AID function in CSR, we tested protein synthesis inhibitors for their ability to block CSR after expression of AID. For this purpose, it is essential to design experiments in which protein synthesis inhibitors neither inhibit AID synthesis nor cause general toxicity to suppress CSR. To avoid prolonged incubation with protein synthesis inhibitors, we used DC-PCR that allows to measure CSR a few hours after AID expression. In addition, we constructed a conditionally active form of AID by the fusion at its C terminus to the hormone-binding domain of the human ER (17) and designated it as AID-ER (Fig. 1a). Inactive AID-ER can be promptly activated by posttranslational conformational change induced by binding to an estrogen analogue, 4-hydroxytamoxifen (OHT).
Figure 1.
AID-ER expression vector and its activity. (a) AID-ER consists of the AID coding sequence fused with flag epitope tag and ER ligand-binding domain. The pAID-ER-GTOP and pAID-ER-BOSbsr contain tet promoter and EF-1α promoter, respectively. The pAID-ER-GTOP and pAID-ER-FBG contain internal ribosomal entry site and enhanced GFP following AID-ER. In the pAID-ER-FBG, AID-ER-IRES-GFP was flanked by LTR. (b) Flow cytometric analysis. AID-ER-expressing cell lines AER and NER7 were stained with anti-IgA or anti-CD8α antibodies 6 or 13 days after OHT stimulation, respectively. Percentages of switching cells are indicated above. (c) Nuclear and cytoplasmic fractions of NER7 cells were separated and monitored by the presence of RNA polymerase II and PI3K p85α, respectively. AID-ER was detected by Western blotting using anti-ER antibodies.
First, AID-ER was stably introduced into a B lymphoma cell line, CH12F3–2, that efficiently undergoes class switching to IgA and NIH 3T3 fibroblast cells that harbor an artificial substrate of CSR. The transfectants of CH12F3–2 and NIH 3T3 were named AER and NER7, respectively. The CSR substrate in NER7 cells is designed to express CD8α on the cell surface after CSR (13). These transfectants were treated with OHT for 6–13 days, and CSR was evaluated by IgA and CD8α surface staining in AER and NER7 cells, respectively. In response to OHT, AID-ER efficiently induced CSR in these cell lines, though there was essentially no switching in the absence of OHT (Fig. 1b).
Intracellular localization of AID-ER was examined by Western blotting after cell fractionation with AID-ER expressing NIH 3T3 cells (NER7). Before OHT addition, AID-ER was almost exclusively in the cytoplasm (Fig. 1c). AID-ER began to increase in nuclei 30 min after addition of OHT. Subsequently, the nuclear localization of AID-ER became more evident with less amounts of cytoplasmic signals. This translocation of AID-ER was not inhibited by cycloheximide (CHX) (data not shown). It has been reported that nuclear localized protein such as Notch1 intracellular domain fused with ER is sequestered in the cytosol by chaperone heat sock protein 90 in the absence OHT. OHT binding releases the ER fusion protein from heat shock protein 90, allows its translocation into nucleus, activation of the fusion partner and elongation of the protein half-life (18, 27). Consistent with this trait of ER, the protein amount of AID-ER increased by the OHT treatment.
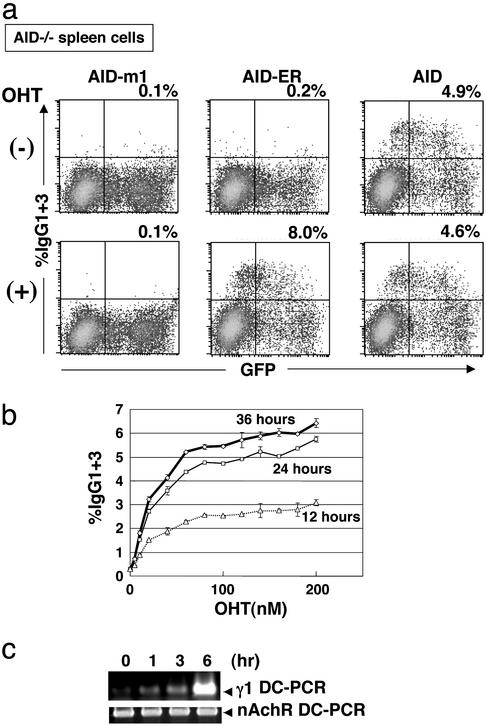
Second, we infected spleen B cells isolated from AID−/− mouse with AID-ER expressing retrovirus. Retroviruses carrying wild-type AID and inactive mutant AID-m1 (28) were also used as controls. OHT was added to the culture 24 h after infection. Two days later, frequencies of IgG1- and IgG3- expressing B cells were measured by FACS (Fig. 2a). In the presence of OHT, AID-ER infected B cells showed efficient switching to IgG-expressing cells. This efficiency was even higher than that observed in wild-type AID-infected cells, which were not affected by the presence of OHT. In AID-ER infected AID−/− spleen cells, percentages of switched cells were elevated by increased concentrations of OHT and with time (Fig. 2b). In the absence of OHT, by contrast, IgG-positive cells were not induced in AID-ER-infected cells. AID-m1 had no activity at all.
Figure 2.
AID-ER induced CSR in AID−/− spleen cells. (a) AID−/− spleen cells were infected with retrovirus containing AID-ER, AID-m1, or wild-type AID in the presence of LPS and IL-4. Twenty-four hours after infection, 1 μM OHT was added. Two days later, cells were harvested and stained with biotinylated anti-IgG1+anti-IgG3 antibodies and streptavidin-phycoerythrin. (b) OHT dose dependency of AID-ER. After infection with retrovirus as above, the concentration of OHT was titrated from 0 to 200 nM. Percentages of IgG1+IgG3 expressing cells were determined by FACS 12, 24, and 36 h after addition of OHT. (c) Sμ-Sγ1 DC-PCR. AID-ER infected AID−/− spleen cells were treated with 1 μM OHT for indicated time intervals in the presence of LPS and IL-4. PCR was carried out for 40 cycles. Gel image of 2% agarose-gel electrophoresis of DC-PCR products with an internal control amplifying the nicotinic acetylcholine receptor gene was visualized by ethidium bromide.
To determine how rapidly AID-ER induces CSR in AID−/− spleen B cells stimulated with LPS and IL-4, we performed DC-PCR that detects CSR to γ1 at the DNA level (21). DC-PCR products were detected already 1 h after OHT addition to AID-ER transfected AID−/− spleen B cells, and continued to increase until 6 h after OHT addition (Fig. 2c). Therefore, γ1 DC-PCR serves as a sensitive assay that allows CSR detection before side-effects of protein synthesis inhibitors may emerge.
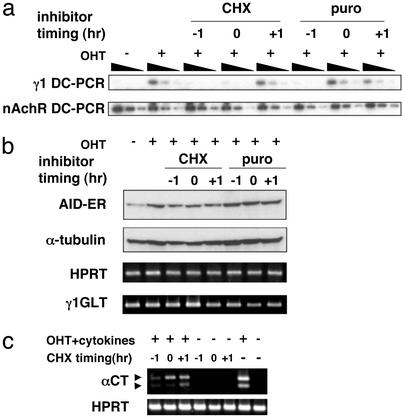
We then asked whether de novo protein synthesis is required for CSR downstream of AID-ER by adding CHX 1 h before OHT stimulation. Because the CHX concentration higher than 1 μg/ml reduced the germ-line transcription activity, we applied 0.2 μg/ml CHX that did not affect levels of γ1 germ-line transcription (Fig. 3b). AID-ER expressing AID−/− B cells were treated with OHT for 6 h in the presence of CHX before cells were harvested for genomic DNA preparation. As shown in Fig. 3a, addition of CHX 1 h before OHT addition (time −1) inhibited formation of DC-PCR γ1 products, namely CSR. Comparison of DC-PCR products from 3-fold serial dilutions of DNA templates indicated that CHX-treatment reduced CSR by at least one order of magnitude. This inhibition was alleviated by delaying timing of CHX addition to 1 h after OHT addition (time +1). Strong inhibition by CHX added at time −1 or simultaneously with OHT (time 0), in contrast to a mild effect by CHX addition at time +1 suggests, that CHX inhibits CSR at a relatively proximal point downstream to AID-ER activation. Puromycin, which inhibits protein synthesis with a different mechanism, suppressed CSR in a similar manner. Although the AID-ER protein levels were slightly decreased by the CHX treatment, they were almost comparable among three conditions where CHX was added at time −1, 0, and +1. Therefore, this slight reduction of the AID-ER protein is not responsible for inhibition of CSR by the CHX pretreatment.
Figure 3.
CSR inhibition by CHX and puromycin. (a) OHT (1 μM) was added to AID-ER-infected AID−/− spleen cells at time 0. Cells were harvested after 6 h. CHX (0.2 μg/ml) and puromycin (1 μM) were added at indicated time points before or after OHT (time −1, 0, and + 1 h). DC-PCR was performed for 30 cycles, and its products were detected by agarose gel electrophoresis followed by Southern blotting. Templates were serially diluted 3-fold. (b) Proteins of AID-ER and α-tubulin were detected by Western blotting with anti-ER and anti-α-tubulin antibodies, respectively. Sample equivalent to 7.5 × 104 cells was applied per lane. Hypoxanthine-guanine phosphoribosyl transferase and γ1 GLT were detected by RT-PCR. (c) OHT and CD40L + IL-4 + transforming growth factor β were added to AER cells. Cells were harvested after 6 h. CHX was added as above. α circle transcripts were amplified by RT-PCR.
A similar experiment was performed with AER cells. Because the nonproductive IgH allele in CH12F3–2 cells have undergone recombination between Sμ and Sα (29), DC-PCR cannot be used for detection of μ-to-α CSR in CH12F3–2-derived AER cells. We used RT-PCR of α circle transcripts (22) to evaluate CSR in AER cells. In agreement with the result from AID-ER expressing AID−/− spleen cells, CHX added 1 h before stimulation with OHT and cytokines strongly inhibited induction of α circle transcripts (Fig. 3b). Therefore, the inhibition of CSR by the protein synthesis inhibitor is not unique to IgG1 switching.
Discussion
We previously reported that CHX inhibits CSR in cytokine-stimulated B cells (10), which led to demonstration of AID requirement in CSR (11, 12). In the present study, CSR was inhibited by CHX, despite the fact that the AID activity was provided by OHT-induced activation of preexisting AID-ER, thus bypassing suppression of AID expression by CHX. Therefore, de novo synthesis of at least an additional protein appears to be essential to the events downstream of AID in CSR. Because CSR was detected within 1 h after the OHT treatment of AID-ER expressing cells, the CSR reaction can be completed within 1 h after activation of AID-ER, whereas CSR products continued to increase at least up to 6 h. Pretreatment of CHX before OHT addition strongly suppressed CSR, whereas the addition of CHX 1 h after OHT addition hardly inhibited CSR. If the new protein synthesis is required for later phases of CSR, CSR should be inhibited similarly by adding CHX after the OHT treatment. Therefore, these results suggest that CHX may inhibit a step relatively proximal to the AID function, most likely synthesis of a recombinase. During the first 1 h of the OHT addition, a sufficient amount of the recombinase appears to be synthesized to accumulate comparable levels of CSR products for the next 5 h. Because the RNA- but not DNA-editing hypothesis for the function of AID postulates de novo protein synthesis immediately downstream of AID expression, the present findings support the RNA-editing hypothesis.
This CHX-sensitive step is unlikely to be the DNA repair phase in CSR because processing of cleaved DNA ends and their ligation are carried out by general DNA repair proteins, which should be ubiquitous to provide against accidental DNA damages by oxygen radicals, UV, x-ray irradiation, and so on. We speculate that the DNA cleavage step downstream of AID may be CHX sensitive. However, we cannot completely exclude the possibility that de novo synthesis of a novel protein is required for processing staggered cleavage ends before their ligation (30). It is also possible that an extremely labile protein is involved in an immediately downstream step of AID. Alternatively, protein synthesis inhibitors superinduce some proteins like p53, giving rise to negative regulation on CSR. If these are the cause, it is likely to have a significant reduction in CSR 5 h after CHX addition at time +1. Because little effect was seen under this condition, we consider that these possibilities failed to explain the mechanism for the CHX inhibition of CSR.
During the progress of this study, it was reported that AID can enhance mutations in Escherichia coli (31). The authors suggested that CSR might be triggered by direct DNA deamination by AID. However, a more recent report shows that Apobec-1, a well established RNA-editing enzyme, can also augment mutations in E. coli more efficiently than AID (32). Although CSR was reduced in B cells deficient in UNG uracil-DNA-glycosylase, a critical enzyme in the base excision repair system (16), similar phenotypes are known in B cells deficient in mismatch repair enzymes (33, 34). It is important to test which step in CSR is affected by UNG uracil-DNA-glycosylase. Such repair enzymes may form a complex with a putative recombinase to hold cleaved DNA ends. It is possible that the deletion of these enzymes inhibits the formation of the recombinase complex (recombinasome). Another interesting possibility is that AID edits a precursor to mRNA coding for a novel DNA cytidine deaminase that directly edits DNA.
Supplementary Material
Supporting Figure
Acknowledgments
We are grateful to Drs. P. Chambon and T. Schroeder for pERT2 plasmid, and to Dr. T. Kitamura for Plat-E packaging cells. We also thank Dr. Shun-ichi Takeda and members of our lab for critical reading of the manuscript and discussion, Mrs. K. Yurimoto, T. Toyoshima, Y. Tabuchi, A. Kawamura, Y. Sasaki, and E. Inoue for their technical assistance, and Ms. K. Saito for preparation of the manuscript. This investigation was supported by the Center of Excellence Grant from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations
AID
activation-induced cytidine deaminase
CSR
class-switch recombination
SHM
somatic hypermutation
GLT
germ-line transcript
OHT
4-hydroxytamoxifen
ER
estrogen receptor
CHX
cycloheximide
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
References
- 1.Kinoshita K, Honjo T. Nat Rev Mol Cell Biol. 2001;2:493–503. doi: 10.1038/35080033. [DOI] [PubMed] [Google Scholar]
- 2.Honjo T, Kinoshita K, Muramatsu M. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 3.Stavnezer-Nordgren J, Sirlin S. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yancopoulos G D, DePinho R A, Zimmerman K A, Lutzker S G, Rosenberg N, Alt F W. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tashiro J, Kinoshita K, Honjo T. Int Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- 6.Rolink A, Melchers F, Andersson J. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- 7.Manis J P, Gu Y, Lansford R, Sonoda E, Ferrini R, Davidson L, Rajewsky K, Alt F W. J Exp Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin X F, Besmer E, Kenter A, Rajewsky K, Nussenzweig M C. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosma G C, Kim J, Urich T, Fath D M, Cotticelli M G, Ruetsch N R, Radic M Z, Bosma M J. J Exp Med. 2002;196:1483–1495. doi: 10.1084/jem.20001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muramatsu M, Sankaranand V S, Anant S, Sugai M, Kinoshita K, Davidson N O, Honjo T. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 11.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 12.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 13.Okazaki I M, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa K, Okazaki I M, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 15.Petersen S, Casellas R, Reina-San-Martin B, Chen H T, Difilippantonio M J, Wilson P C, Hanitsch L, Celeste A, Muramatsu M, Pilch D R, et al. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rada C, Williams G T, Nilsen H, Barnes D E, Lindahl T, Neuberger M S. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 17.Feil R, Wagner J, Metzger D, Chambon P. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 18.Schroeder T, Just U. EMBO J. 2000;19:2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shockett P, Difilippantonio M, Hellman N, Schatz D G. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu C C, Paul W E, Max E E. Proc Natl Acad Sci USA. 1992;89:6978–6982. doi: 10.1073/pnas.89.15.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. Proc Natl Acad Sci USA. 2001;98:12620–12623. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunaga S, Maki K, Komagata Y, Miyazaki J, Ikuta K. J Immunol. 1997;158:4223–4228. [PubMed] [Google Scholar]
- 24.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita S, Kojima T, Kitamura T. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 26.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. J Exp Med. 2002;195:529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wijayaratne A L, McDonnell D P. J Biol Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 28.Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. Nature. 2001;413:639–643. doi: 10.1038/35098100. [DOI] [PubMed] [Google Scholar]
- 29.Ono S J, Zhou G, Tai A K, Inaba M, Kinoshita K, Honjo T. FEBS Lett. 2000;467:268–272. doi: 10.1016/s0014-5793(00)01151-0. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Kinoshita K, Honjo T. Proc Natl Acad Sci USA. 2001;98:13860–13865. doi: 10.1073/pnas.241524898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen-Mahrt S K, Harris R S, Neuberger M S. Nature. 2002;418:99–103. [PubMed] [Google Scholar]
- 32.Harris R S, Petersen-Mahrt S K, Neuberger M S. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 33.Schrader C E, Edelmann W, Kucherlapati R, Stavnezer J. J Exp Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenstein M R, Neuberger M S. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure