Growth Suppression of Pre-T Acute Lymphoblastic Leukemia Cells by Inhibition of Notch Signaling (original) (raw)
Abstract
Constitutive NOTCH signaling in lymphoid progenitors promotes the development of immature T-cell lymphoblastic neoplasms (T-ALLs). Although it is clear that Notch signaling can initiate leukemogenesis, it has not previously been established whether continued NOTCH signaling is required to maintain T-ALL growth. We demonstrate here that the blockade of Notch signaling at two independent steps suppresses the growth and survival of NOTCH1-transformed T-ALL cells. First, inhibitors of presenilin specifically induce growth suppression and apoptosis of a murine T-ALL cell line that requires presenilin-dependent proteolysis of the Notch receptor in order for its intracellular domain to translocate to the nucleus. Second, a 62-amino-acid peptide derived from a NOTCH coactivator, Mastermind-like-1 (MAML1), forms a transcriptionally inert nuclear complex with NOTCH1 and CSL and specifically inhibits the growth of both murine and human NOTCH1-transformed T-ALLs. These studies show that continued growth and survival of NOTCH1-transformed lymphoid cell lines require nuclear access and transcriptional coactivator recruitment by NOTCH1 and identify at least two steps in the Notch signaling pathway as potential targets for chemotherapeutic intervention.
Notch signaling plays an important role in diverse cellular and developmental processes, including differentiation, proliferation, survival, and apoptosis (reviewed in reference 1). For example, the mammalian NOTCH1 gene has an essential role in the development of T cells from common lymphoid progenitors, as NOTCH1 insufficiency leads to intrathymic B-cell development at the expense of T-cell development (43). Conversely, inappropriate increases in NOTCH1 signaling cause ectopic T-cell differentiation within the bone marrow at the expense of B-cell differentiation (42). Enforced NOTCH1 signaling eventually leads to the development of lethal CD4/CD8+/+ T-cell lymphoblastic neoplasms (T-ALLs) (40), indicating NOTCH functions as an oncoprotein in certain contexts.
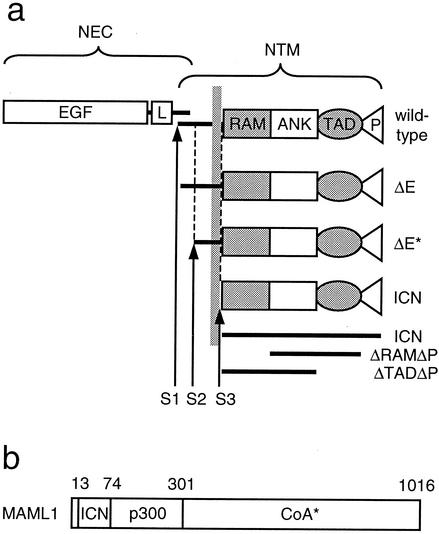
Normal NOTCH1 is a heterodimeric type I transmembrane receptor composed of two polypeptide chains, an extracellular subunit (NEC) and a transmembrane subunit (NTM), which are produced by cleavage (S1 in Fig. 1a) of a single precursor polypeptide by a furin-like convertase (35). The NEC subunit includes 36 iterated epidermal growth factor (EGF)-like repeats that bind ligands of the Delta and Serrate families (45). Although it is very difficult to detect Notch in the nucleus of normal cells, numerous genetic and biochemical studies have converged on a model for signaling in which ligand binding renders the receptor sensitive to at least two successive proteolytic cleavages (reviewed in reference 38). The first cleavage occurs just external to the transmembrane domain (S2 in Fig. 1a) and is mediated by metalloproteases of the ADAM family (7, 35). The second cleavage, which occurs within the inner portion of the lipid bilayer (S3 in Fig. 1a), releases the intracellular domain of NTM (ICN) from its membrane tether. This cleavage requires presenilin 1 or 2 (13, 55), members of a family of novel polytopic transmembrane proteins that likely function as aspartyl proteases (57, 59). Free ICN then translocates to the nucleus, where it interacts with the DNA binding transcription factor CSL [named for its murine, Drosophila, and Caenorhabditis elegans homologs CBF1, Su(H), and Lag-1, respectively] and with conserved transcriptional coactivators of the Mastermind family to form a ternary complex that stimulates the transcription of downstream target genes (15, 41, 60). Although the RAM domain of ICN has been identified as mediating high-affinity interaction with CSL, the ankyrin repeat (ANK) domain also binds weakly (3, 31, 54). The ANK binding site for CSL may be critically important in vivo, as RAM-less forms of ICN1 retain the capability to stimulate transcription from CSL reporters, whereas ANK deletions render ICN1 nonfunctional (3, 4). The ANK domain also serves as the binding site for Mastermind-like coactivators (MAMLs) (15, 41, 60), which interact with ANK through an N-terminal basic domain (Fig. 1b). Structure and leukemogenesis analyses have shown that both ANK and a C-terminal transcriptional activation domain (TAD) are required for induction of T-ALL in a murine model (4).
FIG. 1.
(a) Schematic representation of various forms of NOTCH referred to in this study. The normal mature heterodimeric receptor is produced by cleavage at S1. Cleavages at S2 and S3 are normally regulated by ligand binding to NEC but occur constitutively in the case of the ΔE form. Cleavage of ΔE at S2 by a metalloprotease yields the ΔE* form. Cleavage of ΔE* at S3 by presenilins releases ICN from the membrane. ICN1 constructs utilized in T6E/DFP-AA rescue experiments are indicated (ICN, ΔRAMΔP, and ΔTADΔP). P, PEST domain. (b) Schematic representation of functional domains of MAML1. Amino acid residue positions are indicated; 1016 represents full-length MAML1. ICN, Notch-binding domain; p300, p300 recruitment domain according to Fryer et al. (20); CoA*, recruitment domain for unidentified transcriptional coactivator(s).
Mammalian NOTCH1 was initially identified through analysis of a recurrent (7;9)(q34;q34.3) chromosomal translocation found in sporadic human T-ALL (16). The t(7;9) fuses the 3′ end of NOTCH1 to the T-cell receptor β promoter/enhancer and results in the expression of a series of aberrant mRNAs encoding nuclear forms of NOTCH1 that resemble ICN1, suggesting that T-cell transformation stems from an increase in signals mediated by dysregulated expression of nuclear NOTCH1. While transgenes encoding predominantly nuclear ICN1 induce T-ALL when expressed in murine bone marrow cells (3, 4, 40), the necessity of nuclear localization for T-cell transformation has not been formally proven, and certain observations raise the possibility that alternative signaling mechanisms could be relevant. Most directly, an N-terminally deleted form of membrane-tethered NOTCH1 (termed ΔE) (Fig. 1a) is a potent inducer of T-ALL in the mouse, yet it shows no detectable evidence of processing to a nuclear derivative as judged by immunostaining of cells or cell extracts (3, 40). In addition, some genetic and biochemical data support the existence of CSL-independent NOTCH signaling pathways that might proceed through mechanisms not requiring nuclear access (8, 27, 37, 39, 44, 51, 56).
Another area of uncertainty is whether persistent NOTCH1 signaling is necessary for tumor growth after transformation has occurred. Transgenic expression of active Notch isoforms has been shown both to drive development of immature T cells from lymphoid precursors and to prevent their further maturation (26, 42), suggesting that NOTCH1 signaling contributes to transformation by influencing differentiation. It is not known whether other potential effects of NOTCH signaling, such as inhibition of apoptosis (12, 29) or enhancement of proliferation (6, 21), contribute to Notch-mediated transformation. Determination of whether continued NOTCH1 signaling is required for cell proliferation once tumors have become established is an issue of central importance when considering NOTCH1 as a potential therapeutic target in malignancies such as T-ALL.
By using cell lines derived from NOTCH1-associated T-ALLs, we show here that presenilin inhibitors suppress the growth of ΔE-expressing cells but not ICN1-expressing cells, arguing that NOTCH1 nuclear access is required to maintain tumor cell growth and survival. We then demonstrate that dominant-negative peptides derived from Mastermind-like-1 (MAML1), which prevent recruitment of coactivators to the CSL/ICN1 complex, specifically antagonize the growth of murine ΔE- and ICN1-expressing cell lines as well as cells from a human T-ALL bearing the t(7;9). Together these findings support a model in which recruitment of transcriptional coactivators to ICN1 complexes is necessary for the proliferation and survival of Notch1 leukemia cells.
MATERIALS AND METHODS
Cell lines.
The NOTCH1-associated T-ALL lines used have been described previously (16, 40). All other lines were obtained from the American Type Culture Collection (Manassas, Va.), including the murine T-lymphoblastic cell line BW5147 (ATCC designation BW5147.G.1.4). T-ALL cell lines and BJAB cells were maintained in RPMI (Gibco, Carlsbad, Calif.) supplemented with a solution containing 10% fetal bovine serum (BioWhittaker, Wakersville, Md.), 1 mM sodium pyruvate (Mediatech, Herndon, Va.), 2 mM l-glutamine (Gibco), 100 U of penicillin G/ml, and 100 mg of streptomycin/ml. U2OS and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) with the same supplements. Cells were grown at 37°C under 5% CO2.
Expression plasmids.
Constructs for expression of MAML1 and ICN1 polypeptides were created by PCR amplification with human NOTCH1 (16) and MAML1 (60) cDNAs as templates. Eukaryotic MAML1 expression constructs were engineered in the plasmid vector pcDNA3 (Invitrogen, Carlsbad, Calif.) to produce peptides with a C-terminal three-hemagglutinin tag. To permit stable expression in lymphoid cells, cDNAs encoding MAML1 peptides fused N terminally to green fluorescent protein (GFP) (Clontech, Palo Alto, Calif.) were cloned into the backbone of the retroviral shuttle vector MSCV-IRES-GFP (42), replacing the resident internal ribosome entry sequence (IRES) and GFP sequences. Bacterial MAML1 expression constructs were engineered in the plasmid vector pRSET-A (Invitrogen) to produce peptides with an N-terminal six-His tag followed by a cleavage site for tobacco etch virus (TEV) protease. Constructs were designed such that only a single additional glycine residue remained at the N terminus of encoded MAML1 peptides following cleavage with TEV protease. Expression constructs for NOTCH1 in pcDNA3, MSCV-IRES-GFP (4), pET41 (Novagen, Madison, Wis.), myc-tagged CSL in pcDNA3 (3), and MAML1 in CMV2-FLAG (Sigma, St. Louis, Mo.) (60) have all been described previously.
Presenilin inhibitors.
DFP-AA (also called compound E in the literature) is a benzodiazepine-type compound and was synthesized as described previously (50). DAPT is _N_-[_N_-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine _t_-butyl ester, WPE-III-86 is the unfluorinated counterpart of DAPT, WPE-III-109 is a truncated version of DAPT lacking the phenylglycine residue, WPE-III-18 is the methyl ester variant of DAPT, and WPE-III-141 is the Ala-Leu counterpart to WPE-III-18. All these dipeptide analogues were synthesized as described previously (14). The remaining compounds are all (hydroxyethyl)urea transition-state analogues exemplified by WPE-III-31C (17), with the structure Boc-PheΨPhe-Leu-Val-OMe, where Ψ is the pseudopeptide bond containing the hydroxyethyl group. MW-III-36A is Boc-PheΨAla-Leu-Phe-Ome, MW-III-36B is Boc-PheΨIle-Leu-Phe-Ome, MW-III-36C is Boc-PheΨPhe-Leu-Phe-Ome, MW-III-38A is Boc-PheΨPhe-Ala-Phe-Ome, and MW-III-38B is Boc-PheΨPhe-Val-Phe-OMe. These analogues were all synthesized according to methods described previously (17).
Reporter gene assays.
For the presenilin inhibitor assays, empty pcDNA3 or pcDNA3-ΔE (10 ng/well) was transiently transfected in triplicate into human U2OS cells in 24-well dishes (Falcon 3047) by using Lipofectamine Plus (Invitrogen) along with the firefly luciferase reporter CBF1-luc (24) and an internal Renilla luciferase control plasmid, phRL-TK (Promega, Madison, Wis.). Presenilin inhibitor compounds (1 μM) were added to the cultures immediately posttransfection and were added again in fresh media 1 day posttransfection. To test dominant-negative MAML1 peptides, U2OS cells in 24-well dishes were transfected with empty pcDNA3 or pcDNA3-ICN1 (10 ng/well) plus various pcDNA3 plasmids (50 ng/well) encoding MAML1 or MAML1 peptides with three C-terminal hemagglutinin tags. All dual luciferase assays were performed and analyzed by using cell extracts prepared 40 to 44 h posttransfection, as described previously (4).
Retroviral gene transduction.
Production of pseudotyped MSCV-GFP viruses and retroviral spin infections and flow cytometric detection of GFP-expressing cells were all done as described previously (10, 42). GFP expression appeared by 36 h posttransduction and generally peaked ∼72 h posttransduction.
Cell growth assays.
Cultured cells were treated with a presenilin inhibitor compound (DFPAA, WPE III-18, MW III-36A, or WPE III-109) at concentrations of 100 nM to 5 μM. Viable cell counts were performed daily, either manually by trypan blue exclusion or with an automated clinical hematology analyzer (Advia 120 Hematology System; Bayer Diagnostics, Tarrytown, N.Y.). Viable cells distributed primarily in the lymphocyte and large unstained cell gates and nonviable cells distributed primarily in the platelet and cell debris gates. Two- to 3-ml cultures were initially seeded at 7.5 × 105 viable cells/ml in 12- or 24-well dishes and were reseeded each day at the same density in media with fresh inhibitor. Mock-treated controls were exposed to equivalent concentrations of carrier (up to 0.05% dimethyl sulfoxide). Extrapolated cell counts were calculated each day by using the formula [(current day's cell concentration)/(7.5 × 105 cells/ml)] × (previous day's extrapolated cell count).
Cell cycle and apoptosis analysis.
Cells were stained with either propidium iodide or DRAQ5 for DNA content measurement. For propidium iodide staining, cells were washed once in cold phosphate-buffered saline (PBS), fixed in ice-cold 70% ethanol for at least 24 h at −20°C, and stained with 40 μg of propidium iodide (Sigma)/ml with (100 μg/ml) DNase-free RNase A in PBS for 30 min at 37°C at a concentration of 5 × 105 cells/ml. For DRAQ5 staining, DRAQ5 dye (Biostatus, Leicestershire, United Kingdom) was added to live cells at a final concentration of 10 μM in complete culture medium and was incubated at 37°C for 15 min. Immediately after staining, DNA content was measured by using either a Coulter Epics XL-MCL single-laser 4-color flow cytometer or a Coulter Cytomics FC 500 dual-laser 5-color flow cytometer (Beckman Coulter, Miami, Fla.). Intact, single cells were gated by using propidium iodide and propidium iodide peak fluorescence. For DRAQ5 staining studies, DNA content was measured in separately gated GFP-deficient and GFP+ cell populations. Cell cycle fractions were determined by using MultiCycle software (Phoenix Flow Systems, San Diego, Calif.). Apoptotic cells were identified by staining of PBS-washed, unfixed cells with Annexin-V-phycoerythrin (PE)/7-aminoactinomycin D (7-AAD) according to the manufacturer's recommended conditions (BD Pharmingen, San Diego, Calif.), followed by flow cytometric analysis. Apoptotic cells were measured in the PE+/7-AAD− gate. Dead cells were excluded as 7-AAD+ events.
Immunoprecipitation and Western blotting.
Immunoprecipitates were prepared from whole-cell extracts of 5 × 106 cells with a rabbit polyclonal antibody against NOTCH1, designated α-TC, as described previously (3). Immunoprecipitated proteins on protein A beads (Pharmacia, Piscataway, N.J.) were treated with lambda protein phosphatase (New England Biolabs, Beverly, Mass.) as recommended by the manufacturer prior to electrophoresis in discontinuous sodium dodecyl sulfate polyacrylamide gels. Western blots were stained with α-TC by using a chemiluminescent method (SuperSignal West Pico; Pierce, Rockford, Ill.) as described previously (3).
Northern analysis.
Total RNA was prepared from PBS-washed tissue culture cells by using Trizol reagent (Invitrogen) according to the manufacturer's recommendations. Two micrograms of total RNA from each sample was electrophoresed through 1.2% agarose-2.2 M formaldehyde gels in 1× morpholinepropanesulfonic acid buffer, transferred to Nytran SPC membranes (Schleicher & Schuell, Keene, N.H.) in 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), and UV cross-linked by using a Stratalinker (Stratagene, La Jolla, Calif.). Hybridization was performed according to the membrane manufacturer's recommendations with random hexamer-primed 32P-labeled DNA probes and a final washing with 1× SSPE-0.5% sodium dodecyl sulfate at 37°C. Autoradiography was performed with either a Molecular Dynamics PhosphorImager (Amersham, Arlington Heights, Ill.) or conventional X-ray film. The HES1 probe was described previously (36). The GAPDH probe was a 168-bp fragment spanning bases 201 to 368 (GenBank M32599) derived by PCR from a mouse pre-B cell cDNA library and cloned into pKS+ (Stratagene).
Protein purification.
Bacterial expression plasmids encoding MAML1 or NOTCH1 polypeptides were transformed into BL21(DE3) or BL21(DE3)pLysS Escherichia coli (Invitrogen). Cultures grown in Luria-Bertani broth were induced with 1 mM isopropyl-[exists]-d-thiogalactopyranoside at 37°C for 3 h. After collection by centrifugation, bacterial pellets were resuspended in 1/50th of the original culture volume of a solution containing ice-cold PBS, pH 7.4, containing 5 mM β-mercaptoethanol, 2 μg of aprotinin/ml, 1 μg of leupeptin/ml, and 0.5 mM phenylmethylsulfonyl fluoride. Bacteria were lysed by three cycles of freezing and thawing, each followed by sonication for 90 s (3 times for 30 s each) on ice with a Branson Sonifier (Branson Ultrasonics, Danbury, Conn.).
MAML1 peptides were prepared from inclusion bodies as follows. Triton X-100 (1% [vol/vol]) was added to the lysates, which were mixed for 60 min at 25°C. Inclusion bodies were collected by centrifugation and were solubilized in PBS containing 6 M guanidine-HCl and 5 mM β-mercaptoethanol overnight at 25°C. After removal of insoluble material by centrifugation, soluble proteins were applied to an Ni-nitrilotriacetic acid agarose (Qiagen, Valencia, Calif.) column (bed volume of ∼1/300th of the initial culture volume). After extensive washing, bound proteins were step eluted with PBS solutions containing 6 M guanidine-HCl and 5 mM β-mercaptoethanol equilibrated to pH 6.3, 5.9, and 4.5 and were analyzed by electrophoresis in tricine gels (48). Fractions containing six-His-MAML1 peptides were pooled and adjusted to pH 7.4, reduced by addition of 10 mM dithiothreitol, and dialyzed at 4°C over several days in Spectra/Por membranes (Spectrum Laboratories, Santa Dominguez, Calif.) against PBS, pH 7.4, containing 5 mM β-mercaptoethanol and decreasing concentrations of guanidine-HCl. A precipitate enriched for MAML1 peptides formed during removal of guanidine-HCl that was collected by centrifugation, solubilized in 5% (vol/vol) acetic acid, and freeze dried. Lyophilized peptides were dissolved in 50 mM piperazine sulfonic acid, pH 6.2, containing 0.5 mM EDTA and 1 mM dithiothreitol, and were incubated overnight at 25°C with recombinant TEV protease (Invitrogen). After dialysis against 50 mM Tris, pH 6.0, containing 0.5 mM EDTA and 5 mM β-mercaptoethanol, the cleaved peptides were acidified by addition of 5% (vol/vol) acetic acid and were applied to a C18 preparative-scale high-performance liquid chromatography column, which was developed with a 20 to 40% acetonitrile gradient. Fractions containing eluted peptides (assessed by monitoring optical density at 209 nm) were collected, lyophilized, resuspended in acidified H2O (pH 5.5), and stored at −80°C. Fractions containing purified MAML1 peptides (as judged by the presence of a single species of appropriate size in tricine gels) were used in electrophoretic mobility shift assays (EMSA).
NOTCH1 polypeptides were purified from bacterial lysates as described previously (60). Lysates containing soluble glutathione _S_-transferase-NOTCH1 polypeptides were incubated for 4 h at 4°C with glutathione Sepharose 4B beads (Pharmacia). After extensive washing, the beads were incubated overnight at 25°C in 50 mM Tris, pH 8.0, containing 0.5 mM EDTA, 1 mM dithiothreitol, and recombinant TEV protease. NOTCH1 polypeptides were purified to electrophoretic homogeneity by ion exchange chromatography on Mono-Q resin (Amersham) followed by gel filtration on a Superdex 200 column (Amersham).
CSL was immunopurified from transiently overexpressing 293T cells by using a monoclonal anti-myc antibody (clone 9E10, kindly provided by Jeffrey Parvin) followed by elution with myc peptide (Research Genetics), as described previously (60).
EMSA.
Conditions for EMSA were as described previously (60). Briefly, protein complexes were allowed to form for 30 min at 30°C in a 15-μl volume containing 104 cpm of end-labeled probe (23), 1 μl of immunopurified CSL, 50 ng of ICN1 polypeptides, and/or 10 to 50 ng of MAML1 peptides. Electrophoresis was performed at 175 V in a 10% Tris-glycine-EDTA gel, which was dried and analyzed by autoradiography.
RESULTS
Comparison of presenilin inhibitors in reporter gene assays_._
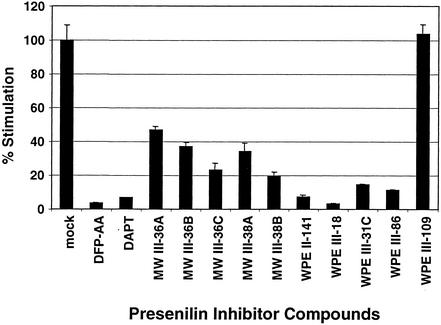
Small-molecule peptidomimetic inhibitors of presenilins block proteolysis and nuclear translocation of membrane-tethered ΔE (5). We used transient expression assays to compare the effects of 12 such inhibitors on ΔE-dependent transactivation of CSL. Our expression construct encodes a form of ΔE consisting of the endogenous NOTCH1 signal peptide fused to the NTM subunit at a position 61 residues external to the transmembrane domain. ΔE thus requires sequential proteolytic cleavage by metalloprotease (S2) and presenilin (S3) for nuclear access (see Fig. 1a). The tested compounds vary widely in inhibitory potency (Fig. 2). The potency of any individual compound in inhibiting ΔE transactivation is highly correlated with its ability to inhibit β-amyloid precursor protein cleavage (M. Wolfe, unpublished data).
FIG. 2.
Potency of presenilin inhibitor compounds in suppression of transcriptional stimulation by ΔE. U2OS cells were transiently transfected with a ΔE expression construct along with a CSL-luciferase reporter. Average normalized luciferase reporter activity (± standard deviations) from triplicate samples is expressed as a percentage of that observed in mock-treated cells.
ΔE-induced T-ALL cell growth is suppressed by presenilin inhibitors.
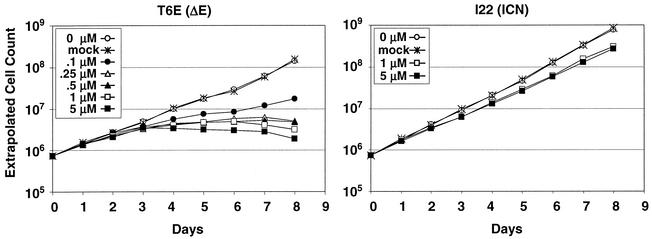
DFP-AA, a potent inhibitor of ΔE-dependent transactivation (Fig. 2), was assessed for its effects on the growth of T6E cells, a murine T-ALL cell line derived from a ΔE-induced tumor (40). DFP-AA causes significant dose-dependent suppression of T6E growth at concentrations as low as 100 nM (Fig. 3, left panel). For three other compounds, WPE III-18, MW III-36A, and WPE III-109 (strong, moderate, and negligible inhibitors of ΔE transactivation, respectively; Fig. 2), growth-suppressive activity also correlates with ΔE inhibition (data not shown).
FIG. 3.
Suppression of T6E cell growth by the presenilin inhibitor compound DFP-AA. T6E (left panel) and I22 (right panel) cells were cultured in the presence of the indicated concentrations of DFP-AA or carrier (mock) and were counted daily. Extrapolated cell counts were calculated as described in Materials and Methods.
To control for NOTCH-independent effects of the inhibitors, we also tested DFP-AA on I22 cells, a murine T-ALL cell line derived from an ICN1-induced tumor (40). At high doses, DFP-AA produces a small decrement in I22 growth, suggesting that any NOTCH-independent effects are minimal (Fig. 3, right panel).
Presenilin inhibitors perturb cell cycle progression and induce apoptosis of T6E cells.
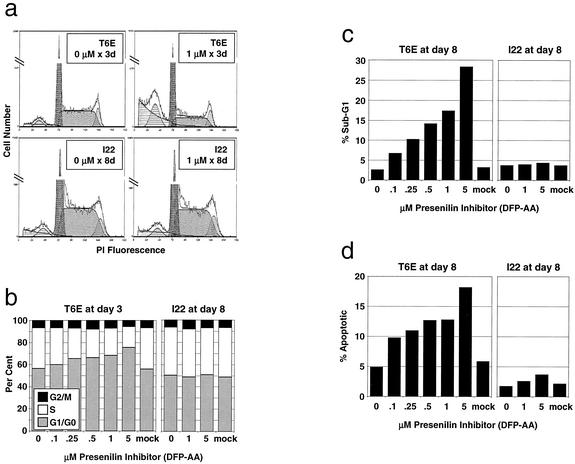
T6E cells demonstrate a dose-dependent increase in G1/G0 fraction and a decrease in S-phase fraction after only 3 days of treatment with DFP-AA, while I22 cells are unaffected by up to 8 days of treatment (Fig. 4a and b). After 8 days, T6E, but not I22, cultures also show a significant accumulation of dead or dying cells, which is reflected by the presence of a large cell fraction with sub-G1 DNA content (Fig. 4c). This is accompanied by dose-dependent induction of apoptosis in T6E, but not I22, cells as judged by an increase in Annexin-V+/7-AAD− cells (Fig. 4d). These data indicate ΔE-expressing T-ALL cells selectively demonstrate altered cell cycle progression and apoptosis in response to DFP-AA treatment.
FIG. 4.
Presenilin inhibitor treatment alters cell cycle progression and induces apoptosis in T6E, but not I22, cells. (a and b) Cell cycle analysis. T6E and I22 cells were treated with the indicated concentrations of DFP-AA for 3 and 8 days, respectively. DNA content was measured by flow cytometry after staining with propidium iodide. (a) Representative propidium iodide (PI) fluorescence histograms with superimposed cell cycle analysis models (including background correction). (b) Cell cycle fractions determined from histograms as for panel a. (c and d) Apoptosis analysis. (c) Sub-G1 fractions determined by using histograms as for panel a. (d) T6E and I22 cells treated with DFP-AA for 8 days were dual-stained with Annexin-V (for apoptotic cells) and 7-ADD (to exclude dead cells) and were analyzed by flow cytometry. Percentages of apoptotic cells (Annexin-V+/7-AAD−) are indicated after excluding cell debris by forward/side scatter gating.
Transforming alleles of ICN1 rescue ΔE-expressing T-ALL cells from presenilin inhibition.
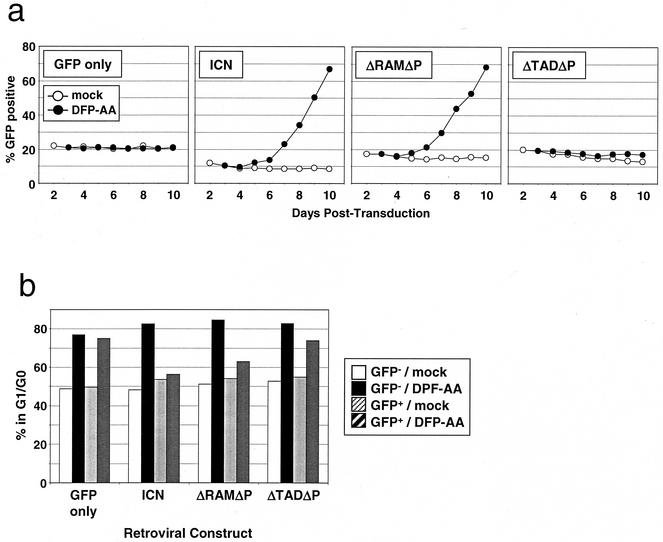
To confirm that the growth suppression of ΔE-expressing cells results from inhibition of NOTCH signaling, we tested whether ICN1 isoforms that do not require presenilin cleavage for nuclear access (ICN1, ΔRAMΔP, and ΔTADΔP; Fig. 1a) prevent presenilin inhibitor-mediated growth suppression. T6E cells were transduced with various ICN1 isoforms along with a marker, GFP, into T6E cells and then were treated with DFP-AA. Retroviral titers were adjusted so that only a subpopulation of cells would be transduced (∼10 to 20% GFP+), allowing the growth of transduced and nontransduced cells to be compared under identical culture conditions. As depicted in Fig. 5a, ICN1 and ΔRAMΔP, but not ΔTADΔP, rescue T6E cells only under conditions of DFP-AA treatment; in mock-treated cells, these isoforms have no effect. Additionally, ICN1, but not ΔTADΔP, prevents the drug-induced increase in G1/G0 fraction (Fig. 5b), while ΔRAMΔP shows an intermediate phenotype. Interestingly, the in vitro phenotypes of these three Notch alleles correlate well with their relative transforming potentials in a murine T-ALL model (4). Specifically, ICN1 induces T-ALL rapidly and activates CSL-dependent transcription strongly, ΔRAMΔP induces T-ALL more slowly and activates CSL-dependent transcription moderately, and ΔTADΔP is nontumorigenic and activates CSL-dependent transcription only weakly. These data show that the effects of presenilin inhibitors are likely mediated through inhibition of ICN1 production and that nuclear access by the ANK and C-terminal TADs of ICN1 is sufficient (and the TAD is necessary) for continued cell growth.
FIG. 5.
Leukemogenic forms of ICN1 rescue T6E cells from presenilin inhibitor-mediated growth suppression. (a) Growth advantage of ICN1-transduced cells under conditions of presenilin inhibitor treatment. T6E cells were transduced with retroviruses encoding various constitutively nuclear forms of NOTCH1 (shown in Fig. 1a) and GFP on a bicistronic mRNA or GFP alone and then were treated with 1 μM DFP-AA or carrier (mock) beginning at day 3 posttransduction and continuing for the duration of the experiment. The percentage of GFP+ cells in each of the cultures was determined daily by flow cytometry, gating for live cells by forward/side scatter criteria. All cultures were repeated in duplicate; a single representative experiment is shown. (b) Cell cycle analysis. T6E cells from the experiment depicted in panel a were harvested after 9 days of exposure to 1 μM DFP-AA or carrier (mock) and were stained with DRAQ5 dye. DNA content was measured by DRAQ5 fluorescence in GFP− and GFP+ subpopulations by flow cytometry.
Presenilin inhibitors result in accumulation of a stable NOTCH1 processing intermediate and downregulate HES1 transcription.
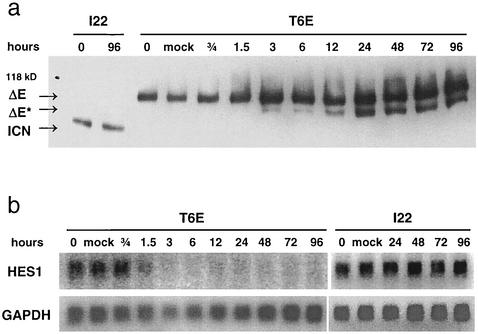
Although proteolytic products derived from ΔE in T6E cells are not detected under normal circumstances, we suspected inhibition of presenilin activity might cause accumulation of ΔE* (Fig. 1a), the product of metalloprotease cleavage of ΔE. Indeed, treatment of T6E cells with DFP-AA permits detection of a new NOTCH1 species of the expected size of ΔE* (Fig. 6a). This polypeptide is first detected after 3 h of treatment and continues to accumulate during the time course of the experiment, suggesting that ΔE* is fairly stable when presenilin is inhibited. No change in NOTCH1 polypeptides occurred in I22 cells treated with DFP-AA or T6E cells treated with dimethyl sulfoxide carrier alone.
FIG. 6.
Presenilin inhibitor treatment blocks NOTCH1 signal transduction in T6E, but not I22, cells. (a) Effects on NOTCH1 polypeptides. T6E and I22 cells were treated with 1 μM DFP-AA for the indicated periods of time. NOTCH1 polypeptides were then immunoprecipitated from whole-cell extracts with an antibody directed against the intracellular domain of NOTCH1 (α-TC) and were analyzed on a Western blot stained with α-TC. ΔE, ΔE*, and ICN are NOTCH1-derived polypeptides diagrammed in Fig. 1. (b) Effects on HES1 transcript level. After T6E and I22 cells were treated with 1 μM DFP-AA for the indicated periods of time, total RNA was collected and analyzed on a Northern blot with probes for HES1 and GAPDH.
To link DFP-AA treatment effects to known NOTCH1 signaling events, we assessed its effects on a well-characterized NOTCH1/CSL target, HES1. HES1 mRNA transcripts decrease rapidly in T6E cells treated with DFP-AA, falling to undetectable levels within 6 h (Fig. 6b). In contrast, DFP-AA has no effect on HES1 expression in I22 cells. These data confirm that DFP-AA treatment inhibits presenilin-dependent cleavage of ΔE in T6E cells and strongly suggest small amounts of ΔE-derived, short-lived ICN1 are essential for maintenance of T6E cell growth and survival.
Mapping and characterization of dominant-negative MAML1peptides.
Recent data suggest that ICN association with CSL is necessary for loading of MAML1 (60) and that both ICN and MAML1 are essential for activation of CSL-dependent transcription (20). Consistent with this view, truncated forms of MAML1 retaining an N-terminal ICN/CSL interaction domain but lacking a C-terminal TAD act as dominant-negative inhibitors of ICN function (20, 60).
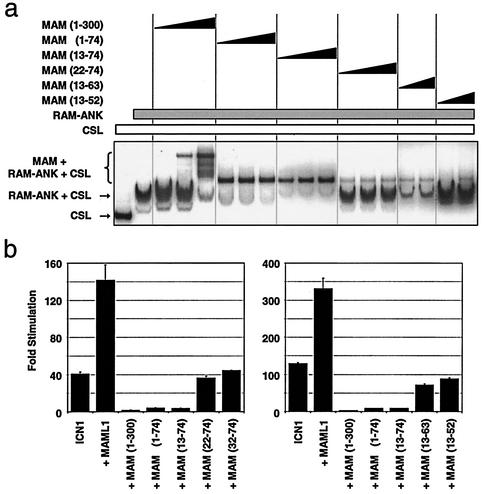
To test the idea that dominant-negative forms of MAML1 might be general inhibitors of NOTCH1-transformed T-ALL cell growth, we first determined the minimal portion of MAML1 needed for ternary complex formation in an EMSA. As part of a parallel study, we also defined a portion of ICN1 spanning the RAM and ANK domains as the minimal domain of NOTCH1 that is needed for stable ternary complex formation in vitro (Y. Nam, unpublished data). This RAM-ANK polypeptide, full-sized CSL, and MAML1 residues 13 to 74 [termed MAML1(13-74)] were sufficient for ternary complex formation on DNA (Fig. 7a), whereas shorter MAML1 peptides (residues 22 to 74, 13 to 63, and 13 to 52) associated weakly with CSL/RAM-ANK. MAML1(13-74) also defined the minimal domain necessary for strong dominant-negative activity in ICN1 gene reporter assays (Fig. 7b). These results indicate that MAML1(13-74) is sufficient for formation of stable, transcriptionally inert ICN1/CSL/MAML1 ternary complexes.
FIG. 7.
Mapping and functional characterization of dominant-negative MAML1 peptides. (a) Mapping MAML1 residues needed for ternary complex formation. Highly purified MAML1 peptides incubated with recombinant ICN1 RAM-ANK and immunopurified CSL were scored for complex formation in an EMSA. MAML1 peptides were included at increasing concentrations of up to a fivefold molar excess over the concentration of RAM-ANK. (b) Dominant-negative activities of MAML1 peptides. Each well of a 24-well plate containing U2OS cells was transiently transfected with pcDNA3 expression constructs for ICN1 (10 ng), various MAML1 peptides (50 ng), a CSL-luciferase reporter (125 ng), and a Renilla luciferase internal control reporter (2.5 ng). Mean normalized luciferase activity (± standard deviations) from triplicate wells is expressed relative to that observed in cells transfected with reporter constructs plus empty pcDNA3 (60 ng).
Dominant-negative MAML1 peptides suppress the growth of NOTCH1-transformed T-ALL cell lines.
To create a readily detectable form of dominant-negative MAML1, we fused the minimal MAML1 dominant-negative peptide [MAML1(13-74)] to GFP. In pilot experiments, MAML1(13-74)-GFP fusion protein expressed from the MSCV retroviral long terminal repeat promoter retained strong dominant-negative activity in reporter gene assays (data not shown).
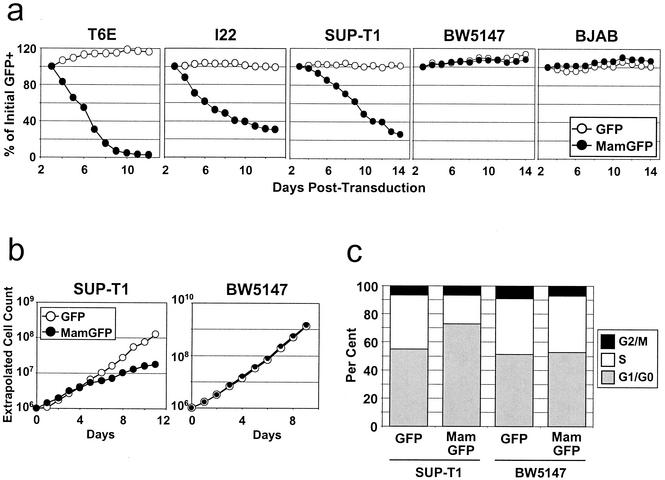
Various lymphoid cell lines were then transduced with dominant-negative MAML1(13-74)-GFP or control GFP retroviruses. In initial experiments, growth of transduced (GFP+) and untransduced (GFP−) cell fractions was compared within unsorted cultures. Growth suppression of the transduced population would thus lead to a decreasing GFP+ percentage over time. As depicted in Fig. 8a, transduction with dominant-negative MAML1(13-74)-GFP virus caused significant growth suppression of murine T6E and I22 cell lines as well as SUP-T1, a human T-ALL cell line with a chromosomal translocation involving NOTCH1 that leads to expression of ICN1-like polypeptides (2). Transduction with GFP-only virus did not inhibit the growth of any of these lines. Human BJAB cells and murine BW5147 cells, both harboring apparently normal NOTCH1 alleles (data not shown), were unaffected by transduction with MAML1(13-74)-GFP virus (Fig. 8a), suggesting the growth-suppressive effects of dominant-negative MAML1 are limited to NOTCH1-transformed cell lines.
FIG. 8.
Dominant-negative MAML1 peptides specifically suppress growth of human and murine NOTCH1-transformed T-cell lines. (a) Growth suppression of cells expressing the dominant-negative MAML1(13-74)-GFP fusion protein. Each cell line was transduced with MAML1(13-74)-GFP or GFP retrovirus at titers such that only a subpopulation of cells (∼40 to 70%) were transduced. The percentage of GFP+ cells was determined daily by flow cytometry, gating for live cells by forward/side scatter criteria. The percentage of GFP+ cells remaining at each day is expressed as a fraction of the initial (day 3 posttransduction) GFP+ percentage. All cell lines except BJAB were used in three independent experiments; a single representative experiment is shown. (b) Absolute growth rates of SUPT-T1 and BW5147 cultures in which >95% of cells had been transduced by the indicated retroviruses. Cell counts were performed daily starting at day 2 postretroviral transduction, and extrapolated cell counts were calculated as described in Materials and Methods. (c) Cell cycle effects of MAML1(13-74)-GFP. DNA content was measured from the SUP-T1 and BW5147 cultures depicted in panel b on day 7 posttransduction (corresponding to day 5 in panel b). Cells were stained with propidium iodide and were analyzed by flow cytometry. MamGFP, MAML1(13-74)-GFP; GFP, GFP only.
To define the growth-suppressive effects of MAML1(13-74)-GFP virus further, we measured absolute growth rates and performed cell cycle analysis on SUP-T1 and BW5147 cultures that contained >95% retrovirally transduced, GFP+ cells. Human SUP-T1 cells transduced with MAML1(13-74)-GFP virus showed a decreased absolute growth rate (Fig. 8b), an increased G1/G0 fraction, and a decreased S-phase fraction (Fig. 8c) compared to those of GFP-only control cells, whereas no difference was seen with BW5147 cells.
DISCUSSION
These studies provide strong evidence that NOTCH1-induced T-ALLs require persistent NOTCH1 signaling for growth and survival. NOTCH signaling was inhibited at two distinct steps. In one set of experiments, presenilin inhibitors prevented cleavage and subsequent nuclear translocation of ICN, leading to growth inhibition and death of a ΔE-expressing murine T-ALL cell line. In a second set of experiments, MAML1 dominant-negative peptides were used to inhibit nuclear NOTCH1, causing growth inhibition and death of Notch1-induced human and murine T-ALL cell lines. Together these findings show that signals transduced by nuclear NOTCH1 are required for growth and survival of Notch1-transformed pre-T cells.
The observation that ΔE proteolysis and nuclear translocation are required for proliferation and survival of T6E cells has several implications. ΔE-expressing T6E cells have levels of nuclear ICN1 that are below the sensitivities of standard antibody-based detection methods (40). Nevertheless, ΔE is a potent activator of CSL-dependent reporter genes (3) and an inducer of T-ALL in our murine model (40), apparently because presenilin-dependent processing leads to inappropriately high levels of nuclear ICN1. In this regard it is noteworthy that, despite convincing evidence that nuclear access is essential for NOTCH function (25, 49, 52, 53), it is difficult or impossible to detect NOTCH in the nucleus of normal cells. It follows that subtle increases in nuclear levels of NOTCH could have an important impact on the behavior of malignant cells.
Other observations also provide support for this possibility. Enforced expression of the NOTCH ligand DELTA-LIKE-4 in bone marrow cells produces T-ALL in mice (61), implying that transformation can occur merely through inappropriate activation of otherwise normal NOTCH receptors. Ligand-mediated NOTCH signaling also stimulates the growth of lymphoid cell lines derived from classical Hodgkin's lymphoma and anaplastic large-cell lymphoma (30). Many mutations producing gain-of-function phenotypes in invertebrates consist of single amino acid substitutions within extracellular portions of NOTCH receptors (11, 22, 34). In each case, relatively small increases in nuclear NOTCH arising through metalloprotease- and presenilin-mediated proteolytic cleavages (akin to the situation in ΔE-induced T-ALLs) likely produce the observed phenotypes. Presenilin inhibitors may be useful in screening cell lines and primary tumors for evidence of ongoing NOTCH processing (on the basis of accumulation of ΔE*, as depicted in Fig. 6a) and dependence on NOTCH nuclear access for growth and survival (58), using the strategy of ICN rescue to control for NOTCH-independent effects. Moreover, NOTCH signals can also transform primary baby hamster kidney cells (9) and murine mammary epithelial cells (46), indicating that screens for Notch activity will have utility in nonhematopoietic tumors as well.
The requirement for very low levels of nuclear ICN1 in ΔE-expressing T6E cells also points out an incongruity in NOTCH signaling relevant to transformation and development. Expression constructs encoding ΔE and ICN1 are equipotent inducers of T-ALL in our murine model, despite large differences in the levels of nuclear ICN1 produced by these two alleles (40). Furthermore, T-ALLs arising from bone marrow cells transduced with ICN1 alleles almost uniformly arise from a GFP-bright, ICN1hi cell population (4), suggesting that high levels of engineered ICN1 are required for efficient transformation. A similar paradox has been observed in the developing Drosophila eye, where ΔE causes more pronounced phenotypes than ICN despite the presence of substantially more nuclear NOTCH in ICN-expressing cells (19). These data suggest ΔE-derived forms of ICN are more potent on a molecule-for-molecule basis in activating downstream signals. Conceivably, during or subsequent to proteolysis ΔE might undergo phosphorylation or other modifying events that augment downstream signaling with greater efficiency than engineered ICN polypeptides. One such modifier could be SEL-5, a serine/threonine kinase that acts upstream of nuclear events to enhance NOTCH signaling in C. elegans (18).
Once ICN1 reaches the nucleus, recruitment of coactivators is important for strong activation of CSL and T-cell transformation. By using a murine model of leukemogenesis, it was previously noted that the minimal transforming portion of ICN1 includes the ANK domain and a C-terminal TAD (4). The same portion of ICN1 is sufficient to rescue T-ALL cells from presenilin inhibition, suggesting that the signals required for growth ex vivo are similar or identical to those required for transformation in vivo.
Likely roles for the ANK domain, which is required for all NOTCH functions, are to form weak contacts with CSL (3, 31, 54) and to recruit MAML1 (60). A critical role for MAML1 binding is supported by growth suppression of NOTCH1-induced T-ALLs by dominant-negative MAML1, a potent inhibitor of CSL activation. These effects could indicate either that MAML recruitment is critical for CSL activation or that multiple, different coactivators are loaded onto ICN through the same contact site. In support of the former possibility, recent work using an in vitro transcription system showed that ICN1 activation of CSL-dependent transcription requires MAML1 (20), which may serve as a docking site for p300 (20) and other uncharacterized coactivators (60). Activation of CSL in vitro also appears to require the ICN1 TAD (20). This same domain is necessary for strong CSL activation in reporter gene assays (4, 32, 33) and may serve to recruit a different class of coactivators (32). Thus, CSL/ICN1/MAML1 is likely an essential subcomplex within a larger multiprotein assembly. In accordance with this prediction, Capobianco's group recently identified and partially purified CSL, ICN1, and MAML1 together in a ∼1.5-MDa complex in SUP-T1 cells (28).
Our findings demonstrate that NOTCH signaling influences the growth potential of transformed T-ALL cells directly, indicating that the mechanism of transformation extends beyond effects on differentiation. NOTCH1 signaling inhibitors suppress the growth of NOTCH1-induced T-ALL lines by perturbing cell cycle progression and inducing apoptosis. NOTCH1 was shown previously to upregulate cyclin D1 in BHK cells (47) and to rescue T-cell lines from glucocorticoid-induced apoptosis (13). It will be of interest to determine the molecular mechanisms underlying growth suppression and apoptosis in NOTCH1-transformed T cells in which both nuclear translocation of ICN1 and recruitment of coactivators are important for transduction of growth and survival signals. Thus, these studies provide proof of principle for the targeting of nuclear Notch complexes in the treatment of NOTCH-induced T-ALL. Furthermore, the Notch-specific inhibitors described herein may prove useful in determining the importance of NOTCH signaling in other forms of T-ALL and in other cancers as well.
Acknowledgments
A.P.W. was supported by an NIH postdoctoral training grant. This work was also supported by Public Health Service grants to J.C.A., S.C.B., J.G., and W.S.P. from the National Cancer Institute. W.S.P. is a Scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284**:**770-776. [DOI] [PubMed] [Google Scholar]
- 2.Aster, J., W. Pear, R. Hasserjian, H. Erba, F. Davi, B. Luo, M. Scott, D. Baltimore, and J. Sklar. 1994. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp. Quant. Biol. 59**:**125-136. [DOI] [PubMed] [Google Scholar]
- 3.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J. Biol. Chem. 272**:**11336-11343. [DOI] [PubMed] [Google Scholar]
- 4.Aster, J. C., L. Xu, F. G. Karnell, V. Patriub, J. C. Pui, and W. S. Pear. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol. Cell. Biol. 20**:**7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berezovska, O., C. Jack, P. McLean, J. C. Aster, C. Hicks, W. Xia, M. S. Wolfe, W. T. Kimberly, G. Weinmaster, D. J. Selkoe, and B. T. Hyman. 2000. Aspartate mutations in presenilin and gamma-secretase inhibitors both impair notch1 proteolysis and nuclear translocation with relative preservation of notch1 signaling. J. Neurochem. 75**:**583-593. [DOI] [PubMed] [Google Scholar]
- 6.Berry, L. W., B. Westlund, and T. Schedl. 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124**:**925-936. [DOI] [PubMed] [Google Scholar]
- 7.Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5**:**207-216. [DOI] [PubMed] [Google Scholar]
- 8.Bush, G., G. diSibio, A. Miyamoto, J. B. Denault, R. Leduc, and G. Weinmaster. 2001. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 229**:**494-502. [DOI] [PubMed] [Google Scholar]
- 9.Capobianco, A. J., P. Zagouras, C. M. Blaumueller, S. Artavanis-Tsakonas, and J. M. Bishop. 1997. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17**:**6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlesso, N., J. C. Aster, J. Sklar, and D. T. Scadden. 1999. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood 93**:**838-848. [PubMed] [Google Scholar]
- 11.De Celis, J. F., and S. J. Bray. 2000. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development 127**:**1291-1302. [DOI] [PubMed] [Google Scholar]
- 12.Deftos, M. L., Y. W. He, E. W. Ojala, and M. J. Bevan. 1998. Correlating notch signaling with thymocyte maturation. Immunity 9**:**777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schruvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent-secretase-like protease mediates release of Notch intracellular domain. Nature 398**:**518-522. [DOI] [PubMed] [Google Scholar]
- 14.Dovey, H. F., V. John, J. P. Anderson, L. Z. Chen, P. de Saint Andrieu, L. Y. Fang, S. B. Freedman, B. Folmer, E. Goldbach, E. J. Holsztynska, K. L. Hu, K. L. Johnson-Wood, S. L. Kennedy, D. Kholodenko, J. E. Knops, L. H. Latimer, M. Lee, Z. Liao, I. M. Lieberburg, R. N. Motter, L. C. Mutter, J. Nietz, K. P. Quinn, K. L. Sacchi, P. A. Seubert, G. M. Shopp, E. D. Thorsett, J. S. Tung, J. Wu, S. Yang, C. T. Yin, D. B. Schenk, P. C. May, L. D. Altstiel, M. H. Bender, L. N. Boggs, T. C. Britton, J. C. Clemens, D. L. Czilli, D. K. Dieckman-McGinty, J. J. Droste, K. S. Fuson, B. D. Gitter, P. A. Hyslop, E. M. Johnstone, W. Y. Li, S. P. Little, T. E. Mabry, F. D. Miller, and J. E. Audia. 2001. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J. Neurochem. 76**:**173-181. [DOI] [PubMed] [Google Scholar]
- 15.Doyle, T. G., C. Wen, and I. Greenwald. 2000. SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97**:**7877-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66**:**649-661. [DOI] [PubMed] [Google Scholar]
- 17.Esler, W. P., W. T. Kimberly, B. L. Ostaszewski, W. Ye, T. S. Diehl, D. J. Selkoe, and M. S. Wolfe. 2002. Activity-dependent isolation of the presenilin-gamma-secretase complex reveals nicastrin and a gamma substrate. Proc. Natl. Acad. Sci. USA 99**:**2720-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fares, H., and I. Greenwald. 1999. SEL-5, a serine/threonine kinase that facilitates lin-12 activity in Caenorhabditis elegans. Genetics 153**:**1641-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortini, M. E., I. Rebay, L. A. Caron, and S. Artavanis-Tsakonas. 1993. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature 365**:**555-557. [DOI] [PubMed] [Google Scholar]
- 20.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16**:**1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Go, M. J., D. S. Eastman, and S. Artavanis-Tsakonas. 1998. Cell proliferation control by Notch signaling in Drosophila development. Development 125**:**2031-2040. [DOI] [PubMed] [Google Scholar]
- 22.Greenwald, I., and G. Seydoux. 1990. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346**:**197-199. [DOI] [PubMed] [Google Scholar]
- 23.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265**:**92-95. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16**:**952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huppert, S. S., A. Le, E. H. Schroeter, J. S. Mumm, M. T. Saxena, L. A. Milner, and R. Kopan. 2000. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 405**:**966-970. [DOI] [PubMed] [Google Scholar]
- 26.Izon, D. J., J. A. Punt, L. Xu, F. G. Karnell, D. Allman, P. S. Myung, N. J. Boerth, J. C. Pui, G. A. Koretzky, and W. S. Pear. 2001. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity 14**:**253-264. [DOI] [PubMed] [Google Scholar]
- 27.Jeffries, S., and A. J. Capobianco. 2000. Neoplastic transformation by Notch requires nuclear localization. Mol. Cell. Biol. 20**:**3928-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffries, S., D. J. Robbins, and A. J. Capobianco. 2002. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol. Cell. Biol. 22**:**3927-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jehn, B. M., W. Bielke, W. S. Pear, and B. A. Osborne. 1999. Protective effects of notch-1 on TCR-induced apoptosis. J. Immunol. 162**:**635-638. [PubMed] [Google Scholar]
- 30.Jundt, F., I. Anagnostopoulos, R. Forster, S. Mathas, H. Stein, and B. Dorken. 2002. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood 99**:**3398-3403. [DOI] [PubMed] [Google Scholar]
- 31.Kato, H., Y. Taniguchi, H. Kurooka, S. Minoguchi, T. Sakai, S. Nomura-Okazaki, K. Tamura, and T. Honjo. 1997. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124**:**4133-4141. [DOI] [PubMed] [Google Scholar]
- 32.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275**:**17211-17220. [DOI] [PubMed] [Google Scholar]
- 33.Kurooka, H., K. Kuroda, and T. Honjo. 1998. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 26**:**5448-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieber, T., S. Kidd, E. Alcamo, V. Corbin, and M. W. Young. 1993. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7**:**1949-1965. [DOI] [PubMed] [Google Scholar]
- 35.Logeat, F., C. Bessia, C. Brou, O. LeBail, S. Jarriault, N. G. Seidah, and A. Israel. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95**:**8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, B., J. C. Aster, R. P. Hasserjian, F. Kuo, and J. Sklar. 1997. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol. Cell. Biol. 17**:**6057-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuno, K., M. J. Go, X. Sun, D. S. Eastman, and S. Artavanis-Tsakonas. 1997. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development 124**:**4265-4273. [DOI] [PubMed] [Google Scholar]
- 38.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228**:**151-165. [DOI] [PubMed] [Google Scholar]
- 39.Nofziger, D., A. Miyamoto, K. M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126**:**1689-1702. [DOI] [PubMed] [Google Scholar]
- 40.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183**:**2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petcherski, A. G., and J. Kimble. 2000. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405**:**364-368. [DOI] [PubMed] [Google Scholar]
- 42.Pui, J. C., D. Allman, L. Xu, S. DeRocco, F. G. Karnell, S. Bakkour, J. Y. Lee, T. Kadesch, R. R. Hardy, J. C. Aster, and W. S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11**:**299-308. [DOI] [PubMed] [Google Scholar]
- 43.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10**:**547-558. [DOI] [PubMed] [Google Scholar]
- 44.Ramain, P., K. Khechumian, L. Seugnet, N. Arbogast, C. Ackermann, and P. Heitzler. 2001. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr. Biol. 11**:**1729-1738. [DOI] [PubMed] [Google Scholar]
- 45.Rebay, I., R. J. Fleming, R. G. Fehon, L. Cherbas, P. Cherbas, and S. Artavanis-Tsakonas. 1991. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67**:**687-699. [DOI] [PubMed] [Google Scholar]
- 46.Robbins, J., B. J. Blondel, D. Gallahan, and R. Callahan. 1992. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J. Virol. 66**:**2594-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronchini, C., and A. J. Capobianco. 2001. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol. Cell. Biol. 21**:**5925-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166**:**368-379. [DOI] [PubMed] [Google Scholar]
- 49.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393**:**382-386. [DOI] [PubMed] [Google Scholar]
- 50.Seiffert, D., J. D. Bradley, C. M. Rominger, D. H. Rominger, F. Yang, J. E. Meredith, Jr., Q. Wang, A. H. Roach, L. A. Thompson, S. M. Spitz, J. N. Higaki, S. R. Prakash, A. P. Combs, R. A. Copeland, S. P. Arneric, P. R. Hartig, D. W. Robertson, B. Cordell, A. M. Stern, R. E. Olson, and R. Zaczek. 2000. Presenilin-1 and -2 are molecular targets for gamma-secretase inhibitors. J. Biol. Chem. 275**:**34086-34091. [DOI] [PubMed] [Google Scholar]
- 51.Shawber, C., D. Nofziger, J. J. Hsieh, C. Lindsell, O. Bogler, D. Hayward, and G. Weinmaster. 1996. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122**:**3765-3773. [DOI] [PubMed] [Google Scholar]
- 52.Struhl, G., and A. Adachi. 1998. Nuclear access and action of notch in vivo. Cell 93**:**649-660. [DOI] [PubMed] [Google Scholar]
- 53.Struhl, G., and I. Greenwald. 2001. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc. Natl. Acad. Sci. USA 98**:**229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tani, S., H. Kurooka, T. Aoki, N. Hashimoto, and T. Honjo. 2001. The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res. 29**:**1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taniguchi, Y., H. Karlstrom, J. Lundkvist, T. Mizutani, A. Otaka, M. Vestling, A. Bernstein, D. Donoviel, U. Lendahl, and T. Honjo. 2002. Notch receptor cleavage depends on but is not directly executed by presenilins. Proc. Natl. Acad. Sci. USA 99**:**4014-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, S., S. Younger-Shepherd, L. Y. Jan, and Y. N. Jan. 1997. Only a subset of the binary cell fate decisions mediated by Numb/Notch signaling in Drosophila sensory organ lineage requires Suppressor of Hairless. Development 124**:**4435-4446. [DOI] [PubMed] [Google Scholar]
- 57.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296**:**2215-2218. [DOI] [PubMed] [Google Scholar]
- 58.Wolfe, M. S. 2001. gamma-Secretase inhibitors as molecular probes of presenilin function. J. Mol. Neurosci. 17**:**199-204. [DOI] [PubMed] [Google Scholar]
- 59.Wolfe, M. S., W. Xia, B. L. Ostaszewski, T. S. Diehl, W. T. Kimberly, and D. J. Selkoe. 1999. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 398**:**513-517. [DOI] [PubMed] [Google Scholar]
- 60.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of drosophila mastermind, is a transcriptional coactivator for NOTCH receptors. Nat. Genet. 26**:**484-489. [DOI] [PubMed] [Google Scholar]
- 61.Yan, X. Q., U. Sarmiento, Y. Sun, G. Huang, J. Guo, T. Juan, G. Van, M. Y. Qi, S. Scully, G. Senaldi, and F. A. Fletcher. 2001. A novel Notch ligand, Dll4, induces T-cell leukemia/lymphoma when overexpressed in mice by retroviral-mediated gene transfer. Blood 98**:**3793-3799. [DOI] [PubMed] [Google Scholar]