The VHL protein recruits a novel KRAB-A domain protein to repress HIF-1α transcriptional activity (original) (raw)
Abstract
The von Hippel-Lindau tumor suppressor (pVHL) is a component of an E3 ubiquitin ligase and targets hypoxia-inducible factor-1α (HIF-1α) for ubiquitylation and degradation under normoxic conditions. pVHL also directly inhibits HIF-1α transactivation by recruiting histone deacetylases. Here, we report a novel pVHL-interacting protein that functions as a negative regulator of HIF-1α transactivation. This protein, generated from the ZnF197 locus by alternative splicing, contains a Kruppel-associated box (KRAB)-A domain and a SCAN domain, but lacks the 22 C2H2-type zinc fingers present in ZnF197. Therefore, we named this protein pVHL-associated KRAB-A domain-containing protein (VHLaK). We demonstrate that the KRAB-A domain in VHLaK mediates pVHL binding and functions as a transcriptional repression module. The SCAN domain mediates VHLaK homo-oligomerization, which enhances VHLaK repressive activity. pVHL can recruit VHLaK to repress HIF-1α transcriptional activity and HIF-1α-induced VEGF expression. Finally, we demonstrate that pVHL, VHLaK and KAP1/TIF-1β can be recruited into a single complex, indicating that KAP1/TIF-1β may participate in pVHL-mediated transcriptional repression of HIF-1α. Our findings provide a novel mechanism for the modulation of HIF-1α transactivation by pVHL.
Keywords: HIF-1α/KRAB domain/VHL/VHLaK/ZnF197
Introduction
von Hippel-Lindau (VHL) disease is a hereditary cancer syndrome that predisposes affected individuals to a variety of tumors including hemangioblastoma of the CNS and retina, clear cell carcinoma of the kidney and pheochromocytoma of the adrenal gland (Maher and Kaelin, 1997). This disease is caused by germline mutations in the VHL tumor suppressor gene (Stolle et al., 1998). The VHL gene is also inactivated in ∼50–80% of sporadic renal clear cell carcinoma (Foster et al., 1994; Gnarra et al., 1994; Shuin et al., 1994). The VHL protein (pVHL) has been demonstrated to be a component of an E3 ubiquitin ligase complex, which also consists of elongin C (EC), elongin B (EB), cullin-2 (CUL-2) and Rbx1 (Pause et al., 1997; Kamura et al., 1999; Skowyra et al., 1999). This multi-protein complex exhibits structural and functional similarity to the SCF (Skp1/Cdc53 or Cullin/F-box) ubiquitin ligase complex (Pause et al., 1997; Iwai et al., 1999; Kamura et al., 1999; Lisztwan et al., 1999; Skowyra et al., 1999; Tyers and Rottapel, 1999). pVHL contains two domains, an α-domain and a β-domain (Stebbins et al., 1999); whereas the α-domain serves as the EC-binding site, the β-domain plays a role in the substrate recognition (Pause et al., 1997; Iwai et al., 1999; Kamura et al., 1999; Lisztwan et al., 1999; Skowyra et al., 1999; Stebbins et al., 1999; Tyers and Rottapel, 1999).
Hypoxia-inducible factor-1 (HIF-1), consisting of an α-subunit and a β-subunit, is a sequence-specific DNA-binding transcriptional complex that regulates genes involved in angiogenesis (Wang et al., 1995; Semenza, 2000). It has been demonstrated that pVHL E3 ligase can ubiquitylate HIF-1α at normoxic conditions via physical interaction with the core of the HIF-1α oxygen-dependent degradation domain (ODDD) (Kaelin, 1999; Maxwell et al., 1999; Cockman et al., 2000; Kamura et al., 2000; Krek, 2000; Ohh et al., 2000). Without functional pVHL, HIF-1α can accumulate in cells and result in over-expression of HIF-1 target genes, such as vascular endothelial growth factor (VEGF) (Tyers and Rottapel, 1999). The C-terminus of HIF-1α contains ODDD sequences that mediate O2-dependent ubiquitylation of HIF-1α (Huang et al., 1998; Sutter et al., 2000). Binding of pVHL is dependent on the hydroxylation of Pro564 within the HIF-1α ODDD sequence via an enzymatic process that requires O2 as well as iron (Ivan et al., 2001; Jaakkola et al., 2001). Recently, several HIF-1α prolyl hydoxylase enzymes have been identified as being responsible for this post-translational modification (Bruick et al., 2001; Epstein et al., 2001). Besides mediating the ubiquitylation and degradation of HIF-1α, pVHL has been shown to function as a transcriptional repressor to directly inhibit the transactivation function of HIF-1α by recruiting histone deacetylases (Mahon et al., 2001). HIF-1α transcriptional activity can also be suppressed under normoxic conditions via hydroxylation of an asparagine residue within its C-terminal transactivation domain by FIH-1, an asparaginyl hydroxylase enzyme (Lando et al., 2002a). This modification blocks HIF-1α association with transcription co-activators, such as p300 (Lando et al., 2002b).
The Kruppel-associated box (KRAB) is a conserved motif found at the N-termini of proteins containing Kruppel-type C2H2 zinc fingers (Bellefroid et al., 1991). It has been suggested that the KRAB domain is a protein–protein interaction motif and possesses transcriptional repression function (Margolin et al., 1994; Witzgall et al., 1994). The KRAB domain spans ∼75 amino acids and is divided into an A and a B box. It has been shown that the KRAB-A domain itself harbors the transcriptional repression activity and the KRAB-B box seems to be dispensable (Margolin et al., 1994; Witzgall et al., 1994; Vissing et al., 1995). This domain is predicted to form an amphipathic helix that may interact with components of the basal transcriptional factor complex or other cellular proteins (Bellefroid et al., 1991). In fact, this has been strongly supported by the finding that the KRAB-A domain can physically interact with a RING domain-containing protein known as KAP1 (KRAB-associated protein-1)/KRIP-1 (KRAB-A interacting protein)/TIF-1β (transcription intermediary factor 1β), and KAP1 can enhance KRAB-A-mediated repression and silence the transcription activity when KAP1–KRAB-domain protein complex directly tethers to DNA (Friedman et al., 1996; Kim et al., 1996; Moosmann et al., 1996). This silencing activity may result from recruitment of histone deacetylase complexes (HDAC), such as N-CoR-1 (Underhill et al., 2000) and Mi-2α (Schultz et al., 2001), and/or from association with members of the heterochromatin protein 1 (HP1) family (Le Douarin et al., 1996; Nielsen et al., 1999; Ryan et al., 1999; Lechner et al., 2000), a family of non-histone heterochromatin-associated proteins with an established gene-silencing function (Eissenberg and Elgin, 2000). Recently, a novel KRAB domain-containing protein (RBaK) has been shown to function as a transcriptional repressor by interacting with RB to repress E2F-dependent genes and prevent DNA synthesis (Skapek et al., 2000).
In this study, we have identified a novel KRAB-A domain-containing protein that can physically interact with pVHL. We designated this protein as VHLaK (pVHL-associated KRAB-A domain-containing protein) in accordance with RBaK. Cloning the full-length cDNA of human VHLaK reveals that this protein contains a SCAN domain and a KRAB-A domain. VHLaK is generated from the ZnF197 locus through alternative splicing and does not contain the 22 C2H2-type zinc finger motifs present in the ZnF197 protein. We have determined that the pVHL-interacting region is located in the KRAB-A domain of the VHLaK protein. The KRAB-A domain is responsible for the transcriptional repression function of VHLaK, and the homo-oligomerization mediated by SCAN domain further enhances this repression function. We show that pVHL can recruit VHLaK to repress the HIF-1α transcriptional activity and HIF-1α-induced VEGF expression. Finally, we demonstrate that VHLaK can recruit both KAP1/TIF-1β and pVHL simultaneously, indicating that KAP1/TIF-1β may participate in pVHL-mediated transcriptional repression of HIF-1α.
Results
Identification of the VHLaK protein as a novel pVHL-interacting protein by yeast two-hybrid screening using pVHL19 as a bait
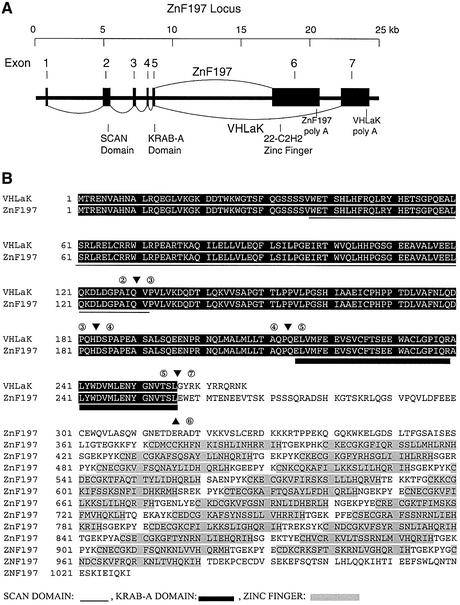
After screening 5.5 × 106 clones, two of our positive cDNA clones were identical and ∼2.3 kb in length. DNA sequencing revealed that these two clones encoded 112 amino acids followed by a 1.9-kb 3′ untranslated region (UTR). The 5′-end coding regions of these two clones were partially matched to a portion of ZnF197 sequence deposited in the DDBJ/EMBL/GenBank database (accession No. NM_006991). A yeast two-hybrid assay using independent co-transformation of the bait vector and these candidate clones, followed by growth selection and a β-galactosidase assay, further confirmed the interaction between pVHL and these candidate clones (data not shown). Using 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR method, we determined the full-length cDNA sequence of this transcript. Multiple in-frame stop codons are present in the 5′-end of this transcript, indicating that the full-length coding region has been obtained. The full-length transcript contains 2975 nucleotides and encodes an open reading frame (ORF) of 267 amino acids with a predicted mol. wt of 30 kDa (DDBJ/EMBL/GenBank accession No. AY074878). Analysis of genomic DNA sequences deposited in the DDBJ/EMBL/GenBank database revealed that this transcript is an alternatively spliced isoform from the ZnF197 locus (Figure 1A). The ZnF197 protein, originally named ZnF20 protein with unknown function, was identified from rapid turnover transcripts over-expressed in thyroid tumors (Gonsky et al., 1997). A SCAN and a KRAB-A domain exist in both the putative protein of this new transcript and the ZnF197 protein. The two proteins differ in their C-termini, with the ZnF197 protein containing 22 C2H2-type zinc finger motifs (Figure 1B). Since the putative protein encoded by this new transcript contains the KRAB-A domain but no zinc fingers, we designated this protein as pVHL-associated KRAB-A domain- containing protein (VHLaK) in accordance with a recently identified RBaK (RB associated KRAB domain- containing protein) (Skapek et al., 2000). The ZnF197 gene spans 23 kb and consists of seven exons. Alternative splicing of exon 6 and exon 7 generates ZnF197 and VHLaK transcripts, respectively (Figure 1A). This gene is located in chromosome 3 clone AC069071, which is mapped to chromosome 3p21, a frequently rearranged region in many cancers including renal cell carcinoma. Northern blot and RT–PCR results and human expressed sequence tag (EST) database search show that both ZnF197 and VHLaK are widely expressed in human tissues (Supplementary data, available at The EMBO Journal Online). Like the ZnF197 mRNA, the 3′-UTR of the VHLaK mRNA also contains three copies of the AUUUA motif, which indicates that the VHLaK transcript is also subjected to rapid turnover (Gonsky et al., 1997). A polyclonal antibody raised against the full-length VHLaK protein recognizes two bands on a western blot of HeLa cell extract. One 120-kDa band corresponds to the size of ZnF197, while another 35-kDa band is consistent with the size of VHLaK (Supplementary data).
Fig. 1. Gene structure and alternative splicing of the ZnF197 locus, amino acid sequence alignment of VHLaK and ZnF197 proteins. (A) Exon–intron structure and alternative splicing of the ZnF197 gene. The ZnF197 gene consists of seven exons and generates VHLaK and ZnF197 transcripts by alternative splicing of exons 1–5 then exon 7, and exons 1–6, respectively. The corresponding coding regions for the SCAN domain, KRAB-A domain, and 22 C2H2 zinc finger motifs are indicated. (B) Amino acid sequence alignment of VHLaK and ZnF197 proteins. The arrowheads and numbers above and below the amino acids indicate the corresponding splicing junctions and exon numbers. The SCAN domain and KRAB-A domain are underlined using different lines. The 22 C2H2 zinc finger motifs of ZnF197 are shaded. Note that the VHLaK protein contains a short C-terminus and lacks all 22 C2H2 zinc finger motifs present in the ZnF197 protein.
The VHLaK protein interacts with pVHL through its KRAB-A domain
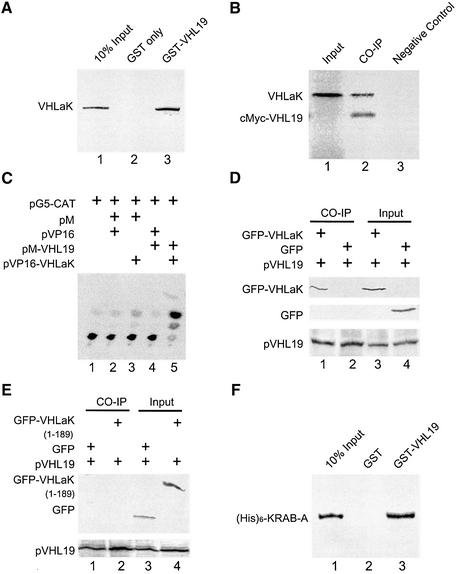
The association of full-length VHLaK with pVHL was tested in vitro using a GST pull-down assay. The 35S-labeled VHLaK specifically bound to the GST–pVHL19 fusion protein, but not to GST alone (Figure 2A), indicating a physical association between these two proteins in vitro. The association of the VHLaK protein with pVHL was also tested by in vitro co-immunoprecipitation. The 35S-labeled VHLaK and c-Myc-fused pVHL19 (residues 54–213) were transcribed and translated separately in vitro. After interacting with c-Myc-fused pVHL, VHLaK was precipitated by an anti-c-Myc antibody (Figure 2B). To determine whether pVHL and VHLaK could interact in mammalian cells, a mammalian two-hybrid assay was performed. As shown in Figure 2C, the expression of chloramphenicol acetyl transferase enzyme was stimulated by the interaction between these two proteins. The interaction between VHLaK and pVHL was also tested by co-immunoprecipitation from protein extracts prepared from COS-7 cells co-transfected with pEGFP-C3-VHLaK and pCMX-pVHL19. GFP–VHLaK could be co-immunoprecipitated by an anti-VHL antibody (Figure 2D). These results strongly support our conclusion that the VHLaK protein is a pVHL-interacting partner.
Fig. 2. The VHLaK protein interacts with pVHL through its KRAB-A domain. (A) Analysis of the interaction between the VHLaK protein and pVHL by GST pull-down assay. Five microliters of in vitro translated [35S]methionine-labeled VHLaK protein were incubated with GST–pVHL19 or GST-coated beads. The eluted solutions and 10% of input protein were resolved on a 10% SDS–PAGE gel. (B) In vitro co-immunoprecipitation of c-Myc-pVHL19 with the VHLaK protein. Five microliters of in vitro translated c-Myc-pVHL19 and VHLaK (lane 2) or 5 µl of in vitro translated VHLaK protein alone (lane 3) were incubated at 4°C. The proteins were immunoprecipitated with anti-c-Myc antibody and analyzed on a 10% SDS–PAGE gel. In lane 1, 0.5 µl of in vitro translated VHLaK protein were loaded into the gel as a marker. (C) Analysis of the interaction between the VHLaK protein and pVHL in a mammalian two-hybrid assay. In lane 5, pM-pVHL19 and pVP16-VHLaK were co-transfected into COS-7 cells together with reporter plasmid pG5CAT and cells were lysed for chloramphenicol acetyltransferase (CAT) assay. CAT activity was measured using a thin-layer chromatogram and 14C-labeled chloramphenicol as substrate. Other lanes served as negative controls. (D) In vivo co-immunoprecipitation of pVHL with GFP–VHLaK fusion protein. Lysates prepared from COS-7 cells transfected with pCMX-VHL19 and pEGFP-C3 or pEGFP-C3-VHLaK were subjected to immunoprecipitation (IP) with an anti-VHL antibody followed by anti-GFP or anti-pVHL immunoblotting. (E) Mapping pVHL-interacting region in the VHLaK protein by in vivo co-immunoprecipitation. Lysates prepared from COS-7 cells transfected with pCMX-VHL19 and pEGFP-C3 or pEGFP-C3-VHLaK (amino acids 1–189) were subjected to immunoprecipitation (IP) with an anti-VHL antibody followed by anti-GFP or anti-VHL immunoblotting. (F) Mapping pVHL-interacting region in VHLaK by GST pull-down assay. Five microliters of in vitro translated [35S]methionine- labeled His6-KRAB-A protein were incubated with GST–pVHL19 or GST-coated beads. The eluted solutions and 10% of input protein were resolved on a 15% SDS–PAGE gel.
Since the initially isolated pVHL-interacting clones from yeast two-hybrid screening contain the KRAB-A domain, we examined whether this domain was responsible for the pVHL-binding. In contrast to Figure 2D, GFP-fused truncated VHLaK (amino acids 1–189) without the KRAB-A domain could not be immunoprecipitated by an anti-pVHL antibody (Figure 2E). A GST pull-down assay result further demonstrated that the 35S-labeled His6-KRAB-A specifically bound to the GST–pVHL19 fusion protein, but not to GST alone (Figure 2F), indicating a physical association in vitro. Taken together, we conclude that the KRAB-A domain is responsible for the interaction with pVHL.
The KRAB-A domain of VHLaK exhibits transcriptional repression activity independent of HDAC activity and the oligomerization mediated by the SCAN domain further enhances this repression activity
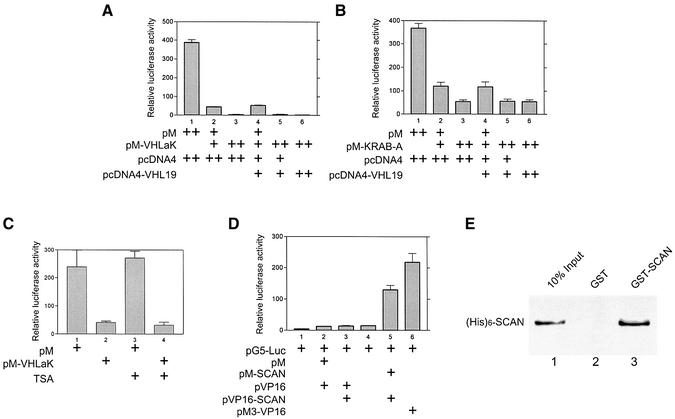
Since KRAB domains in other zinc-finger proteins have been suggested to function as a transcription repressor module, we examined whether the KRAB-A domain of VHLaK possesses transcriptional repression activity. The full-length VHLaK or its KRAB-A domain were fused in-frame to the GAL4 DNA-binding domain and were expressed together with reporter plasmid pGAL4tkLUC, which contains five GAL4 binding sites upstream of the luciferase start site. Co-expression of the full-length VHLaK or the KRAB-A domain with pGAL4tkLUC potently decreased the expression level of the reporter gene in a dose-dependent manner, confirming the repressive activity of the KRAB-A domain of VHLaK (Figure 3A and B). More interestingly, the full-length VHLaK exhibits much stronger repression than the KRAB-A domain alone, indicating that domains other than the KRAB-A domain may regulate this repression function (Figure 3A and B). Moreover, the transcriptional repression activity is independent of HDAC activity, as demonstrated by the fact that trichostatin A, which can inhibit all histone deacetylases, has no effect on the transcriptional repression of VHLaK (Figure 3C).
Fig. 3. The KRAB-A domain of VHLaK exhibits transcriptional repression activity independent of HDAC activity and the homo-oligomerization mediated by the SCAN domain enhances this repressive activity. (A and B) Transcriptional repression activity of the VHLaK protein and its KRAB-A domain. Various doses of GAL4-DBD-VHLaK (pM-VHLaK) (A) and of GAL4-DBD-KRAB-A (pM-KRAB) (B), along with pVHL expression plasmid (pcDNA4-VHL19), 300 ng of pGAL4-tk-LUC and 10 ng pRLSV40-LUC, were transfected into COS-7 cells. Twenty-four hours after transfection, cells were harvested and measured for firefly and Renilla activity. The firefly luciferase activity was normalized against Renilla luciferase activity for each sample, and mean values of relative luciferase activity were calculated from triplicate wells. The experiments were repeated three times to ensure consistency. (C) Transcriptional repression activity of VHLaK protein is independent of HDAC activity. Lanes 3 and 4, 18 h after transfection, the cells were treated with 300 nM of trichostatin A (TSA) for another 18 h and then the cells were lysed for luciferase activity assay. (D) Analysis of self-interaction of the SCAN domain in mammalian two-hybrid assay. Lane 5, pM-SCAN and pVP16-SCAN were co-transfected into COS-7 cells together with reporter plasmid pG5-LUC and cells were lysed for luciferase activity assay. Lane 6, pM3-VP16 served as a positive control. (E) Analysis of the self-interaction of the SCAN domain by GST pull-down assay. Five microliters of in vitro translated [35S]methionine-labeled His6-SCAN protein were incubated with GST–SCAN or GST-coated beads. The eluted solutions and 10% of input protein were resolved on a 15% SDS–PAGE gel.
The SCAN domain located upstream of the KRAB-A domain is a highly conserved, leucine-rich motif that can mediate the protein–protein interactions based on its predicted amphipathic secondary structure. Recent studies have illustrated that the SCAN domain mediates a selective homotypic or heterotypic oligomerization and may represent a regulatory mechanism of transcriptional activity (Williams et al., 1999; Schumacher et al., 2000). To determine whether the SCAN domain of VHLaK could mediate homo-oligomerization of VHLaK, we fused the SCAN domain of VHLaK to GAL4-DBD and VP16-AD, and performed a mammalian two-hybrid assay. Co-expression of GAL4-DBD-SCAN and VP16-AD-SCAN significantly stimulates the expression of the reporter gene pG5LUC (Figure 3D). To further demonstrate the self-interaction of the SCAN domain, a GST pull-down assay was performed. The 35S-labeled His6-SCAN specifically bound to the GST–SCAN fusion protein, but not to GST alone (Figure 3E), indicating a physical association in vitro. These results confirmed that the SCAN domain of VHLaK could mediate the homo-oligomerization of VHLaK. Together with the above results, this suggests that the homo-oligomerization of VHLaK could further enhance the transcriptional repression activity of VHLaK.
The VHLaK protein enhances pVHL-mediated suppression of HIF-1α transcriptional activity
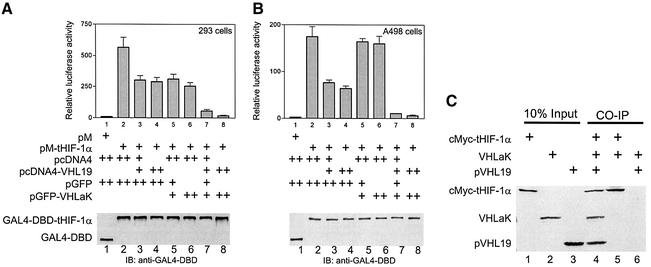
To determine whether VHLaK could function as a negative regulator with pVHL to suppress HIF-1α transactivation, we examined the effects of VHLaK and pVHL on the transactivation activity of HIF-1α. GAL4-DBD fused to the transactivation region of HIF-1α (amino acids 530–826) and reporter plasmid pG5LUC were transfected into HEK 293 cells or human renal cell carcinoma A498 cells, which harbor VHL mutation, with or without pVHL or/and VHLaK. The results showed that pVHL exhibits repression effects on the HIF-1α transactivation in both cell lines (Figure 4A and B, lanes 3 and 4). VHLaK alone exhibited no significant effect on the transactivation activity of HIF-1α in A498 cells (without wild-type pVHL) (Figure 4B, lanes 5 and 6). In contrast, VHLaK alone showed an inhibitory effect in HEK 293 cells (with wild-type pVHL) (Figure 4A, lanes 5 and 6). More strikingly, co-expression of pVHL and VHLaK resulted in a much more significant repression on HIF-1α transactivation (Figure 4A and B, lanes 7 and 8). As demonstrated by immunoblot analysis (immunoblot panels in Figure 4A and B), the protein levels of GAL4-DBD-fused truncated HIF-1α (amino acids 530–826), which does not contain the entire domain (amino acids 429–608) required for O2-dependent ubiquitylation and degradation (Huang et al., 1998; Sutter et al., 2000), was not affected by over-expression of VHLaK and/or pVHL in these experiments. This indicated that the transcriptional repression demonstrated in our experiments was not caused by decreased HIF-1α level. To illustrate more directly that pVHL could recruit VHLaK to HIF-1α, an in vitro co-immunoprecipitation assay was performed. As shown in Figure 4C, VHLaK could be immunoprecipitated with the c-Myc-fused truncated HIF-1α (amino acids 530–826) in the presence of pVHL by an anti-c-Myc antibody. In contrast, VHLaK could not be precipitated with the c-Myc-fused truncated HIF-1α in the absence of pVHL (Figure 4C). Taken together, our experiments suggest that the VHLaK protein and pVHL repress HIF-1α transactivation and that pVHL-mediated recruitment of VHLaK to HIF-1α is required for the transcriptional repression.
Fig. 4. VHLaK enhances pVHL-mediated repression of HIF-1α transcriptional activity. (A and B) GAL4-DBD-tHIF-1α expression plasmid (pM-tHIF-1α) together with various doses of pVHL expression plasmid (pcDNA4-VHL19) and/or VHLaK expression plasmids were transfected into HEK 293 cells (A) and A498 cells (B) along with 300 ng of pG5-LUC and 10 ng pRLSV40-LUC. Twenty-four hours after transfection, cells were harvested and measured for luciferase activity. The experiments were repeated three times to ensure consistency. (C) VHLaK can be recruited into pVHL–HIF-1α complex. VHLaK, pVHL19 and cMyc-tHIF-1α (amino acids 530–826) were expressed in the presence of [35S]methionine. Co-immunoprecipitation was performed using an anti-c-Myc antibody. Note that the VHLaK protein can only be co-immunoprecipitated with cMyc- tHIF-1α in the presence of pVHL.
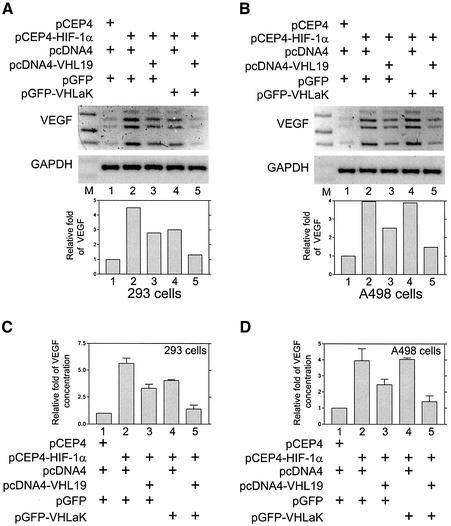
The VHLaK protein inhibits VEGF mRNA and protein expression in the presence of pVHL
Since the transcription factor HIF-1α activates VEGF transcription, we investigated whether VHLaK possesses any inhibitory effect on endogenous VEGF expression in HEK 293 cells and pVHL-null renal cell carcinoma cell line, A498 cells. Our results demonstrated that over-expression of HIF-1α could significantly increase the VEGF mRNA expression (Figure 5A and B, lanes 1) and VEGF secretion (Figure 5C and D, lanes 1) and the co-transfection of pVHL and VHLaK significantly decreased the VEGF mRNA expression (Figure 5A and B, lanes 5) and VEGF protein secretion levels (Figure 5C and D, lanes 5). In contrast, pVHL alone had some inhibitory effect on the VEGF mRNA and protein levels in both HEK 293 cells and A498 cells (Figure 5A–D, lanes 3). While VHLaK alone exhibited an inhibitory effect on the VEGF mRNA and protein level in HEK 293 cells (Figure 5A and C, lanes 4), in pVHL-null A498 cells, VHLaK alone did not show any inhibitory effect (Figure 5B and D, lanes 4). These results suggest that VHLaK participates in the down-regulation of VEGF expression and that this activity requires the presence of the wild-type pVHL.
Fig. 5. The VHLaK protein inhibits VEGF mRNA and protein expression in the presence of pVHL. (A and B) the VHLaK protein inhibits HIF-1α- induced VEGF mRNA expression in the presence of pVHL. HEK 293 cells (A) and A498 cells (B) were transfected with the plasmids as indicated. Thirty-six hours after transfection, total RNA was extracted and semi-quantitative RT–PCR was performed using primers specific for VEGF or GAPDH. Note that four bands were observed representing four VEGF isoforms (VEGF 121, 165, 189 and 206). The densities of VEGF bands were quantified and normalized with the density of GAPDH bands. Molecular size markers were loaded in lane M. (C and D) The VHLaK protein inhibits HIF-1α-induced VEGF protein expression in the presence of pVHL. HEK 293 cells (C) and A498 cells (D) were transfected with the plasmids as indicated. Thirty-six hours after transfection, aliquots of medium were collected for VEGF ELISA and results were normalized for cell numbers. Histograms represent the results of three independent experiments.
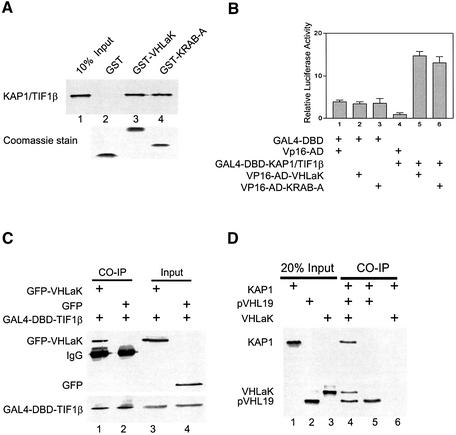
The VHLaK protein interacts with KAP1/TIF-1β through its KRAB-A domain and the VHLaK protein can recruit pVHL and KAP1 simultaneously
It has been reported that the KRAB-A domain in several other KRAB domain-containing proteins can physically interact with KAP1/KRIP-1/TIF-1β and enhance KRAB-A-mediated repression (Friedman et al., 1996; Kim et al., 1996; Moosmann et al., 1996). We further examined whether the repressive activity of pVHL and VHLaK is associated with KAP1/TIF-1β. Our results showed that GST-fused VHLaK or KRAB-A proteins could bind to the in vitro translated KAP1/TIF-1β protein (Figure 6A), suggesting that VHLaK can interact with KAP1/TIF-1β and the KRAB-A domain of VHLaK is responsible for this interaction. Our mammalian two-hybrid assay further demonstrated that VHLaK and KAP1/TIF-1β could interact with each other in vivo (Figure 6B). In the in vivo co-immunoprecipitation assay, GFP–VHLaK fusion protein could be co-precipitated with GAL4-DBD-TIF-1β fusion protein by anti-GAL4-DBD antibody, but not GFP alone (Figure 6C). To investigate whether VHLaK can interact with both pVHL and KAP1 simultaneously, an in vitro co-immunoprecipitation assay was performed. As shown in Figure 6D, KAP1 could be co-immunoprecipitated with pVHL19 in the presence of VHLaK by an anti-VHL antibody. In contrast, KAP1 could not be precipitated with pVHL19 in the absence of VHLaK (Figure 4D). Further analysis using all endogenous proteins demonstrated that both endogenous VHL and KAP1 proteins can be precipitated by anti-VHLaK antibody (Figure 6E). Together, these results demonstrate that the VHLaK protein can recruit KAP1/TIF-1β or vice versa, indicating that KAP1/TIF-1β is involved in pVHL-mediated suppression of HIF transactivation function through VHLaK.
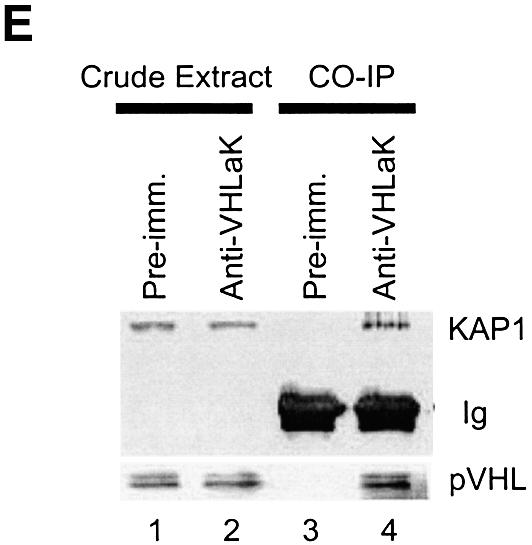
Fig. 6. The VHLaK protein can recruit KAP1/TIF-1β through its KRAB-A domain and both endogenous VHL and KAP1/TIF-1β proteins interact VHLaK simultaneously. (A) Analysis of the interaction between VHLaK and KAP1/TIF-1β by GST pull-down assay. Five microliters of in vitro translated [35S]methionine-labeled KAP1/TIF-1β protein were incubated with GST–VHLaK, GST–KRAB-A or GST-coated beads. The eluted solutions and 10% of input protein were resolved on an 8% SDS–PAGE gel. (B) The interaction between VHLaK and KAP1/TIF-1β in mammalian two-hybrid assay. Lanes 4 and 5, GAL4-DBD-KAP1/TIF-1β and pVP16-VHLaK or pVP16-KRAB-A were co-transfected into COS-7 cells, respectively. Thirty-six hours after transfection, cells were harvested and lysed for luciferase assay. (C) In vivo co-immunoprecipitation of KAP1/TIF-1β with VHLaK. Lysates prepared from COS-7 cells transfected with GAL4-DBD-KAP1/TIF-1β and pEGFP-C3 or pEGFP-C3-VHLaK were subjected to immunoprecipitation (IP) with an anti-GAL4-DBD antibody followed by anti-GFP or anti-GAL4-DBD immunoblotting. (D) Both pVHL and KAP1 can interact with VHLaK simultaneously. VHLaK, pVHL19, and KAP1 were expressed in the presence of [35S]methionine. Co-immunoprecipitation was performed using an anti-VHL antibody. Note that the KAP1 protein can only be co-immunoprecipitated with pVHL in the presence of VHLaK. (E) The endogenous VHLaK protein can recruit endogenous VHL and KAP1 proteins. The top panel shows a western blot analysis of crude extracts (lanes 1 and 2), control immunoprecipitates with the pre-immune serum (lane 3) or immunoprecipitates with anti-VHLaK antibody (lane 4) from HeLa cells immunoblotted with anti-KAP1 antibody; the bottom panel shows similar analysis of same crude extracts and immunoprecipitates immunoblotted with anti-VHL antibody.
Discussion
For most cells cultured under hypoxic conditions, there is a dramatic increase in the level of HIF-1α protein, which is achieved by stabilizing HIF-α subunits at low oxygen tensions (Huang et al., 1998; Sutter et al., 2000). Under normoxic conditions, HIF-α subunits are efficiently ubiquitylated by pVHL-E3 ubiquitin ligase and degraded by the proteasome (Maxwell et al., 1999; Cockman et al., 2000; Kamura et al., 2000; Ohh et al., 2000). It has been demonstrated that re-expression of pVHL in renal carcinoma cells carrying mutant pVHL is sufficient for HIF-1α destruction under normal oxygen conditions. The fact that loss of pVHL function is sufficient to activate the HIF system in the mouse liver, in Caenorhabditis elegans and in cultured Chinese hamster ovary-K1 cells suggests that this pVHL-dependent regulation of HIF activation is widely employed by different cells (Epstein et al., 2001; Haase et al., 2001; Vaux et al., 2001). Furthermore, loss of pVHL function leads to full, rather than partial, activation of HIF system, suggesting that pVHL could be involved in other regulatory aspects of the HIF system in response to change in oxygen tension. In fact, recent reports have demonstrated that FIH-1 is an asparaginyl hydroxylase that regulates HIF-1α transcriptional activity by modifying an asparagine residue in the C-terminal activation domain of HIF-1α (Lando et al., 2002a,b). These reports provided a unifying mechanism for the modulation of HIF-α protein stabilization and transcriptional activation in response to changes in cellular O2 concentration.
In this report, we add another new component to the complex regulatory system of HIF-1α function. In the present study, we have identified a novel pVHL-interacting protein, pVHL-associated KRAB-A domain-containing protein (VHLaK), which contains a SCAN domain and a KRAB-A domain. These structure motifs prompted us to investigate transcriptional regulation of VHLaK in association with pVHL. As demonstrated in our experiments, the KRAB-A domain of VHLaK exhibits transcriptional repression activity and this repressive activity is consistent with the function of KRAB-A domain described previously (Margolin et al., 1994; Witzgall et al., 1994; Vissing et al., 1995). We have demonstrated that SCAN domain-mediated oligomerization could further enhance this repressive activity. The transcriptional repression function of VHLaK in association with pVHL is probably mediated through recruiting KAP1 to pVHL by VHLaK.
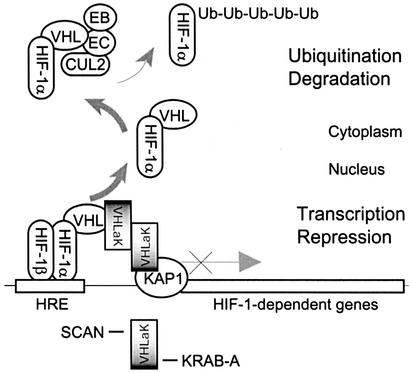
The KRAB-A domain is predicted to form an amphipathic helix, which may interact with components of the basal transcriptional factor complex or other cellular proteins (Bellefroid et al., 1991). The KRAB-A domain can physically interact with KAP1/KRIP-1/TIF-1β and KAP1 can enhance KRAB-A-mediated repression and silence transcription when directly tethered to DNA. The data presented in this study demonstrate that VHLaK can recruit KAP1 and that the transcriptional repression of VHLaK is independent of HDAC activity; this result is consistent with the report that the transcriptional repression mediated by KRAB-A-KAP1 may result from association with members of the HP1 family (Le Douarin et al., 1996; Nielsen et al., 1999; Ryan et al., 1999; Lechner et al., 2000), a family of non-histone heterochromatin-associated proteins with an established gene-silencing function (Eissenberg and Elgin, 2000). Based on the data presented in this study and previous findings, we propose that pVHL recruits VHLaK to HIF-1 transcription factor complex by interacting with the KRAB-A domain of VHLaK. VHLaK recruits more VHLaKs by its SCAN domain-mediated homo-oligomerization, and subsequently these VHLaKs are able to recruit KAP-1/KRIP-1/TIF-1β and HP1 family members to repress the transactivation activity of HIF-1α (Figure 7).
Fig. 7. Schematic diagram of the proposed mechanism that represses the HIF-1α transactivation by pVHL and the VHLaK protein. Based on the data presented in this study and previously published findings, we propose that pVHL not only mediates the ubiquitylation and degradation of HIF-1α in the cytoplasm, but also recruits the VHLaK protein, through interaction with the KRAB-A domain, to HIF-1α transcription factor complex. The VHLaK protein forms homo-oligomerization via its SCAN domain and can recruit KAP-1/KRIP-1/TIF-1β, a well- characterized transcriptional repressor, to repress the transcriptional activity of HIF-1α.
Based on genomic sequence analyses, VHLaK is an alternatively spliced isoform from the ZnF197 locus (Gonsky et al., 1997). ZnF197 contains the same SCAN domain and the KRAB-A domain, but differs from VHLaK at its C-terminus, which contains 22 C2H2-type zinc fingers. Although we did not provide any experimental data in this report, we expect that ZnF197 would interact with pVHL and KAP1/TIF-1β in the same or a similar manner, due to the same molecular structure in their N-termini. ZnF197 may bind to DNA directly through its zinc fingers and function as a transcription factor. Its higher molecular weight and 22 zinc fingers may also provide multiple interfaces for interacting with other proteins. Since VHLaK can form homo-oligomers through its SCAN domain, VHLaK may well be able to form a heterodimer with ZnF197 though the same mechanism. It will be of interest to determine whether VHLaK and ZnF197 can also suppress HIF-2α and HIF-3α transactivation activities.
Combining the fact that VHLaK functions as a repressor of HIF-1α transactivation function in a pVHL-dependent manner and, more interestingly, that the ZnF197 locus is located in chromosome 3p21, a region known frequently to be rearranged in renal cell carcinomas as well as other cancers (van den Berg and Buys, 1997), it is conceivable that VHLaK might be a potential tumor suppressor. VHLaK loss of function may contribute to the increase of HIF-1α transactivation activity, and therefore lead to the overproduction of HIF-1α-regulated downstream target genes such as VEGF in cancer cells. The VHL gene is inactivated in ∼50–80% of the sporadic form of renal clear cell carcinomas (Foster et al., 1994; Gnarra et al., 1994; Shuin et al., 1994). Whether VHLaK loss of function plays a role in the carcinogenesis of clear cell renal cell cancinomas or other cancer cells remains to be explored. Taken together, our findings provide a novel mechanism for the modulation of HIF-1α transactivation by pVHL. Further investigations are warranted in order to understand the complex functional relationship between VHLaK and other pVHL interacting partners, and whether VHLaK is involved in _VHL_-related tissue specific tumorigenesis.
Materials and methods
Yeast two-hybrid screening
Yeast two-hybrid system 3 (Clontech) was used according to the manufacturer’s protocol. The detailed method has been described previously (Li et al., 2002a,b).
RACE, PCR and sequence analysis
The 5′ and 3′ RACE experiments were performed using Marathon kidney cDNA Amplification kit (Clontech) according to the manufacturer’s protocol. The PCR products were subcloned into pCR2.1 vector (Invitrogen) and selected clones were sequenced to assemble the full-length cDNA. The putative protein sequences were analyzed by MacDNASIS software (Hitachi) and NCBI database searches.
Plasmid construction
Construction of pGBKT7-VHL19, pCMX-VHL19, pCDNA4-VHL19, pM-VHL19 and pGEX6p1-VHL19 was as described previously, and all these plasmids encode VHL protein residues from 54 to 213 (Li et al., 2002a, b). pSG5-VHLaK was prepared for expressing native VHLaK. pVP16-VHLaK, pM-VHLaK and pM-KRAB-A(217–257) was prepared for the luciferase activity assay. pEGFP-C3-VHLaK and pEGFP-C3-VHLaK(1–189) were constructed to express GFP–VHLaK or truncated VHLaK (amino acids 1–189) fusion protein. pGEX6p1-VHLaK and pGEX6p1-KRAB-A(217–257) were constructed to express GST fusion proteins in bacteria. pcDNA4-KRAB-A(217–257) was constructed for in vitro expression of His6-KRAB-A. pM-SCAN, pVP16-SCAN, pGEX-6p1-SCAN and pcDNA4-SCAN were constructed for oligomerization studies. _pM-tHIF-1_α (amino acids 530–826) and _pGBKT7-tHIF-1_α (amino acids 530–826) were constructed by inserting PCR-amplified HIF-1α fragment into the corresponding vectors. _pCEP4/HIF-1_α was kindly provided by Dr Gregg L.Semenza. _pcDNA3-KAP1/TIF-1_β and _GAL4-DBD-KAP1/TIF-1_β were kindly provided by Dr Frank J.Rauscher III and Dr Walter Schaffner, respectively. All sequences amplified by PCR were confirmed by complete sequencing.
Mammalian cell transfection and luciferase activity assay
COS-7 cells, HEK 293 and human renal cell carcinoma A498 cells were maintained in DMEM, RPMI and EMEM containing penicillin (25 U/ml), streptomycin (25 µg/ml) and 10% fetal bovine serum, respectively. Transient transfection was performed using SuperFect (Qiagen) transfection solution according to the manufacturer’s protocol. For the luciferase assay, cells seeded in 12-well plates were transfected with recombinant plasmids, internal control plasmid pRLSV40-LUC, and reporter plasmid pGAL4tkLUC or pG5LUC, both of which contain five consensus GAL4 binding sites and different promoters. Analyses of luciferase activity were performed according to the protocol of Dual Luciferase Assay System (Promega) and relative light units were measured using a luminometer.
Antibodies and immunoblotting
Anti-cMyc, anti-GAL4-DBD and anti-GFP monoclonal antibodies were obtained from Santa Cruz Co. Anti-VHL monoclonal, anti-KAP1 polyclonal and anti-HIF-1α monoclonal antibodies were purchased from PharMingen, Affinity Bioreagents and Novus Biologicals, respectively. Anti-VHLaK antisera were raised in rabbits against the purified GST–VHLaK full-length fusion protein (Covance), and further affinity-purified on a protein A column. The pre-bleed serum from the same rabbit was used as a pre-immune negative control. Samples for western blot analysis were resolved on SDS–PAGE gels and transferred to PVDF membrane (Millipore) for subsequent immunoblotting.
GST pull-down assay and co-immunoprecipitation
The detailed methods for GST pull-down assay and co-immunoprecipitation have been described previously (Li et al., 2002a,b).
Mammalian two-hybrid assay
Different combinations of plasmids together with reporter plasmid pG5CAT or pG5-Luc, were co-transfected into COS-7 cells with a constant amount of a second reporter, pCMV-GAL4 or pRLSV40-LUC, for normalizing the transfection efficiency. After 24 h, cells were harvested and lysed. The lysates were used for radioactive chloramphenicol acetyltransferase (CAT) assay or luciferase activity assay.
RNA isolation and RT–PCR
Total RNA was isolated using TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol. One microgram of total RNA from each sample was reverse transcribed using 200 U of MMLV reverse transcriptase (Promega) and 0.5 µg of oligo(dT)15 primer. The reactions were incubated at 42°C for 90 min. Five microliters of each RT product were used for PCR. The VEGF primers, 5′-CGAAGTGGTGAA GTTCATGGATG-3′ (sense) and 5′-TTCTGTATCAGTCTTTCCTGG TGA-3′ (antisense) and GAPDH primers, 5′-CCAAAACTGGCAATT CCATGGCA-3′ (sense) and 5′-TCTAGACGGCAGGTCAGGTCC ACC-3′ (antisense) were used in our experiments. The PCR program was 94°C for 5 min, 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min, followed by a final extension at 72°C for 7 min.
Determination of VEGF protein levels in cell culture medium by ELISA
Equal numbers of cells were plated on 6-well plates and grown overnight. Cells were transfected with different combinations of plasmids. Thirty-six hours after transfection, cell supernatants were collected, clarified by centrifugation at 500 g for 5 min, and stored at –70°C. Concomitantly, the total cell numbers in each well were determined after trypsinization. VEGF concentrations in medium were determined in duplicate by ELISA using the reagents and the protocol supplied with the Quantikine Human VEGF Immunoassay kit (R&D Systems). Differences in VEGF concentrations in different groups of cell medium were normalized for the total numbers of cells.
Accession number
The nucleotide sequence reported in this paper has been submitted to the DDBJ/EMBL/GenBank database under accession No. AY074878.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Gregg L.Semenza (McKusick-Nathans Institute of Genetic Medicine, The Johns Hopkins University School of Medicine) for his generous gift of the full-length HIF-1α expression plasmid. We thank Dr Frank J.Rauscher III (The Wistar Institute, PA) and Dr Walter Schaffner (Institut für Molekularbiologie der Universität Zurich-Irchel, Switzerland) for their generous gifts of the plasmids pcDNA3-KAP1 and GAL4-DBD-TIF-1β, respectively. We also thank Dr Pierre Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale/ULP/Collège de France, France) for his generous gift of the plasmid pASV3-TIF-1β.
References
- Bellefroid E.J., Poncelet,D.A., Lecocq,P.J., Revelant,O. and Martial,J.A. (1991) The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc. Natl Acad. Sci. USA, 88, 3608–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick R.K. and McKnight,S.L. (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science, 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- Cockman M.E. et al. (2000) Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem., 275, 25733–25741. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C. and Elgin,S.C. (2000) The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev., 10, 204–210. [DOI] [PubMed] [Google Scholar]
- Epstein A.C. et al. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell, 107, 43–54. [DOI] [PubMed] [Google Scholar]
- Foster K. et al. (1994) Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum. Mol. Genet., 3, 2169–2173. [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Fredericks,W.J., Jensen,D.E., Speicher,D.W., Huang,X.P., Neilson,E.G. and Rauscher,F.J.,III (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev., 10, 2067–2078. [DOI] [PubMed] [Google Scholar]
- Gnarra J.R. et al. (1994) Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet., 7, 85–90. [DOI] [PubMed] [Google Scholar]
- Gonsky R., Knauf,J.A., Elisei,R., Wang,J.W., Su,S. and Fagin,J.A. (1997) Identification of rapid turnover transcripts overexpressed in thyroid tumors and thyroid cancer cell lines: use of a targeted differential RNA display method to select for mRNA subsets. Nucleic Acids Res., 25, 3823–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase V.H., Glickman,J.N., Socolovsky,M. and Jaenisch,R. (2001) Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl Acad. Sci. USA, 98, 1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.E., Gu,J., Schau,M. and Bunn,H.F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin–proteasome pathway. Proc. Natl Acad. Sci. USA, 95, 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M. et al. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science, 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Iwai K., Yamanaka,K., Kamura,T., Minato,N., Conaway,R.C., Conaway,J.W., Klausner,R.D. and Pause,A. (1999) Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA, 96, 12436–12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P. et al. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science, 292, 468–472. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. Jr (1999) Cancer. Many vessels, faulty gene. Nature, 399, 203–204. [DOI] [PubMed] [Google Scholar]
- Kamura T. et al. (1999) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science, 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato,S., Iwai,K., Czyzyk-Krzeska,M., Conaway,R.C. and Conaway,J.W. (2000) Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl Acad. Sci. USA, 97, 10430–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.S., Chen,Y.M., O’Leary,E., Witzgall,R., Vidal,M. and Bonventre,J.V. (1996) A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc. Natl Acad. Sci. USA, 93, 15299–15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W. (2000) VHL takes HIF’s breath away. Nat. Cell Biol., 2, E121–E123. [DOI] [PubMed] [Google Scholar]
- Lando D., Peet,D.J., Gorman,J.J., Whelan,D.A., Whitelaw,M.L. and Bruick,R.K. (2002a) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev., 16, 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Peet,D.J., Whelan,D.A., Gorman,J.J. and Whitelaw,M.L. (2002b) Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science, 295, 858–861. [DOI] [PubMed] [Google Scholar]
- Lechner M.S., Begg,G.E., Speicher,D.W. and Rauscher,F.J.,III (2000) Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol., 20, 6449–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen,A.L., Garnier,J.M., Ichinose,H., Jeanmougin,F., Losson,R. and Chambon,P. (1996) A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J., 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Li Z., Na,X., Wang,D., Schoen,S.R., Messing,E.M. and Wu,G. (2002a) Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J. Biol. Chem., 277, 4656–4662. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang,D., Na,X., Schoen,S.R., Messing,E.M. and Wu,G. (2002b) Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem. Biophys. Res. Commun., 294, 700–709. [DOI] [PubMed] [Google Scholar]
- Lisztwan J., Imbert,G., Wirbelauer,C., Gstaiger,M. and Krek,W. (1999) The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin–protein ligase activity. Genes Dev., 13, 1822–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher E.R. and Kaelin,W.G.,Jr (1997) von Hippel-Lindau disease. Medicine (Baltimore), 76, 381–391. [DOI] [PubMed] [Google Scholar]
- Mahon P.C., Hirota,K. and Semenza,G.L. (2001) FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev., 15, 2675–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin J.F., Friedman,J.R., Meyer,W.K., Vissing,H., Thiesen,H.J. and Rauscher,F.J.,III (1994) Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl Acad. Sci. USA, 91, 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.H. et al. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–275. [DOI] [PubMed] [Google Scholar]
- Moosmann P., Georgiev,O., Le Douarin,B., Bourquin,J.P. and Schaffner,W. (1996) Transcriptional repression by RING finger protein TIF1β that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res., 24, 4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz,J.A., You,J., Oulad-Abdelghani,M., Khechumian,R., Gansmuller,A., Chambon,P. and Losson,R. (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J., 18, 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M., Park,C.W., Ivan,M., Hoffman,M.A., Kim,T.Y., Huang,L.E., Pavletich,N., Chau,V. and Kaelin,W.G. (2000) Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat. Cell Biol., 2, 423–427. [DOI] [PubMed] [Google Scholar]
- Pause A., Lee,S., Worrell,R.A., Chen,D.Y., Burgess,W.H., Linehan,W.M. and Klausner,R.D. (1997) The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl Acad. Sci. USA, 94, 2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R.F., Schultz,D.C., Ayyanathan,K., Singh,P.B., Friedman,J.R., Fredericks,W.J. and Rauscher,F.J.,III (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol., 19, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.C., Friedman,J.R. and Rauscher,F.J.,III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev., 15, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher C. et al. (2000) The SCAN domain mediates selective oligomerization. J. Biol. Chem., 275, 17173–17179. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. (2000) HIF-1 and human disease: one highly involved factor. Genes Dev., 14, 1983–1991. [PubMed] [Google Scholar]
- Shuin T. et al. (1994) Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res., 54, 2852–2855. [PubMed] [Google Scholar]
- Skapek S.X., Jansen,D., Wei,T.F., McDermott,T., Huang,W., Olson,E.N. and Lee,E.Y. (2000) Cloning and characterization of a novel Kruppel-associated box family transcriptional repressor that interacts with the retinoblastoma gene product, RB. J. Biol. Chem., 275, 7212–7223. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Koepp,D.M., Kamura,T., Conrad,M.N., Conaway,R.C., Conaway,J.W., Elledge,S.J. and Harper,J.W. (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science, 284, 662–665. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Kaelin,W.G.,Jr and Pavletich,N.P. (1999) Structure of the VHL–ElonginC–ElonginB complex: implications for VHL tumor suppressor function. Science, 284, 455–461. [DOI] [PubMed] [Google Scholar]
- Stolle C. et al. (1998) Improved detection of germline mutations in the von Hippel-Lindau disease tumor suppressor gene. Hum. Mutat., 12, 417–423. [DOI] [PubMed] [Google Scholar]
- Sutter C.H., Laughner,E. and Semenza,G.L. (2000) Hypoxia-inducible factor 1α protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc. Natl Acad. Sci. USA, 97, 4748–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. and Rottapel,R. (1999) VHL: a very hip ligase. Proc. Natl Acad. Sci. USA, 96, 12230–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C., Qutob,M.S., Yee,S.P. and Torchia,J. (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem., 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- van den Berg A. and Buys,C.H. (1997) Involvement of multiple loci on chromosome 3 in renal cell cancer development. Genes Chromosomes Cancer, 19, 59–76. [DOI] [PubMed] [Google Scholar]
- Vaux E.C., Wood,S.M., Cockman,M.E., Nicholls,L.G., Yeates,K.M., Pugh,C.W., Maxwell,P.H. and Ratcliffe,P.J. (2001) Selection of mutant CHO cells with constitutive activation of the HIF system and inactivation of the von Hippel-Lindau tumor suppressor. J. Biol. Chem., 276, 44323–44330. [DOI] [PubMed] [Google Scholar]
- Vissing H., Meyer,W.K., Aagaard,L., Tommerup,N. and Thiesen,H.J. (1995) Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett., 369, 153–157. [DOI] [PubMed] [Google Scholar]
- Wang G.L., Jiang,B.H., Rue,E.A. and Semenza,G.L. (1995) Hypoxia-inducible factor 1 is a basic-helix–loop–helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA, 92, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.J., Blacklow,S.C. and Collins,T. (1999) The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell. Biol., 19, 8526–8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzgall R., O’Leary,E., Leaf,A., Onaldi,D. and Bonventre,J.V. (1994) The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl Acad. Sci. USA, 91, 4514–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]