HIV reproducibly establishes a latent infection after acute infection of T cells in vitro (original) (raw)
Abstract
The presence of latent reservoirs has prevented the eradication of human immunodeficiency virus (HIV) from infected patients successfully treated with anti-retroviral therapy. The mechanism of postintegration latency is poorly understood, partly because of the lack of an in vitro model. We have used an HIV retroviral vector or a full-length HIV genome expressing green fluorescent protein to infect a T lymphocyte cell line in vitro and highly enrich for latently infected cells. HIV latency occurred reproducibly, albeit with low frequency, during an acute infection. Clonal cell lines derived from latent populations showed no detectable basal expression, but could be transcriptionally activated after treatment with phorbol esters or tumor necrosis factor α. Direct sequencing of integration sites demonstrated that latent clones frequently contain HIV integrated in or close to alphoid repeat elements in heterochromatin. This is in contrast to a productive infection where integration in or near heterochromatin is disfavored. These observations demonstrate that HIV can reproducibly establish a latent infection as a consequence of integration in or near heterochromatin.
Keywords: chromatin/HIV/latent infection/Tat/transcriptional silencing
Introduction
Combination anti-retroviral therapy can control HIV-1 replication and delay disease progression. However, despite the complete suppression of detectable viremia in many patients, viremia re-emerges rapidly after interruption of treatment, consistent with the existence of a latent viral reservoir (Finzi et al., 1997; Wong et al., 1997). This reservoir is thought to consist mainly of latently infected resting memory CD4+ T cells (recently reviewed by Chun and Fauci, 1999; Butera, 2000; Pierson et al., 2000). Due to the long half-life of this reservoir (44 months), it has been estimated that its total eradication with current treatment would require over 60 years (Finzi et al., 1999).
Latently infected cells contain replication-competent integrated HIV-1 genomes that are blocked at the transcriptional level, resulting in the absence of viral protein expression. HIV depends on both cellular and viral factors for efficient transcription of its genome, and the activity of the HIV promoter is tightly linked to the level of activation of its host cell. HIV transcription is characterized by two temporally distinct phases. The early phase occurs immediately after integration and relies solely on cellular transcription factors. Because of a transcriptional elongation defect in the basal HIV promoter, most transcripts cannot elongate efficiently and terminate rapidly after initiation. However, a few transcripts elongate throughout the genome, resulting in transcription of the viral transactivator Tat. The late phase of transcription occurs when enough Tat protein has accumulated. Tat binds to TAR, recruits the pTEFb complex and causes the hyperphosphorylation of RNA polymerase II, dramatically increasing its ability to elongate (Karn, 1999).
To understand how postintegration latency is established, an in vitro cell system reflecting the state of HIV-1 latency is required. While several latently infected cell lines have been established after HIV infection, the proviruses integrated in these cell lines harbored mutations in their Tat–TAR transcriptional axis (Emiliani et al., 1996, 1998) or in NF-κB binding sites in the HIV promoter (Antoni et al., 1994). The presence of mutations in HIV-infected latent cell lines has raised questions about the significance of these lines to latency in vivo. However, the very presence of these mutations has also strengthened the concept that transcription inhibition is critical to the establishment and maintenance of HIV latency.
We reported previously that the site of integration of the HIV-1 provirus into the cell genome affects its basal Tat-independent transcriptional activity (Jordan et al., 2001). We therefore predicted that a small fraction of integration sites might lead to such low basal promoter activity that no Tat mRNA would accumulate. Such a provirus would be locked in the early phase of transcription, resulting in functional latency. In this manuscript, we have used recombinant viruses that express green fluorescent protein (GFP) to selectively enrich for rare cells blocked at the transcriptional level during an acute infection. Using this approach, we have isolated a large number of clonal cell lines harboring HIV in a latent state. The latent provirus can be reactivated at the transcriptional level in these cell lines by a variety of agents. This study documents that HIV reproducibly establishes a latent infection with low frequency after acute infection of T cells in vitro.
Results
Establishment of an in vitro model of HIV-1 latency
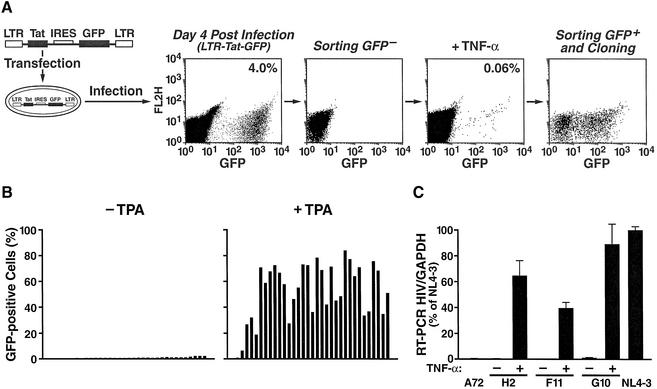
To determine whether unique integration events can lead to latency, we used an HIV-based retroviral vector containing the Tat and GFP open reading frames both under the control of the HIV promoter in the 5′ long terminal repeat (LTR). We infected a culture of the lymphocytic cell line Jurkat with viral particles containing this vector and used differential fluorescence-activated cell sorting (FACS) based on GFP expression (Figure 1A). First, we infected Jurkat cells with the LTR–Tat– IRES–GFP virus at a low m.o.i. and isolated GFP-negative cells by FACS 4 days after infection (Figure 1A). This population presumably harbored both uninfected cells and cells with transcriptionally silenced proviruses. To activate HIV expression, we treated this population with TPA or tumor necrosis factor α (TNF-α) and purified GFP-positive cells by FACS (Figure 1A). These cells, representing <0.06% of the original population, corresponded to the latent phenotype: GFP-negative under basal conditions and GFP-positive after activation with TPA or TNF-α. By comparing the proportions of productively infected cells (4%) and cells exhibiting a latent phenotype (0.06%), we calculate that ∼1.5% of infections (1 in 66) resulted in a latent state in this system. These cells were both grown as a group and individually sorted for further characterization. Re-analysis 17 days after sorting showed that a significant proportion of the cells had no GFP expression, indicating transcriptional silencing in the absence of TPA (Figure 1A).
Fig. 1. Enrichment and purification of HIV-latently infected cells by FACS. (A) A schematic representation of our enrichment protocol is shown. See text for details. The percentage of GFP-positive cells obtained after infection (4%) or after TNF-α treatment of GFP-negative cells (0.06%) is shown. Similar data were obtained with TPA. (B) Clonal cell lines isolated using the procedure described above were analyzed for GFP expression under basal and stimulated conditions (24 h treatment with TNF-α). (C) mRNA levels were measured for HIV and for GAPDH using TaqMan PCR in untreated and TNF-α-treated cell lines. Results are expressed as a percentage of mRNA levels measured in Jurkat cells acutely infected with HIVNL4-3 (day 6 post-infection). Clone A72 is infected with an LTR–GFP construct (Jordan et al., 2001), clone H2 is infected with the LTR–Tat–IRES–GFP vector while clones F11 and G10 are infected with HIV-R7/E–/GFP.
Individual cells exhibiting a latent phenotype were cloned and clonal cell lines were further characterized. Flow cytometry analysis of individual clones 4 weeks after their isolation showed low basal GFP expression (Figure 1B, left panel). After TPA treatment, all clones were activated, and GFP levels reached a maximum that was relatively independent of basal GFP activity (Figure 1B, right panel). Similar results were obtained after stimulation with TNF-α (data not shown).
Examination of mRNA levels in these cell lines under repressed (–TNF-α) and activated (+TNF-α) conditions confirmed the presence of very low transcriptional levels under basal conditions and the activation of HIV transcription by TNF-α (Figure 1C, see clone H2). Importantly, HIV-specific transcript levels after TNF-α treatment were of the same order of magnitude as mRNA levels measured in Jurkat cells acutely infected with wild-type HIV (NL4-3; Figure 1C).
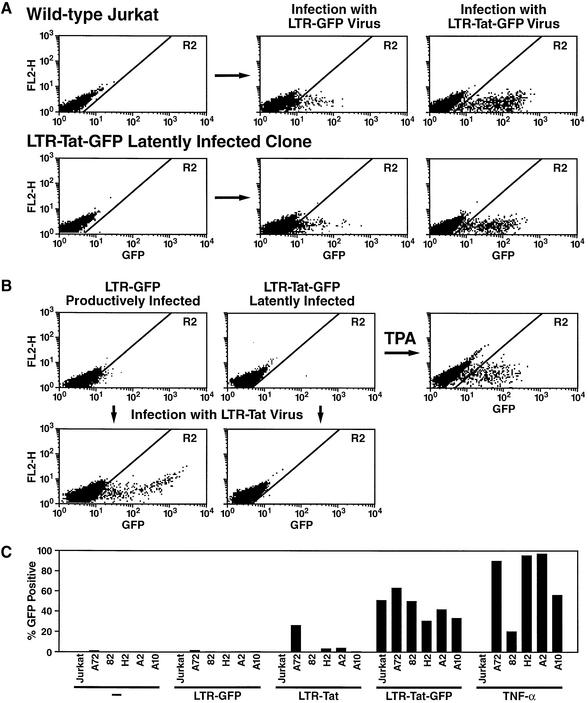
Mutations in a cellular factor important for HIV transcription, such as CDK9 or cyclin T1 for example, could be responsible for a latent phenotype. To test this possibility, we infected one of our latent cell lines (clone 82) and control Jurkat cells with an LTR–GFP virus or with the LTR–Tat–IRES–GFP virus (Figure 2A). Both cell lines produced similar amounts of GFP after infection with the LTR–GFP–virus (Figure 2A). GFP levels were higher but also comparable in both cell lines after infection with the LTR–Tat–GFP virus (Figure 2A). Similar results were observed in three other latent cell lines (clones H2, A2 and A10; Figure 2C).
Fig. 2. Characterization of latently infected clones. (A) Clone 82 is competent for basal and Tat-dependent HIV promoter activity. Clone 82 was infected with viral particles containing HIV-derived vectors LTR–GFP or LTR–Tat–IRES–GFP, and GFP expression was measured by flow cytometry. As a control, Jurkat cells were infected in parallel. (B) The integrated latent HIV promoter in clone 82 is unresponsive to Tat stimulation. Clone 82 was infected with a Tat-expression retroviral vector, or treated with TPA, as a positive control. As a control of the infection process and Tat expression, a clone containing a single integration of the HIV-derived LTR–GFP vector was infected in parallel. (C) HIV promoter activity (percentage of GFP-positive cells in R2 gate) was measured in four latent clonal cell lines (82, H2, A2 and A10) 24 h after infection with retroviral particles containing the following vectors: LTR–GFP, LTR–Tat, LTR–Tat–GFP or after treatment with TNF-α. Control Jurkat cells and a cell line containing a stably integrated LTR–GFP retroviral construct (Jordan et al., 2001) were used as controls.
This experiment demonstrates that the cellular environment in latent cell lines can support HIV transcription and Tat transactivation with the same efficiency as control Jurkat cells and is therefore not responsible for the latent phenotype.
HIV transcription is characterized by an early, Tat-independent phase and a late, Tat-dependent phase. Since no GFP was produced by our latent cell lines, it is likely that Tat was not expressed since both proteins are located on a polycistronic mRNA. To test whether Tat alone was sufficient to reactivate latent HIV gene expression, we infected clone 82 with a HIV-derived retroviral vector expressing the Tat protein (LTR–Tat virus; Figure 2B). Infection of a Jurkat clone containing a single copy of an integrated LTR–GFP retroviral vector (clone A, described in Jordan et al., 2001) with the same LTR–Tat virus stock showed high levels of GFP expression but had no effect in the latently infected cell line (Figure 2B). Similar results were obtained after transfection of a Tat-expression plasmid into these cell lines (data not shown). Similar results were also observed in three other latent cell lines (clones H2, A2 and A10) although weak stimulation of GFP expression was noted in clones H2 and A2 (Figure 1C). We conclude that the latent HIV promoter is relatively unresponsive to Tat stimulation, suggesting that the blockage of transcription in this clone lies primarily at the level of transcription initiation.
Preferential HIV integration in or near alphoid DNA in latently infected cells
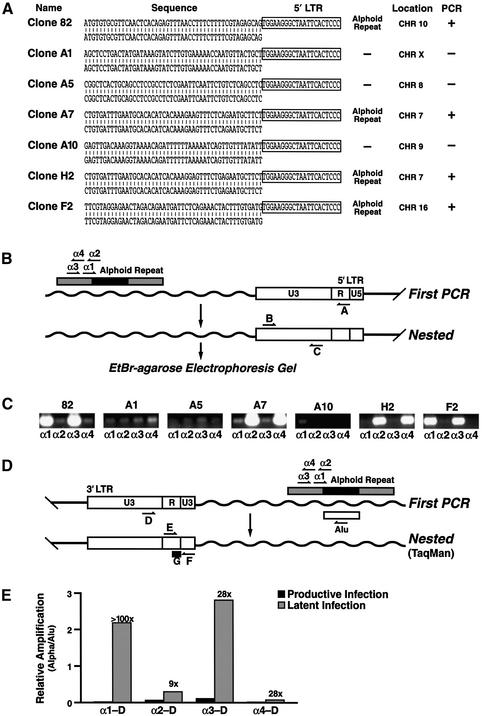
The basal activity of the HIV promoter is determined by a combination of _cis_- and _trans_-acting variables. The observations described above indicate that the cellular environment in each latent cell line is fully capable of supporting the transcription of the HIV genome and therefore points to the role of the site of integration of the provirus as a likely cause of low basal transcription. Accordingly, we isolated and sequenced the provirus integration sites in eight distinct clonal cell lines (Figure 3A) Mapping of the integration site on the human genome using BLAST showed that the retroviral vector had integrated in an alphoid repeat element in four out of eight clones on chromosomes 7, 10 and 16 (Figure 3A). The site of integration of three clones was identified in the human genome in non-alphoid repeat DNA while the site of integration in one clone did not yield significant homology to any region of the human genome (Figure 3A).
Fig. 3. Latency is associated with preferred integration in or near alphoid repeats. (A) The sequence of integration site of the HIV vector is shown for seven clonal cell lines aligned with the corresponding genomic sequence. The match in GenBank for each clone corresponded to DDBJ/EMBL/GenBank accession number M93288 for clone 82, AL591625 for clone A1, AC023948 for clone A5, M16037 for clone A7, Al354920 for clone A10, AC019063 for clone H2 and AC079801 for clone F2. The sequence corresponding to the HIV promoter is indicated by a closed box. The chromosomal location of each integration site is shown and integration sites into alphoid repeat elements are indicated. (B) A nested PCR assay designed to quantify integration in or near alphoid repeats is schematically represented. The position of primers in the two sequential PCRs are shown aligned with the HIV 5′ LTR and a putative alphoid element in the genome. (C) Four PCRs were conducted for each cell line using either primer α1, α2, α3 or α4 in combination with primer A for the first reaction and the primer pair B+C for the second PCR. PCR products amplified according to this scheme are shown for each latent cell line. (D) Real-time PCR analysis of integration in or near alphoid repeats. Primer G represents an internal fluorescent primer used for the quantification of the TaqMan reaction. (E) Quantification of PCR products after TaqMan PCR analysis as indicated in (D). Productively infected cells (GFP-positive cells in Figure 1A, panel 1; productive infection) are compared with latently infected cells (Figure 1A, panel 4; latent infection) using the Alu and alphoid PCR assay. Data is expressed as the relative signal intensity (alphoid/Alu). Numbers on top of bar indicate the ratio between latent and productive infection.
To confirm this observation, we developed a novel PCR assay in which degenerate primers matching alphoid DNA consensus sequence (α1, α2, α3, α4) are used in combination with an LTR specific primer (primer A; Figure 3A). Amplified products were detected with two nested primers in the LTR (primers B and C; Figure 3B). Since primers α1 and α3 are orientated in the same way in the alphoid repeat, a positive signal with one primer should be accompanied by a positive signal for the other (Figure 3B). The same holds true for primers α2 and α4 (Figure 3B). Similarly, a positive response with primers α1 and α3 would be accompanied by a negative signal for primers α2 and α4. Examination of the PCRs showed perfect concordance with the sequencing data: clones 82, A7, H2 and F2 showed a positive PCR and had a documented alphoid repeat integration (Figure 3C) while clones A1, A5, A10 (Figure 3C) and A11 (not shown) tested negative in the PCR assay and did not integrate in an alphoid element.
For comparison, we subjected DNA obtained from a library of 34 random integrations (not sorted for a latent phenotype) to the same procedure (Jordan et al., 2001). None of these 34 clones showed a pattern consistent with alphoid repeat integration, in agreement with previous reports that HIV integration in or near alphoid repeat elements is disfavored (Carteau et al., 1998).
To examine the relative distribution of alphoid integration in latently versus productively infected cells at a population level, we adapted the alphoid PCR assay to TaqMan PCR. Because Alu elements are randomly distributed within the genome, this analysis included a control using primers specific for the Alu element and the HIV LTR. The product amplified by this primer pair is assumed to represent random integration events within the genome and has been used as a reliable marker of HIV integration (Minami et al., 1995; Chun et al., 1997b; Butler et al., 2001). Results are therefore presented as the ratio of the alphoid–LTR product to the _Alu_–LTR product as an indication of the frequency of integration in or near alphoid DNA (Figure 3E). When cell populations corresponding to productively infected cells (GFP-positive in first panel of Figure 1A) were compared to latently infected cell population (GFP-positive in third panel of Figure 1A), we observed preferential integration (9-fold to >100-fold enrichment) in or near alphoid repeats in latent cells in comparison to productively infected cells (Figure 3E). Similar results were obtained after infection of human peripheral blood mononuclear cells (PBMCs) with the HIV-derived vector (data not shown). Cloning and sequencing of several PCR products obtained after amplification with the alphoid specific primers confirmed that these contained HIV integration events into alphoid DNA (data not shown).
Transcriptional activation of the HIV promoter in latently infected cells
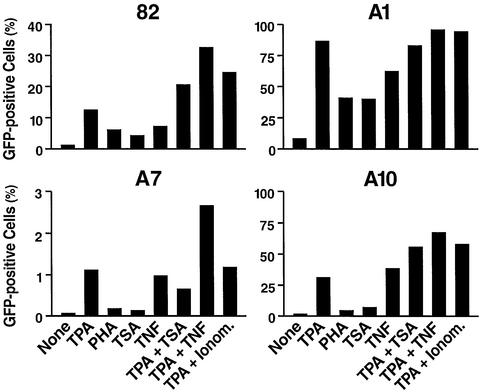
We have tested a number of biological and chemical agents for their ability to reactivate latent HIV expression. Several NF-κB activators, including TPA, TNF-α and PHA, independently induced HIV expression, as expected, since NF-κB strongly induces the HIV promoter activity. TPA was the strongest inducer tested (Figure 4). All treatments increased the number of GFP-positive cells and the mean fluorescence intensity reflective of GFP levels (data not shown). Incubation with anti-CD3 antibodies, which crosslink the surface T-cell receptors, an alternative pathway leading to NF-κB activation, also induced GFP expression (data not shown). These results suggest that activation of the NF-κB pathway may boost HIV transcription initiation, the production of Tat, and transition to Tat-dependent transcriptional activation. Trichostatin A (TSA), an inhibitor of histone deacetylases also activated expression but to a lesser extent than NF-κB activators and only in some cell lines (see clones A1 as an example, Figure 4). Treatment with 5-axa-2-deoxycytidine (aza-dC), an inhibitor of DNA methylation, had little effect on the fraction of cells induced to transcribe HIV alone or in combination with a histone deacetylase inhibitor (Groudine et al., 1981; Chen et al., 1997; data not shown).
Fig. 4. Transcriptional activation of the HIV promoter in latently infected cells. Cells from clones 82, A1, A7 and A10 were treated as described in Materials and methods with several indicated agents and LTR expression was measured by flow cytometry. Data are expressed as percentage of cells becoming GFP-positive after a 24 h treatment.
Latent cell lines containing a full-length integrated HIV genome
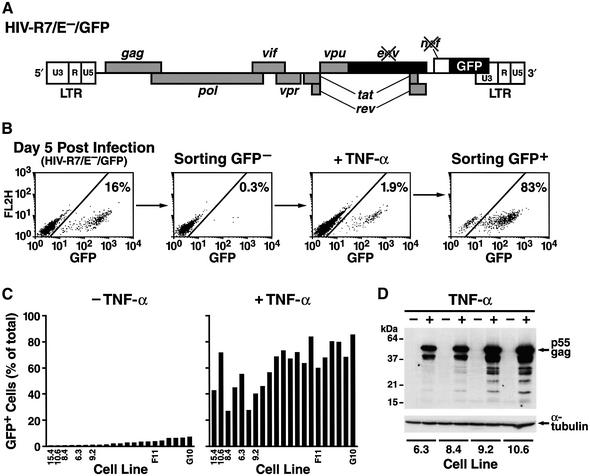
To confirm that HIV latency can be established in the context of a full-length provirus, we used a recombinant HIV molecular clone containing the GFP open reading frame in place of the Nef gene (Bieniasz and Cullen, 2000) (HIV-R7/E–/GFP; Figure 5A). To restrict our analysis to a single infection cycle, the env gene was suppressed by introduction of a frameshift mutation. This defect was complemented by coexpression of a VSV-G envelope protein to generate pseudotyped viral particles. We infected a culture of the lymphocytic cell line Jurkat with viral particles containing this HIV genome and used differential FACS based on GFP expression to isolate GFP-negative cells by FACS 4 days after infection (Figure 5B). Based on our previous experiments, we predicted that this population harbored both uninfected cells and cells with transcriptionally silenced proviruses. To activate HIV expression, we treated this population with TNF-α and observed that a small fraction of the cell population (1.9%) became positive. These activated cells were purified by FACS based on GFP expression levels (Figure 5B). These cells were both grown as a group and individually sorted for further characterization. Re-analysis after sorting showed that a small proportion of the cells had no GFP expression, indicating transcriptional silencing has occurred after withdrawal of TNF-α (Figure 5B). Flow cytometry analysis of individual clones showed low basal GFP expression (Figure 5C). After TNF-α treatment, HIV expression was increased in all clones both in terms of the fraction of cells that became GFP-positive (Figure 5C) but also in terms of mean fluorescence intensity (data not shown). Measurement of virus-specific mRNA showed that the mechanism of latency in these clones was controlled at the transcriptional level (Figure 1C, clones F11 and G10). Levels of viral mRNA after activation were similar to those measured after a productive HIV infection (compare clones F11 and G10 with NL4-3, Figure 1C). Similar results were obtained using northern blot analysis (data not shown). Analysis of HIV-specific protein expression in several clones by western blotting using an antiserum from an HIV-infected individual showed no detectable expression of HIV proteins under basal conditions (Figure 5D). Treatment of the same clones with TNF-α led to a dramatic increase in HIV protein expression, particularly the gag p55 precursor (Figure 5D). When the culture supernatants of the same clones were examined for HIV-specific p24 expression, no or low picogram amounts could be detected under basal conditions (Table I). Treatment with TNF-α led to a >1000-fold increase in p24 measurement for several representative clones (Table I). These observations demonstrate that transcriptional latency can also be established in the context of a full-length HIV infection.
Fig. 5. Establishment of latently infected cell lines with a full-length HIV provirus. (A) Genome organization of a molecular clone of HIV encoding GFP and containing a frameshift mutation in env. (B) Schematic representation of protocol for enrichment of latently infected cells after infection of Jurkat cells with HIV-R7/E–/GFP (see text for details). (C) Clonal cell lines isolated using the procedure described above were analyzed for GFP expression under basal and stimulated conditions (24 h treatment with TNF-α). (D) Western blot analysis of four representative Jurkat clones latently infected with HIV-R7/E–/GFP. Clones were treated for 24 h with TNF-α (10 ng/ml) and cell lysates were analyzed by western blotting using an antiserum from an HIV-infected individual (provided by the NIH AIDS Research and Reagent Reference Program). A predominant band of 55 kDa corresponds to the Gag precursor protein. The same samples were analyzed using an antiserum specific for α-tubulin to ensure equal loading.
Table I. Low to undetectable HIV protein expression in several latently infected clonal cell lines under basal conditions.
| | GFP-positive (%) | GFP signal (MFI) | p24 (pg/ml) | | | | | | ------------------- | ---------------- | ----------- | ------- | -------- | ------- | --------------- | | | TNF-α – | TNF-α + | TNF-α – | TNF-α + | TNF-α – | TNF-α + | | | Clone 15.4 | <1 | 46 ± 6 | 6 ± 0.5 | 188 ± 34 | 7 ± 12 | 6066 ± 1960 | | Clone 6.3 | <1 | 27 ± 9 | 5 ± 0.2 | 135 ± 43 | 0 | 10 100 ± 4573 | | Clone 8.4 | <1 | 77 ± 6 | 5 ± 0.3 | 488 ± 74 | 0 | 32 967 ± 10 537 | | Clone 9.2 | <1 | 75 ± 7 | 7 ± 0.5 | 522 ± 61 | 23 ± 6 | 41 067 ± 9100 | | Clone 10.6 | <1 | 96 ± 1 | 5 ± 1.4 | 645 ± 45 | 14 ± 3 | 85 500 ± 5981 |
Discussion
We present evidence that HIV can reproducibly lead to a state of transcriptional silencing and true postintegration latency. We have used a FACS-based protocol to highly enrich for latently infected cells and show that HIV integration leads with low frequency to integration sites that cannot support basal transcription of the HIV promoter. The low frequency of integration events leading to latency (∼1%) is likely to be the reason why this phenomenon has eluded discovery until now.
Integration into alphoid repetitive DNA, a component of centromeric heterochromatin, occurred frequently in our latent clone population. A recent comprehensive analysis of the site of integration of HIV during a productive infection showed that HIV integrates preferentially in actively transcribed genes (69% of integrations sites; Schroder et al., 2002). In the same study, α satellite integration represented <1% of all integration events (Schroder et al., 2002). In contrast, in our small sample of sequenced integration sites from latent infections, we observed 50% (four of eight) of integration sites in alphoid repeats. Analysis of a larger sample using the alphoid repeat PCR assay indicated that the true frequency of alphoid repeat integration is probably lower, between 20 and 30% in latent cells (data not shown).
Heterochromatin is found in transcriptionally inactive regions of the genome and is associated with specialized chromosome structures, such as centromeres and telomeres. It is characterized by a strongly condensed nucleosomal structure that impairs access to the underlying DNA for transcription factors and the basal transcriptional machinery. The unique properties of hetero chromatin are associated with the incorporation of particular histone variants, unique post-translational modifications of the histone tails and, heterochromatin-specific proteins (Wallrath, 1998; Jenuwein, 2001; Jenuwein and Allis, 2001). The transcriptional silencing effect of heterochromatin is not restricted to the region packaged into heterochromatin itself but extends to several kilobases of adjoining DNA.
Heterochromatin-mediated transcriptional silencing has been characterized extensively in Saccharomyces cerevisiae, where the integration of a retrotransposon in or near heterochromatin leads to transcriptional repression and, in Drosophila, where chromosomal rearrangements (inversions, translocation), occurring as a result of X-ray irradiation, placed euchromatic genes close to a heterochromatic breakpoint. This chromosomal rearrangement led to a position-dependent inactivation of the transposed euchromatic gene. Because the expression of the rearranged euchromatic gene or of the transposon is often characterized by a variegated phenotype, transcriptional repression by integration in or near heterochromatin has been referred to as position effect variegation (Tartof, 1994). Interestingly, transcription of the HIV promoter in latent clones also shows a variegated phenotype after activation, i.e. only a fraction of the population becomes reactivated in response to a global signal. Variegated expression of the HIV promoter was also observed during progressive repression after withdrawal of the activation signal. Different clones reverted to the silenced phenotype at different rates, possibly indicating that different chromatin environments lead to transcriptional repression with different efficiencies.
We propose as a working hypothesis that a repressive chromatin environment is the common underlying mechanism for transcriptional repression in all of our clones. As discussed above, we estimate that 20–30% of integration events in latent cells occurred in or near alphoid repeat elements. It is likely that other chromatin environments, such as proximity to a transcriptional silencer or heterochromatin islands not associated with alphoid elements, could account for transcriptional silencing in the rest of the clones. Since silencing by heterochromatin can occur over genomic distances of several kilobases, our PCR-based assay might also have missed a significant number of integrations near alphoid repeat elements. In addition, alphoid elements can differ significantly at the level of their DNA sequence, a factor that may account for negative PCR for integration events near specific alphoid repeats despite our use of consensus sequence primers.
The occurrence of integration into heterochromatin raises a number of interesting questions since this DNA is usually considered invisible to nuclear multiprotein complexes. One possibility could be that integrase, through its association with INI1, a component of the chromatin remodeling complex SWI/SNF, transiently remodels chromatin to make DNA more accessible (Kalpana et al., 1994; Miller and Bushman, 1995). Alternatively, heterochromatin is known to accumulate at the periphery of nuclei during interphase and could potentially become the primary target of the preintegration complex after it crosses the nuclear membrane.
HIV-1 latency occurs with extremely low frequency in HIV-infected individuals (106–107 cells per infected individual, 0.1–1 cell per million lymphocytes; Chun et al., 1997a). Since this rate is significantly lower than the frequency measured in the present study (one latent infection per 100 productive infections), it can be assumed that other factors contribute to latency in vivo. First, the virus that we have used does not express a functional Nef protein. Since Nef activity contributes to T-cell activation, it is possible that latency would occur with a lesser frequency in the context of a Nef-competent virus. The role of T-cell activation in the establishment of latency has also been considered independently of Nef (Pierson et al., 2000). In particular, it was proposed that withdrawal of T-cell activation signals might reduce transcriptional activity and ultimately lead to latency. Our data support this model and show that HIV transcription reverts to a latent phenotype at different rates depending on the site of integration after withdrawal of the activation signal (TPA in our study, T-cell activation signals in vivo).
According to this model, latency could be established in vivo when two events occur simultaneously in an infected T cell: integration in or near heterochromatin and withdrawal of T-cell activation signals. It will be important in the future to validate this model in HIV-infected individuals. However, the extremely low frequency of latently infected cells in blood samples from patients makes quantification of integration events into alphoid repeats unfeasible. Nevertheless, we were able to amplify the integration site of provirus integrated into alphoid repeats from PBMCs from HIV-1-infected individuals treated with highly active anti-retroviral therapy (see Supplementary data, available at The EMBO Journal Online).
The pool of latently infected cells represents a significant barrier to the eradication of HIV infection in an infected individual. In theory, the problem of the latent pool could be solved if HIV expression could be reactivated uniformly and effectively in all latently infected cells. Reactivated cells would be eliminated by a combination of host immune response and virally mediated cytopathic effects (Chun and Fauci, 1999; Pierson et al., 2000). Previous studies have reported the use of biological and pharmacological agents to reactivate latent HIV. For example, TPA, histone deacetylase inhibitors and various combinations of cytokines, including TNF-α, can activate HIV expression from latently infected cell lines (Poli and Fauci, 1993; Verdin et al., 1993; Van Lint et al., 1996; Chun et al., 1998, 1999; Butera, 2000).
The cell lines described here represent valuable tools for further studies of the molecular mechanism of HIV transcriptional silencing via heterochromatin and for the identification of small molecules that can reactivate latent HIV.
Materials and methods
Viral production and cell infections
For the production of viral particles containing the HIV-derived vector LTR–Tat–IRES–GFP, 5 × 106 293T cells were transfected with plasmids pEV731 (Jordan et al., 2001) (10 µg), pCMV-R8.91 (6.5 µg), and pMD.G (3.5 µg) in 10-cm dishes. After 16 h, the medium was replaced, and supernatants containing viral particles were harvested 24 h later. The number of infective particles per ml was established by infecting 2 × 105 Jurkat cells with different amounts of viral suspension. The titer of the virus stock was measured by flow cytometry analysis of GFP expression 96 h after infection. To obtain a random library of clones containing the vector LTR–Tat–IRES–GFP integrated, Jurkat cells were infected at an m.o.i. of 0.1 followed by serial dilution and plating in 96-well plates. Individual clones obtained after 3 weeks were analyzed by flow cytometry for GFP expression. For the purification of latently infected cells, Jurkat cells were infected with the LTR–Tat–IRES–GFP viral stock at an m.o.i. of 0.1 and kept in culture for at least 96 h. GFP-negative (GFP–) and GFP-positive (GFP+) cells were separated by FACS and further cultured. One week later, GFP– cells were incubated with 10 nM TPA or 10 ng/ml TNF-α for 17 h. The resulting GFP+ cells were sorted as a population and kept in culture or sorted into 96-well plates at one cell per well to generate clonal cell lines. Viral particles harboring an LTR–Tat vector were obtained as described above by transfecting 293T cells with plasmids pEV695, pCMV-R8.91 and pMD.G (Jordan et al., 2001).
Cell culture, transfections and cell treatments
Jurkat cells were grown in RPMI 1640 medium (Mediatech Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM l-glutamine at 37°C under a 95% air/5% CO2 atmosphere. PBMCs were cultivated in the same medium supplemented with 100 U/ml human interleukin (hIL)-2 and were PHA-stimulated (5 µg/ml) once every 2 weeks. 293T cells were grown under the same conditions as Jurkat cells in Dulbecco’s modification of Eagle’s medium (Mediatech Cellgro). 293T cells were routinely transfected with calcium phosphate. Jurkat cells (107 cells/0.4 ml serum-free medium) were electroporated in 0.4-cm gap cuvettes at 250 V and 950 mA (Gene Pulser II; Bio-Rad, Hercules, CA). Plasmid DNA for transfections was purified with the Qiagen Plasmid Maxi kit, followed by phenol extraction and ethanol precipitation. To test the HIV promoter inducibility, Jurkat-derived clones were incubated for 17 h with 10 nM TPA, 10 ng/ml TNF-α, 5 µg/ml PHA, 1 µM ionomycin, 5 µg/ml anti-CD3 antibody, 400 nM TSA. Incubations with 5 µM aza-dC lasted for 48 h.
Flow cytometry analysis and sorting
Cells were washed in PBS and resuspended in PBS containing 1% paraformaldehyde. GFP fluorescence was measured with a FACScalibur machine (Becton Dickinson, San Jose, CA). A two-parameter analysis to distinguish GFP-derived fluorescence from background fluorescence was used: GFP was measured in FL1 and cellular autofluorescence was monitored in FL2. Electronic compensation was applied during analysis. Analysis was gated on live cells according to forward and side scatter. A gate (R2) containing GFP-positive cells was drawn compared to an uninfected control, and the data shown refer to the percentage of cells in R2 or mean fluorescence intensity (MFI) of those cells. Results shown throughout the manuscript are representative of three independent experiments, except when libraries of clones were analyzed. Cell sorting was carried out with a FACSVantage (Becton Dickinson).
Southern blotting and colony hybridizations
Genomic DNA from infected Jurkat cells was extracted with the DNeasy Tissue kit (Qiagen, Valencia, CA). Southern hybridization was performed on digested DNA with [α-32P]dCTP-labeled probes (Multiprime DNA labeling system, Amersham Pharmacia Biotech, Piscataway, NJ) as described previously (Jordan et al., 2001). For probes, DNA fragments internal to the pEV731 retroviral vector were generated by PCR amplification: a 1.4-kb fragment extending between the 5′ LTR and Tat was generated with primers EV1048 (5′-GTGGCGCCCGAACAGG GACC-3′) and EV1049 (antisense, 5′-CCGTCGAGATCCGTTCACTA-3′); a 171-bp fragment corresponding to the 5′ end of the LTRs was produced with primers EV976 (5′-GCTAATTCACTCCCAACGAAG AC-3′) and EV1333 (antisense, 5′-gcttcttctaccttctcttgctc-3′); a 70-bp fragments for the 3′ end of the LTRs was generated with primers EV984 (5′-GCCCGTCTGTTGTATGACTCTG-3′) and primer EV934 (antisense, 5′-CGCCACTGCTAGAGATTTTCCAC-3′).
Sequencing of flanking genomic regions
A variety of strategies were used to clone genomic DNA at the integration site of latent clones. Inverse PCR was used to obtain the genomic region flanking the 3′ LTR on the integrated provirus of clone 82. Briefly, genomic DNA was digested with _Nco_I (cleavage site between IRES and GFP), and the resulting products were circularized by incubating with T4 DNA ligase (New England Biolabs, Beverly, MA). A nested series of three inverse PCRs were performed with three primers for GFP [EV1253, EV1335 (5′-ggtcttgtagttgccgtcgtc-3′) and EV1336 (5′-gaagaagatggtg cgctcc-3′); antisense] and 3 primers for LTR [EV933, EV996 (5′-TT GCCTGTACTGGGTCTCTCTG-3′) and EV984]. Before cloning the amplification products, the presence of LTR-containing products was confirmed by Southern hybridization with the probe EV984/EV934 described above.
Once a particular clone was identified as containing the retroviral vector integrated close to alphoid or Alu repeats, a series of two or three nested PCR amplifications were carried out with alphoid (α1–4) or Alu (EV1255) primers and with primers for the HIV LTR EV977 (5′-ATT CCATGCAGGCTCACAGG-3′), EV1332 (5′-gtgtaacaagcgggtgttctctc-3′) and EV1333 for the 5′ LTR, or EV933, EV996 and EV984 for the 3χ LTR.
To clone the integration site from clones that were not flanked by alphoid repeats, we used ligation-mediated PCR (LM-PCR) (Schmidt et al., 2001). Briefly, genomic DNA was digested with _Nla_III (New England Biolabs) and ligated to 100 pmol of annealed linker cassette (oligonucleotides EV1534 5′-GACCCGGGAGATCTGAATTCAGTG GCACAGCAGTTAGG-3′, and EV1535 5′-CCTAACTGCTGTGCCAC TGAATTCAG-3′). The ligation products were used as a template in a PCR amplification with retroviral primer EV996 and linker-specific primer EV1532 (5′-GACCCGGGAGATCTGAATTC-3′), that amplifies the 3′ LTR and flanking genomic region. Next, an aliquot of the first PCR was used on a nested PCR with the retroviral primer EV984 (LTR) and linker-specific primer EV1533 (5′-AGTGGCACAGCAGTTAGG-3′) to increase specificity of amplification.
In all cases, amplification products were cloned into pCR-Blunt vector and colony hybridization was used to screen for colonies harboring an LTR-containing fragment with probe EV976/EV1333 or EV984/EV934 described above. Selected clones were sequenced as described. BLAST (National Center for Biotechnology Information) was used to compare sequences with the human genome draft sequence and nucleotide database.
Alphoid PCR amplifications and quantitative PCR
To quantify the occurrence of integration close to alphoid repeats, we developed a PCR assay based on previously reported methods for Alu elements (Minami et al., 1995; Chun et al., 1997a). We used a human α satellite monomer consensus sequence derived from 293 cloned monomers of diverse chromosomal origins (Choo et al., 1991) to design oligonucleotides α1 (5′-AGACAGAAGCATTCTSAGAA-3′), α2 (5′-ATCACAAAGNAGTTTCTSAGAAT-3′), α3 (5′-TTTSATWGAGCA GNTTKGAAAC-3′) and α4 (5′-AAAGAGTGTTTCMAANCTGCT CW-3′). During the first PCR, genomic DNA was amplified with primer A (EV1371, 5′-AGGCAAGCTTTATTGAGGCTTAAGC-3′; antisense LTR) and either primer α1, α2, α3, α4, Alu (EV1255, 5′-TCCC AGCTACTCGGGAGGCTGAGG-3′) or B (EV1372, 5′-CACACA CAAGGCTACTTCCCT-3′; LTR) as a positive control for the presence of the LTR. As negative controls, amplifications with no primer or with primer A alone were performed. Taq DNA polymerase (0.75 U/25 µl reaction; Life Technologies, Rockville, MD), 200 nM each dNTP and 500 nM each primer were used, and the reaction was run with the following program: (i) 3 min at 94°C; (ii) 30 cycles of 30 s at 94°C, 30 s at 53°C and 4 min at 72°C; and (iii) 10 min at 72°C. A second nested PCR amplification was carried out by using 1 µl of the first reaction with primers B (EV1372) and C (EV1373, 5′-GCCACTCCCCIGTCCC GCCC-3′; antisense LTR), which allows amplification of a fragment of the LTR. This second PCR was done using the same conditions as the first one, but the extension time was 1 min and amplification was run for 25 cycles. PCR products were analyzed by ethidium bromide–agarose gel electrophoresis and DNA bands were quantified with the EagleSight software (Stratagene, La Jolla, CA).
Real-time PCR (TaqMan) was also used to quantify integration of HIV-derived vectors close to alphoid or Alu repeats, using a modification of a published protocol (Butler et al., 2001). A first PCR was carried out for 25 cycles as described above but, instead of primer A, the LTR primer D (EV933, 5′-GAGCCCTCAGATGCTGCATATAAG-3′) was used in combination with primers α1–4 or Alu to amplify genomic regions downstream of the 3′ LTR. For the nested real-time PCR, internal LTR primers E (EV1441, 5′-AACTAGGGAACCCACTGCTTAAG-3′) and F (EV934, 5′-CGCCACTGCTAGAGATTTTCCAC-3′; antisense) were used. An aliquot (1 µl) of the first amplification product was amplified with 200 nmol of each of the specific primers and 100 nmol of the LTR-specific TaqMan probe G (EV1444, 5′-6 FAM-ACACTACTTGAAG CACTCAAGGCAAGCTTT-TAMRA-3′) using the TaqMan Universal PCR Master mix (Perkin Elmer Applied Biosystems, Foster City, CA). The reaction was run for 40 cycles (15 s at 95°C plus 1 min at 60°C), in an AbiPrism 7700 Sequence Detector (Perkin Elmer Applied Biosystems). Quantification was performed as recommended by Perkin Elmer.
Sequencing of integrated provirus
To check for the occurrence of mutations, the 5′ LTR from selected clones was amplified (823-bp fragment) from genomic DNA with primers EV976 (5′-GCTAATTCACTCCCAACGAAGAC-3′; 5′ end of LTR) and EV987 (antisense, 5′-TCGCTTTCAGGTCCCTGTTCG-3′; gag region downstream 5′ LTR). A Tat–IRES–GFP′ fragment (1307 bp) was amplified with primers EV1140 (5′-CCATCGATGCCACCATGGA GCCAGTAGA-3′; 5′ end of Tat) and EV1253 (antisense, 5′-AGGGT GTCGCCCTCGAA-3′; internal to GFP). Pfu DNA polymerase (Stratagene) was used and the reaction was run with the following program: (i) 45 s at 94°C; (ii) 30 cycles of 45 s at 94°C, 45 s at 58°C and 1.5 min at 72°C; and (iii) 10 min at 72°C. The amplified product was purified from ethidium bromide-containing 1%-agarose gel with the GenClean Spin kit (Qbiogene, Carlsbad, CA) and cloned in the pCR-Blunt vector provided in the Zero Blunt PCR Cloning kit (Invitrogen, Carlsbad, CA). Two to four recombinant clones containing the expected DNA insert were sequenced with primers M13 Forward and M13 Reverse and the Big Dye d-Rhodamine Terminator Ready Reaction kit (Perkin Elmer Applied Biosystems).
HIV-specific mRNA measurements
RNA was isolated using TRIzol (Invitrogen) followed by digestion with RQ1 DNase (Promega). First strand cDNA was synthesized using Superscript II (Invitrogen). TaqMan PCR was performed on an ABIprism 7700 detector using the following primer/probe set: primer 1: 5′-GTG TGCCCGTCTGTTGTGTGA-3′, primer 2: 5′-GCCACTGCTAGAGAT TTTCCA-3′, probe 5′-CTGGTAACTAGAGATCCC-3′. The GAPDH primer/probe set was purchased from Applied Biosystems.
Construction of HIV-R7/E–/GFP molecular clone
The HIV molecular clone (HIV-R7/E–/GFP) was constructed by introducing a frameshift mutation in the env gene (by filling-in _Nde_I site with T4 polymerase) in the backbone of HIV-R7/3/GFP (a generous gift from Mark Muesing) (Bieniasz and Cullen, 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank John Carroll for graphics, Heather Gravois for manuscript preparation, Stephen Ordway and Gary Howard for editorial assistance, Melanie Ott for advice on the manuscript. We thank Didier Trono for plasmids, and Veronique Kiermer and other members of the Verdin laboratory for discussions. We thank Mark Muesing (Aaron Diamond AIDS Research Center) for providing the R7/3/GFP HIV molecular clone. We thank the AIDS Research and Reagent Reference Program at NIH for reagents. We thank the Gladstone Institute Flow Cytometry core for technical assistance. This work was supported in part by a Public Health Service grant from the NIH (GM 51671-05A2). A.J. was the recipient of a fellowship from the Human Frontier Science Program Organization. D.B. was supported by the Alberta Heritage Foundation for Medical Research.
References
- Antoni B.A., Rabson,A.B., Kinter,A., Bodkin,M. and Poli,G. (1994) NF-κB-dependent and -independent pathways of HIV activation in a chronically infected T cell line. Virology, 202, 684–694. [DOI] [PubMed] [Google Scholar]
- Bieniasz P.D. and Cullen,B.R. (2000) Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol., 74, 9868–9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butera S.T. (2000) Therapeutic targeting of human immunodeficiency virus type-1 latency: Current clinical realities and future scientific possibilities. Antiviral Res., 48, 143–176. [DOI] [PubMed] [Google Scholar]
- Butler S.L., Hansen,M.S. and Bushman,F.D. (2001) A quantitative assay for HIV DNA integration in vivo. Nat. Med., 7, 631–634. [DOI] [PubMed] [Google Scholar]
- Carteau S., Hoffmann,C. and Bushman,F. (1998) Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol., 72, 4005–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Y., Bailey,E.C., McCune,S.L., Dong,J.Y. and Townes,T.M. (1997) Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc. Natl Acad. Sci. USA, 94, 5798–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K.H., Vissel,B., Nagy,A., Earle,E. and Kalitsis,P. (1991) A survey of the genomic distribution of α satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res., 19, 1179–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W. and Fauci,A.S. (1999) Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl Acad. Sci. USA, 96, 10958–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W. et al. (1997a) Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature, 387, 183–188. [DOI] [PubMed] [Google Scholar]
- Chun T.W., Stuyver,L., Mizell,S.B., Ehler,L.A., Mican,J.A., Baseler,M., Lloyd,A.L., Nowak,M.A. and Fauci,A.S. (1997b) Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl Acad. Sci. USA, 94, 13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W., Engel,D., Mizell,S.B., Ehler,L.A. and Fauci,A.S. (1998) Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med., 188, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W. et al. (1999) Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med., 5, 651–655. [DOI] [PubMed] [Google Scholar]
- Emiliani S., Van Lint,C., Fischle,W., Paras,P.,Jr, Ott,M., Brady,J. and Verdin,E. (1996) A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl Acad. Sci. USA, 93, 6377–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S., Fischle,W., Ott,M., Van Lint,C., Amella,C.A. and Verdin,E. (1998) Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol., 72, 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D. et al. (1997) Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science, 278, 1295–1300. [DOI] [PubMed] [Google Scholar]
- Finzi D. et al. (1999) Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med., 5, 512–517. [DOI] [PubMed] [Google Scholar]
- Groudine M., Eisenman,R. and Weintraub,H. (1981) Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature, 292, 311–317. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol., 11, 266–273. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jordan A., Defechereux,P. and Verdin,E. (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J., 20, 1726–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana G.V., Marmon,S., Wang,W., Crabtree,G.R. and Goff,S.P. (1994) Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science, 266, 2002–2006. [DOI] [PubMed] [Google Scholar]
- Karn J. (1999) Tackling Tat. J. Mol. Biol., 293, 235–254. [DOI] [PubMed] [Google Scholar]
- Miller M.D. and Bushman,F.D. (1995) HIV integration. Ini1 for integration? Curr. Biol., 5, 368–370. [DOI] [PubMed] [Google Scholar]
- Minami M., Poussin,K., Bréchot,C. and Paterlini,P. (1995) A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics, 29, 403–408. [DOI] [PubMed] [Google Scholar]
- Pierson T., McArthur,J. and Siliciano,R.F. (2000) Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol., 18, 665–708. [DOI] [PubMed] [Google Scholar]
- Poli G. and Fauci,A.S. (1993) Cytokine modulation of HIV expression. Semin. Immunol., 5, 165–173. [DOI] [PubMed] [Google Scholar]
- Schmidt M. et al. (2001) Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther., 12, 743–749. [DOI] [PubMed] [Google Scholar]
- Schroder A.R., Shinn,P., Chen,H., Berry,C., Ecker,J.R. and Bushman,F. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell, 110, 521–529. [DOI] [PubMed] [Google Scholar]
- Tartof K.D. (1994) Position effect variegation in yeast. BioEssays, 16, 713–714. [DOI] [PubMed] [Google Scholar]
- VanLint C., Emiliani,S., Ott,M. and Verdin,E. (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J., 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Paras,P.,Jr and Van Lint,C. (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J., 12, 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L.L. (1998) Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev., 8, 147–153. [DOI] [PubMed] [Google Scholar]
- Wong J.K., Hezareh,M., Günthard,H.F., Havlir,D.V., Ignacio,C.C., Spina,C.A. and Richman,D.D. (1997) Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science, 278, 1291–1295. [DOI] [PubMed] [Google Scholar]