Cellular Gene Expression Survey of Vaccinia Virus Infection of Human HeLa Cells (original) (raw)
Abstract
Vaccinia virus (VV) is a cytocidal virus that causes major changes in host cell machinery shortly after infecting cells. To define the consequences of virus infection on host gene expression, we used microarrays of approximately 15,000 human cDNAs to examine expression levels of mRNAs isolated at 2, 6, and 16 h postinfection from cultures of infected HeLa cells. The majority of profiling changes during VV infection corresponded to downregulation of genes at 16 h postinfection. Differentially expressed genes were clustered into seven groups to identify common regulatory pathways, with most of them (90%) belonging to clusters 6 and 7, which represent genes whose expression was repressed after infection. Cluster 1, however, contained 37 transcripts (2.81%) showing a robust pattern of induction that was maintained during the course of infection. Genes in cluster 1 included those for Wiskott-Aldrich syndrome protein (WASP) family member WASF1, thymosine, adenosine A2a receptor, glutamate decarboxylase 2, CD-80 antigen, KIAA0888 protein, selenophosphate synthetase, pericentrin, and attractin as well as several expressed sequence tags. We analyzed in more detail the fate of WASP protein in VV-infected cells, because a related family member, N-WASP, is involved in viral motility. WASP protein accumulated in the course of infection; its increase required viral DNA replication and de novo protein synthesis, and it localized in cytoplasmic structures distinct from uninfected cells. This study is the first quantitative analysis of host gene expression following VV infection of cultured human cells, demonstrating global changes in the expression profile, and identifies upregulated genes with potential roles in the virus replication cycle.
Vaccinia virus (VV) is a member of the Poxviridae family, a group of large, double-stranded DNA viruses that reproduce exclusively in the cytoplasm of the infected host cell (28). The VV genome is subdivided into three temporally expressed classes, termed early, intermediate, and late genes, which encode over 200 proteins, including most of the enzymes and factors required for transcription, genome replication, and virion assembly (28). Immediately after infection, early VV genes are transcribed, but the first steps of viral DNA replication are required to transcribe late and intermediate genes (44). The genomes of a number of poxviruses have been sequenced and found to contain virulence genes (20, 24; www.poxviruses.org).
Poxvirus infection induces a number of changes in cell function, metabolism, and cellular morphology (7). The cytopathic effect observed early after vaccinia virus infection is linked to a strong rearrangement of the cell cytoskeleton (2, 18, 31). Moreover, actin fibers are actively mobilized by nascent viral particles to form tails that are induced by intracellular envelope virus particles (10, 35). In addition, cell motility is also modified after infection (36).
Vaccinia virus infection also interferes with multiple steps in host gene expression. First, VV induces an immediate and rapid inhibition of HeLa RNA synthesis and processing (5, 30). In addition, virus-induced degradation of actin and tubulin mRNAs has been well documented in early reports (33). Concurrent with the degradation of cellular mRNA, the virus proceeds in orderly temporal expression of its own genetic information (33). Finally, host translation is drastically impaired, while viral protein synthesis takes place at maximal efficiency (3, 4).
Although many aspects of the biology of this virus have been described, little is known of its impact on host gene expression. The recent development of high-density cDNA microarrays allows simultaneous expression profiling of thousand of genes and represents a powerful approach with which to study the impact of viral infections on gene expression (6, 9, 15, 21, 29, 32, 43, 45). To obtain a more comprehensive view of the global effects of VV infection on human cells at the mRNA level, we performed cDNA microarray analysis of mRNAs obtained from VV-infected HeLa cells at various times postinfection and evaluated about 15,000 human cDNAs. We identified several mRNAs from a variety of cellular mechanisms whose levels were altered by VV infection. Genes involved in known cellular pathways and expressed sequence tags (ESTs) were defined as being differentially regulated by VV infection.
(Part of this work was presented at the XIVth International Poxvirus and Iridovirus Meeting, 20-25 September 2002, Lake Placid, N.Y.)
MATERIALS AND METHODS
Cells, viruses, and infection conditions.
HeLa cells (ATCC) were cultured in Dulbecco's medium supplemented with 10% newborn bovine serum and antibiotics. VV wild-type, Western Reserve strain (WR), was grown in spinner HeLa cells and purified by banding on sucrose gradients as described previously (11) and titrated in monkey BSC-40 cells. All infections were performed at a multiplicity of infection of 5 PFU/cell with VV.
Microarray fabrication.
To generate cDNA arrays, we used the Research Genetics 40K clone human cDNA library sequence verified (http://www.resgen.com/products/SVHcDNA.php3). Forty plates containing 15,360 clones (most of known genes) were selected and grown in Luria-Bertani (LB) plus 7.5% glycerol medium (overnight, 37°C). PCRs were performed with 3 μl of each bacterial culture as the template and 5 U of Gibco Taq polymerase (Invitrogen) in the following buffer: 20 mM Tris-HCl (pH 9.0), 8.5 mM NaCl, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.4 mM deoxynucleoside triphosphate mix, 0.5 μM forward primer (5′-CTG CAA GGC GAT TAA GTT GGG TAA C-3′), and 0.5 μM reverse primer (5′-GTG AGC GGA TAA CAA TTT CAC ACA GGA AAC AGC-3′). Amplification was performed in the following conditions: 1 min of initial denaturation, 40 cycles of 30 s at 94°C, 45 s at 60°C, and 4 min at 72°C, and a final extension of 10 min at 72°C. cDNAs were confirmed with MultiScreen PCR 96-well (Millipore) and confirmed by agarose gel. Finally, PCR products were reformatted to 384-well plates, dried, and resuspended in 50% dimethyl sulfoxide. Printing was performed on CMT-GAPS II slides (Corning) with a Total Array System (BioRobotics) at 22°C and 40 to 45% relative humidity.
Microarray hybridization.
Total RNA from VV-infected (5 PFU/cell) or mock-infected HeLa cells cultured in 10-cm plates was isolated with RNAwiz (Ambion) following the manufacturer's instructions. Each RNA (40 μg) was labeled with dUTP-indodicarbocyanine (Cy5) or dUTP-indocarbocyanine (Cy3) (Pharmacia-Biotech) by direct labeling during the reverse transcription. Mock-infected sample was labeled with Cy3, and the infected sample was labeled with Cy5. This combination was changed in one experiment (mock/Cy5 and VV-infected/Cy3) to abolish differences in labeling and hybridization due to the specific characteristics of each Cy-labeled dUTP. Briefly, a mixture containing 40 μg of RNA, 150 pmol of oligo(dT)20,, 0.5 mM each dATP, dGTP, and dCTP, 0.1 mM dTTP, 0.05 mM Cy3/Cy5-dUTP, 1× first-strand reaction buffer (Invitrogen), and 10 mM dithiothreitol in a volume of 38 μl was heated (65°C, 5 min) and preincubated (42°C, 5 min), after which 400 U of SuperScript II (Invitrogen) and 40 U of RNase Inhibitor (Roche) were added and the mixture was incubated (42°C, 2.5 h).
The reaction was terminated with EDTA, and the starting RNA template was removed by adding 2 μl of 10 N NaOH, followed by incubation (65°C, 20 min). The reaction was then neutralized by adding 4 μl of 5 M acetic acid. Cy5 and Cy3 probes were mixed, and unincorporated dye was removed by isopropanol precipitation. Probes were resuspended in deionized water, and blocking reagents were added to increase specificity: poly(A) (20 μg; Sigma), tRNA (20 μg; Sigma), and human Cot-1 DNA (20 μg; Invitrogen). While the probes were drying in a Speed-Vac, microarray slides were prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate (SDS)-1% bovine serum albumin (42°C, 1 h). Slides were finally rinsed five times with water and dried by centrifugation (563 × g, 1 min). Probes were resuspended in 40 μl of hybridization buffer (50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt's solution) and incubated with slides (42°C, 16 h) in hybridization chambers (Array-It) in a water bath in the dark. After incubation, slides were washed twice in 0.1× SSC-0.1% SDS for 5 min each and three times in 0.1× SSC for 5 min each. Finally, the slides were dried by centrifugation as before and scanned on a ScanArray 4000 (Packard Biosciences) with ScanArray 3.1 software.
Images from Cy5 and Cy3 were equilibrated, and spots were quantified with QuantArray 3.0 software (Packard Biosciences). Raw data were normalized based on the signal of genes that did not undergo variation after infection, as confirmed by Northern blot (genes APEXL2 and FLJ20643). Data from a single hybridization were viewed as log2-ratio (Cy3/Cy5) in which significant variations from 1 (no change) were indicative of increased (>2) or decreased (<−2) levels of gene expression relative to the mock-infected sample. The data shown were obtained from three independent experiments. Experiments 1 and 2 corresponded to mRNA obtained at 2, 6, and 16 h postinfection, and experiment 3 corresponded to mRNA obtained at 6 and 16 h postinfection.
Gene expression analysis.
All samples were prepared from HeLa cells grown in monolayers and infected with sucrose-purified VV. Hybridization was performed in triplicate, and only those genes whose log2-ratio values were consistently repeated in all the replicates were included in the analysis. For this purpose, we removed inconsistent replicates with quite restrictive criteria based on maximum distance to the median value: for each replicated gene, we calculated the log2-ratio median and removed those values that were beyond 1 from the median value. In this way, we assured the consistency of the resulting data set.
A variation filter was then used to eliminate genes that did not change significantly across samples (flat patterns). In this case, genes that did not show an abundance ratio of 2.0 in at least one experimental condition were eliminated from the data set. The resulting data set after the different preprocessing steps was composed of 1,318 genes. The length of each resulting expression vector was normalized to 1 previous to clustering.
To classify gene expression profiles into groups according to their behavior patterns, cluster analysis was carried out by self-organizing maps (22, 42). Since the experimental data contain only three conditions (2 h, 6 h, and 16 h), a simple three-dimensional scatter plot would be sufficient for visual clustering of the data set. Nonetheless, as visual inspection might fail to detect subtle variation patterns hidden in the data set, a more sophisticated technique (self-organizing maps) was used in this study for clustering purposes. Self-organizing maps is a powerful technique that allows nonlinear projection of the original data onto a two-dimensional grid so that the resulting map is ordered according to certain data features, allowing a simple, intuitive visualization of the clustering structure. To calculate the self-organizing maps, we used the Engene software package (www.engene.cnb.uam.es) (14).
The expression patterns were projected onto a 10 by 5 hexagonal grid with the self-organizing maps algorithm. Each node in the map is an expression profile representing a subset of the expression patterns under analysis. The self-organizing maps algorithm was trained in two phases, a global ordering and a fine adjustment (22). In the first stage, the algorithm was iterated 100,000 times, linearly decreasing the learning rate from 0.3 to 0. The neighborhood radius was also linearly decreased from 5 to 1. In the second fine-adjustment phase, the algorithm was iterated 500,000 times, varying the learning rate linearly from 0.1 to 0 and decreasing the neighborhood radius linearly from 2 to 1 (22).
Northern blot.
Total RNA (20 μg) was fractionated by electrophoresis through 1% agarose gels containing formaldehyde and blotted in 20× SSC onto Nytran nylon membranes (Schleicher and Schuell). cDNA fragments were labeled with [32P]dCTP with the ReadyPrime system (Amersham). Hybridization was performed in Church buffer (0.2 M sodium phosphate [pH 7], 1 mM EDTA, 1% bovine serum albumin, 10% SDS) at 68°C overnight. Membranes were washed twice in 2× SSC-0.1× SDS (65°C, 1 h) and twice in 0.1× SSC-0.1× SDS (65°C, 1 h) and exposed for 12 h to a Storm phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Quantitative real-time RT-PCR.
One microgram of RNA was reverse transcribed according to the Superscript first-strand synthesis system for reverse transcription-PCR (RT-PCR) protocol (Invitrogen). A 1:40 dilution of the RT reaction mixture was used in the quantitative PCR. Primers, probe set, and protocols used for the amplification of PCNT2, WASF1, and IL-6ST were performed with the instructions of the manufacturer (Assay-on-Demand of Applied Biosystems). Gene expression quantification with Assay-on-Demand is optimized to work with TaqMan Universal PCR MasterMix, No AmpErase UNG. We used the human housekeeping gene hypoxanthine ribosyltransferase (HPRT) for internal calibration. The thermal cycler conditions were 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C followed by 1 min at 60°C. The amplification of cDNA was carried out in a 96-well reaction plate. All the samples were assayed in duplicate. The threshold cycle (Ct) values were used to plot a standard curve in which Ct decreased in linear proportion of the log of the template copy number. The correlation values of standard curves were always more than 99%.
Western blot.
HeLa cells were infected at a multiplicity of infection of 5 PFU/cell with VV, collected, and lysed at 2, 6, and 16 h postinfection in lysis buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 10% NP-40, 1% SDS) maintained on ice for 5 min. Equal amounts of protein lysates were separated by SDS-polyacrylamide gel electrophoresis on 14% or 8% gels, transferred to nitrocellulose membranes, and reacted with primary antiactin (Sigma), antitubulin (Sigma), and anti-Wiskott-Aldrich syndrome (WAS) family member antibodies (kindly provided by Antonio Bernad) and with secondary antibodies (mouse and rabbit immunoglobulin-peroxidase conjugates). Protein expression was detected with ECL Western blotting reagents (Amersham).
Immunofluorescence.
HeLa cells cultured on coverslips were infected at 5 PFU/cell with VV. At 2, 6, and 16 h postinfection, cells were washed with phosphate-buffered saline, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (room temperature, 10 min). Cells were incubated with antibodies to the A27L viral protein (monoclonal antibody C3α14k) together with anti-WAS family member antibody and the DNA-staining reagent ToPro (Molecular Probes). Images were obtained with a Bio-Rad Radiance 2100 confocal laser microscope.
RESULTS
Analysis of clusters.
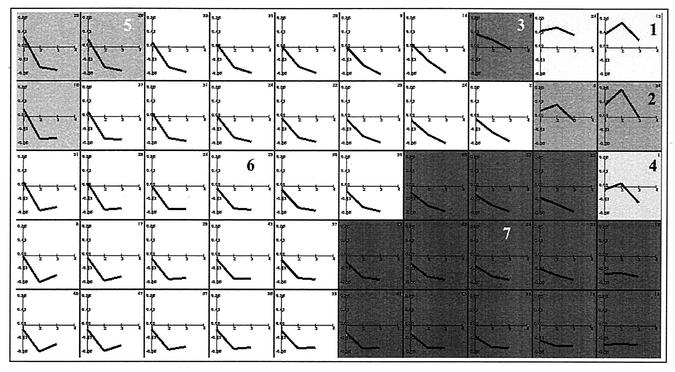
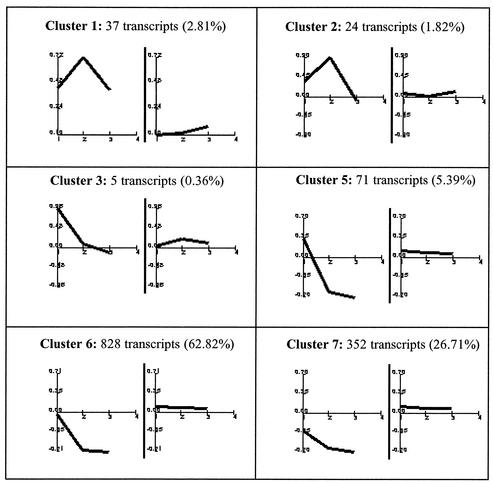
We used human cDNA microarrays to study the global transcriptional response of HeLa cells infected with VV (WR strain). We chose HeLa cells because they are highly susceptible to VV infection and many fundamental studies of VV biology have been performed with this cell line. To study the cellular transcriptional response during VV infection, we compared the relative abundance of a specific mRNA in infected cells with the same specific mRNA from mock-infected cells in cDNA microarrays. The gene expression clusters of the resulting 1,318 genes that passed the filtering conditions are depicted in Fig. 1. In global terms, expression of most of these genes (90%) was downregulated at 6 to 16 h after VV infection. However, a more detailed analysis of profiles led us to group the genes into seven main clusters according to their behavior over the three time points of VV infection. These clusters seem to represent specific regulation patterns, and the average profiles for each of the clusters depicted are shown in Fig. 2.
FIG. 1.
Representation of 10 by 5 map obtained by the self-organizing maps algorithm, showing the gene expression clusters for VV-infected HeLa cells. Experimental points on the x axis are indicated as 1 for 2 h, 2 for 6 h, and 3 for 16 h postinfection. The y axis shows normalized expression values. Each cluster depicted was numbered from 1 to 7.
FIG. 2.
Characteristic expression patterns represented in each cluster. Mean values (left) and standard deviations (right) of the expression profiles of genes assigned to each cluster. Experimental points on the x axis are as in Fig. 1. The y axis shows normalized expression values. Cluster 4 was not included because it contains only one transcript (0.08%).
Clusters 1 to 3 correspond roughly to three different kinetics of gene activation, whereas clusters 4 to 7 show different repression kinetics. Cluster 1 contained 37 transcripts (2.81%) that were upregulated during the infection; it is the only cluster that showed a generalized induction pattern maintained from 2 to 16 h postinfection. The members of cluster 1 are detailed in Table 1. Cluster 2 contained 24 transcripts (1.82%), including genes whose induction was maintained from 2 to 6 h postinfection and returned to basal levels at 16 h postinfection; cluster 2 members are represented in Table 2. Cluster 3 contained five transcripts (0.36%) with an upregulation pattern at 2 h postinfection that returned to basal levels at 6 h postinfection. Cluster 4 contained one transcript, gamma interferon-inducible protein 16, which was slightly upregulated at 6 h postinfection. Cluster 5 contained 71 transcripts (5.39%) that were upregulated at 2 h postinfection and downregulated at 6 and 16 h postinfection. Cluster 6 was the largest group, containing 828 transcripts (62.82%), which were completely repressed at 6 and 16 h postinfection. This cluster included genes involved in transcription, translation, subcellular trafficking, apoptosis, metabolic pathways, and other vital cell processes. Cluster 7 contained 352 transcripts (26.71%), with a pattern of decreasing repression at 2, 6, and 16 h postinfection (Table 3).
TABLE 1.
Genes in cluster 1
| Function and gene name | Accession no. | Change (fold) at h postinfection: | ||
|---|---|---|---|---|
| 2 | 6 | 16 | ||
| WAS protein family member, WASF1 | N59851 | 2.35 | 2.43 | 2.99 |
| KIAA0136 protein | N73634 | 3.92 | 4.23 | 3.81 |
| Diacylglycerol kinase delta (130 kDa), DGKD | AA280692 | 1.68 | 2.41 | 2.55 |
| KIAA0671 gene product | R33011 | 1.39 | 1.87 | 2.20 |
| Claudin 3, CLDN3 | AA039323 | 1.78 | 2.13 | 2.30 |
| Neurotrophic tyrosine kinase receptor type 2, NTRK2 | N63949 | 2.04 | 2.17 | 1.83 |
| Thymosin beta 4 X chromosome, TMSB4X | AA634103 | 3.81 | 5.70 | 2.91 |
| Adenosine A2a receptor, ADORA2A | N57553 | 3.89 | 4.14 | 4.11 |
| PRO2047 protein | N66208 | 2.28 | 2.75 | 1.77 |
| Glutamate decarboxylase 2 (pancreatic islets and brain, 65 kDa), GAD2 | R44005 | 3.56 | 2.38 | 3.14 |
| Golgi resident protein GCP60 | AA255954 | 1.41 | 2.00 | 1.79 |
| CD80 antigen (CD28 antigen ligand 1, B7-1 antigen), CD80 | AA983817 | 1.75 | 2.58 | 2.06 |
| KIAA0888 protein | AI376113 | 4.69 | 16.22 | 22.78 |
| Homo sapiens normal mucosa of esophagus specific 1 (NMES1) mRNA, complete coding sequence | AA620995 | 1.64 | 2.53 | 1.28 |
| Selenophosphate synthetase, SPS | AA488081 | 3.73 | 7.11 | 2.17 |
| Prostaglandin-endoperoxide synthase 1, PTGS1 | AA454668 | 1.51 | 2.38 | 1.21 |
| Pericentrin, PCNT | N45326 | 3.20 | 7.11 | 1.58 |
| Attractin, ATRN | AA683500 | 2.16 | 2.60 | 1.41 |
| Melanoma antigen family C 1, MAGEC | AI126114 | 1.40 | 2.04 | 1.26 |
| ATPase aminophospholipid transporter (APLT) class I type 8A member 1, ATP8A1 | AI218581 | 1.46 | 2.69 | 1.57 |
| ESTs | T86959 | 3.63 | 19.84 | 9.45 |
| N34895 | 3.18 | 5.35 | 2.85 | |
| N47089 | 5.78 | 9.06 | 7.16 | |
| N62132 | 5.74 | 13.55 | 3.71 | |
| H78999 | 2.64 | 11.79 | 8.28 | |
| N40165 | 3.84 | 6.87 | 4.63 | |
| N47431 | 3.2 | 5.24 | 3.18 | |
| N63062 | 3.78 | 5.5 | 3.05 | |
| N64688 | 8.17 | 10.34 | 8.69 | |
| N64662 | 2.35 | 2.58 | 2.16 | |
| AA884618 | 1.74 | 1.64 | 2 | |
| AA005135 | 1.67 | 5.28 | 2.23 | |
| N62516 | 2.57 | 11.08 | 1.72 | |
| N52149 | 4.50 | 12.30 | 1.46 | |
| N59137 | 3.27 | 5.28 | 1.73 | |
| N59214 | 3.76 | 8.88 | 1.80 | |
| N51223 | 2.81 | 4.92 | 1.83 |
TABLE 2.
Genes in cluster 2
| Function and gene name | Accession no. | Change (fold) at h postinfection: | ||
|---|---|---|---|---|
| 2 | 6 | 16 | ||
| Suppression of tumorigenicity 14, ST14 | AA489246 | 1.8 | 4.56 | 0.93 |
| Solute carrier family 4 anion exchanger member 3, SLC4A3 | AA609880 | 1.58 | 2.10 | 0.95 |
| Postmeiotic segregation increased (Saccharomyces cerevisiae) 1, PMS | AA504838 | 1.49 | 3.14 | 0.81 |
| KIAA1288 protein | AA621202 | 1.43 | 2.36 | 0.87 |
| KIAA0082 protein | AA504460 | 1.39 | 2.33 | 0.89 |
| Hypothetical protein FLJ23018 | T83664 | 1.43 | 2.43 | 0.85 |
| Hypothetical protein FLJ10748 | AA398335 | 1.07 | 2.08 | 0.93 |
| Homo sapiens mRNA from chromosome 5q21-22, clone:357Ex | AA481425 | 1.44 | 2.81 | 1.08 |
| Homo sapiens cDNA FLJ13569 fis, clone PLACE1008369 | AA478962 | 1.15 | 2.01 | 0.79 |
| GT198 complete ORF HUMGT198A | AA134555 | 1.62 | 2.25 | 0.79 |
| ESTs | N48707 | 2.35 | 11.79 | 1.58 |
| AA461091 | 1.41 | 2.03 | 0.75 | |
| AA101833 | 1.55 | 2.17 | 0.88 | |
| AA446019 | 1.57 | 2.93 | 0.87 | |
| AA399260 | 1.45 | 2.51 | 0.84 | |
| AA905171 | 1.49 | 2.27 | 0.92 | |
| N51444 | 3.16 | 8.40 | 1.36 | |
| N63609 | 2.79 | 7.94 | 1.37 | |
| AA621192 | 1.57 | 3.20 | 0.74 | |
| AA620632 | 2.64 | 3.61 | 1.06 | |
| AA620746 | 1.32 | 3.01 | 0.84 | |
| DKFZP434D193 protein | N63452 | 2.25 | 6.87 | 0.81 |
| Chromosome 16 open reading frame 5 | AA401341 | 1.40 | 3.12 | 1.11 |
TABLE 3.
Genes in clusters 3 to 7
| Cluster | Function and gene name | Accession no. | Change (fold) at h postinfection: | ||
|---|---|---|---|---|---|
| 2 | 6 | 16 | |||
| 3 | Diacylglycerol kinase, alpha (80 kDa) | AA456830 | 2.01 | 1.17 | 0.90 |
| Golgi autoantigen golgin subfamily a2 | AA424786 | 2.41 | 0.97 | 1.01 | |
| H2B histone family member B | N33927 | 5.94 | 0.78 | 0.61 | |
| H3 histone family member B | AI399887 | 4.92 | 1.12 | 1.11 | |
| Hypothetical protein FLJ10569 | AA195648 | 2.00 | 1.41 | 0.85 | |
| 4 | Interferon gamma protein 16 | AA490996 | 0.84 | 1.20 | 0.33 |
| 5 | Adhesion and migration | ||||
| Exportin1 (CRM1 yeast homolog) | T59055 | 2.10 | 0.20 | 0.15 | |
| Fibrillarin | AA663986 | 1.47 | 0.50 | 0.49 | |
| Karyopherin (importin) beta 1 | AA251527 | 2.01 | 0.20 | 0.18 | |
| Kinesin family member 5B | AA046690 | 2.31 | 0.17 | 0.17 | |
| Matrin 3 | AA075307 | 1.38 | 0.40 | 0.48 | |
| Splicing factors | |||||
| Splicing factor arginine/serine-rich (transformer 2 Drosophila homolog) 10 | H11720 | 1.97 | 0.20 | 0.15 | |
| Splicing factor arginine/serine-rich 2 | AA454585 | 1.95 | 0.16 | 0.12 | |
| Splicing factor arginine/serine-rich 3 | AA598400 | 1.41 | 0.43 | 0.31 | |
| Splicing factor (CC1.3) | H47069 | 1.60 | 0.32 | 0.20 | |
| Transcription | |||||
| Metal-regulatory transcription factor 1 | AA448256 | 1.47 | 0.49 | 0.50 | |
| Translation | |||||
| Eukaryotic translation initiation factor 1A | AA281733 | 2.25 | 0.72 | 0.50 | |
| Eukaryotic translation initiation factor 1A Y chromosome | AA047039 | 2.30 | 0.29 | 0.29 | |
| Eukaryotic translation initiation factor 5A | AA878570 | 2.19 | 0.43 | 0.35 | |
| Eukaryotic translation initiation factor 3 subunit 10 (theta 150/170 kDa) | R60031 | 2.93 | 0.21 | 0.14 | |
| 6 | ATPases | ||||
| ATP binding protein associated with cell differentiation | N80741 | 1.46 | 0.21 | 0.11 | |
| ATP synthase H+ transporting mitochondrial F0 complex subunit b isoform 1 | AA453765 | 1.39 | 0.18 | 0.12 | |
| ATP synthase H+ transporting mitochondrial F0 complex subunit c (subunit 9) isoform 2 | AA455126 | 1.43 | 0.20 | 0.09 | |
| ATPase H+ transporting lysosomal (vacuolar proton pump) member J | AA608567 | 1.37 | 0.17 | 0.11 | |
| ATPase Na+/K+ transporting beta 3 polypeptide | AA486417 | 1.23 | 0.20 | 0.13 | |
| Apoptosis | |||||
| Caspase 3 apoptosis-related cysteine protease | R42530 | 1.56 | 0.12 | 0.04 | |
| Caspase 8 apoptosis-related cysteine protease | AA448468 | 1.17 | 0.30 | 0.21 | |
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 1 | AA425687 | 1.38 | 0.21 | 0.20 | |
| Death-associated protein 3 | AA488173 | 1.59 | 0.22 | 0.21 | |
| Programmed cell death 5 | AA156940 | 0.79 | 0.26 | 0.23 | |
| Death-associated protein 6 | AA487253 | 1.74 | 0.13 | 0.12 | |
| Cell cycle | |||||
| Cyclin A2 | AA608568 | 1.21 | 0.57 | 0.39 | |
| Cyclin C | AA451686 | 1.85 | 0.39 | 0.14 | |
| Cyclin D1 | AA487486 | 1.06 | 1.00 | 0.35 | |
| Cyclin G1 | AA082943 | 1.24 | 0.62 | 0.40 | |
| CDC16 (cell division cycle 6 S. cerevisiae homolog) | AA410604 | 1.18 | 0.13 | 0.08 | |
| Cell division cycle 2 G1 to S and G2 to M | AA598974 | 1.03 | 0.25 | 0.18 | |
| Cyclin H | AA454146 | 1.27 | 0.73 | 0.45 | |
| Cell division cycle 25A | H59260 | 1.00 | 0.56 | 0.47 | |
| Cytochrome c oxidase subunit VIa polypeptide 1 | AA482243 | 0.68 | 0.07 | 0.38 | |
| Cytochrome c oxidase subunit VIb | N71160 | 0.68 | 0.09 | 0.10 | |
| Cytochrome c oxidase ubunit VIIa polypeptide 2 (liver) | A1002403 | 0.54 | 0.08 | 0.06 | |
| Cytochrome c oxidase subunit VIIc | AA629719 | 1.11 | 0.38 | 0.36 | |
| Cytochrome P450 subfamily IIIA (niphedipine oxidase) polypeptide 4 | R91078 | 0.91 | 0.45 | 0.41 | |
| Cytochrome _c_-1 | AA447774 | 0.30 | 0.21 | 0.04 | |
| Cytoskeleton | |||||
| Actin gamma 2 smooth muscle enteric | T60048 | 1.62 | 0.22 | 0.13 | |
| Actin-related protein 2/3 complex subunit 3 (21 kDa) | H73961 | 1.58 | 0.19 | 0.12 | |
| Actin-related protein 2/3 complex subunit 5 (16 kDa) | W55964 | 1.74 | 0.13 | 0.09 | |
| Calnexin | AA126265 | 1.28 | 0.22 | 0.15 | |
| Calumenin | R78586/R78585 | 1.37 | 0.19 | 0.11 | |
| Caveolin 1 caveolae protein 22 kDa | AA055835 | 1.48 | 0.09 | 0.03 | |
| Caveolin 3 | AA425319 | 1.59 | 0.06 | 0.03 | |
| Collagen type IX alpha 1 | N69335 | 0.97 | 0.36 | 0.19 | |
| Calmegin | AA778675 | 1.39 | 0.21 | 0.11 | |
| Destrin (actin depolymerizing factor) | AA424824 | 1.37 | 0.22 | 0.21 | |
| Karyopherin (importin) beta 2 | AA481067 | 0.90 | 0.24 | 0.33 | |
| Karyopherin alpha 2 (RAG cohort 1 importin alpha 1) | AA676460 | 0.86 | 0.16 | 0.17 | |
| Myosin heavy polypeptide 9 nonmuscle | T69926 | 0.79 | 0.19 | 0.20 | |
| Myosin light polypeptide 1 alkali skeletal fast | T52894 | 0.84 | 0.15 | 0.15 | |
| Myosin VA (heavy polypeptide 12 myoxin) | AA025850 | 0.68 | 0.11 | 0.11 | |
| Procollagen-proline 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase) beta | AA426212 | 0.79 | 0.28 | 0.26 | |
| Tubulin alpha brain-specific | AA865469 | 0.71 | 0.20 | 0.24/PICK> | |
| Tubulin beta 2 | AA888148 | 0.73 | 0.22 | 0.26 | |
| Tubulin beta polypeptide | H38210 | 0.83 | 0.27 | 0.31 | |
| Tubulin-specific chaperone a | N93021 | 0.66 | 0.10 | 0.14 | |
| Tubulin-specific chaperone d | A1668870 | 0.78 | 0.17 | 0.22 | |
| Microtubule-associated protein RP/EB family member 1 | AA001749 | 0.82 | 0.11 | 0.11 | |
| Microtubule-associated protein RP/EB family member 3 | AA019549 | 0.80 | 0.19 | 0.18 | |
| Immune response | |||||
| CD9 antigen (p24) | AA412053 | 1.15 | 0.12 | 0.06 | |
| Interleukin enhancer binding factor 2 45 kDa | AA894687 | 0.87 | 0.21 | 0.26 | |
| Nuclear factor interleukin-3 regulated | AA633811 | 0.81 | 0.29 | 0.27 | |
| Proteasoma and ubiquitin | |||||
| Ubiquitin carrier protein | AA464729 | 0.75 | 0.18 | 0.19 | |
| Ubiquitin carrier protein E2-C | AA430504 | 0.78 | 0.24 | 0.26 | |
| Ubiquitin-conjugating enzyme E2 variant 2 | AA448676 | 0.76 | 0.24 | 0.27 | |
| Ubiquitin-conjugating enzyme E2B (RAD6 homolog) | AA598492 | 0.80 | 0.23 | 0.26 | |
| Ubiquitin-conjugating enzyme E2D 3 (homologous to yeast UBC4/5) | AA017200 | 0.78 | 0.22 | 0.25 | |
| Ubiquitin-like 1 (sentrin) | AA488626 | 0.78 | 0.25 | 0.26 | |
| Proteasome (prosome macropain) 26S ubunit ATPase 2 | AA251770 | 0.85 | 0.20 | 0.31 | |
| Proteasome (prosome macropain) 26S subunit ATPase 3 | AA282230 | 0.82 | 0.22 | 0.33 | |
| Proteasome (prosome macropain) 26S subunit ATPase 6 | AA424503 | 0.81 | 0.27 | 0.42 | |
| Proteasome (prosome macropain) 26S subunit non-ATPase 4 | AA450227 | 0.74 | 0.17 | 0.30 | |
| Proteasome (prosome macropain) activator subunit 3 (PA28 gamma Ki) | AA486324 | 0.80 | 0.21 | 0.37 | |
| Proteasome (prosome macropain) subunit alpha type 2 | R71913 | 0.71 | 0.20 | 0.32 | |
| Proteasome (prosome macropain) subunit alpha type 3 | AA465237 | 0.82 | 0.17 | 0.29 | |
| Proteasome (prosome macropain) subunit alpha type 5 | AA598815 | 0.70 | 0.21 | 0.32 | |
| Proteasome (prosome macropain) subunit alpha type 6 | AA047338 | 0.82 | 0.50 | 0.66 | |
| Proteasome (prosome macropain) subunit beta type 2 | N53065 | 0.72 | 0.31 | 0.47 | |
| Protein folding | |||||
| Chaperonin-containing TCP1 subunit 2 (beta) | N38959 | 1.01 | 0.49 | 0.37 | |
| Chaperonin-containing CP1 subunit 4 (delta) | AA598637 | 0.99 | 0.26 | 0.17 | |
| Chaperonin-containing TCP1 subunit 6A (zeta 1) | AA872690 | 1.01 | 0.21 | 0.13 | |
| Chaperonin-containing TCP1 subunit 7 (eta) | AA676588 | 1.04 | 0.12 | 0.07 | |
| Heat shock 70-kDa protein 8 | AA629567 | 0.81 | 0.27 | 0.22 | |
| Heat shock factor binding protein 1 | AA664067 | 0.84 | 0.27 | 0.22 | |
| Ribosomal protein | |||||
| 40S ribosomal protein S27 isoform | AA156054 | 1.43 | 0.28 | 0.23 | |
| 60S ribosomal protein L30 isolog | AA063398 | 1.61 | 0.23 | 0.17 | |
| EST moderately similar to A31233 ribosomal protein RS 40K cytosolic (H. sapiens) | T48293 | 0.97 | 0.14 | 0.14 | |
| EST moderately similar to RL1 human 60S ribosomal protein L18A (H. sapiens) | W81118 | 1.12 | 0.21 | 0.15 | |
| Ribosomal protein L18 | T49327 | 0.71 | 0.14 | 0.21 | |
| Ribosomal protein L19 | AA083485 | 0.66 | 0.20 | 0.28 | |
| Ribosomal protein L21 | AA464743 | 0.74 | 0.20 | 0.27 | |
| Ribosomal protein L27a | AA599178 | 0.71 | 0.13 | 0.20 | |
| Ribosomal protein L28 | AA486746 | 0.81 | 0.23 | 0.31 | |
| Ribosomal protein L29 | A1018613 | 0.78 | 0.22 | 0.31 | |
| Ribosomal protein L35 | AA625634 | 0.75 | 0.24 | 0.33 | |
| Ribosomal protein L36a | AA669359 | 0.81 | 0.22 | 0.31 | |
| Ribosomal protein L39 | N54526 | 0.69 | 0.19 | 0.27 | |
| Ribosomal protein 5 | AA496880 | 0.72 | 0.20 | 0.29 | |
| Ribosomal protein L6 | AA629808 | 0.78 | 0.19 | 0.28 | |
| Ribosomal protein S10 | A1611010 | 0.71 | 0.25 | 0.34 | |
| Ribosomal protein S13 | AA629641 | 0.74 | 0.15 | 0.23 | |
| Ribosomal protein S16 | AA668301 | 0.72 | 0.16 | 0.23 | |
| Ribosomal protein S19 | T72208 | 0.80 | 0.15 | 0.23 | |
| Ribosomal protein S23 | N73091 | 0.79 | 0.14 | 0.23 | |
| Ribosomal protein S24 | N27154 | 0.74 | 0.18 | 0.27 | |
| Ribosomal protein S25 | T98662 | 0.81 | 0.14 | 0.22 | |
| Ribosomal protein S27a | AA625632 | 0.84 | 0.34 | 0.43 | |
| Ribosomal protein S4 Y-linked | T69468 | 0.76 | 0.22 | 0.29 | |
| Ribosomal protein S6 | N91584 | 0.78 | 0.24 | 0.33 | |
| Ribosomal protein S7 | AI675707 | 0.80 | 0.16 | 0.24 | |
| Ribosomal protein S8 | AA683050 | 0.72 | 0.14 | 0.22 | |
| EST weakly similar to RL2 human 60S ribosomal protein L22 (H. sapiens) | AA188378 | 1.06 | 0.42 | 0.40 | |
| Riboproteins | |||||
| Heterogeneous nuclear ribonucleoprotein A1 | AA126911 | 0.79 | 0.13 | 0.11 | |
| Heterogeneous nuclear ribonucleoprotein A2/B1 | W02101 | 0.85 | 0.26 | 0.21 | |
| Heterogeneous nuclear ribonucleoprotein D-like | AA598578 | 0.84 | 0.29 | 0.22 | |
| Heterogeneous nuclear ribonucleoprotein H1 (H) | W96058 | 0.77 | 0.22 | 0.18 | |
| Heterogeneous nuclear ribonucleoprotein M | AA504272 | 0.77 | 0.16 | 0.13 | |
| Heterogeneous nuclear ribonucleoprotein H2 (H′) | AA679345 | 0.77 | 0.20 | 0.17 | |
| Heterogeneous nuclear ribonucleoprotein H3 (2H9) | R02069 | 0.84 | 0.22 | 0.17 | |
| Heterogeneous nuclear ribonucleoprotein L | AA398352 | 0.84 | 0.20 | 0.16 | |
| Transcription | |||||
| Polymerase (RNA) II (DNA directed) polypeptide H | A1554561 | 0.59 | 0.09 | 0.07 | |
| Nuclear transcription factor Y, beta | AA130846 | 0.77 | 0.25 | 0.25 | |
| Polymerase (RNA) II | AA458646 | 0.74 | 0.14 | 0.12 | |
| Translation | |||||
| Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | AA489523 | 1.04 | 0.22 | 0.26 | |
| Eukaryotic translation elongation factor 2 | R43766 | 1.00 | 0.17 | 0.20 | |
| Eukaryotic translation initiation factor 2 subunit 3 (gamma, 52 kDa) | AA448301 | 1.06 | 0.36 | 0.35 | |
| Eukaryotic translation initiation factor 3 subunit 4 (delta, 44 kDa) | AA668703 | 1.08 | 0.14 | 0.14 | |
| Eukaryotic translation initiation factor 4E | AA194246 | 0.93 | 0.29 | 0.28 | |
| Eukaryotic translation initiation factor 3 subunit 6 (48 kDa) | AA669674 | 0.92 | 0.33 | 0.30 | |
| Splicing | |||||
| Splicing factor arginine/serine-rich 1 (splicing factor 2 alternate splicing factor) | T65786 | 0.74 | 0.17 | 0.21 | |
| Splicing factor arginine/serine-rich 11 | H56944 | 0.73 | 0.21 | 0.27 | |
| Splicing factor arginine/serine-rich 5 | AA598965 | 0.73 | 0.25 | 0.32 | |
| Splicing factor arginine/serine-rich 9 | AA491213 | 0.74 | 0.21 | 0.25 | |
| Splicing factor proline/glutamine-rich | AA425853 | 0.79 | 0.16 | 0.22 | |
| Capping | |||||
| Capping protein (actin filament) muscle Z-line alpha 2 | AA083228 | 1.16 | 0.36 | 0.27 | |
| Capping protein (actin filament) muscle Z-line beta | W45165 | 1.51 | 0.10 | 0.04 | |
| 7 | Apoptosis | ||||
| CGI-39 protein cell death-regulatory protein GRIM19 | W32281 | 0.39 | 0.13 | 0.15 | |
| P75NTR-associated cell death executor ovarian granulosa cell protein (13 kDa) | R63543 | 0.43 | 0.22 | 0.05 | |
| ATP synthase | |||||
| ATP synthase H+ transporting mitochondrial F0 complex subunit c (subunit 9) isoform 1 | AA046701 | 0.43 | 0.12 | 0.05 | |
| ATP synthase H+ transporting mitochondrial F0 complex subunit e | AA431433 | 0.42 | 0.18 | 0.08 | |
| ATP synthase H+ transporting mitochondrial F0 complex subunit f isoform 2 | AA938704 | 0.51 | 0.14 | 0.14 | |
| ATP synthase H+ transporting mitochondrial F1 complex beta polypeptide | AA708298 | 0.31 | 0.05 | 0.08 | |
| ATP synthase H+ transporting mitochondrial F1 complex delta subunit | AA669314 | 0.28 | 0.33 | 0.04 | |
| ATP synthase H+ transporting mitochondrial F1 complex O subunit | AA873577 | 0.63 | 0.11 | 0.43 | |
| ATPase H+ transporting lysosomal (vacuolar proton pump) 21 kDa | AA457717 | 0.43 | 0.19 | 0.31 | |
| ATPase H+ transporting lysosomal (vacuolar proton pump) 16 kDa | AA486080 | 0.55 | 0.24 | 0.14 | |
| Cell cycle | |||||
| Cell cycle progression 2 protein | AA676387 | 0.56 | 0.22 | 0.13 | |
| CDC37 (cell division cycle 37 S. cerevisiae homolog) | AA458870 | 0.60 | 0.54 | 0.17 | |
| Cell division cycle 27 | T81764 | 0.82 | 0.19 | 0.45 | |
| Cytochrome | |||||
| Cytochrome _b_-245 alpha polypeptide | AA876021 | 0.22 | 0.22 | 0.14 | |
| Cytochrome _b_5 outer mitochondrial membrane precursor | AA406485 | 0.59 | 0.07 | 0.18 | |
| Cytochrome c oxidase subunit Vb | AI688757 | 0.27 | 0.24 | 0.09 | |
| Cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) | AA872125 | 0.46 | 0.08 | 0.28 | |
| Cytochrome coxidase subunit VIII | AA862813 | 0.39 | 0.17 | 0.24 | |
| Cytoskeleton | |||||
| Tubulin alpha 2 | AA626698 | 0.69 | 0.15 | 0.16 | |
| Tubulin gamma 1 | T77733 | 0.51 | 0.18 | 0.13 | |
| Tubulin gamma 2 | AA126760 | 0.71 | 0.23 | 0.15 | |
| Actin-related protein 2/3 complex subunit 4 (20 kDa) | AA865878 | 0.48 | 0.20 | 0.17 | |
| Cofilin 1 (non-muscle) | AI203139 | 0.33 | 0.45 | 0.16 | |
| Collagen type IV alpha 5 (Alport syndrome) | AA029997 | 0.59 | 0.15 | 0.18 | |
| Integrin alpha E (antigen CD103 human mucosal lymphocyt antigen 1 alpha polypeptide) | AA425451 | 0.53 | 0.09 | 0.19 | |
| Integrin beta 4 binding protein | AI017019 | 0.43 | 0.19 | 0.10 | |
| Keratin 16 (focal nonepidermolytic palmoplantar keratoderma) | AA928454 | 0.30 | 0.49 | 0.18 | |
| Keratin 7 | AA485959 | 0.44 | 0.08 | 0.06 | |
| Keratin 8 | AA598517 | 0.19 | 0.18 | 0.04 | |
| Kinesin family member 5A | AA984728 | 0.36 | 0.22 | 0.38 | |
| Kinesin-like 4 | AA430503 | 0.60 | 0.12 | 0.16 | |
| Microtubule-associated protein RP/EB family member 2 | AA608576 | 0.74 | 0.15 | 0.33 | |
| Microtubule-associated proteins 1A/1B light chain 3 | AA460003 | 0.53 | 0.25 | 0.13 | |
| Myogenin (myogenic factor 4) | AI291603 | 0.18 | 0.39 | 0.35 | |
| Myosin light polypeptide 4 alkali atrial embryonic | AA705225 | 0.57 | 0.28 | 0.21 | |
| Myosin light polypeptide 6 alkali smooth muscle and nonmuscle | AA488346 | 0.67 | 0.18 | 0.29 | |
| Myosin phosphatase target subunit 2 | AA463926 | 0.59 | 0.19 | 0.10 | |
| Myosin regulatory light chain 2 smooth muscle isoform | AA877166 | 0.38 | 0.17 | 0.18 | |
| Immunity | |||||
| Interleukin-1 receptor antagonist | T72877 | 0.15 | 0.21 | 0.18 | |
| CD151 antigen | AA443118 | 0.58 | 0.69 | 0.30 | |
| CD84 antigen (leukocyte antigen) | H65107 | 0.76 | 0.19 | 0.30 | |
| Delta sleep-inducing peptide immunoreactor | AA775091 | 0.50 | 0.18 | 0.23 | |
| Macrophage-stimulating 1 (hepatocyte growth factor-like) | T47813 | 0.55 | 0.16 | 0.11 | |
| Major histocompatibility complex class 1 F | AA455292 | 0.71 | 0.34 | 0.20 | |
| Secretory leukocyte protease inhibitor (antileukoproteinase) | AA683520 | 0.35 | 0.34 | 0.10 | |
| Small inducible cytokine subfamily A (Cys-Cys) member 8 (monocyte chemotactic protein 2) | AI268937 | 0.40 | 0.37 | 0.32 | |
| Metabolism | |||||
| Acetyl-coenzyme A acetyltransferase 2 (acetoacetyl coenzyme A thiolase) | R46821 | 0.52 | 0.21 | 0.16 | |
| Adenine phosphoribosyltransferase | AA598510 | 0.45 | 0.18 | 0.08 | |
| Adenylate kinase 1 | W23690 | 0.58 | 0.20 | 0.19 | |
| Carbamoyl-phosphate synthetase 1 (mitochondrial) | T61078 | 0.52 | 0.74 | 0.17 | |
| Carboxypeptidase A2 (pancreatic) | AA844831 | 0.43 | 0.31 | 0.44 | |
| Creatinine kinase (brain) | AA894557 | 0.50 | 0.24 | 0.37 | |
| Galactose-1-phosphate uridylyltransferase | AA857212 | 0.65 | 0.56 | 0.27 | |
| Glucose phosphate isomerase | AA401111 | 0.59 | 0.50 | 0.47 | |
| Glucose-regulated protein 58 kDa | R33030 | 0.52 | 0.29 | 0.69 | |
| Glucose-6-phosphatase transport (glucose-6-phosphate) protein 1 | AA490159 | 0.55 | 0.09 | 0.23 | |
| Glutathione peroxidase 4 (phospholipid hydroperoxidase) | AA454856 | 0.39 | 0.30 | 0.06 | |
| Glutathione _S_-transferase M3 (brain) | R63065 | 0.42 | 0.15 | 0.19 | |
| Glutathione _S_-transferase pi | R33642 | 0.32 | 0.16 | 0.11 | |
| Glutathione synthetase | AA463458 | 0.51 | 0.19 | 0.15 | |
| Guanine nucleotide binding protein (G protein) beta polypeptide 2 | N68166 | 0.57 | 0.19 | 0.13 | |
| Guanylate kinase 1 | AA490902 | 0.53 | 0.15 | 0.10 | |
| Hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A | AA916323 | 0.48 | 0.16 | 0.16 | |
| Insulin-induced gene 1 | H59620 | 0.31 | 0.10 | 0.22 | |
| Isocitrate dehydrogenase 3 (NAD+) gamma | AA459380 | 0.29 | 0.18 | 0.17 | |
| Isopentenyl-diphosphate delta isomerase | H08820 | 0.49 | 0.40 | 0.17 | |
| Phosphofructokinase, liver | W72140 | 0.43 | 0.11 | 0.16 | |
| Phosphogluconate dehydrogenase | AA598759 | 0.43 | 0.19 | 0.09 | |
| Phosphoglycerate kinase 1 | AA599187 | 0.45 | 0.18 | 0.15 | |
| Phosphoribosylglycinamide formyltransferase phosphoribosylglycinamide synthetase | AA598487 | 0.44 | 0.07 | 0.13 | |
| Pyruvate dehydrogenase (lipoamide) beta | AA521401 | 0.57 | 0.53 | 0.18 | |
| Transaldolase 1 | AA955007 | 0.51 | 0.15 | 0.30 | |
| Aldehyde dehydrogenase 3 family member B1 | N93686 | 0.61 | 0.59 | 0.25 | |
| Aldehyde dehydrogenase 7 family member A1 | AA102646 | 0.66 | 0.09 | 0.47 | |
| Aldolase A fructose-bisphosphate | AA775241 | 0.42 | 0.26 | 0.06 | |
| Aldolase C fructose-bisphosphate | R39463 | 0.38 | 0.25 | 0.18 | |
| Alkaline phosphatase intestinal | AA190871 | 0.62 | 0.13 | 0.24 | |
| Alkylation repair alkB homolog | AA609609 | 0.46 | 0.19 | 0.10 | |
| Aminoacylase 1 | AA402915 | 0.59 | 0.08 | 0.26 | |
| Argininosuccinate synthetase | AA676466 | 0.35 | 0.21 | 0.04 | |
| Succinate dehydrogenase complex subunit A flavoprotein (Fp) | T70043 | 0.48 | 0.20 | 0.10 | |
| Succinate dehydrogenase complex subunit B iron sulfur (Ip) | AA463510 | 0.45 | 0.28 | 0.18 | |
| NADH dehydrogenase (ubiquinone) 1 subcomplex unknown 1 (6 kDa KFYI) | AA460251 | 0.63 | 0.14 | 0.10 | |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 1 (7.5 kDa MWFE) | AA111999 | 0.57 | 0.29 | 0.10 | |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 2 (8 kDa B8) | AA425211 | 0.54 | 0.17 | 0.16 | |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 7 (14.5 kDa B14.5a) | AA022627 | 0.53 | 0.16 | 0.15 | |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 9 (39 kDa) | AA598884 | 0.34 | 0.08 | 0.12 | |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 (16 kDa SGDH) | N93053 | 0.51 | 0.16 | 0.08 | |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex 7 (18 kDa B18) | AA428058 | 0.43 | 0.07 | 0.19 | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 4 (18 kDa) (NADH-coenzyme Q reductase) | AA055102 | 0.24 | 0.13 | 0.08 | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 8 (23 kDa) (NADH-coenzyme Q reductase) | AA127014 | 0.34 | 0.10 | 0.15 | |
| B-cell CLL/lymphoma 7A | AA281583 | 0.46 | 0.43 | 0.11 | |
| Proteosome | |||||
| Proteasome (prosome macropain) 26S subunit ATPase 5 | AI348774 | 0.51 | 0.20 | 0.08 | |
| Proteasome (prosome macropain) 26S subunit non-ATPase 2 | AA454852 | 0.66 | 0.08 | 0.17 | |
| Proteasome (prosome macropain) 26S subunit non-ATPase 8 | AA464557 | 0.41 | 0.09 | 0.07 | |
| Proteasome (prosome macropain) 26S subunit non-ATPase 9 | AA403126 | 0.49 | 0.19 | 0.10 | |
| Proteasome (prosome macropain) activator subunit 2 (PA28 beta) | H65395 | 0.61 | 0.19 | 0.22 | |
| Proteasome (prosome macropain) subunit alpha type 7 | AA863149 | 0.54 | 0.20 | 0.19 | |
| Proteasome (prosome macropain) subunit beta type 10 | T54166 | 0.50 | 0.16 | 0.19 | |
| Proteasome (prosome macropain) subunit beta type 3 | AA620580 | 0.50 | 0.17 | 0.17 | |
| Ubiquinol-cytochrome c reductase (6.4 kDa) subunit | R46837 | 0.60 | 0.29 | 0.14 | |
| Ribosomal protein | |||||
| Ribosomal protein L36 | AI475653 | 0.65 | 0.07 | 0.29 | |
| Ribosomal protein S28 | AA856556 | 0.43 | 0.20 | 0.09 | |
| Ribosomal protein S29 | N93715 | 0.46 | 0.25 | 0.26 | |
| Ribosomal protein S5 | AA456616 | 0.39 | 0.26 | 0.32 | |
| Ubiquitin-conjugating enzyme E2M (homologous to yeast UBC12) | AA449119 | 0.62 | 0.19 | 0.36 | |
| Transcription | |||||
| Transcription elongation factor B (SIII) polypeptide 2 (18 kDa elongin B) | AA630017 | 0.58 | 0.08 | 0.14 | |
| Transcription factor CP2 | AA488618 | 0.35 | 0.28 | 0.09 | |
| Activating transcription factor 4 (_tax_-responsive enhancer element B67) | AA600217 | 0.38 | 0.32 | 0.24 | |
| Splicing factor 3a subunit 3 60 kDa | R43015 | 0.46 | 0.11 | 0.18 | |
| Polymerase (RNA) II (DNA-directed) polypeptide L (7.6 kDa) | AA873691 | 0.49 | 0.11 | 0.20 | |
| Polymerase (RNA) mitochondrial (DNA directed) | R31115 | 0.31 | 0.17 | 0.13 | |
| TATA box binding protein (TBP)-associated factor RNA polymerase II I 28 kDa | N92711 | 0.57 | 0.35 | 0.15 | |
| Translation | |||||
| Eukaryotic translation elongation factor 1 gamma | AA620477 | 0.57 | 0.12 | 0.06 | |
| Eukaryotic translation initiation factor 3 subunit 2 beta (36 kDa) | AA936783 | 0.54 | 0.10 | 0.15 | |
| Eukaryotic translation initiation factor 3 subunit 8 (110 kDa) | AA598863 | 0.38 | 0.19 | 0.14 |
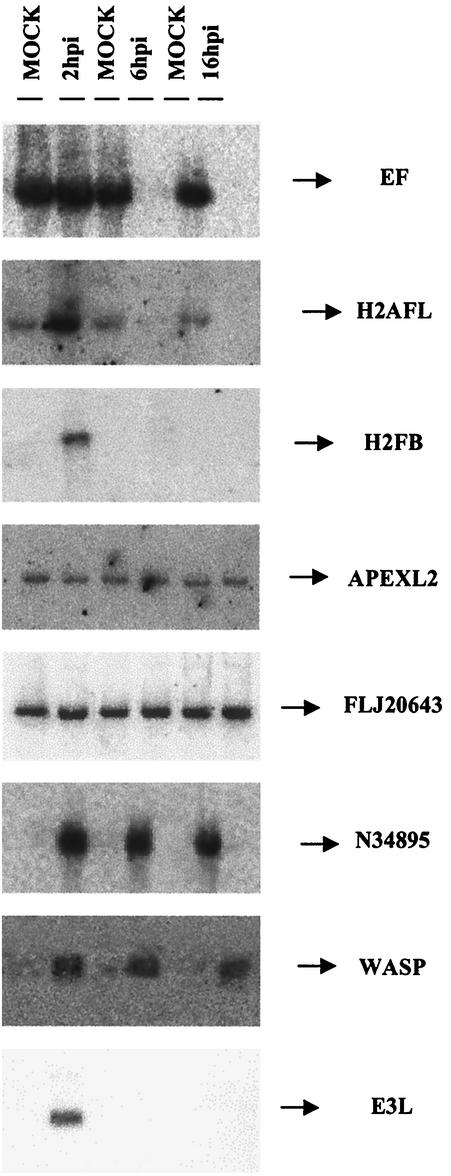
Target verification by Northern blot analysis and real-time RT-PCR of representative genes.
Selected genes with different post-VV infection expression patterns, as derived from microarray results, were chosen for target verification by Northern blot analysis. Total RNA was purified from uninfected or VV-infected cells at 2, 6, and 16 h postinfection, fractionated by gel electrophoresis, blotted, and probed with 32P-labeled PCR products that were spotted on the microarray. The RNA preparation used for this analysis was the same as that used in the microarray. The amount of RNA spotted on the blot was normalized based on rRNA content. In all cases, the Northern blot confirmed the microarray results (Fig. 3). Whereas the expression pattern of the elongation factor gene (EF) was still detected at 2 h postinfection, strong inhibition was observed at 6 and 16 h postinfection compared to the control sample. A peak of histone L gene H2AFL mRNA expression was reached at 2 h postinfection; a similar expression pattern was shown by the histone H2FB gene. A constitutive expression pattern of the apurinic/apyridiminic endonuclease gene (APEXL2) and the EST FLJ20643 was observed in the presence and absence of VV. EST N34895 expression was detected only following infection. Strong upregulation of WASP expression was also observed during VV infection. The expression of the VV E3L early gene was analyzed as a control for virus infection.
FIG.3.
Validation of microarray data by Northern blot. Total RNA (20 μg) purified from uninfected and VV-infected cells at 2, 6, and 16 h postinfection was hybridized with probes derived from PCR products that were spotted on the microarray. The genes included in the autoradiogram are N34895 (EST), WASP (Wiskott-Aldrich syndrome protein), H2AFL (histone L), H2FB (histone B), EF (elongation factor), APEXL2 (apurinic/apyridiminic endonuclease), FLJ20643 (EST), and E3L (VV protein).
As an alternative to the Northern blot data, real-time RT-PCR was used to verify the transcriptional change in selected genes, as detected by microarray. Three genes were chosen for analysis: two were upregulated (PCNT2 and WASP) and one was not altered (IL-6ST). HPRT was used as an internal control. As shown in Table 4, the RT-PCR data confirmed the microarray results, showing the same relative regulation of transcription of the selected genes and hence validating the Northern and the microarray findings. The absolute values are not identical between the microarray and RT-PCR data, but this is probably due to the intrinsic differences between the techniques.
TABLE 4.
Confirmation of microarray data by real-time RT-PCR
| Gene product | Changea (fold) | |||||
|---|---|---|---|---|---|---|
| Microarray | RT-PCR | |||||
| 2 hpi | 6 hpi | 16 hpi | 2 hpi | 6 hpi | 16 hpi | |
| PCNT2 | 3.20 | 7.11 | 1.56 | 3.82 | 8.54 | 2.21 |
| WASF1 | 2.35 | 2.43 | 3.81 | 2.05 | 3.30 | 4.27 |
| IL-6ST | 1.02 | 1.35 | 0.98 | 1.08 | 1.53 | 1.07 |
Target verification by Western blot and immunofluorescence of representative cellular proteins.
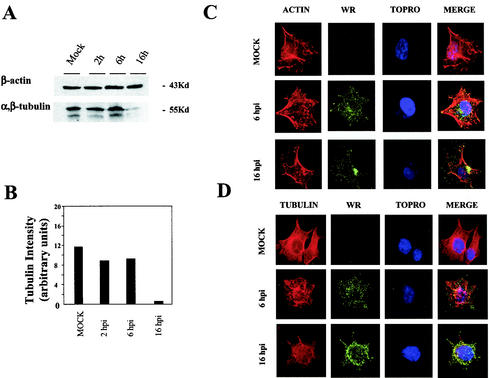
To correlate changes in transcription with protein levels, we studied the effect of infection on some cytoskeletal components by analyzing actin and tubulin expression levels by immunoblot. Although actin gene expression was downregulated at 6 h postinfection in microarray analysis (see Table 3), the protein was present in equal amounts in mock- and VV-infected cells at 2, 6, and 16 h postinfection (Fig. 4A). This is probably due to the high stability of actin. After virus infection, the microarray data indicated that tubulin expression was downregulated (Table 3), and this was confirmed by Western blot analysis of tubulin protein levels (Fig. 4A). Tubulin degradation was quantified by densitometry, which showed that tubulin expression decreased 10-fold at 16 h postinfection compared to mock-infected cells (Fig. 4B). These data were confirmed by immunofluorescence staining with fluorescein-labeled phalloidin and antitubulin antibodies (Fig. 4C and D). These findings show that the correlation between levels of mRNAs and protein depends on the stability of a given protein.
FIG. 4.
Changes in cytoskeletal components after VV infection. (A) Western blot comparison of actin and tubulin protein levels in lysates of mock- and VV-infected cells. Total proteins (100 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-β-actin and anti-β-tubulin antibodies. (B) Densitometric quantification of tubulin protein. (C) Immunofluorescence analysis of the effect of VV infection on actin fibers in HeLa cells. Mock- and VV-infected cells at 6 and 16 h postinfection (hpi) were labeled with monoclonal antibody C3α14k to detect the A27L viral p14 protein, followed by the appropriate phalloidin-conjugated secondary antibody and ToPro reagent. The samples were analyzed by confocal immunofluorescence microscopy. (D) Immunofluorescence analysis of the effect of VV infection on tubulin.
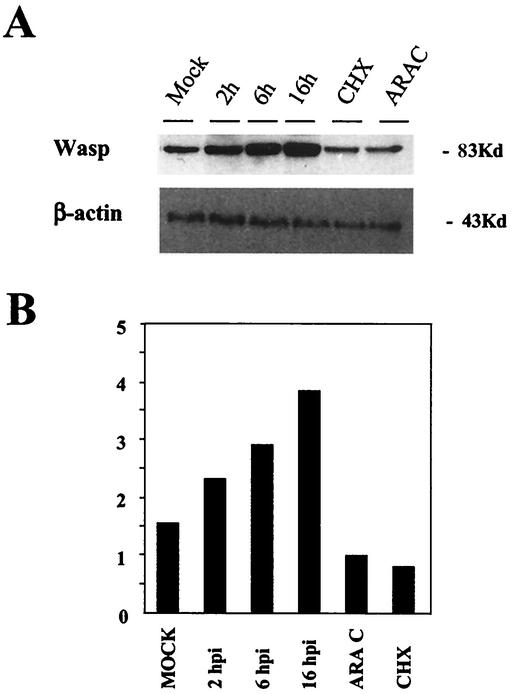
Since N-WASP has been shown to play an important role in the actin-mediated motility of VV as a mechanism to viral spread between cells (12-13, 19, 34), we next focused our work on the WASP protein family member WASF1, which was upregulated more than twofold in VV-infected cells compared to uninfected control cells (Table 1). In agreement with the Northern, microarray, and real-time RT-PCR data, a clear increase in WASP protein levels was evident in VV-infected cells by Western blot analysis (Fig. 5). The WASP protein signal increased early in infection and reached a maximal level at 16 h postinfection. At this time, the amount of WASP in VV-infected cells was threefold higher than in control cells. The increase in WASP protein required viral DNA replication, as observed by infecting cells in the presence of adenosine arabinosidede, an inhibitor of VV DNA synthesis. Moreover, the increase in the WASP protein level required de novo protein synthesis, because its accumulation was prevented by cycloheximide treatment. This result discards the possibility that VV infection might be increasing the WASP protein level by enhancing protein stability.
FIG. 5.
Upregulation of WASP protein after VV infection. (A) Comparison of WASP protein levels by Western blot from lysates of mock- and VV-infected cells at 2, 6, and 16 h postinfection (hpi) and from cells treated at infection with cycloheximide (CHX; 100 μg/ml) and adenosine arabinoside (ARAC; 50 μg/ml) for 16 h. Total proteins (100 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-WASP and anti-β-actin antibodies. (B) Densitometric quantification of WASP protein.
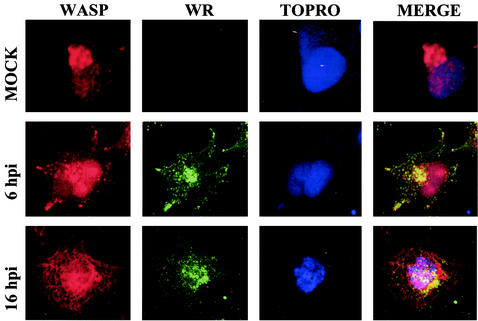
Given the known involvement of N-WASP in VV-mediated cytoskeletal changes (19, 34), we next analyzed the subcellular localization of WASP. To this end, we performed comparative immunofluorescence analysis of intracellular WASP distribution between VV-infected cells at 6 and 16 h postinfection and uninfected cells. While in uninfected cells WASP appeared in defined regions of the cytoplasm, with time of VV infection, there was a clear increase in WASP protein intensity, with a subcellular distribution distinct from that of uninfected cells (Fig. 6). The subcellular distribution of WASP was compared with that of the VV membrane protein p14 (A27L gene). At late times postinfection, p14 localized as a soluble component but mostly as part of intracellular mature virus, giving a single-dot appearance. Image merging indicated some colocalization of WASP with p14 (Fig. 6).
FIG. 6.
Immunofluorescence analysis showing WASP redistribution in VV-infected HeLa cells. Mock- and VV-infected cells at 6 and 16 h postinfection (hpi) were double-labeled with monoclonal antibody C3α14k to detect the A27L viral p14 protein and anti-WASP antibody, followed by the appropriate fluorescent secondary antibody and ToPro reagent. Cells were visualized by confocal immunofluorescence microscopy.
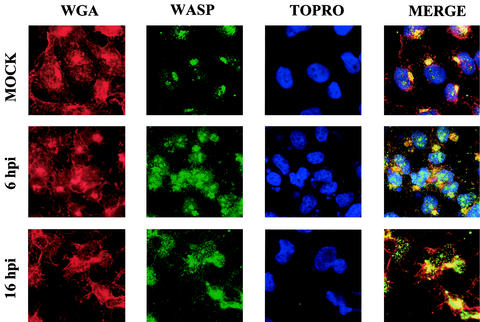
To analyze further the intracellular localization of WASP in infected cells, we used antibodies to the wheat germ antigen, a specific marker for Golgi structures. In uninfected cells, WASP protein was present mainly in the Golgi region, whereas in infected cells, WASP protein localized in cytoplasmic structures that lacked Golgi organization (Fig. 7). Since a significant fraction of mature virions colocalized with WASP protein (compare the dots in Fig. 6 and 7, merge panel), these observations suggest a possible functional and/or biochemical interaction between VV proteins and WASP.
FIG. 7.
VV infection changes the subcellular localization of WASP from the Golgi complex. HeLa cells cultured on coverslips were infected with VV for 6 and 16 h. Cells were treated with anti-WASP antibody, followed by an appropriate fluorescent secondary antibody, ToPro, and fluorescent anti-wheat germ antigen (WGA).
DISCUSSION
With cDNA microarrays, we analyzed the expression of approximately 15,000 human cDNAs during VV infection of HeLa cells and identified genes that are differentially expressed and belong to a variety of cellular pathways. This work describes for the first time the global change in the cellular mRNA content in response to a poxvirus infection. Our results show that VV infection has a major impact on the expression pattern of specific cellular genes. We performed our analysis at 2, 6, and 16 h postinfection and observed drastic changes in the amount of mRNA in VV-infected compared to uninfected cells. This led us to analyze the temporal behavior of host genes over infection that could bring new clues on the role that these genes play in the context of the VV-infected cell.
The majority (90%) of the differentially expressed genes showed expression patterns characterized by diminished levels compared to mock-infected controls. Expression of host genes that govern vital cell processes such as replication, transcription, and translation was downregulated during the course of infection. The expression of other genes involved in apoptosis or the proteasome-ubiquitin degradation pathway was also repressed. These findings support the notion that cytocidal viruses often lead to profound changes in host gene expression (6, 30). Notably, our results indicate that VV infection upregulates expression of a discrete number of genes with known functions and several ESTs with unknown functions, not described previously as being upregulated by other viruses (9, 15, 29).
The global reduction in mRNA content observed in VV-infected cells might be due to a general repression of host transcription or increased degradation rates of cellular mRNAs. At this time we cannot distinguish between these two possibilities. However, since VV infection affects both the synthesis and the stability of cellular mRNAs, the data obtained here probably reflect the interference of VV with multiple steps in host gene expression (5, 30, 33).
More interesting is the fact that VV infection increased the level of some specific host mRNAs. This could be due to a general response of the cell to stress situations or may be a more specific response of the cell to poxvirus infection. Notably, some of these genes encode structural components of the cell, such as Golgi resident protein 60 (GCP-60), claudin 3, and pericentrin. Although the roles that these proteins play in the cell are well established (1, 38-39) the biological meaning of their upregulation by VV is not immediately clear. VV could benefit from increased expression of host genes encoding structural components of the cell, since production of progeny virus requires the integrity of some cellular structures. For instance, formation of extracellular envelope virus requires the Golgi apparatus for intracellular mature virus acquisition of an extra double membrane (17).
Another gene that underwent upregulation after VV infection was EST N34895, a homolog of coxsackievirus and adenovirus receptors. One can speculate that N34895 might be involved in some step of VV entry, although further experiments will be necessary to test this possibility.
Among the genes upregulated by VV infection, perhaps the function of WASP has the most obvious biological meaning in VV-infected cells. VV infection triggers transduction pathways, and WASP is an effector molecule involved in signal transmission from tyrosine kinase receptors and small GTPases to the actin cytoskeleton (8, 26, 40-41). N-WASP, a related member of the WASP family, is implicated in the actin-mediated motility of VV as a mechanism to spread virus between cells (12-13, 19, 34). The molecular mechanism of this process involves the Src family kinase-dependent tyrosine phosphorylation of tyrosine residues 112 and 132 of viral transmembrane protein A36R, the tail nucleator (19, 27, 37). These posttranslational modifications regulate a direct recruitment of Nck, Grb2, WAS-interacting protein, and N-WASP to the virus particle and actin polymerization via the Arp2/3 complex (16, 23, 25).
We observed a clear upregulation of WASP expression during VV infection, with a correlation between WASP mRNA and protein levels. The increase in WASP protein required viral DNA replication, shown by infecting cells in the presence of adenosine arabinoside, an inhibitor of VV DNA synthesis. Moreover, the increase in WASP protein level required de novo protein synthesis because it was prevented by cycloheximide treatment. Although the biological relevance of this upregulation remains to be established, it can be speculated in view of the functions assigned to N-WASP that WASP protein might be involved in some step of virus budding or maturation by regulating the organization of actin-containing cytoskeleton, thus facilitating the spread of the virus. Our preliminary observations in tissue-specific knockouts of WAS in mice suggest a role for WAS family members in the release of VV from infected cells (unpublished data).
In conclusion, this study is the first quantitative analysis of host cell gene expression following VV infection in cultured human cells. The results identify a number of cellular genes whose expression levels are markedly modified by VV infection and suggest potential roles as regulators of viral infection. We demonstrate the usefulness of microarray technology in the study of poxvirus-host interactions. Additional studies with a variety of virus strains, mutants, cell lines, and in vivo animal model systems will be required to obtain a comprehensive pattern of host cell gene expression and biological effects following poxvirus infection. The impact of VV on host gene expression is important considering the potential application of VV as a recombinant vaccine and the threat of bioterrorism with variola virus and related poxviruses.
Acknowledgments
We are indebted to to R. Conde for helpful quantitative PCR assistance, to A. Bernad for kindly providing the WASP antibody, to J. M. Fernández and J. C. Oliveros for bioinformatic assistance, to I. Ventoso, S. Masciotra, and C. Mark for critical review of the manuscript, and to V. Jiménez for expert technical assistance.
This work was supported by the Spanish Ministry of Science and Technology BIO2000-0340P4 and BIO2001-2269 (to M.E.). The Department of Immunology and Oncology was founded and is supported by the Spanish Council (CSIC) and Pharmacia.
REFERENCES
- 1.Adams, C., P. Diadori, L. Schoenroth, and M. Fritzler. 2000. Autoantibodies in childhood post-varicella acute cerebellar ataxia. Can. J. Neurol. Sci. 4**:**316-320. [DOI] [PubMed] [Google Scholar]
- 2.Bablanian, R., B. Baxt, J. A. Sonnabend, and M. Esteban. 1978. Studies on the mechanism of vaccinia virus cytopathic effects. II. Early cell rounding is associated with virus polypeptide synthesis. J. Gen. Virol. 39:403-413. [DOI] [PubMed] [Google Scholar]
- 3.Bablanian, R., G. Coppola, S. Scribani, and M. Esteban. 1981. Inhibition of protein synthesis by vaccinia virus. IV. The role of low-molecular-weight viral RNA in the inhibition of protein synthesis. Virology 112**:**13-24. [DOI] [PubMed] [Google Scholar]
- 4.Bablanian, R., M. Esteban, B. Bax, and J. A. Sonnabend. 1978. Studies on the mechanisms of vaccinia virus cytopathic effects. I. Inhibition of protein synthesis in infected cells is associated with virus-induced RNA synthesis. J. Gen. Virol. 39:391-402. [DOI] [PubMed] [Google Scholar]
- 5.Becker, Y., and W. K. Joklik. 1964. Messenger RNA in cells infected with vaccinia virus. Proc. Natl. Acad. Sci. USA 51**:**577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolt, G., K. Berg, and M. Blixenkrone-Moller. 2002. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 83**:**1157-1165. [DOI] [PubMed] [Google Scholar]
- 7.Buller, R. M. L., and G. J. Palumbo. 1991. Poxvirus pathogenesis. Microbiol. Rev. 55**:**80-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron, E. 2002. Regulation of Wiskott-Aldrich syndrome protein and related molecules. Curr. Opin. Cell Biol. 14**:**82-87. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional target of human papillomavirus type 31. J. Virol. 74**:**4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378**:**636-638. [DOI] [PubMed] [Google Scholar]
- 11.Esteban, M. 1984. Defective vaccinia virus particles in interferon-treated infected cells. Virology l33:220-227. [DOI] [PubMed]
- 12.Frischknecht, F., S. Cudmore, V. Moreau, I. Reckmann, S. Rottger, and M. Way. 2001. Tyrosine phosphorylation is required for actin based motility of vaccinia virus but not for Listeria or Shigella. Curr. Biol. 9**:**89-92. [DOI] [PubMed] [Google Scholar]
- 13.Frischknecht, F., V. Moreau, S. Rottger, I. Reckmann, C. Superti-Furga, and M. Way. 1999. Actin based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 404**:**1007-1011. [DOI] [PubMed] [Google Scholar]
- 14.García de la Nava, J., Franco D. Santaella, Cuenca J. Alba, J. M. Carazo, O. Trelles, and A. Pascual-Montano. 2002. Engene: the processing and exploratory analysis of gene expression. Bioinformatics 19**:**657-658. [DOI] [PubMed] [Google Scholar]
- 15.Geiss, G. K., R. E. Bumgarner, M. C. An, A. B. Agy, A. B. van't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2001. Large-scale monitoring of host gene expression during HIV-1 infection with cDNA microarrays. Virology 266**:**8-16. [DOI] [PubMed] [Google Scholar]
- 16.Higgs, H. N., and T. D. Pollard. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70**:**649-676. [DOI] [PubMed] [Google Scholar]
- 17.Hiller, G., and K. Weber. 1982. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55**:**651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiller, G., K. Weber, L. Schneider, C. Parajsz, and C. Jungwirth. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98**:**142-153. [DOI] [PubMed] [Google Scholar]
- 19.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154**:**389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, G. P., S. J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus genome. Virology 196**:**196., 381-401. [DOI] [PubMed]
- 21.Jones, J., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77**:**1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohonen, T. 1997. Self-organizing maps, 2nd ed. Springer-Verlag, Berlin, Germany.
- 23.Martinez-Quiles, N., R. I. M. Rohatgi, M. Medina, S. P. Saville, H. Miki, H. Yamaguchi, T. Takenawa, J. H. Hartwig, R. S. Geha, and N. Ramesh. 2001. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat. Cell Biol. 3:484-491. [DOI] [PubMed] [Google Scholar]
- 24.Massung, R. F., J. J. Esposito, L. Liu, J. Qi, T. R. Utterback, J. C. Knight, L. Aubin, L., T. E. Yuran, J. M. Parson, V. N. Loparev, et al. 1993. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature 224**:**1280-1284. [DOI] [PubMed] [Google Scholar]
- 25.McKarty, J. H. 1998. The Nck SH2/SH3 adaptador protein: a regulator of multiple intracellular signal transduction events. Bioassays 20**:**913-921. [DOI] [PubMed] [Google Scholar]
- 26.Miki, H., K. Minura, and T. Takenawa. 1996. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream or tyrosine kinases. EMBO J. 15**:**5326-5335. [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau, V., F. Frischknecht, I. Reckmann, G. Superti-Furga, and M. Way. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2**:**441-448. [DOI] [PubMed] [Google Scholar]
- 28.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2649-2885. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Press, New York, N.Y.
- 29.Nees, N., J. M. Geoghegan, T. Hyman, S. Frank, L. Miller, and C. I. Woodworth. 2001. Papillomavirus type 16 oncogenes downregulate expression interferon-responsive genes and upregulate proliferation-associated and NFκB-responsive genes in cervical keratinocytes. J. Virol. 75**:**4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedley, S., and R. J. Cooper. 1984. The inhibition of HeLa cell RNA synthesis following infection with vaccinia virus. J. Gen. Virol. 65**:**1687-1697. [DOI] [PubMed] [Google Scholar]
- 31.Ploubidou, A., V. Moreau, K. Ashman, I. Reckmann, C. González, and M. Way. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19**:**3932-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portis, T., and R. Longnecker. 2003. Epstein-Barr virus LMP2A interferes with global transcription factor regulation when expressed during B-lymphocyte development. J. Virol. 77:105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, A. P., and B. E. Roberts. 1983. Vaccinia virus induces cellular mRNA degradation. J. Virol. 47**:**529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3**:**992-1000. [DOI] [PubMed] [Google Scholar]
- 35.Rottger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus intracellular envelope virus membrane proteins and their roles in intracellular envelope virus assembly and actin tail formation. J. Virol. 73**:**2863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanderson, C. M., M. Way, and G. L. Smith. 1998. Virus-induced cell motility. J. Virol. 72**:**1235-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scaplehorn, N., A. Holmstrom, V. Moreau, F. Frischknecht, I. Reckmann, and M. Way. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12:740-745. [DOI] [PubMed] [Google Scholar]
- 38.Sharon, H., A. Purohit, J. Doxey, and R. B. Vallee. 2000. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 42**:**32763-32768. [DOI] [PubMed] [Google Scholar]
- 39.Sohda, M., Y. Misumi, A. Yamamoto, A. Yano, N. Nakamura, and Y. Ikehara. 2001. Identification and characterization of a novel Golgi protein, GC60, that interacts with the integral membrane protein giantin. J. Biol. Chem. 48**:**45298-45306. [DOI] [PubMed] [Google Scholar]
- 40.Symons, M., J. M. Derry, B. Karlak, S. Jiang, V. Lemahieu, F. McCormick, U. Francke, and A. Abo. 1996. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase Cdc42Hs, is implicated in actin polymerization. Cell 84**:**723-734. [DOI] [PubMed] [Google Scholar]
- 41.Takenawa, T., and H. Miki. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114**:**1801-1809. [DOI] [PubMed] [Google Scholar]
- 42.Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, E. Dmitrovsky, E. S. Lander, and T. R. Golub. 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96**:**2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+ T-cell lines. J. Virol. 77**:**1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vos, J. C., and H. G. Stunnenberg. 1988. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 7**:**3487-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, H., J. P. Cong, G. Mantora, T. Gineras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95**:**14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]