C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis (original) (raw)
Abstract
Chromosome segregation and X-chromosome gene regulation in Caenorhabditis elegans share the component MIX-1, a mitotic protein that also represses X-linked genes during dosage compensation. MIX-1 achieves its dual roles through interactions with different protein partners. To repress gene expression, MIX-1 acts in an X-chromosome complex that resembles the mitotic condensin complex yet lacks chromosome segregation function. Here we show that MIX-1 interacts with a mitotic condensin subunit, SMC-4, to achieve chromosome segregation. The SMC-4/MIX-1 complex positively supercoils DNA in vitro and is required for mitotic chromosome structure and segregation in vivo. Thus, C. elegans has two condensin complexes, one conserved for mitosis and another specialized for gene regulation. SMC-4 and MIX-1 colocalize with centromere proteins on condensed mitotic chromosomes and are required for the restricted orientation of centromeres toward spindle poles. This cell cycle-dependent localization requires AIR-2/AuroraB kinase. Depletion of SMC-4/MIX-1 causes aberrant mitotic chromosome structure and segregation, but not dramatic decondensation at metaphase. Moreover, SMC-4/MIX-1 depletion disrupts sister chromatid segregation during meiosis II but not homologous chromosome segregation during meiosis I, although both processes require chromosome condensation. These results imply that condensin is not simply required for compaction, but plays a more complex role in chromosome architecture that is essential for mitotic and meiotic sister chromatid segregation.
Keywords: Mitosis, meiosis, condensin, chromosome segregation, centromere, dosage compensation
Chromosomes must undergo extensive structural reorganization, compaction, and untangling during mitosis to ensure their faithful segregation to daughter cells. Mitotic chromosome condensation begins after DNA replication, with compaction of two connected sister chromatids at prophase. Condensation continues as chromosomes attach to the mitotic spindle during prometaphase and is maximal once chromosomes align along the spindle equator at metaphase. The condensed sisters resolve their entanglements and segregate during anaphase. Chromosomes decondense as the cycle progresses to telophase and cell division occurs. Condensation serves several purposes. Interphase chromosomes are much longer than the length of the cell, making reduction of chromosome length essential to pull chromosomes into discrete compartments. Condensation also facilitates the decatenation of tangles between intertwined sister chromatids (Holmes and Cozzarelli 2000) and provides extra mechanical strength to withstand the forces of the mitotic spindle.
Critical to mitotic chromosome condensation is the conserved five-protein condensin complex (Hirano 2000). Condensin includes members of an ATPase superfamily, the SMC (structural maintenance of chromosomes) proteins, and non-SMC proteins. SMC proteins play important roles in chromosome mechanics other than condensation, including sister chromatid cohesion and recombination-mediated repair (Cobbe and Heck 2000). To carry out these different functions, the four classes of eukaryotic SMC proteins associate with each other in heterodimeric pairs. Cohesin, the complex required for sister chromatid cohesion, includes an SMC1–SMC3 heterodimer pair, whereas condensin includes an SMC2–SMC4 heterodimeric pair.
Two lines of evidence implicated condensin in chromosome condensation. In Xenopus, sperm DNA added to mitotic egg extracts assembled into condensed mitotic chromosomes, and depletion of condensin disrupted condensation (Hirano and Mitchison 1994; Hirano et al. 1997). In yeast, mutational inactivation of condensin subunits increased the average distance between two fluorescently labeled loci on a mitotic chromosome (Strunnikov et al. 1995; Lavoie et al. 2000; Ouspenski et al. 2000). However, a Drosophila condensin mutation disrupted sister chromatid resolution without disrupting the longitudinal shortening of chromosomes, implying that some compaction occurs without condensin activity (Steffensen et al. 2001). Condensin is also essential for chromosome segregation. Mitotic chromosomes often fail to separate in fly and fission yeast mutants defective in condensin components, leaving chromatin bridges between sister chromatids at anaphase (Saka et al. 1994; Bhat et al. 1996; Sutani et al. 1999; Steffensen et al. 2001).
In vitro assays suggested a molecular mechanism by which condensin modifies chromosome structure. In the presence of a topoisomerase, the Xenopus condensin complex introduced ATP-dependent topological changes in naked plasmid DNA. On the basis of these changes, condensin was proposed to organize DNA by introducing global positive writhe, which promotes condensation by stabilizing positively supercoiled chromatin loops (Kimura and Hirano 1997; Kimura et al. 1999). The relationship, however, between the in vitro activities on naked DNA and condensin function on chromosomes in living cells is not known.
Condensin activity must be temporally restricted, so that chromosomes are condensed at mitosis for segregation but decondensed at interphase for transcription and replication. Mitotic kinases control the localization of condensin to fly and fission yeast chromosomes (Sutani et al. 1999; Giet and Glover 2001) and control the positive supercoiling and condensation activities of purified frog and human condensins (Kimura et al. 1998, 2001).
Although mitotic chromosome condensation may seem mechanistically unrelated to the regulation of gene expression, a molecular component unites these two processes in Caenorhabditis elegans. MIX-1, a homolog of the condensin subunit SMC2, is required both to segregate mitotic chromosomes and to represses transcription of X-linked genes during dosage compensation (Lieb et al. 1998). MIX-1 performs dosage compensation as part of a complex that assembles onto both X chromosomes of hermaphrodites to down-regulate X-linked gene expression by half, thereby equalizing X expression between males (XO) and hermaphrodites (XX). The C. elegans dosage compensation complex resembles condensin, containing both conserved non-SMC components (Lieb et al. 1996) as well as the SMC2 and SMC4 proteins MIX-1 and DPY-27 (Dumpy-27), respectively (Chuang et al. 1994; Lieb et al. 1998). DPY-27 is required for the X localization and dosage compensation function of MIX-1, but has no role in mitosis, suggesting that MIX-1 has a different SMC4 partner for mitosis.
In this study, we addressed how MIX-1 is commissioned for its roles in two distinct cellular processes, dosage compensation and mitosis. We identified a second C. elegans SMC4 protein, SMC-4, and demonstrated its interaction with MIX-1 in a complex that is the functional homolog of mitotic condensin. The complex introduces positive supercoils into plasmid DNA in vitro, a conserved condensin function. It also associates with condensed mitotic chromosomes in a restricted pattern coincident with centromere proteins and is required for the orientation of centromere proteins toward the spindle poles, a role not described previously for condensin. Analysis of chromosome segregation in live embryos depleted for SMC-4 or MIX-1 revealed normal homologous chromosome segregation in meiosis I, but defective sister chromatid segregation in mitosis and meiosis II, even though all three processes require chromosome condensation. Moreover, depletion of SMC-4 or MIX-1 caused aberrant chromosome morphology throughout mitosis and dramatic chromosome decondensation at prometaphase but not metaphase. Thus, condensin is critical for overall chromosome structure and segregation, but is not likely the sole complex required for compaction.
Results
SMC-4 interacts with MIX-1 in a protein complex distinct from the dosage compensation complex
To achieve dosage compensation, MIX-1 interacts with proteins that lack mitotic functions, suggesting that MIX-1 uses other protein partners for mitosis (Lieb et al. 1998). A search of the C. elegans genome revealed proteins with hallmarks of SMC protein structure, including an amino-terminal nucleotide-binding motif (the Walker A motif), two coiled-coil regions separated by a central hinge, and a conserved carboxy-terminal region containing the Walker B motif (Cobbe and Heck 2000). The F35G12.8 protein on chromosome III had the greatest similarity to the SMC4 class of proteins, which include DPY-27 (Fig. 1A), making it a possible mitotic partner for MIX-1. The F35G12.8 gene was named smc-4. Most organisms have single smc-2 and smc-4 genes, but C. elegans has one smc-2 gene and two smc-4 genes.
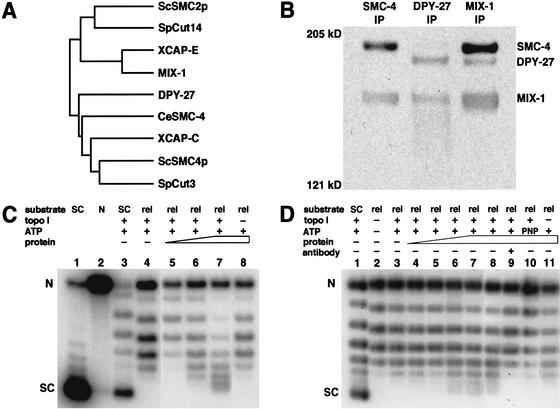
Figure 1.
MIX-1 associates with SMC-4 in a complex that supercoils DNA in vitro. (A) Dendrogram of SMC2 and SMC4 proteins in S. cerevisiae, S. pombe, X. laevis, and C. elegans. Unlike other organisms, C. elegans has two SMC4 proteins, DPY-27 and SMC-4. (B) Western blot of immunoprecipitations (IPs) from embryonic extracts using antibodies against SMC-4 (left), DPY-27 (middle), and MIX-1(right), probed with all three antibodies. (C) Supercoiling assay. Plasmid DNA that is supercoiled (SC, lane 1), nicked (N, lane 2), or supercoiled but partially relaxed with topoisomerase I (lane 3) are markers for the distribution of topoisomers. Relaxed plasmid substrate (rel, lane 4) incubated with ATP, topoisomerase I, and increasing amounts of protein immunoprecipitated by SMC-4 antibody showed a shift in topoisomer distribution toward more supercoiled forms (lanes 5_–_7). Supercoiling activity was not observed without topoisomerase I to cancel the compensatory supercoils (lane 8). (D) Supercoiling assay. A separate experiment shows the topoisomer distribution of both the supercoiled plasmid partially relaxed with topoisomerase I (lane 1) and the relaxed substrate for the supercoiling reaction (lane 2). Supercoiling activity was observed with the addition of increasing amounts of SMC-4 immunoprecipitate (lanes 3_–_8). Activity was blocked by preincubation of the immunoprecipitated protein with SMC-4 antibody (lane 9) or by replacing ATP with a non-hydrolyzable analog, AMP-PNP (lane 10). Supercoiling activity was not observed in the absence of topoisomerase I (lane 11).
SMC-4 could interact with MIX-1 in a mitotic complex, the dosage compensation complex, or could perform an unrelated function. To discriminate among these possibilities, we asked whether affinity-purified antibodies to SMC-4 could immunoprecipitate DPY-27 or MIX-1 from embryonic extracts. As shown in the Western blot of Figure 1B, SMC-4 antibodies coimmunoprecipitated MIX-1, but not DPY-27, indicating that SMC-4 interacts with MIX-1 but is not a component of the dosage compensation complex. Likewise, DPY-27 antibody coimmunoprecipitated MIX-1, but not SMC-4 (Fig. 1B, middle lane). Finally, MIX-1 antibody immunoprecipitated both DPY-27 and SMC-4 (Fig. 1B, right lane). Thus, MIX-1, the sole SMC2 protein in C. elegans, associates with either DPY-27 or SMC-4, and its association with SMC-4 occurs within a complex distinct from the dosage compensation complex.
The complex containing MIX-1 and SMC-4 drives positive DNA supercoiling in vitro, a conserved condensin activity
If MIX-1 and SMC-4 are members of a C. elegans mitotic condensin complex, the complex might alter DNA topology in the same manner as Xenopus and human condensins by introducing positive supercoils into circular DNA in the presence of a type I topoisomerase (Kimura and Hirano 1997; Kimura et al. 1999, 2001). To test the complex containing SMC-4 and MIX-1 for this activity, we immunoaffinity purified the complex by large-scale immunoprecipitation using an SMC-4 peptide antibody (Materials and Methods).
For the supercoiling assay, the complex was incubated with relaxed plasmid DNA and topoisomerase I. Because the plasmid was covalently closed, any changes in twist or writhe introduced by condensin binding would result in compensatory changes elsewhere in the DNA. Only the compensatory supercoils would be relieved by topoisomerase I; any supercoils introduced and protected by condensin would remain. Supercoils could then be detected by deproteinizing the plasmid and separating the topoisomers by gel electrophoresis in the presence of a low concentration of chloroquine.
Although the level of supercoiling activity by the SMC-4-containing complex varied somewhat between protein preparations, positive DNA supercoiling was observed in several different experiments. The two experiments in Figure 1 showed that increasing amounts of SMC-4 immunoprecipitate shifted the distribution of topoisomers toward more supercoiled forms (Fig. 1C, lanes 5–7, and D, lanes 4–8) relative to those of the starting relaxed substrate (Fig. 1C, lane 4, and D, lane 3). Two observations showed that these supercoils are positive. First, chloroquine makes relaxed DNA migrate as positively supercoiled, and condensin makes such DNA migrate even faster. Second, the topoisomers produced by incubating DNA with the SMC-4-containing complex ran slightly faster than the negative supercoils of a control plasmid partially relaxed with topoisomerase I (Fig. 1C, lane 3, and D, lane 1). Supercoiling activity was not observed without topoisomerase I to cancel the compensatory negative supercoils (Fig. 1C, lane 8 and D, lane 11). Moreover, the supercoiling activity was blocked by pre-incubation with SMC-4 antibody (Fig. 1D, lane 9) and by replacing ATP with a nonhydrolyzable analog, AMP-PNP (Fig. 1D, lane 10), indicating that the activity is dependent on the SMC-4 immunocomplex and ATP hydrolysis. Thus, SMC-4, MIX-1, and their associated proteins possess positive ATP-dependent DNA supercoiling activity, as found for Xenopus and human condensins.
SMC-4 and MIX-1 localize to the poleward face of chromosomes at times of the cell cycle when chromosomes are condensed
SMC-4 antibody staining showed that SMC-4 localized to mitotic chromosomes in a dynamic pattern with two striking features. First, SMC-4 localized to chromosomes only at times of the cell cycle when they were condensed. Little or no SMC-4 was observed on decondensed chromosomes during interphase (Fig. 2A,F). SMC-4 localized to chromosomes at prometaphase (Fig. 2B,G), when the six C. elegans chromosomes, but not their sister chromatids, become visibly distinct. SMC-4 persisted through metaphase (Fig. 2C,H) and anaphase (Fig. 2D,I), then disappeared again as chromosomes decondensed and entered telophase (Fig. 2E,J).
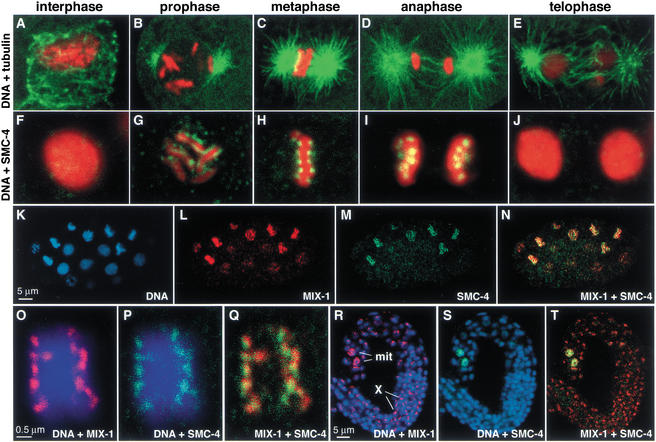
Figure 2.
SMC-4 colocalizes with MIX-1 at the poleward face of condensed mitotic chromosomes but not on X chromosomes. (A_–_E) The mitotic chromosome condensation cycle. Merged confocal images of wild-type embryonic nuclei costained with DNA dye (red) and tubulin antibody (green) show C. elegans chromosomes and microtubules throughout mitosis. (F_–_J) Merged confocal images of wild-type embryonic nuclei costained with SMC-4 antibody (green) and DNA dye (red); overlap appears yellow. (K_–_N) Early wild-type embryo costained with DNA dye (blue, K), MIX-1 antibody (red, L), and SMC-4 antibody (green, M). MIX-1 and SMC-4 colocalize in nuclei with condensed chromosomes (yellow when merged, N). At this early stage, dosage compensation is not active; MIX-1 and other dosage compensation proteins are not localized to the X chromosome. (O_–_Q) Metaphase chromosomes costained with DNA dye (blue), MIX-1 antibody (red, O), and SMC-4 antibody (green, P) show colocalization of MIX-1 and SMC-4 at the poleward face of condensed chromosomes (yellow when merged, Q). (R_–_T) Wild-type larva costained for DNA (blue), MIX-1 (red), and SMC-4 (green). MIX-1 and SMC-4 both stain nuclei of two mitotic cells (mit), but only MIX-1 shows the punctate subnuclear staining characteristic of dosage compensation proteins bound to hermaphrodite X chromosomes (X).
Second, SMC-4 associated only with certain regions of the chromosomes. SMC-4 appeared in a faint speckled pattern during prophase, then appeared in two stripes outlining each chromosome during prometaphase (Fig. 2G). When chromosomes were aligned at metaphase, SMC-4 localized to distinct foci on the poleward sides of the metaphase plate (Fig. 2H). The foci appear as paired spots of staining in cross-section and may represent the poleward sides of paired sister chromatids. This distinctive pattern of localization is remarkably similar to that of proteins bound to the centromere and will be discussed later. C. elegans chromosomes are holocentric, meaning that the centromere, and thus spindle attachment, extends along their entire length. Antibodies against centromere proteins outline the poleward facing length of mitotic chromosomes (Buchwitz et al. 1999; Howe et al. 2001; Moore and Roth 2001; Oegema et al. 2001).
The mitotic localization pattern of MIX-1 was identical to that of SMC-4 throughout the cell cycle. For example, in a wild-type embryo that had not yet implemented dosage compensation, SMC-4 and MIX-1 were both absent from decondensed chromosomes and coincident on condensed chromosomes (Fig. 2K–N). During metaphase, MIX-1 and SMC-4 decorated the chromosomes in the same distinctive poleward pattern (Fig. 2O–Q). Unlike MIX-1, however, SMC-4 did not localize to hermaphrodite X chromosomes, further confirming that SMC-4 is not part of the dosage compensation complex (Fig. 2R–T). The wild-type larva of Figure 2 (R–T) contains two mitotic cells brightly stained by both MIX-1 and SMC-4, but only MIX-1 shows the punctate nuclear staining characteristic of dosage compensation proteins bound to X chromosomes. These results are consistent with SMC-4 interacting with MIX-1 for its role in mitosis, but not for its role in dosage compensation.
Chromosomes depleted of SMC-4 retain a high degree of condensation during metaphase, but have altered morphology and fail to segregate
To assess SMC-4 function, we depleted SMC-4 by double-stranded RNA interference (RNAi) and examined chromosomes in both fixed and living embryos. Analysis of chromosome structure and movement in real time eliminates the difficulty in inferring the progression of a dynamic behavior from static images. This powerful approach was made possible by a strain carrying a GFP-tagged histone H2B transgenic protein (H2B–GFP) that fluorescently labels chromosomes (Praitis et al. 2001). Movies of cell division were made by merging a time-lapse series of GFP images with the corresponding differential interference contrast (DIC) images, in which structures such as the nuclear envelope, spindle, and cytokinesis furrow were readily visualized. Relevant individual images from wild-type and smc-4(RNAi) movies (see Supplemental Material movies 1 and 2 at http://www.genesdev.org) are shown in Figure 3, A–J. In this and all subsequent RNAi experiments, antibody staining was used to show that the targeted protein had been reduced to levels undetectable by immunofluorescence.
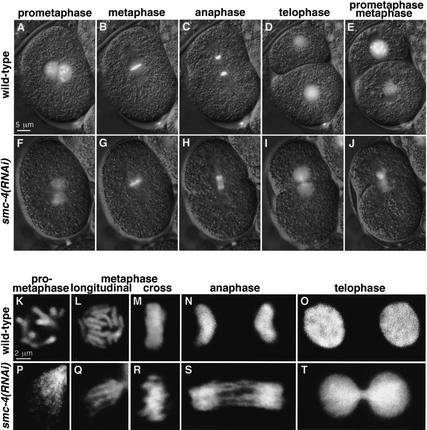
Figure 3.
Depletion of SMC-4 severely disrupted chromosome segregation and prometaphase chromosome organization but not metaphase chromosome compaction. (A_–_J) Merged DIC (gray) and GFP (white) frames from time-lapse movies of wild-type (A_–_E) and smc-4(RNAi) (F–J) embryos carrying histone H2B–GFP. Chromosomes depleted of SMC-4 were stringy rather than rod-shaped at prometaphase (A,F), disorganized but fairly well compacted and aligned at metaphase (B,G), and connected by chromatin bridges rather than separated at both anaphase (C,H) and telophase (D,I). Despite chromosome segregation failure, cytokinesis occurs (D,I) and embryos proceed to the next cell division, at which defects in chromosome morphology are also observed (E,J). (K_–_T) Confocal images of wild-type (K_–_O) and smc-4(RNAi) (P_–_T) nuclei stained for DNA (white). smc-4(RNAi) caused defects in prometaphase chromosome condensation (K,P). Wild-type chromosomes compact and align at metaphase, appearing as a disk of 12 rods when viewed from the spindle pole (L) and as a bar when viewed in cross-section (M). SMC-4-depleted chromosomes appear as fairly compacted, rodlike structures at metaphase, although somewhat abnormal and not precisely aligned (Q,R). Not all 12 chromosomes are in the focal plane (Q). Chromosomes failed to separate at anaphase (N,S) and telophase (O,T).
Severe defects in mitotic chromosome morphology and segregation were observed when SMC-4 was depleted from embryos. The analysis of Figure 3 began after the fertilized embryo had completed meiosis, the maternal and paternal pronuclei had migrated toward one another, and chromosomes were condensing in preparation for the first cell division. In wild-type embryos, prometaphase chromosomes are distinct and rod-shaped (Fig. 3A). However, in smc-4(RNAi) embryos, prometaphase chromosomes have an aberrant wispy morphology, and multiple thin fibers are observed (Fig. 3F). Nevertheless, by metaphase, the chromosomes of RNAi-treated embryos achieve a fairly high degree of compaction compared with chromosomes in wild-type embryos (Fig. 3B,G). Anaphase chromosomes of wild-type embryos separate suddenly and completely. In contrast, anaphase chromosomes of smc-4(RNAi) embryos fail to resolve their connections and remain attached by chromatin bridges (Fig. 3C,H). Telophase chromosomes of wild-type embryos decondense, and two daughter cells with equal genetic material are separated by cytokinesis. Telophase chromosomes of smc-4(RNAi) embryos decondense without fully separating, yet cytokinesis still occurs (Fig. 3D,I). Despite having unresolved chromosome attachments, RNAi-treated embryos attempt division and have similar defects during the next mitosis (Fig. 2E,J). Chromosomes again fail to segregate, but spindle attachment and cytokinesis appear to proceed normally (see Supplemental Material movie 2). These defects in mitotic chromosome structure and segregation indicate a critical role for SMC-4 in mitosis and support the view that SMC-4 is part of a mitotic condensin complex in C. elegans.
A more detailed view of aberrant chromosome morphology in SMC-4-depleted cells was evident from confocal microscopy. Prometaphase chromosomes in RNAi-treated embryos formed into multiple thin fibers instead of individual thick rods, showing that SMC-4 is required for proper chromosome condensation (Fig. 3K,P). Yet rod-like structures and significant compaction were achieved by metaphase (Fig. 3L,Q), although chromosome structure and alignment within the metaphase plate were somewhat disorganized (Fig. 3M,R). Chromatin bridges at anaphase (Fig. 3N,S) and telophase (Fig. 3O,T) indicate that SMC-4 is critical for resolving chromosome linkages. Thus, depletion of a C. elegans condensin component does not cause severe compaction defects at metaphase, as might have been predicted on the basis of studies in frogs and yeast. Instead, the observed defects suggest a more complex role for condensin in chromosome organization and sister chromatid segregation.
SMC-4 and MIX-1 require each other for chromosome localization, and loss of either causes identical phenotypes, consistent with function in the same complex
Depletion of the MIX-1 protein by RNAi resulted in chromosome abnormalities identical to those seen in smc-4(RNAi) embryos (Fig. 4A–C). In mix-1(RNAi) embryos, prometaphase chromosome morphology was altered, and chromosomes remained connected by chromatin bridges at anaphase (Fig. 4C and Supplemental Material movie 3). Giant nuclei were observed in both smc-4(RNAi) and mix-1(RNAi) embryos, presumably resulting from repeated rounds of DNA replication and attempted division without chromosome segregation (Fig. 4B,C). Fluorescent in situ hybridization (FISH) to a single locus showed that individual nuclei from smc-4(RNAi) embryos had either many signals or no signals, whereas nuclei from wild-type embryos had only the expected two signals (Fig. 4E,D). Thus, cells depleted of either SMC-4 or MIX-1 undergo multiple rounds of DNA replication despite the failure of chromosome segregation, leading to aneuploid nuclei.
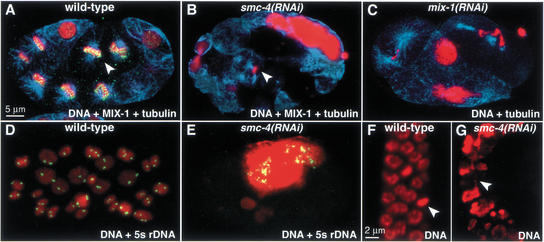
Figure 4.
Depletion of SMC-4 or MIX-1 causes aneuploidy and chromosome segregation defects in cell divisions throughout development. (A,B) Early embryos stained with DNA dye (red), tubulin antibody (blue), and MIX-1 antibody (green). (A) MIX-1 associates with mitotic chromosomes in wild-type embryos (yellow, MIX-1 and DNA overlap). (B) MIX-1 is not observed on mitotic chromosomes in smc-4(RNAi) embryos. (Arrowheads) Metaphase chromosomes within a mitotic spindle. (C) mix-1(RNAi) embryo stained with DNA dye (red) and tubulin antibody (blue) shows phenotypes similar to those of smc-4(RNAi) embryos. (D,E) Early embryos stained with DNA dye (red) and a FISH probe against the 5S rDNA locus (green) show normal diploid nuclei in wild-type (D) but aneuploid nuclei in smc-4(RNAi) embryos (E). (F,G) Mitotic cells of adult gonad stained with DNA dye. Normal mitotic figures are seen in a wild-type gonad (F, arrowhead marks metaphase plate), but large nuclei and chromatin bridges are observed in gonads partially depleted of SMC-4 by RNAi (G, arrowhead marks chromatin bridge).
In smc-4(RNAi) embryos, MIX-1 antibody staining was reduced to undetectable levels (Fig. 4B), as was SMC-4 antibody staining in mix-1(RNAi) embryos (data not shown). The identical mitotic phenotypes of smc-4(RNAi) and mix-1(RNAi) embryos and the dependence of SMC-4 and MIX-1 on each other for stability and/or localization provides further evidence that these two SMC proteins act together in a mitotic complex.
SMC-4 and MIX-1 are required for chromosome segregation throughout development
In addition to their role in embryonic mitosis, SMC-4 and MIX-1 are required for mitosis at all stages of development and in all tissues examined. In the experiments described above, progeny from RNAi-treated hermaphrodites were examined 24 h post injection. At that time, protein levels were undetectable by antibody staining, and 100% embryonic lethality was observed, presumably from aneuploidy. In contrast, embryos produced only 4–12 h after injection developed into adults, presumably because interference was incomplete, allowing us to assess the role of SMC-4 and MIX-1 later in development. Many animals were sterile and lacked coordinated movement in adulthood, phenotypes characteristic of mix-1 and other chromosome-segregation-defective mutants (Lieb et al. 1998; O'Connell et al. 1998).
Analysis of the defective progeny revealed chromosome segregation defects in cells that divided postembryonically, such as the ventral nerve cord precursors, intestinal cells, and germ-line cells. For example, mitotic cells in the gonad were unevenly sized and often connected by chromatin bridges (Fig. 4G), explaining the observed sterility. Consistent with this finding, both SMC-4 and MIX-1 localize to condensed mitotic chromosomes in the gonad (data not shown). Thus, partial depletion of SMC-4 and MIX-1 caused pleiotropic defects consistent with a requirement for these proteins in postembryonic cell divisions.
Condensin localization to mitotic chromosomes requires AIR-2, a kinase that phosphorylates histone H3 during mitosis
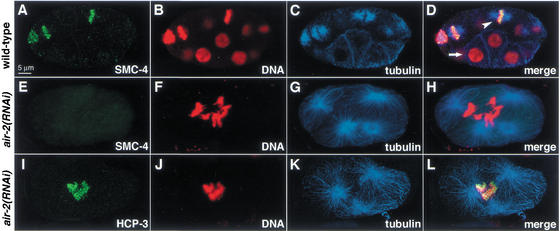
Mitotic kinases are known to restrict condensin activity to mitosis. Therefore, we asked whether the conserved mitotic kinase AIR-2 was essential for the cell-cycle-dependent localization of SMC-4 and MIX-1 to chromosomes. We focused on AIR-2, the C. elegans homolog of Aurora-B/Ipl1, because it is required for chromosome segregation (Schumacher et al. 1998) and is responsible for the mitotic-specific phoshorylation of histone H3 at serine 10, a modification correlated with chromosome condensation (Hsu et al. 2000).
Depletion of the AIR-2 kinase prevented the association of SMC-4 and MIX-1 with mitotic chromosomes, as assessed by costaining of air-2(RNAi) embryos with antibodies against tubulin to identify cells in mitosis, AIR-2 to monitor its depletion, and SMC-4 or MIX-1. Histone H3 phosphorylated at serine 10 was also undetectable by antibody staining, confirming that AIR-2 had been depleted (data not shown). In the air-2(RNAi) embryos (Fig. 5E–H) but not wild-type embryos (Fig. 5A–D), both cytokinesis and chromosome segregation failed, causing aneuploid nuclei. When these aneuploid nuclei attempted mitosis, as indicated by their mitotic spindle structure, no staining was observed with SMC-4 or MIX-1 antibodies (Fig. 5E–H). The effect on SMC-4 and MIX-1 was relatively specific, as other proteins with similar mitotic localization, such as the centromere protein HCP-3 (Fig. 5I–L), and the kinetochore protein CeMCAK (data not shown), retained their mitotic chromosome association in air-2(RNAi) embryos. Thus, AIR-2 is a kinase important for the cell-cycle-dependent localization of C. elegans condensin components.
Figure 5.
AIR-2 kinase is required for localization of condensin proteins during mitosis. (A_–_D) Wild-type embryo costained with SMC-4 antibody (green, A), DNA dye (red, B), and tubulin antibody (blue, C; merge in D). SMC-4 associates with condensed mitotic chromosomes within mitotic spindles, but not with interphase chromosomes. (Arrowhead) Mitotic nucleus; (arrow) interphase nucleus. (E_–_H) air-2(RNAi) embryo, stained as above. SMC-4 (E) does not associate with condensed mitotic chromosomes (F) within a tetrapolar mitotic spindle (G; merge in H). (I_–_L) air-2(RNAi) embryo, costained with HCP-3 antibody (green, I), DNA dye (red, J), and tubulin antibody (blue, K; merge in L). HCP-3 associates with mitotic chromosomes when AIR-2 is depleted.
C. elegans condensin components colocalize with centromere proteins on mitotic chromosomes and appear necessary for orientation of the centromere toward the spindle poles
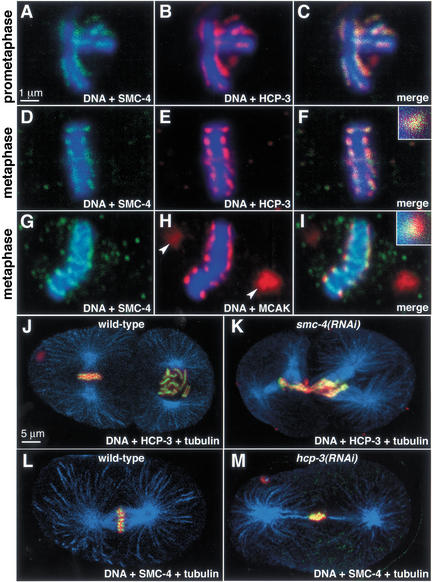
In addition to their temporally restricted localization to chromosomes, C. elegans condensin components are spatially restricted on chromosomes (Fig. 2). Unlike condensins of other organisms, which associate with a central chromosome axis (Hirano and Mitchison 1994; Schmiesing et al. 1998; Steen et al. 2000; Steffensen et al. 2001), SMC-4 and MIX-1 outline condensing prophase chromosomes along their length, then localize to the poleward faces of the metaphase plate. This dynamic subchromosomal staining pattern resembles that of C. elegans centromere and kinetochore proteins (Buchwitz et al. 1999; Howe et al. 2001; Moore and Roth 2001; Oegema et al. 2001).
Double staining experiments showed that condensin colocalized with the centromere protein HCP-3, the C. elegans homolog of CENP-A, a conserved histone H3-like protein incorporated specifically into centromeric chromatin (Buchwitz et al. 1999). Both SMC-4 and HCP-3 outlined prometaphase chromosomes (Fig. 6A–C) and were restricted to poleward-facing foci on cross sections of the metaphase plate (Fig. 6D–F). SMC-4 and the conserved kinetochore-associated protein CeMCAK also overlapped in a punctate, poleward-facing pattern on metaphase cross-sections, with CeMCAK staining extending further toward the poles (Fig. 6G–I). Thus, C. elegans condensin components appear coincident with a protein that marks centromeric DNA and internal to a protein that marks the kinetochore.
Figure 6.
Condensin components colocalize with centromere components and are required for the restricted distribution of centromere proteins toward the spindle poles during mitosis. (A_–_F) Wild-type prometaphase (A_–_C) and metaphase (D_–_F) chromosomes stained with DNA dye (blue), and antibodies against SMC-4 (green) and centromere protein HCP-3 (red). Merged images (C, F) show that SMC-4 and HCP-3 colocalize. The magnified focus of staining from the metaphase plate shows overlap of the two proteins (F, inset). (G_–_I) Metaphase chromosome stained with DNA dye (blue), and antibodies to SMC-4 (green) and the kinetochore–centrosome-associated protein CeMCAK (red). In the merged image and magnified inset (I), CeMCAK partially overlaps (yellow) SMC-4 and extends more poleward (red) than SMC-4 on metaphase chromosomes. Unlike SMC-4, CeMCAK stains the centrosomes (H, arrowheads). (J) Wild-type and (K) smc-4(RNAi) two-cell embryos stained with tubulin antibody (blue), DNA dye (red), and HCP-3 antibody (green; yellow represents overlap between HCP-3 and DNA). HCP-3 associates with mitotic chromosomes when SMC-4 is depleted, but is not present in a restricted pattern facing the spindle poles. (L) Wild-type and (M) hcp-3(RNAi) one-cell embryos stained with tubulin antibody (blue), DNA dye (red), and SMC-4 antibody (green; yellow represents overlap between SMC-4 and DNA). When HCP-3 is depleted, SMC-4 associates with DNA but in a disorganized pattern.
These results raised two questions. First, is condensin required for the localization of centromere and kinetochore proteins to chromosomes? Second, is the condensin-mediated folding of chromosomes required to organize the centromere? To address the first question, we examined the localization of HCP-3, CeMCAK, and HIM-10 in the absence of SMC-4. Both the centromere protein HCP-3 (Fig. 6J,K) and the kinetochore proteins CeMCAK and HIM-10 (data not shown) associated with chromosomes in embryos depleted for SMC-4. The reciprocal was also true; SMC-4 and MIX-1 associated with mitotic chromosomes in both wild-type (Fig. 6L) and HCP-3-depleted (Fig. 6M) embryos. Thus, SMC-4/MIX-1 and HCP-3 are not codependent for their chromosome association.
In contrast, depletion of either a condensin or centromere component altered the distribution of the other component on the chromosome. In the absence of HCP-3, kinetochore proteins fail to load onto mitotic chromosomes, the mitotic spindle is mechanically unstable, and chromosomes fail to segregate (Moore and Roth 2001; Oegema et al. 2001). We confirmed these phenotypes by observing the loss of chromosome-associated kinetochore proteins in hcp-3(RNAi) animals (data not shown) and by observing aberrant chromosome and spindle dynamics in live hcp-3(RNAi) embryos carrying both histone–GFP and tubulin–GFP (Strome et al. 2001; see Supplemental Material movies 4 and 5). In hcp-3(RNAi) embryos (Fig. 6M), SMC-4 and MIX-1 retained their mitotic chromosome localization but were not restricted to the poleward foci typical of wild-type embryos (Fig. 6L).
In reciprocal experiments involving smc-4(RNAi), HCP-3 localized to mitotic chromosomes, but in a disorganized pattern (Fig. 6K) that contrasted with the wild-type, poleward-facing pattern (Fig. 6J). In smc-4(RNAi) embryos, chromosome morphology and segregation were altered, but, nonetheless, kinetochore function appeared intact. Spindle attachment and stability appeared robust by tubulin antibody staining (Fig. 6J,K) and by tubulin–GFP movies (see Supplemental Material movie 6). Thus, condensin components do not require the centromere chromatin structure specified by HCP-3 to load onto mitotic chromosomes, but their proper restricted distribution does depend on HCP-3. Likewise, HCP-3 does not require condensin to load onto mitotic chromosomes, but it does require the mitotic chromosome organization promoted by SMC-4/MIX-1 to maintain its normal orientation toward the mitotic spindle.
C. elegans condensin components co-localize with centromere proteins on meiotic chromosomes, and their depletion impairs chromosome segregation in meiosis II, but not in meiosis I
Similarities between mitosis and meiosis prompted us to ask whether condensin localizes to meiotic chromosomes and is required for segregation of homologous chromosomes during meiosis I and/or sister chromatids during meiosis II. Chromosome condensation is required for both meiotic divisions, but chromosome segregation in meiosis II, but not in meiosis I, resembles mitotic chromosome segregation. In immunofluorescence experiments, both SMC-4 and MIX-1 localized to meiotic chromosomes in metaphase of meiosis I and of meiosis II (Fig. 7A–D). In confocal sections, SMC-4 appeared somewhat more concentrated around the periphery of condensed chromosomes in meiosis I (Fig. 7B) and meiosis II, in a pattern that resembled centromere proteins. The meiotic centromere of C. elegans chromosomes is unusual in that it completely surrounds and encases the chromosomes (Howe et al. 2001). In costaining experiments with HCP-3 and SMC-4 antibodies, both proteins were coincident at metaphase of meiosis I (Fig. 7A–D) and meiosis II (data not shown), consistent with localization of SMC-4 to the meiotic centromere.
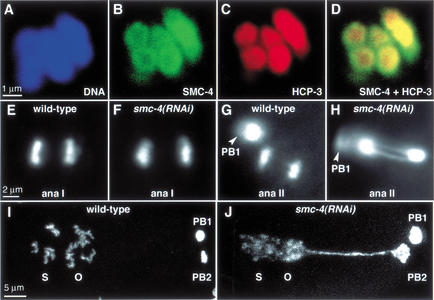
Figure 7.
SMC-4 and MIX-1 co-localize with centromere proteins on meiotic chromosomes, and their depletion impairs chromosome segregation in meiosis II but not meiosis I. (A_–_D) Meiosis I bivalents stained with a DNA dye (blue, A), SMC-4 antibody (green, B), and an antibody to centromere protein HCP-3 (red, C). SMC-4 is concentrated at the periphery of the chromosomes and colocalizes with HCP-3. (D) SMC-4 and HCP-3 overlap (yellow). (E_–_H) Images of anaphase I (ana I) and anaphase II (ana II) from time-lapse movies of meiosis in a strain carrying histone H2B–GFP. Normal segregation of homologs at anaphase of meiosis I is observed in wild-type (E) and smc-4(RNAi) embryos (F). Sister chromatids separate normally at anaphase of meiosis II in wild-type embryos (G; PB1 is first polar body from meiosis I), but anaphase II chromatin bridges are observed in smc-4(RNAi) embryos (H; PB1 is out of focus). (I_–_J) DNA-stained wild-type (I) and smc-4(RNAi) (J) embryos after formation of the first and second polar bodies (PB1 and PB2) and migration and fusion of the oocyte (O) and sperm (S) pronuclei. In smc-4(RNAi) embryos, the oocyte pronucleus remains connected to the second polar body by a long chromatin bridge; condensation of prophase chromosomes in the pronuclei is abnormal (J).
The role of SMC-4 and MIX-1 in meiosis was assessed by use of RNAi to deplete the proteins from germ lines of injected hermaphrodites. Because SMC-4 and MIX-1 are required for germ-line mitosis, we analyzed meiotic phenotypes 48 h after RNA injection. This regimen allowed sufficient time for germ-line protein stores to be depleted, but insufficient time for nuclei in mitosis at the time of injection to have progressed to metaphase of meiosis I. Thus, any defects in meiotic chromosome segregation could not result from an earlier defect in germ-line mitosis.
Depletion of either SMC-4 or MIX-1 caused chromosome segregation defects during meiosis II, but not during meiosis I. Using histone H2B–GFP to follow meiotic chromosome segregation in real time, we found no obvious differences in the segregation of homologous chromosomes at meiosis I between wild-type and RNAi-treated animals (Fig. 7E,F; see Supplemental Material movies 7 and 8). Also, no obvious morphological changes were apparent in the meiosis I chromosomes, although their small size precludes a precise assessment. These results were not due to a general inability of RNAi treatment to affect meiosis I, as depletion of the AIR-2 kinase by a similar RNAi regimen prevented chromosome segregation during both meiosis I and II (data not shown). The presence of the first polar body, one product of the reductional meiosis I division, was further evidence that meiosis I proceeded normally in RNAi-treated embryos.
Although wild-type chromosomes segregated completely at anaphase of meiosis II, resulting in the second polar body and the maternal pronucleus (Fig. 7G; see Supplemental Material movie 9), chromosomes depleted of SMC-4 failed to segregate at anaphase II (Fig. 7H; see Supplemental Material movie 10). This failed segregation was clearly manifested by a long chromatin connection between the second polar body and the maternal pronucleus as it migrated toward the paternal pronucleus (Fig. 7I,J). Because of this connection, DNA of the second polar body was sometimes pulled into the embryo and incorporated into one of the embryonic nuclei (see Supplemental Material movie 10). These results provide the first evidence for condensin localization and function during meiosis. Moreover, the apparent requirement for condensin during meiosis II when sister chromatids separate, but not meiosis I when homologs separate, implies that condensin may play a special role in sister chromatid segregation.
Discussion
C. elegans has two condensin-like complexes, with both shared and specialized components
We have uncovered the C. elegans mitotic condensin complex and described both conserved and previously uncharacterized condensin activities. This complex was discovered in a search for the mitosis-specific partner of MIX-1, a protein with dual functions. Within the same hermaphrodite cell, MIX-1 can either assemble onto X chromosomes to down-regulate their gene expression or assemble onto mitotic chromosomes to promote their folding and segregation. MIX-1 carries out these distinct roles through its physical interaction with two different SMC4 paralogs, the dosage compensation-specific protein DPY-27 for regulation of X gene expression (Lieb et al. 1998) and the mitotic-specific protein SMC-4 for chromosome segregation. SMC-4 and MIX-1 are co-dependent for function: Depletion of either causes identical mitotic chromosome segregation defects and reduction of the other. However, SMC-4 does not associate with dosage compensation proteins, localize to the X chromosome, or cause dosage compensation phenotypes, indicating that it is not involved in dosage compensation. Thus, C. elegans has two separate condensin complexes, one for chromosome segregation and one for X-linked gene regulation.
Because the role of SMC proteins in chromosome segregation is conserved from bacteria to humans, the segregation function is likely the ancestral function and the role in gene regulation more recently evolved. The need for X-chromosome dosage compensation would have occurred as the X-chromosome counting mechanism of sex determination arose, resulting in males and hermaphrodites with different doses of X chromosomes. As dosage compensation evolved, proteins like MIX-1 were apparently recruited for this new role and shared between the mitotic and dosage compensation complexes. Other proteins like DPY-27 may have arisen from gene duplication and divergence, resulting in a new paralog specialized for dosage compensation. The shared MIX-1 protein and the similar composition of dosage compensation and condensin complexes suggest that the dosage compensation mechanism may be related to the function of condensin on mitotic chromosomes.
Regulating condensin activity through the cell cycle
Condensin activity must be restricted to the mitotic stage of the cell cycle, a feat achieved in many organisms by mitotic kinases. In Xenopus and human cells, the mitotic kinase cdc2 phosphorylates the non-SMC subunits of condensin, thereby stimulating supercoiling and condensation activities (Kimura et al. 1998, 2001). In Schizosaccharomyces pombe, condensin activity is regulated at the level of nuclear import, and phosphorylation of the SMC4 homolog by cdc2 is required for this nuclear localization (Sutani et al. 1999). In C. elegans, the role of the cdc2 homolog NCC-1 (Boxem et al. 1999) in localizing condensin was difficult to assess, because the cell cycle arrests prior to condensin loading when NCC-1 is depleted. Instead, AIR-2 was shown here to be required for condensin localization to C. elegans mitotic chromosomes.
Coincident with our study, localization of the Barren condensin subunit to mitotic chromosomes was shown to require the Drosophila homolog of AIR-2, Aurora B (Giet and Glover 2001). Thus, regulation of condensin by AIR-2 appears to be conserved. It is not yet known whether AIR-2 directly phosphorylates one of the condensin subunits to promote chromosome association of the complex, or whether its action on a different substrate is a prerequisite for condensin targeting. One proposal is that the phosphorylation of histone H3 by AIR-2 may serve to mark mitotic chromosomes and recruit condensin (Wei et al. 1999; Schmiesing et al. 2000; Giet and Glover 2001), but evidence that mitotic phosphorylation of histone H3 is neither necessary nor sufficient to target condensin contradicts this view (Kimura and Hirano 2000; Schmiesing et al. 2000; Murnion et al. 2001).
Condensin is required for chromosome condensation and sister chromatid separation, but is not likely the sole factor required for compaction
Condensin is so named for its effects on chromosome condensation at mitosis. Indeed, we found that chromosomes depleted of SMC-4 or MIX-1 formed multiple thin fibers instead of thick rods at prometaphase, confirming that condensin is required for the transition from a decondensed chromosome structure at interphase to a more condensed structure at prometaphase. However, in live smc-4(RNAi) embryos, the stringy prometaphase chromosomes compacted substantially to form rodlike structures at metaphase. Thus, in C. elegans, condensin components appear not to be the sole proteins involved in compaction in vivo. The degree of metaphase condensation was at first surprising, given the effects of condensin depletion in frogs and yeast (Hirano and Mitchison 1994; Strunnikov et al. 1995). However, even in these organisms, condensin depletion does not cause complete decondensation. Moreover, mutation of the Drosophila SMC4 gene disrupts sister chromatid resolution at prophase, but not shortening of the longitudinal chromosome axis at metaphase (Steffensen et al. 2001).
The most dramatic consequence of condensin depletion in C. elegans is not a condensation defect, but rather the failure to segregate sister chromatids at anaphase. Chromosomes depleted of SMC-4 and MIX-1 remained attached after each attempted cell division, ultimately forming giant aneuploid nuclei. Such failure of chromosome separation occurs in both prokaryotes and eukaryotes when SMC proteins are altered (for review, see Hirano 2000). Rather than simply compact mitotic chromosomes, condensin appears to play a more complex role in chromosome organization and folding that is essential for sister chromatid segregation.
For example, condensin might facilitate the action of topoisomerase II in decatenating intertwined sisters. Because unambiguous data supporting a physical interaction and enzymatic cooperation between condensin and topoisomerase II are lacking (Bhat et al. 1996; Hirano et al. 1997; Lavoie et al. 2000), other models must be considered. Most plausibly, the supercoiling and condensation activities of condensin directly facilitate chromosome untangling. The link between SMC proteins, supercoiling, and decatenation in prokaryotes suggest this idea. Mutations in the Escherichia coli SMC-like gene mukB cause partitioning defects rescuable by mutations that increase overall DNA supercoiling (Sawitzke and Austin 2000). These results suggest that MukB and supercoiling cooperate in condensing DNA, which drives partitioning (for review, see Holmes and Cozzarelli 2000). Consistent with increased supercoiling promoting partitioning, supercoiled plasmids are much more efficiently decatenated by topoisomerase than relaxed plasmids (Ullsperger and Cozzarelli 1996). By analogy, the supercoiling activity of eukaryotic condensin, extended to C. elegans in this study, may promote decatenation. Condensin activity could also provide directionality to topoisomerase II. Because topoisomerase II does not know whether it breaks and passes strands within or between sisters, it is capable of either catenating or decatenating sisters. The action of condensin in folding each sister into itself and away from its opposite sister may help drive topoisomerase II equilibrium toward decatenation.
Condensin is required for sister separation at meiosis II, but apparently not homolog separation at meiosis I
Chromosome segregation during meiosis II and mitosis are similar in that sister chromatids catenated during replication must untangle and separate. Meiosis I differs in that the connection created by recombination between sister chromatids of opposing homologs must be resolved but cohesion between sister chromatids of a single homolog must be maintained. For both meiotic divisions, chromosome compaction must occur. The differential effect of SMC-4 RNAi on meiosis II suggests two possibilities, assuming that depletion of condensin by RNAi was complete, as suggested but not proven by antibody staining. First, condensation and segregation of chromosomes in meiosis I might require proteins other than condensins. Alternatively, condensin might facilitate the decatenation and segregation of sister chromatids intertwined by replication but not by recombination. Whatever the mechanism, our work provides the first demonstration of a role for condensin in meiosis.
Condensin and the centromere
Condensin components colocalize with centromere proteins on C. elegans mitotic chromosomes. Moreover, depletion of condensins prevents the restricted orientation of centromere proteins toward the spindle poles. Even at meiosis, condensin components appear to colocalize with the centromere, which encircles each chromosome. These findings were unexpected, given that condensin is not restricted to the centromere in other organisms. Although human, frog, and fly condensins are concentrated at the centromere, they also extend along the length of each sister chromatid, appearing as a narrow, patchy line of staining along the internal axis, rather than facing out toward the spindle poles (Hirano and Mitchison 1994; Schmiesing et al. 1998; Steen et al. 2000; Steffensen et al. 2001). Characterization of the Saccharomyces cerevisiae condensin subunit BRN1 suggests that the functional connection between condensins and centromeres may extend beyond C. elegans. In one study, chromosomes were displaced away from the spindle in brn1 mutants (Lavoie et al. 2000), and in the other, brn1 mutations caused a discontinuous spindle (Ouspenski et al. 2000), phenotypes that may reflect a failure in spindle microtubule attachment to the centromere.
What is the significance of condensin localization to C. elegans centromeres? It has been proposed that assembly of centromeres on holocentric chromosomes occurs by bringing dispersed sequences of interphase chromosomes together into a single structure during mitotic condensation. This model is based in part on the progression of HCP-3 staining foci from dispersed speckles at early prophase, to parallel stripes at prometaphase, to a compact, poleward structure at metaphase (Buchwitz et al. 1999; Moore and Roth 2001). It is an extension of the repeat subunit model, in which centromeres of monocentric chromosomes are formed by folding together centromeric repeat sequences (Zinkowski et al. 1991). The proposal for centromere formation on holocentric chromosomes resembles the proposal that condensin promotes global writhe and thus brings dispersed sequences together by reeling in DNA and stabilizing higher order chromosome structure (Kimura et al. 1999). Although depletion of C. elegans condensin components did not prevent centromere assembly or spindle attachment, it did prevent the restricted centromere orientation toward the spindle poles. These results are consistent with condensin being a driving force to coalesce separated sequences and form proper centromere structure as it condenses mitotic chromosomes. Moreover, the orderly folding promoted by condensin may influence the orientation of the DNA within each sister, so that centromeric regions become positioned toward the spindle pole.
Materials and methods
RNAi
RNAi was achieved by injection of wild-type hermaphrodites with doubled-stranded RNA (1–2 mg/mL) corresponding to the full genomic regions of hcp-3 or air-2 and to the central region of smc-4 (amino acids 343–724) or the amino-terminal region of mix-1 (amino acids 8–148) according to Fire et al. (1998). RNAi effectiveness was monitored by antibody staining for the protein targeted for depletion. RNAi was considered successful if protein levels were undetectable. Embryos produced 24 or 48 h after injection were used for analysis of mitosis or meiosis, respectively. Mitotic phenotypes described for each RNA were consistently observed in >10 different experiments analyzing 100 total embryos. Meiotic phenotypes were observed in >5 different experiments analyzing 20 total embryos.
Antibodies
SMC-4
Rabbit and rat antibodies were made to fusion proteins covering amino-terminal or central epitopes (provided by D. Pasqualone, University of California, Berkeley and M. Albrecht, University of Southern California, Los Angeles) and to amino- or carboxy-terminal peptides (synthesized by D. King, HHMI and University of California, Berkeley). Affinity-purified, but not preimmune, sera of all four recognized the same ∼176-kD band in Western blots of embryonic extracts, showed the same immunofluorescent staining pattern, and failed to stain smc-4(RNAi) embryos. Most experiments used the carboxy-terminal peptide antibody.
MIX-1
Two different MIX-1 antibodies showed the same immunoprecipitation and immunostaining results. Most experiments used the carboxy-terminal peptide antibody provided by R. Chan (HHMI and University of California, Berkeley) against a carboxy-terminal peptide of MIX-1.
Other antibodies
Phospho-histone H3 (Upstate Biotechnology), DPY-27 (P.-T. Chuang, University of California, San Francisco [rabbit] and R. Chan [rat]), CeMCAK (M. Albrecht), α-tubulin (D. Asai, Purdue University, West Lafayette, IN), HIM-10 (Howe et al. 2001), HCP-3 (Buchwitz et al. 1999), and AIR-2 (Schumacher et al. 1998).
Antibody staining
The antibody staining protocol was devised by A. Chan (University of California, Berkeley) and performed as described (Howe et al. 2001). Wild-type staining patterns are representative of >100 nuclei examined in at least 15 different experiments and were obtained using both paraformaldehyde (protocol above) and paraformaldehyde/methanol fix. To assess the lack of staining in RNAi experiments, a positive staining antibody such as tubulin was used on the same slide, and wild-type controls were stained in parallel.
Immunoprecipitation reactions and supercoiling assays
Immunoprecipitations were performed on whole-cell extracts prepared by sonication of wild-type embryos as described in Chu et al. (2002) except that the homogenization buffer was 50 mM HEPES-KOH (pH 7.6), 200 mM KCl, 1 mM EDTA, 0.5% NP-40, and 10% glycerol. Large-scale immunoprecipitations were performed by incubation of 6 mL of 20 mg/mL extract pre-cleared with IgGsorb (The Enzyme Center), with 6 mL of homogenization buffer and 1 mg of affinity-purified carboxy-terminal peptide SMC-4 antibody for 2 h at 4°C. Protein A beads were added and incubated 45 min, spun down, washed 3 times with 35 mL of homogenization buffer, then eluted 2 times with 250 μL of 1 mg/mL carboxy-terminal peptide in buffer (20 mM HEPES-KOH at pH 7.6, 50 mM KCl, 5 mM MgCl2, 10% glycerol). Samples were concentrated and washed with the above buffer in Microcon concentrators (Amicon). Extracts were made from embryos with mitotic and nonmitotic cells, suggesting that only some of the purified complexes were active. Supercoiling reactions were performed on relaxed Bluescript plasmid DNA (Strategene) that had been made by incubation of purified supercoiled DNA with topoisomerase I for 30 min at 25°C. Assays were essentially as described previously (Kimura and Hirano 1997). Relaxed plasmid DNA (5 ng) was incubated with 3 μL of eluted protein (1–3 μg) and wheat germ topoisomerase I in a total volume of 5 μL, containing 10 mM HEPES-KOH (pH 7.6), 50 mM KCl, 5 mM MgCl2, 0.1 mM CaCl2, 5 mM ATP, 1 mM DTT, 0.1 mg/mL BSA. After 15 min of incubation, 45 μL of stop buffer was added (20 mM EDTA at pH 8, 0.1%SDS, 0.1 mg/mL proteinase K) and the reactions were incubated 20 min at 55°C. DNA was phenol extracted, ethanol precipitated, electrophoresed in 1% agarose with 0.2 μg/mL chloroquine, and visualized by Southern blotting. For antibody blocking, the carboxy-terminal SMC-4 antibody was used. For testing of dependence on ATP hydrolysis, 5 mM AMP-PNP was used in place of ATP.
Fluorescence in situ hybridization
A 950-bp repeat from the 5S rDNA locus on chromosome V was labeled and used for FISH as described in (Howe et al. 2001).
Time-lapse microscopy
Gravid hermaphrodites were dissected in standard M9 buffer and mounted on a 2% agarose pad under a coverslip. Embryos were analyzed by widefield microscopy with a motorized microscope (Zeiss Axioplan II) and 12-bit camera (Hamamatsu Orca 100) controlled with Openlab software (Improvision). A histone H2B–GFP transgene (Praitis et al. 2001) allowed chromosome visualization; a β-tubulin–GFP transgene (Strome et al. 2001) allowed spindle visualization. DIC and GFP images were collected every 10 sec. Quicktime movies can be viewed as Supplemental Material at http://www.genesdev.org.
Acknowledgments
We thank D. Pasqualone for the original SMC-4 antibody and RNAi observations; R. Chan for the immunoprecipitation protocol and MIX-1 peptide antibody; A. Chan for the antibody staining protocol and microscopy assistance; M. Albrecht for the original SMC-4 and CeMCAK antibodies; D. King for peptide synthesis; M. Roth for the HCP-3 antibody; J. Schumacher for the AIR-2 antibody; J. Austin for the H2B–GFP strain; C. Mallone and J. White for the tubulin–GFP strain; J. Harb for movie assistance; and R. Heald, D. Reiner, and C. Tsai for critical reading of the manuscript. K.H. was a fellow of the Damon Runyon Cancer Research Fund and is a research associate of the Howard Hughes Medical Institute. V.H. was a predoctoral fellow of the Howard Hughes Medical Institute. This work was supported by NIH grants GM30702 (to B.J.M.) and GM31655 (to N.R.C.). B.J.M. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bjmeyer@uclink4.berkeley.edu; FAX (510) 643-5584.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.968302.
References
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Boxem M, Srinivasan DG, van den Heuvel S. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 1999;126:2227–2239. doi: 10.1242/dev.126.10.2227. [DOI] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Chu, D.S., Dawes, H.E., Lieb, J.D., Chan, C.C., Kuo, A.F., and Meyer, B.J. 2002. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes & Dev (in press). [DOI] [PMC free article] [PubMed]
- Chuang PT, Albertson DG, Meyer BJ. DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. SMCs in the world of chromosome biology—From prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Holmes VF, Cozzarelli NR. Closing the ring: Links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M, McDonald KL, Albertson DG, Meyer BJ. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- ————— Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci. 2000;97:11972–11977. doi: 10.1073/pnas.220326097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Cuvier O, Hirano T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J Biol Chem. 2001;276:5417–5420. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: Implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol Biol Cell. 2000;11:1293–1304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Capowski EE, Meneely P, Meyer BJ. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science. 1996;274:1732–1736. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- Moore LL, Roth MB. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnion ME, Adams RR, Callister DM, Allis CD, Earnshaw WC, Swedlow JR. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J Biol Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- O'Connell KF, Leys CM, White JG. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics. 1998;149:1303–1321. doi: 10.1093/genetics/149.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenski II, Cabello OA, Brinkley BR. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol Biol Cell. 2000;11:1305–1313. doi: 10.1091/mbc.11.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci. 2000;97:1671–1676. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing JA, Ball AR, Jr, Gregson HC, Alderton JM, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing JA, Gregson HC, Zhou S, Yokomori K. A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, co-localizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol Cell Biol. 2000;20:6996–7006. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RL, Cubizolles F, Le Guellec K, Collas P. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J Cell Biol. 2000;149:531–536. doi: 10.1083/jcb.149.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MM, Sunkel CE. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr Biol. 2001;11:295–307. doi: 10.1016/s0960-9822(01)00096-3. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Spindle dynamics and the role of γ-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: A repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]