Conjugative Plasmid Transfer in Gram-Positive Bacteria (original) (raw)
Abstract
Conjugative transfer of bacterial plasmids is the most efficient way of horizontal gene spread, and it is therefore considered one of the major reasons for the increase in the number of bacteria exhibiting multiple-antibiotic resistance. Thus, conjugation and spread of antibiotic resistance represents a severe problem in antibiotic treatment, especially of immunosuppressed patients and in intensive care units. While conjugation in gram-negative bacteria has been studied in great detail over the last decades, the transfer mechanisms of antibiotic resistance plasmids in gram-positive bacteria remained obscure. In the last few years, the entire nucleotide sequences of several large conjugative plasmids from gram-positive bacteria have been determined. Sequence analyses and data bank comparisons of their putative transfer (tra) regions have revealed significant similarities to tra regions of plasmids from gram-negative bacteria with regard to the respective DNA relaxases and their targets, the origins of transfer (oriT), and putative nucleoside triphosphatases NTP-ases with homologies to type IV secretion systems. In contrast, a single gene encoding a septal DNA translocator protein is involved in plasmid transfer between micelle-forming streptomycetes. Based on these clues, we propose the existence of two fundamentally different plasmid-mediated conjugative mechanisms in gram-positive microorganisms, namely, the mechanism taking place in unicellular gram-positive bacteria, which is functionally similar to that in gram-negative bacteria, and a second type that occurs in multicellular gram-positive bacteria, which seems to be characterized by double-stranded DNA transfer.
INTRODUCTION
Conjugative Transfer of Resistance Determinants from Antibiotic Producers into Pathogens
Soon after the successful introduction of antibiotics as therapeutic agents of infectious diseases, resistant bacteria emerged. Pathogenic bacteria have developed numerous strategies to resist the action of antibiotics, including modification and inactivation of the drug, exclusion of the antibiotic, and alteration of the target. The increased prevalence of antibiotic resistance in pathogenic bacteria is an outcome of evolution and selective pressure due to the widespread use of antibiotics in medicine, veterinary medicine, animal feeding, and agriculture. The origin of antibiotic resistance genes in pathogenic bacteria is unclear. The period from the beginning of antibiotic treatment (50 to 60 years ago) to the emergence of bacteria expressing effective resistance mechanisms is too short to explain the development of resistance factors from other proteins by spontaneous mutation. In particular, if a resistance mechanism requires the cooperative action of several proteins (e.g., vancomycin resistance) the de novo generation of such a resistance mechanism in the pathogen is very unlikely.
Most of the antimicrobial drugs currently in use are derived from metabolites of soil organisms, mainly fungi and actinomycetes. All resistance mechanisms that have been identified in pathogenic bacteria, including RNA methylases, ATP-binding cassette transporters, aminoglycoside phosphotransferases, and β-lactamases, already exist in the respective antibiotic producers. In Streptomyces coelicolor (http://www.sanger.ac.uk/Projects/S_coelicolor/), as well as in the glycopeptide producers of the genus Amycolatopsis, even the vancomycin resistance determinants vanH (d-Ala dehydrogenase), vanA (d-Ala-d-Lac ligase), and vanX (d,d-dipeptidase) are present in the very same gene organization as found in the enterococcal conjugative transposon Tn_1549_ (79).
The resistance genes probably evolved in the antibiotic producers as part of the biosynthetic gene cluster to protect the producing organism from the detrimental action of its own antibiotic. Subsequent gene transfer events might have spread the resistance determinants to other bacteria. Whereas the ability of broad-host-range plasmids from gram-negative bacteria in intergeneric and transkingdom transfer is well documented (9, 53, 216), the role of the gram-positive transfer systems in the dissemination of resistance determinants needs further evaluation.
Conjugative Transfer in Gram-Negative Bacteria as a Paradigm for Key Steps in Conjugative Plasmid Transfer
Bacterial conjugation is a highly specific process whereby DNA is transferred from donor to recipient bacteria by a specialized multiprotein complex, termed the conjugation apparatus. An important prerequisite for conjugative transfer is an intimate association between the cell surfaces of the interacting donor and recipient cells. In gram-negative bacteria, this physical contact is established by complex extracellular filaments, designated sex pili. For the majority of gram-positive bacteria, the means to achieve this intimate cell-cell contact have not yet been identified.
To facilitate homology studies with gram-negative systems and to develop a transfer model for gram-positive unicellular bacteria, the current model for conjugative transfer in gram-negative bacteria is briefly presented here. We restrict our overview to the fundamental findings of one of the best-studied conjugative systems, the IncP transfer (tra) system of the broad-host-range plasmid RP4. The IncP transfer system consists of two regions, Tra1 and Tra2, including 30 transfer functions, 20 of which are essential for intraspecies Escherichia coli matings. The central question in bacterial conjugation is how the DNA traverses the cell envelopes of the mating cells. The current model is that two protein complexes exist, namely, the relaxosome and the mating-pair formation (mpf) complex, which are connected via interaction with a TraG-like coupling protein. The relaxosome has been defined as a multiprotein-DNA complex that is generated at the plasmid origin of transfer, oriT. Plasmid-encoded and chromosomally encoded proteins participate in this complex (77, 120). The mpf complex is a plasmid-encoded multiprotein complex that is involved in the traffic of the donor DNA strand from the donor to the recipient cell (124).
The RP4 relaxosome was localized in the cytoplasm and found to be associated with the cytoplasmic membrane independent of the membrane-spanning mpf complex (89, 123). DNA relaxases are the key enzymes in the initiation of conjugative transfer and operate by catalyzing the cleavage of a specific phosphodiester bond in the nic site within oriT in a strand- and site-specific manner. In all systems encoded by self-transmissible and mobilizable plasmids studied so far, the DNA cleavage reaction is a strand transfer reaction involving a covalent DNA-relaxase adduct as an intermediate. This intermediate is proposed to be a prerequisite for the recircularization of the cleaved plasmid after completion of transfer by a joining reaction between the free 3′ hydroxyl and the 5′ terminus of the covalently bound relaxase. An exception is plasmid CloDF13, for which data suggest that nic cleavage possibly results in a free nicked-DNA intermediate (152).
IncP-type relaxases seem to be the most widely distributed among different gram-positive and gram-negative conjugative plasmids, conjugative transposons, mobilizable elements, and the agrobacterial T-DNA transfer system (226). All conjugative DNA relaxases have common domains in which the N-terminal moiety seems to contain the catalytic activity whereas the C-terminal moiety may be involved in interactions with other components of the transfer machinery. The enzymatic properties of DNA relaxases are discussed in more detail below.
Biochemical, genetic, and electron microscopic data imply the existence of complicated structures of the mpf complex. Eleven mpf components (trbB to trbL) and traF are required for IncP pilus formation in the absence of any DNA-processing factors (92), and these components are also required to establish conjugative junctions (181). The mpf system of RP4 was localized in the cell membrane (89) and was suggested to form a complex that connects the cytoplasmic and the outer membrane. These data agree with a role of the mpf complex in protein transport. Experimental evidence for interaction of the complex with DNA has been recently obtained, since nonspecific DNA binding activity of TrbE was shown (11).
The _tra1_-encoded TraG protein is also associated with the cytoplasmic membrane independent of the presence of the Tra2 region. The results also suggest a connection of TraG with the mpf complex, thereby supporting its proposed role as a potential interface between the mpf system and the relaxosome (89).
Gram-negative bacteria possess two very efficient barriers which have to be traversed by macromolecules during export from and import into the cell: the outer membrane and the inner membrane, which are separated by a cellular compartment, the periplasm. From this point of view, it is evident that macromolecules such as plasmid DNA and prepilin subunits (the building blocks of the pili) need a transport channel to cross the two membranes and the periplasmic space.
Conjugative plasmids have evolved systems of regulation that minimize the metabolic and phenotypic load exerted by the maintenance of a conjugative transfer apparatus while optimizing the adaptive advantages of self-transmission. For instance, IncP plasmids transfer at high frequencies under optimal conditions, so that the transfer frequencies can approach one transfer event during a 5-min mating on nutrient agar. However, IncP transfer genes are not expressed constitutively. In fact, their expression is regulated by complex local autoregulatory circuits as well as by global regulators, resulting in the coordinated expression of transfer genes with other plasmid functions (52, 225).
CONJUGATIVE TRANSFER IN UNICELLULAR GRAM-POSITIVE BACTERIA
Conjugative transfer systems have been detected in many different gram-positive species. The available information suggests that the major differences between conjugation in gram-negative and gram-positive bacteria lie in the mechanisms that have evolved to establish cell-cell contact in order to initiate conjugal transfer. Determination of the nucleotide sequence of the tra regions or whole genomes of several large conjugative plasmids from gram-positive bacteria (15, 59, 143, 186) has revealed homologies to proteins belonging to the TraG/TrwB/VirD4 family of coupling proteins (for recent reviews and meeting reports on type IV secretion systems, see references 11, 30, 33, 36, 37, 51, 119, and 193), to the conjugative transfer ATPase VirB4 and its homologues (51, 52) involved in substrate translocation processes during T-DNA and plasmid transfer in gram-negative bacteria, and to the VirB1 family of lytic transglycosylases. These interesting homologies to components of type IV secretion systems are discussed below. Some representatives of conjugative plasmids and transposons of unicellular gram-positive bacterial origin discussed here are listed in Table 1.
TABLE 1.
Conjugative elements from unicellular gram-positive bacteria
| Plasmid or transposon | Original host | Size (kb) | Antibiotic resistancea | Host range/induction of transfer |
|---|---|---|---|---|
| pIP501 | S. agalactiae | 30.2 | Cm, MLS | Broad/? |
| pAMβ1 | E. faecalis | 26.5 | MLS | Broad/? |
| pRE25 | E. faecalis | 50.2 | Cm, MLS | Broad/? |
| Tn_916_ | E. faecalis | 18.0 | Tc | Broad/? |
| Tn_1545_ | S. pneumoniae | 25 | Em, Km, Tc | Broad/? |
| pSK41 | S. aureus | 46.4 | Bm, Gm, Km, Nm, Tm | Staphylococcus/? |
| pGO1 | S. aureus | 52.0 | Bm, Gm, Km, Nm, Tm, Tmp | Staphylococcus/? |
| pMRC01 | L. lactis | 60.2 | Noneb | Lactococcus/? |
| pAD1 | E. faecalis | 59.3 | Nonec | Enterococcus/sex pheromone |
| pCF10 | E. faecalis | 65 | Tc | Enterococcus/sex pheromone |
| pPD1 | E. faecalis | 56 | Noneb | Enterococcus/sex pheromone |
| pXO16 | B. thuringiensis | 200 | Unknown | B. thuringiensis/aggregation |
| pRS01 | L. lactis | 48.4 | Unknown | Lactococcus/aggregation |
Conservation of Conjugative DNA Relaxases
DNA relaxases are the main players in the initiation of conjugative plasmid transfer (for reviews, see references 31, 54, 161, and 226). Relaxases from several transfer systems have been studied so far, and the best characterized are those for which in vitro systems with purified relaxosome components have been developed (reference 226 and references therein).
Three conserved motifs (I to III) were first identified in IncP-like relaxases (160, 163). Motif I contains a conserved Tyr residue (Tyr-22 in Tra1 from RP4) that reversibly attacks the DNA backbone in the relaxase-catalyzed reaction. A Ser residue within motif II was shown to be involved in tight binding of the 3′ terminus generated in the DNA cleavage reaction. Motif III contains two His residues thought to be involved in activating Tyr-22 for its nucleophilic attack at the nic site (161, 163). Interestingly, these conserved His residue are found not only in conjugative relaxases but also in several rolling-circle replicating (RCR) initiator proteins, and they have been proposed to participate in the binding and coordination of the metal cation (Mg2+ or Mn2+) needed for cleavage of the DNA substrate (101, 114). Motifs I and III are found in all conjugative DNA relaxases, which were divided into four distinct DNA relaxase families, the IncP, the IncF/IncW, the IncQ, and the RCR (pMV158)-type family, on the basis of to overall homology (226).
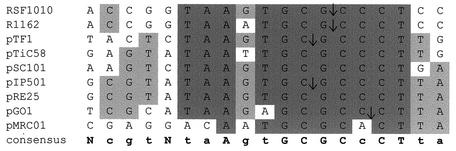
The relaxases of conjugative and mobilizable plasmids from gram-positive bacteria mainly belong to two families, the IncQ-type family and the pMV158-type family (91, 226). The relaxases encoded by pIP501, pRE25, pSK41, pMRC01, and pGO1 belong to the IncQ-type family. An alignment of the IncQ-type relaxases, including three proteins of plasmids from gram-negative bacteria (RSF1010, pTF1, and pSC101) and five relaxases of gram-positive bacterial origin, is shown in Fig. 1. It shows conservation of motif I, characterized by the Tyr residue (Tyr-26 in pIP501-encoded TraA), and of motif III, specified by the two His residues (His-134 and -136 in TraA) in all members of the family. Mutation of these His residues in TraI of RP4 resulted in strong reduction of relaxase-mediated cleavage activity (162, 163).
FIG. 1.
Alignment of conjugative DNA relaxases of the IncQ-type family. Amino acid positions that are conserved throughout are shown in pink. Green letters mark positions that are conserved in at least five of the eight proteins. The delimitations of two conserved motifs that were identified first in IncP-like relaxases are indicated by lines above the sequence block. The active tyrosine in motif I and the two histidines in motif III that are conserved in all conjugative DNA relaxases are marked with asterisks. GenBank/EMBL accession numbers: MobA (RSF1010), X04830; MobL (pTF1), S12190; Mob (pSC101), P14492; Nes (pGO1), U50629; Nes (pSK41), AF051917; TraA (pIP501), L39769; Orf24 (pRE25), X92945; TraA (pMRC01), NC_001949.
The first of the IncQ-type DNA relaxases of gram-positive bacterial origin, for which some enzymatic properties have been determined, is the pIP501 relaxase TraA. traA was cloned, overexpressed in E. coli, and purified as a fusion with glutathione _S_-transferase via affinity chromatography. Full-length TraA, as well as a C-terminally truncated version, TraA*, encompassing the first 293 amino acids of a total 660 amino acids, showed specific cleavage activity on supercoiled DNA containing _oriT_pIP501. The site- and strand-specific cleavage required the presence of Mg2+ or Mn2+, which could not be substituted by Ca2+ or Zn2+ ions, and was highest at temperatures between 42 and 45°C. Interestingly, the N-terminal portion of TraA, TraA*, also cleaves supercoiled DNA containing _oriT_pIP501, although less efficiently than the full-length protein (25% conversion of _oriT_pIP501 supercoiled DNA to open circular forms in comparison to a maximum conversion rate of 55% exerted by TraA [118]). These data agree with the results obtained for the MobA protein encoded by the gram-negative host plasmid RSF1010 (183). The minimal functional domain of MobA, the DNA relaxase encoded by R1162, which is virtually identical to RSF1010, was determined recently. It comprises the N-terminal 184 to 188 amino acids (containing motifs I and III). It is the smallest fragment capable of strong binding to oriT DNA and the smallest fragment that cleaves this DNA (13).
Another family of DNA relaxases is made up of the mobilization (Mob) proteins encoded by many RCR plasmids isolated from a variety of gram-positive bacteria. Interestingly, these proteins were first described as participating in the generation of cointegrates between staphylococcal plasmids, so that they were termed plasmid recombination enzymes (Pre) (151). Indications that these proteins were involved in mobilization were provided later by methods showing that the mobM gene of the streptococcal plasmid pMV158 was required for its mobilization by pIP501 between strains of Streptococcus pneumoniae (180), and that two regions of the staphylococcal plasmid pC221 are involved in transfer (174), as well as by sequence similarity analyses (159, 209). Comparative studies of staphylococcal plasmids related to pT181 showed that these plasmids also carry a Pre function (175). In addition, the region where the plasmid cointegration occurred (named recombination site a [RSa]) was identified as the plasmid oriT (91, 173). The only Mob protein of this category of plasmids that has been characterized so far is the pMV158-MobM protein (91), but there are nearly 50 Mob proteins that show a high degree of similarity to MobM, and they include Mob proteins from well-characterized staphylococcal plasmids like pUB110, pE194, pT181, and pC221; curiously enough, staphylococcal plasmid closely related to pUB110, pC194, does not appear to carry a mobilization cassette (91).
Using the Pfam algorithms from the Sanger Institute (http://www.sanger.ac.uk/cgi-bin) and from the Swiss Institute for Experimental Cancer Research (http://hits.isb-sib.ch/cgi-bin/PFSCAN), all these proteins, especially those from RCR plasmids, can be grouped within a single family, termed the Mob-Pre family of proteins, and they have been found in a wide variety of bacteria from the family Bacteroidaceae to firmicutes (Bacillus-Clostridium-Staphylococcus groups) and proteobacteria. The family can be extended to other proteins that show homology to the Mob proteins at the C-terminal end, like two hypothetical proteins from Lactobacillus lactis (36.1 kDa; accession number Q9L973) and from Moraxella catharralis (80.7 kDa; accession number Q9L973).
Conservation of nic Regions
In virtually all conjugative transfer systems studied so far, a DNA single strand is thought to be transferred between donor and recipient cell (70, 120, 125, 228). The only locus required in cis for generation of the single-stranded plasmid intermediate is the oriT, where the DNA relaxase exerts its cleavage within the phosphodiester bond of a specific dinucleotide, the nic site. In several cases, comparisons of components of conjugative transfer systems with vegetative replication systems of the rolling-circle type revealed significant similarities which indicate not only functional but also phylogenetic relatedness. These include leading-strand replication and transfer origins as well as conjugative DNA relaxases and replication initiator proteins of RCR plasmids and single-stranded coliphages (101, 114, 218). Five families of oriT core sequences have been defined through comparisons of a wide range of transfer origins (91, 120, 226). A close inspection of the oriT sequences of these five families revealed a core consensus sequence that is common even among apparently phylogenetically distant oriT families (226).
The RSF1010-oriT family (118, 226) includes the prototype IncQ plasmids RSF1010 (55) and R1162 (25), the Thiobacillus ferrooxidans plasmid pTF1 (60), the A. tumefaciens Ti plasmid pTiC58 (49), the Salmonella plasmid pSC101 (138), and four plasmids from gram-positive hosts, namely, pRE25, pIP501, pGO1, and pMRC01. The 5′ end of the nick site was mapped for five of these nine plasmids and showed identical nic sites for RSF1010 and R1162 (55, 60) and for pTF1 and pIP501 (60, 215), respectively, while the dinucleotide cleaved in _oriT_pGO1 (48) is different.
A consensus sequence for the RSF1010-oriT family was deduced: 5′-NcgtNtaAgtGCGCcCTta-3 (Fig. 2). An additional similarity is the presence of inverted repeats directly adjacent to the nic site. These inverted repeats have the potential to generate hairpin structures, so that their generation would allow the specific recognition of the oriT region by the cognate DNA relaxase and the cleavage reaction, which would take place in an unpaired region. Although all conjugative and mobilizable plasmids analyzed thus far show different inverted repeats within their oriT regions, their location relative to the nick is similar. Experimentally determined nick sites in the DNAs of plasmids of the RSF1010-oriT family mapped between 7 and 11 nucleotides upstream of the inverted repeats. In addition, the inverted repeats of RSF1010, pTF1, R1162, pIP501, pRE25, and pGO1 are all centered on the nucleotide sequence GAA.
FIG. 2.
Alignment of oriT nick regions. Nucleotides conserved in the nick regions of at least eight of nine plasmids of the RSF1010 oriT family are indicated by dark grey shading. Nucleotides that are conserved in at least five of the nine plasmids are indicated by light grey shading. A consensus sequence is also shown. Nucleotides conserved in all oriT regions are shown in capital letters; positions conserved in at least five of the nine sequences are indicated in lowercase letters. The cleavage sites determined experimentally are indicated by arrows. GenBank/EMBL accession numbers: RSF1010, M28829; R1162, M13380; pTF1, X52699; pTiC58, M95646; pSC101, X01654; pIP501, L39769; pRE25, X92945; pGO1, U50629; pMRC01, NC_001949. Modified from reference 118.
Conjugative Transfer of Broad-Host-Range Plasmids
Transfer of broad-host-range plasmids occurs at a variable frequency (generally in the range of 10−3 to 10−6) depending on the plasmid and the mating-pair genotype, and mating requires cocultivation of donor and recipient cells on a solid surface. Most conjugative plasmids identified to date in streptococci and enterococci actually show a broad host range (and hence are referred to as broad-host-range plasmids [38, 182]), while those found in staphylococci seem to be limited to the genus Staphylococcus. Both groups of conjugative plasmids confer a broad spectrum of antibiotic resistance, and their lower size limit is 15 to 20 kb (132).
Most of the broad-host-range conjugative streptococcal plasmids encode resistance to macrolides, lincosamides, and the streptogramin B antibiotics (MLSr). This resistance determinant (erm) is found in a wide variety of gram-positive cocci and bacilli and has also been found in species of the gram-negative genus Bacteroides (153). Some streptococcal plasmids, like pIP501, also carry a resistance determinant against chloramphenicol. Broad-host-range plasmids of the MLSr type have been found in various clinically important streptococci worldwide (reference 132 and references therein). Comparisons of the nucleotide and amino acid sequence of the streptococcal MLSr gene with those of different gram-positive bacteria suggest that the MLSr determinants are ancestrally related (61, 135, 219). Transfer of the streptococcal broad-host-range plasmids to a wide range of gram-positive species, including Enterococcus, Lactococcus, Staphylococcus, Clostridium, Pediococcus, and Listeria, has been demonstrated.
The appearance of conjugative plasmids in staphylococci coincided with reports of the emergence of gentamicin resistance in U.S. hospitals in the mid-1970s. However, the first demonstrations of true conjugative transfer of antibiotic resistance plasmids in staphylococci were made much later as a consequence of new outbreaks of infections due to gentamicin-resistant staphylococci in several hospitals (6, 73, 136). In these early reports, interspecies conjugative transfer between Staphylococcus epidermidis and S. aureus was demonstrated to occur on human skin (102, 208). The presence of conjugative resistance plasmids with identical restriction patterns in hospital isolates of S. aureus and S. epidermidis from the same patient (6) confirmed the epidemiological importance of bacterial conjugation.
Staphylococcal plasmids seem to be remarkably stable, since the plasmids which were detected in hospitals in the early 1980s were still the main carriers of gentamicin resistance genes in S. aureus 10 years later (7).
The Transfer Regions of Plasmids pIP501, pRE25, pSK41, pGO1, and pMRC01
The complete nucleotide sequences of the staphylococcal plasmid pSK41 (15, 71), the lactococcal plasmid pMRC01 (59), and the enterococcal plasmid pRE25 (186) have been determined. A 30.5-kb segment of pRE25 was found to be highly similar to pIP501. The chloramphenicol acetyltransferase gene, the 23S RNA methylase gene, and part of the putative conjugative unit of pRE25 (oriT and orf24 to orf29) showed 100% identity to the pIP501 49-bp oriT region and the first six genes of the pIP501 transfer operon (186). The 30.5-kb segment of pRE25 is flanked by two IS_1216V_ elements and is also highly similar to the two other plasmids, pSM19035 (34) and pAMβ1 (28). Together with pIP501, they constitute incompatibility group Inc18 of streptococcal and enterococcal plasmids replicating unidirectionally by a novel theta mechanism (24, 27, 28). The restriction map of pIP501 also exhibits a considerable degree of identity to the corresponding 30.5-kb fragment of pRE25. These data led to the hypothesis that pRE25 is pIP501 enlarged by an IS_1216V_-induced insertion (186).
The entire transfer region of the staphylococcal self-transmissible plasmid pGO1 has also been sequenced (143), and very recently we determined the 3′ part of the pIP501 tra region encompassing orf7-15 (accession number AJ505823).
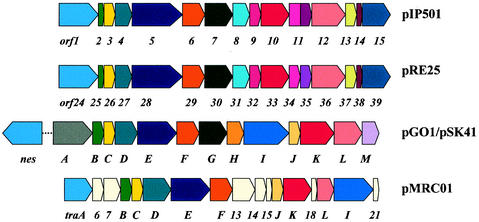
Sequence comparisons revealed interesting similarities extending the known homologies of the first six orf genes carried on pSK41, pMRC01, pGO1, pIP501, and pRE25 (15, 59, 68, 186). The modular organization of these tra regions is shown in Fig. 3. The arrangement of the first seven genes is well conserved among all compared tra regions, with the exception of an insertion of two genes of unknown function between the putative relaxase gene traA and gene traB in pMRC01. In pSK41 and pGO1, the nicking activity is encoded not by the first gene of the tra region but by the nes gene (for pSK41, nes/oriT are located approximately 11 kb upstream of the tra region, [15]). The pMRC01 tra region is the most distantly related and contains seven unique genes. Interestingly, the traG gene in pMRC01 is missing, while traK and traL homologues are present in all five plasmids (Fig. 3).
FIG. 3.
Comparison of the transfer regions of pIP501, pRE25, pGO1, pSK41, and pMRC01. Similar gene products are shown in the same color. Cream-colored boxes represent tra genes unique to pMRC01. The putative transfer proteins of pRE25 and pIP501 show a high degree of identity (between 80 and 100%). Orf1 to Orf6, Orf8 to Orf9, and Orf14 are 100% identical to the corresponding pRE25 gene products. Orf13 (262 amino acids) is significantly larger than the corresponding Orf37 (231 amino acids) encoded by pRE25. In pIP501, one big Orf (Orf11, 306 amino acids) comprises the regions of the corresponding Orf34 and Orf35 in pRE25. The gene products of the transfer region of pGO1 (trsA to trsM) and pSK41 (traA to traM) also exhibit a very high degree of similarity (between 97 and 98% identity). Tra proteins of pMRC01 show 25 to 42% identity to the corresponding proteins of pGO1 (59). pGO1 and pSK41 encode at least one additional tra gene, nes, located outside the transfer region. In pSK41, the distance between nes and the _tra_-region is approximately 11 kb. Specific single-strand nicking mediated by Nes at the respective oriT site was demonstrated for pGO1 (48).
With the exception of the sex pheromone-responding plasmids, information about the regulatory processes involved in gene transfer of conjugative plasmids in gram-positive bacteria is scarce. Only TrsN, a 7.2-kDa protein encoded by pGO1, was shown to repress the expression of essential transfer genes (196). This occurs by binding of TrsN to promoter-like sequences upstream of trsA, the first gene of the conjugative gene cluster, trs.
The operon organization of the major part of the pIP501 transfer genes was elucidated recently. Reverse transcription-PCR studies of mRNA isolated from Enterococcus faecalis JH2-2 cultures harboring pIP501 revealed cotranscription of the first 11 genes of the pIP501 tra region. The tra genes orf1 to orf11 are transcribed as a single operon of 11.3 kb (118). The compact organization of the pIP501 oriT region makes autoregulation of the tra operon by the TraA protein likely. The −10 region of the P_tra_ promoter overlaps half of an inverted repeat structure, proposed to represent the binding site for the TraA relaxase, the product of orf1 (118). This assumption is currently under investigation (B. Kurenbach and E. Grohmann, unpublished data).
Homologies to Type IV Secretion Systems
Macromolecular transfer systems ancestrally related to the conjugal mpf complexes are called type IV secretion systems, as originally proposed by Salmond (179). This nomenclature distinguishes the conjugation-related systems from other bacterial secretion pathways, such as the type I or ATP-binding cassette transporter superfamily and the type II, III, and V secretion systems. The unifying mechanistic feature among the type IV secretion systems is the capacity to transfer protein substrates intercellularly. Conjugation systems appear to be a subgroup of type IV systems that have evolved the additional capacity to translocate DNA-protein complexes (37).
Type IV systems include conjugative transfer apparatus, filamentous bacteriophage secretion, protein secretion systems of several pathogens, and natural transformation systems. Several reviews of type IV secretion in gram-negative bacteria have been published recently (30, 33, 36, 37, 51, 119, 193). A list of sequenced members of the type IV secretory pathway (IVSP) family is available at http://www-biology.ucsd.edu/∼msaier/align/align_table/VirB_Table_S4_.html. We briefly describe homologues found on conjugative elements of gram-positive bacterial origin (Table 2).
TABLE 2.
Type IV homologues encoded by conjugative elements from unicellular gram-positive bacteria
| Vir protein | Homologous Orf protein | Size (amino acids) | Reference |
|---|---|---|---|
| VirB1 | Orf41 of pAD1 | 423 | 75 |
| Orf50 of pAD1 | 830 | 75 | |
| Orf7 of pIP501 | 369 | 118 | |
| Orf30 of pRE25 | 368 | 186 | |
| TrsG of pGO1 | 358 | 143 | |
| TraG of pSK41 | 358 | 15 | |
| Orf16 of Tn_916_ | 816 | 72 | |
| VirB4 | Orf5 of pIP501 | 653 | 215 |
| Orf28 of pRE25 | 653 | 186 | |
| TrsE of pGO1 | 672 | 143 | |
| TraE of pSK41 | 672 | 15 | |
| TraE of pMRC01 | 672 | 59 | |
| Orf20 of Tn_1549_ | 800 | 79 | |
| VirB11 | Orf59 of pXO1 | 477 | 154 |
| Orf25 of pXO2 | 443 | 155 | |
| VirD4 | Orf10 of pIP501 | 551 | 118 |
| Orf33 of pRE25 | 551 | 186 | |
| TrsK of pGO1 | 546 | 143 | |
| TraK of pSK41 | 546 | 15 | |
| TraK of pMRC01 | 530 | 59 | |
| Orf16 of Tn_1549_ | 564 | 79 | |
| Orf53 of pAD1 | 747 | 76 |
ATPases.
Two gene products, Orf59 of the large Bacillus anthracis anthrax toxin-encoding plasmid pXO1 (154, 155), and Orf25 of pXO2 (155), show homologies to the VirB11 ATPase of the Agrobacterium tumefaciens T-DNA transfer system. However, the role of Orf59 and Orf25 in the B. anthracis host is not known.
VirB11, TrbB (RP4), TrwD (R388), and HPO525 of the Helicobacter pylori cag pathogenicity island belong to a family of ATPases with members present among all type II and IV secretion systems characterized so far (37). ATP hydrolysis activity of purified VirB11 has been demonstrated (35). These ATPases generally associate tightly, but peripherally, with the cytoplasmic membrane. Genetic and biochemical studies have supplied evidence for the formation of homo-oligomers of these ATPases. Recently, the TrbB, TrwD, and HPO525 ATPases have been shown to assemble as homohexameric rings with a ∼12-nm diameter, as visualized by electron microscopy (116, 117). The rings were stabilized by the addition of ATP. ATP hydrolysis was increased by the addition of phospholipids, thus indicating that interaction of these proteins with the cell membrane is likely. The crystal structure of a binary complex of HPO525 bound to ADP has been solved at a resolution of 2.5 Å (223). In the hexamer, the N- and C-terminal domains build two rings, which together form a chamber open on one side and closed on the other. The crystal structure led to a model in which the VirB11-type ATPases function as GroEL-like chaperones in translocation of unfolded proteins across the cytoplasmic membrane (117, 223). For the TrwD ATPase of R388, association with membrane vesicles that was independent of ATP hydrolysis was demonstrated, so that the protein could indeed act as a chaperone involved in the translocation of transfer components across the membranous system (131).
Three putative gene products with homologies to type IV secretion systems are encoded by plasmids pIP501, pRE25, pSK41, pGO1, and pMRC01 (Table 2). pIP501-Orf5, pRE25-Orf28, pGO1-TrsE, pMRC01-TraE, and pSK41-TraE have homologies, albeit weak, to the IncP/TrbE IncF/TraC Ti/VirB4 family of conjugative ATPases (sequenced VirB4 homologuesat http://www-biology.ucsd.edu/∼msaier/align/align_table/VirB_Table_S4_.html). Orf5 (pIP501) shows a score of 71.2 and E value of 3 × 10−13 as a member of the VirB4 family of intracellular trafficking and secretion proteins (COG3451). Orf20, encoded by Tn_1549_, a VanB-type conjugative transposon of the Tn_916_ family, has significant similarity to TrsE, the VirB4 homologue encoded by pGO1 (27% identity in 437 of a total of 800 amino acids [79]).
VirB4-type proteins are ubiquitous among the type IV systems and are sometimes present in two or more copies. Experimental evidence for VirB4 self-association and a structural contribution to channel formation that is independent of the VirB4 ATPase activity has been provided (for a review, see reference 37). Based on these features, this family of ATPases might transduce information, possibly in the form of ATP-induced conformational changes, across the cytoplasmic membrane to extracytoplasmic subunits (52).
Mating-channel proteins.
Interestingly, Orf7 encoded by pIP501 and its homologues (Table 2) show weak similarities to the family of lytic transglycosylases (pfam01464; score for Orf7, 36.1; E value, 0.007; http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid = pfam01464&version = v1.54) encoded by bacteriophages and type III and type IV secretion systems. For Orf7, the membrane localization was predicted by the PSORT program. The family of lytic transglycosylases includes the pilT gene of conjugative plasmids R64 and ColIb-P9 (AB021078 [178]), the p19 gene of the conjugative resistance plasmid R1 (P14499 [12]), trbN of RP4 (M93696), the traL gene of pKM101 (AAA86448), and virB1 of the T-DNA transfer machinery of A. tumefaciens (P17791). Although the contribution of a functional lytic transglycosylase to pathogenicity could be established only for VirB1 (17, 127, 144), it is tempting to speculate that all these transglycosylases aid the DNA and/or protein(s) to cross the cell envelope by locally opening the peptidoglycan (12, 57).
Recently, determination of the nucleotide sequence of the E. faecalis conjugative sex pheromone plasmid pAD1 was completed (75). By sequence analysis, two Orf proteins with significant similarity to lytic transglycosylases were identified: (i) Orf41, which has 61% similarity to TraG of S. aureus plasmid pSK41, and (ii) Orf50, another potential VirB1 homologue, which has 243% similarity to Orf16 of Tn_916_.
Coupling proteins.
Coupling proteins are thought to link the DNA transfer intermediate to, and perhaps lead it through, the mating channel. This family of proteins includes TraG (RP4 and Ti), TrwB (R388), TraD (F), and VirD4 of the T-DNA transfer system.
Gomis-Rüth et al. (85, 86) proposed an elegant model based on the crystal structure of the coupling protein TrwB of plasmid R388. TrwB was shown to be a large multimeric DNA-binding integral membrane protein that participates in the transfer of the single DNA strand during the mating process. The three-dimensional structure of TrwB was shown to be a homohexamer. TrwB revealed an almost spherical quaternary structure with striking similarity to F1-ATPase. A central channel with a diameter of 20 Å traverses the hexamer, although this channel may be too narrow in the cytoplasmic extreme to accommodate a single DNA strand appropriately if there is no further modification (86). The TrwB structure also shows high similarity to DNA helicases, which use the energy from nucleoside triphosphate (NTP) hydrolysis to unwind double-stranded DNA. The strong structural resemblance of TrwB to ring helicases suggests that the transferred DNA single strand might pass through the central channel of the TrwB hexamer, thereby entering the translocation apparatus connecting the donor and recipient cells. ATP hydrolysis would provide the energy to pump the single-stranded DNA through the TrwB channel, in much the same way as it does in helicases for their processive movement along the DNA (86). In fact, conformational changes have been observed in TrwB crystals after binding and putative ATP hydrolysis (85).
Topology analysis of TraG (the coupling protein of plasmid RP4) revealed that it is a multimeric transmembrane protein with cytosolic N and C termini and a short periplasmic domain close to the N terminus. It has been suggested that TraG forms a pore and that the relaxosome binds to the TraG pore via a TraG-DNA complex and that TraG interactions with the TraI relaxase (185).
Orf10 of the pIP501 tra region belongs to the pfam02534 TraG/TraD family of coupling proteins (score, 291; E value, 1e − 79). These proteins contain a P-loop and a Walker B site for nucleotide binding. For pIP501, the most closely related putative type IV secretion proteins are all encoded by pRE25 (e.g., Orf10 is 99% identical to Orf33 of pRE25). The pIP501 orf10 product is also 27% identical to the orf16 product encoded by the E. faecalis conjugative transposon Tn_1549_ (79). On the recently completed E. faecalis V583 genome sequence, another putative VirD4 homologue (20% identity) was detected. Putative homologues of coupling proteins have been detected on the chromosomes of many recently sequenced gram-positive and gram-negative bacteria as well as on transposons harbored by them.
In summary, the broad-host-range plasmids, pIP501 and pRE25, as well as pSK41, pGO1, and pMRC01, encode at least one protein homologue of most of the protein families involved in T-DNA transfer and in gram-negative bacterial plasmid transfer (37). These provide substrate presentation (VirD4 homologue), energetics of the translocation process (VirB4 homologue), and formation of the mating channel (putative VirB1 homologue). Homologues for contact formation between donor and recipient cells and for the major components of the mating channel (VirB1 is not an essential transfer protein) were not yet detected. These homologies to type IV secretion systems make a similar mechanism perhaps simpler, because only one membrane per mating partner has to be crossed for the conjugative DNA transport of plasmids from gram-positive hosts to be likely. However, the most important questions still remain unanswered: how the cell-cell contact between donor and recipient cells is established and how the DNA-protein complex is transported through the cell envelope.
Conjugative Transposons
Conjugative transposons are mobile DNA elements that encode all the necessary functions for intracellular transposition and intercellular conjugation. They are present in a wide variety of gram-positive and gram-negative bacteria and are important for the spread of antibiotic resistance genes (for reviews, see references 42, 137, 180, and 188). The transfer frequency of the investigated conjugative transposons is between 10−4 and 10−9 (76, 180). The first conjugative transposon identified was the 18-kb Tn_916_ transposon from E. faecalis (76). Tn_916_ and the closely related element Tn_1545_ from S. pneumoniae (32, 50) form the basis of a family of conjugative transposons with an extremely broad host range (177). All members of this family encode a tetracycline determinant of the TetM type (29), and many of them also carry further genes encoding resistance to antimicrobial agents. Members of the Tn_916_-Tn_1545_ family have been found naturally in, or have been introduced into, more than 50 different species and more than 20 genera of bacteria (42).
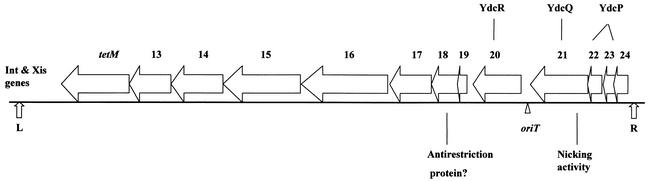
Conjugative transfer of Tn_916_ requires a series of genes located at the right end of the transposon (189). A map of Tn_916_ with open reading frames thought to be related to conjugation is shown in Fig. 4. Genetic data suggest that a single DNA strand is transferred from the donor cell to a recipient cell during conjugative transfer of Tn_916_ (187). Tn_916-oriT_ was identified as a segment of DNA that, when cloned onto a plasmid, causes mobilization of the plasmid by Tn_916_ (103). Tn_916-oriT_ is presumably the site where the DNA is nicked to initiate the transfer of a single-stranded DNA molecule, although this has not been demonstrated directly. Furthermore, definite experimental evidence for a DNA relaxase encoded by Tn_916_ exerting this site- and strand-specific nick at oriT has not yet been obtained.
FIG. 4.
Map of the conjugative transposon Tn_916_ with open reading frames thought to be related to conjugation. The genes related to excision of Tn_916, int_ and xis, are shown near the left end of the transposon. tetM is the inducible tetracycline resistance determinant of the transposon. Orf13 and Orf14 have homologues (near identity) within Tn_5397_, a conjugative transposon in Clostridium difficile. Orf15 and Orf16 each have homologues on both pAD1 and pAM373. Orf18 has similarity to the bacterial antirestriction proteins, Ard of plasmid Coll-b-P9 and ArdA of pKM101 (72). Orf20 is a homologue of the hypothetical YdcR in B. subtilis; Orf22 and Orf23, which have some similarity to each other, are both homologues of the hypothetical YdcP. Orf21 has nonspecific nicking activity that is likely to be related to an origin-involved transfer event (44). Orf21 has strong homology to a B. subtilis hypothetical protein (YdcQ), and an internal segment resembles the FtsK/SpoIIIE family (14). Modified from reference 44.
Tn_4555_ is a tetracycline resistance mobile element from the gram-negative genus Bacteroides, whose oriT shows significant homology to the nic sites of the pMV158-oriT family (see below), which are present in many RCR plasmids of gram-positive bacteria (90, 202).
Sex Pheromone Plasmids
Pheromone-responding conjugative plasmids seem to be confined to enterococci. They encode antibiotic resistance, bacteriocins, and hemolysins (reviewed in references 39, 40, 43, and 63). In this unique transfer system, recipient cells secrete a family of heat-stable, protease-sensitive peptide pheromones with specificities for donors carrying various conjugative plasmids. Bacteria harboring a particular plasmid respond by synthesis of an adhesin which facilitates the formation of mating aggregates with nearby recipients. The mating cells appear to make contacts by random collisions, and chemotaxis does not seem to be involved. Pheromones increase the transfer frequency of a plasmid by 5 to 6 orders of magnitude. The induced surface material on donor cells is named aggregation substance, while its counterpart (receptor) on the recipient surface is referred to as binding substance. When one copy of the transferred plasmid has entered the recipient, the production of the corresponding pheromone is stopped while synthesis and secretion of pheromones specific for other plasmids continues. All the pheromones characterized to date are hydrophobic peptides of 7 or 8 amino acids. With the recent availability of the E. faecalis genome sequence data, it was recognized that the sex pheromones in general correspond to parts of signal sequences of precursors of certain lipoproteins (45). Very recently, the cAD1 sex pheromone precursor in E. faecalis FA2-2 was identified. The gene, cad was found to encode a 309-amino-acid lipoprotein precursor with the last 8 residues of its 22-amino-acid signal sequence representing the cAD1 moiety (2). The sex pheromones display biological activity at very low concentrations. In pCF10, the pheromone induces transfer at concentrations of <5 pM, which corresponds to one to five molecules per donor cell under the assay conditions (142). Each plasmid, in turn, encodes an inhibitor peptide, which is secreted and acts as a specific inhibitor of the corresponding pheromone. These competitive inhibitors can block self-induction of donors and desensitize donors to pheromone secreted by recipients too far away from the donors to be encountered by random collisions. When equal numbers of recipients and donors are present, pheromone usually outcompetes the inhibitor (39).
The best-studied pheromone-induced plasmid transfer systems are pAD1, pCF10, and pPD1. pAD1 is a 59.3-kb hemolysin/bacteriocin plasmid that responds to the pheromone cAD1. The 65-kb pCF10 encodes tetraycline resistance and confers response to the pheromone cCF10. pPD1 is a 56-kb plasmid encoding bacteriocin production (Bac21), and its conjugative response depends on the pheromone cPD1. For pAD1, two oriT sequences have been identified (1, 75), with oriT1 being located within the repA determinant whereas the more efficiently utilized oriT2 is located between orf53 and orf57, two genes found to be essential for conjugation (74). oriT2 of pAD1 contains a large inverted repeat (about 140 nucleotides) adjacent to a series of short direct repeats. The orf57 gene product, the TraX relaxase, nicks within the inverted repeat (46, 74). Orf53 exhibits certain structural similarities to TraG-like proteins, although there is little overall homology (74).
Pheromone internalization is essential for induction of the pheromone response (122). This import is achieved by pheromone-binding lipoproteins, the products of traC for pAD1 and pPD1 and of prgZ for pCF10, which act as surface receptors that bind to the exogenous pheromone peptide. The pheromone is then internalized, making use of a host-encoded peptide transport system (for a recent detailed review, see reference 44). A functional analysis of TraA, the intracellular sex pheromone receptor encoded by pPD1, was recently performed (99). When cPD1 is taken up by a pPD1 donor cell, it binds to an intracellular receptor, TraA. Once a recipient cell acquires pPD1, it starts to produce an inhibitor of cPD1, termed iPD1, which functions as a TraA antagonist and blocks self-induction in donor cells. Horii et al. (99) discussed how TraA transduces the signal of cPD1 to the mating response. For pAD1, Fujimoto and Clewell (78) presented evidence that after transport into the bacterial cell, the primary target of pheromone is the pAD1-encoded TraA protein and that a conformational shift leads to induction of conjugation functions via an alteration of TraA DNA-binding activity at the iad promoter. The available information relating to the complex regulation of the pheromone response has been generated most extensively with pAD1 and pCF10 (for recent reviews, see references 43, 44, and 64).
The response system developed by conjugative pheromone plasmids to sense whether a potential host harbors the same plasmid is unique among plasmids studied so far. However, parallels to the Ti plasmids appear to exist, insofar as many of the components of the pheromone-mediated conjugation system also seem to be involved in host-parasite interactions of enterococci (95, 217, 226). Hirt et al. (95) demonstrated that E. faecalis cells harboring pCF10 showed significantly increased virulence in a rabbit endocarditis model. Their results confirmed in vivo induction of the normally highly controlled plasmid-encoded aggregation substance. Host plasma induction was dependent on the presence of the pCF10-encoded pheromone receptor protein PrgZ, indicating the requirement of the pheromone-sensing system in the induction process (95).
Interestingly, the traH gene of the conjugative S. aureus plasmid pSK41 has been reported to encode a lipoprotein precursor bearing a signal sequence whose carboxyl-terminal region consists of seven or eight contiguous amino acid residues identical to cAD1 (16, 69). A cAD1 activity could even be detected in supernatants of pSK41-carrying staphylococci but not in plasmid-free cells. Thus far, the involvement of recipient-produced pheromones as mating signals related to plasmid transfer has been observed only in E. faecalis. However, a few other bacterial species secrete peptides with a cAM373-like activity, the pAM373-specific pheromone. These include Enterococcus hirae, S. aureus, and Streptococcus gordonii (41).
A 65.1-kb conjugative plasmid, pMG1, that transfers efficiently in broth matings from E. faecalis to E. faecium strains and vice versa was isolated from a gentamicin-resistant E. faecium clinical isolate (100). Interspecies transfer of pMG1 occurs at a frequency of approximately 10−4 per donor cell in broth matings and appears to proceed independently of the presence of pheromone-like signal molecules in the culture supernatants. Interestingly, Southern hybridization of pMG1 DNA did not show any homology to pheromone-responsive plasmids and the broad-host-range plasmids pAMβ1 and pIP501. These results indicate that another efficient broth transfer system might exist in E. faecium which differs from the sex pheromone-mediated transfer system in E. faecalis (100).
Aggregation-Mediated Plasmid Transfer in Bacillus thuringienis and in Lactic Acid Bacteria
Bacillus thuringiensis subsp. israelensis is a gram-positive pathogen that is highly toxic to larvae of several dipteran aquatic insects. It contains up to 10 plasmids. One of them, a 114-kb plasmid, carries the toxin genes, while the plasmid transfer factors are ascribed to the conjugative 200-kb plasmid pXO16 (5, 87, 104).
Mobilization of small plasmids between strains of B. thuringiensis subsp. israelensis is accompanied by non-pheromone-induced and protease-sensitive coaggregation between donor and recipient cells (4). Two aggregation phenotypes (Agr+ and Agr−) were identified. They are characterized by macroscopically visible aggregates when exponentially growing cells of the Agr+ and the Agr− types are mixed in broth. The mobilization of small plasmids was found to occur unidirectionally, from Agr+ to Agr− cells. The Agr+ phenotype is transferred at a high frequency (∼100%) to Agr− cells in broth matings (4). Loci essential for the Agr+ phenotype have been localized on plasmid pXO16 (104), and it is supposed that the pXO16-mediated B. thuringiensis subsp. israelensis plasmid transfer system mobilizes plasmids of distinct replication types independent of the presence of oriT and mob functions on the mobilized plasmids (5). The fact that all plasmids tested so far (theta-replicating plasmid pAMβ1; ori43-, ori44-, and _ori60_-containing plasmids; and Bacillus cereus plasmid pBC16) could be mobilized by the pXO16-encoded conjugation system suggests that pXO16 possesses an exceptional and, so far, unique system (5), whose molecular basis remains to be elucidated.
Transfer of plasmid-encoded genes for lactose catabolism by a conjugation-like mechanism in lactic acid bacteria was described early (80, 109). Subsequently, conjugal transfer of lactose utilization genes has been reported for various Lactococcus lactis strains and for a lactose plasmid in Lactobacillus casei (210). Cell aggregation is mediated by the interaction of two cell surface components. One seems to be active only after molecular rearrangements of the lactose plasmid with the sex factor, and its clu gene(s) is encoded by the sex factor on the enlarged lactose plasmid. The second component is constitutively expressed and is encoded by a chromosomal agg gene(s). High-frequency transfer and cell aggregation occur only when a pair of strains includes both the agg and clu genes. These genes can be present in the donor, in which case it aggregates, or clu can be in the donor with agg in the recipient, in which case the mating mixture aggregates but the individual strains do not (210).
The conjugative transfer systems encoded by the sex factor of L. lactis subsp. lactis 712 and the conjugative plasmid pRS01 of L. lactis subsp. lactis ML3 (3, 81) have features in common with the aggregation-mediated plasmid transfer system in E. faecalis. In both systems, donor-recipient aggregation is associated with efficient plasmid transfer, but there is no evidence for a sex-pheromone-like induction system in L. lactis (210). Plasmid pRS01 and the sex factor from L. lactis subsp. lactis 712 are prototype mobile elements in lactococci (140). Both elements mediate high-frequency transfer of genes encoding lactose utilization (Lac+) by insertion sequence-directed cointegration with nonconjugative Lac+ plasmids (82, 171). The clu genes have been shown to be associated within an inversion region (3, 83; J.-J. Godon, C. Pillidge, K. Jury, C. A. Shearman, and M. J. Gasson, Proc. 4th Int. Conf. Streptococcal Genet., p. 43, 1994). Mapping of pRS01 identified four distinct regions (Tra1, Tra2, Tra3, and Tra4) involved in conjugative transfer. Sequence analysis of the Tra1 region revealed a gene, ltrB, with extensive homology to replicative and conjugative relaxases (139). oriT of pRS01 was localized and shown to reside within the Tra1 region upstream of the ltrB gene (141). Conjugative transfer of pRS01 requires splicing of a group II intron, LI.ltrB, for accurate translation of the mRNA for the exon gene ltrB (140). The protein product of ltrB was shown to be a conjugative relaxase, essential for pRS01 transfer. A functional promoter within LI.ltrB was identified upstream from the ltrA gene. LtrA is required for efficient splicing of LI.ltrB in vivo. Zhou et al. (227) showed that the major source of ltrA mRNA in the LI.ltrB system is from this promoter within the intron and that the promoter activity is essential for normal expression of LtrA protein, LI.ltrB splicing, and pRS01 conjugation functions in L. lactis.
It has been shown that an autoaggregating strain of L. plantarum was able to act as a donor of, or recipient for, the broad-host-range Inc18 plasmid pAMβ1 with high efficiency of plasmid transfer when mated on solid surfaces and at a low rate in broth matings. It was suggested that cell aggregation and high frequency of conjugation are associated with a secreted protein of 32 kDa, which recognizes and specifically binds to lipoteichoic acids or substitutions in teichoic acids in the cell membrane (176). The molecular mechanism of this plasmid transfer system remains to be elucidated.
RCR Mobilizable Plasmids: the pMV158 Family
The mobilization protein MobM, encoded by the broad-host-range plasmid pMV158, was the first DNA relaxase from an RCR plasmid from gram-positive bacteria that has been purified and characterized (91). The protein was shown to cleave supercoiled (but not linear) pMV158 DNA at the plasmid oriT region in a reaction that requires Mg2+ ions. On cleavage, MobM remained tightly associated with the 5′ end of the DNA, whereas the 3′ end was accessible to labeling (91). Analytical ultracentrifugation analyses have shown that MobM is a dimer in solution, with a high content of α-helices as determined by circular dichroism experiments (C. de Antonio, M. García de Lacoba, M. E. Farías, and M. Espinosa, unpublished results). Gel retardation assays showed that the protein was specifically bound to a linear double-stranded DNA segment containing two inverted repeat sequences, termed IR-1 and IR-2, which partially overlap (Fig. 5A). IR-2 has the potential to generate a secondary structure that would leave the MobM nic site unpaired and exposed in a single-stranded configuration. This would explain why MobM is able to cleave supercoiled DNA and single-stranded oligonucleotides harboring the IR but not linear double-stranded DNA because, in the latter case, the nic site would be buried within the DNA helix (90, 91). DNase I footprinting assays showed that purified MobM protein protected the IR-2 sequence (90). Since the −10 extended region of the promoter that directs synthesis of the mobM mRNA (at least in lactococcal cells) is also included within the IR-2 (66), it would appear that the protein regulates its own synthesis, similarly to the Mob protein of plasmid pBBR1 (205), a hypothesis that is currently under investigation (C. de Antonio and M. Espinosa, personal communication). Consequently, this pMV158-DNA region was defined as the oriT plasmid in vitro (90, 91) and was later shown to promote transfer of pSC101 when MobM was provided in trans (65). An identical, or nearly identical, sequence and structure of the oriT region of pMV158 can be found in various RCR plasmids from different origins (Fig. 5A); curiously, _oriT_-like sequences were found in plasmids like pCI411 and pA1, which lack a mob gene (91). Whether this genetic situation reflects a remnant of an ancient mobilization cassette present in these plasmids and/or, in addition to mobilization, these sequences play a role in plasmid cointegration when incompatibility processes take place (e.g., a plasmid-bearing host being colonized by an incompatible replicon under selection conditions) is presently unkown.
FIG. 5.
The oriT region of the pMV158 family of plasmids. (A) Features of the pMV158 oriT and its conservation among RCR-plasmids. The inverted repeats (IR) are indicated by arrows above and below the sequence. Nucleotides protected from DNase I cleavage by MobM are indicated in boldface italics, whereas the extended −10 promoter region of the mobM gene is underlined. The last nucleotide depicted (G, underlined) is the mobM transcription initiation site in L. lactis. The MobM-mediated nick (l) and the changes in the nucleotide sequence (boldface) are indicated. (B) Characterized oriT regions of three plasmids with similar mobilization cassette and the same nick site (l). Differences are underlined. DNA from plasmid pVA380-1 is a substrate of pMV158 MobM.
The Mob protein from the streptococcal plasmid pVA380-1 (121) has about 90% identity to pMV158 MobM at its N-terminal moiety, and divergences are found at its C terminus. Although the origins of transfer of the two plasmids exhibit dissimilarities (Fig. 5B), MobM protein was able to cleave supercoiled DNA of pVA380-1 in vitro (90). Based on the conservation of the N-terminal but not of the C-terminal moiety, it was proposed that the former region of MobM could be involved in the nicking reaction, a hypothesis supported by the conservation of a Tyr residue among the Mob proteins of RCR plasmids (91). For the Mob protein of plasmid pBBR1 (a broad-host-range theta replicating plasmid from the gram-negative host Bordetella bronchiseptica), it was shown that the protein is related to pMV158 MobM (205) and the pBBR1 oriT region has homologies to the pMV158 oriT (Fig. 5B). In spite of these similarities, mutation of each of the seven Tyr residues of the pBBR1-Mob protein did not affect the frequency of conjugation of the plasmid, whereas changes at two conserved residues, Asp120 and Glu121, totally abolished plasmid transfer (205). These results leave open the question whether this family of Mob proteins cleaves its target DNA through a Tyr-mediated transesterification reaction or by a water-mediated nucleophilic attack performed by a Glu residue, as proposed for the closing reaction carried out by the replication initiator protein of plasmid pC194 (150).
Based on homology analyses, it has been proposed that the DNA-binding domain of the Mob proteins is located within their C-terminal moieties, a region that in MobM contains a putative coiled-coil region that could be involved in protein dimerization (de Antonio, personal communication). No indication of DNA-binding motifs, such as helix-turn-helix or ribbon-helix-helix motifs, has been obtained.
CONJUGATIVE PLASMID TRANSFER IN MYCELIUM- FORMING STREPTOMYCETES
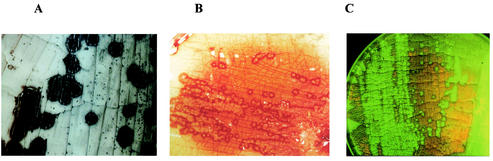
About 50 years ago, conjugation was shown to be implicated in the generation of prototrophic recombinants in mixed cultures of auxotrophic Streptomyces strains (96, 190). As in other bacteria, the fertility was ascribed to the presence of plasmids (21, 97, 211). However, conjugation in Streptomyces is a very distinct process from genetic exchange in other bacteria not only in its molecular mechanism but also in its phenotypic appearance. If a plasmid-carrying donor is plated onto agar plates together with an excess of plasmid-free recipient, plasmid transfer is associated with the formation of characteristic pock structures due to macroscopically visible growth inhibition zones of 1 to 3 mm (20). Within this pock structure, the morphological differentiation of the recipient mycelium which has newly acquired a plasmid is temporarily retarded (Fig. 6A). This feature makes Streptomyces the only microorganism where genetic exchange is visible with the naked eye.
FIG. 6.
Pock formation, indicating the intramycelial plasmid spreading. (A) If spores of a donor strain carrying a self-transmissible plasmid are mixed with an excess of plasmid-free recipient spores, characteristic growth retardation zones (pocks) are formed, indicating the area where the recipient mycelium has acquired a plasmid. The size of the pocks depends on the action of the spd genes. (B) In S. lividans pock formation is also associated with the induction of the red-pigmented antibiotic actinorhodin. (C) A pIJ101-carrying donor was streaked on a lawn of a recipient expressing gfp from Aequorea victoria. Since the aerial mycelium and spores of S. lividans show red autofluorescence, only the pock regions, where morphological differentiation is retarded, light up green.
Different Types of Conjugative Streptomyces Plasmids
A wide variety of different plasmids, most of them conjugative, have been characterized from mycelial actinomycetes. In general, these plasmids do not encode any resistance genes or other traits beside replication and fertility. The Streptomyces plasmids include huge linear plasmids of several hundreds of kilobases. Plasmid SCP1 from S. coelicolor was the first Streptomyces plasmid shown to confer fertility (211). SCP1-containing strains that transferred chromosomal markers with enhanced frequency have been isolated (97). This allowed a genetic linkage map of the S. coelicolor chromosome to be established (112). The huge linear plasmids often encode antibiotic biosynthetic pathways (113) and are the only Streptomyces plasmids that encode any phenotypic traits. The linear plasmids replicate from a centrally located origin and carry proteins bound to the repetitive ends (10, 222). These linear plasmids seem to recombine frequently with the linear chromosome (18), resulting in the exchange of a plasmid end and a chromosomal end. Since the chromosomal ends of Streptomyces do not contain essential genes, the loss of a chromosomal end usually does not interfere with the viability of the Streptomyces strain. However, the exchange of plasmid and chromosomal DNA fragments could be a very efficient route for the distribution of chromosomal genes by horizontal gene transfer.
The Streptomyces plasmids also include integrative plasmids, such as pSAM2 (21, 156, 166) or the Amycolatopsis methanolica plasmid pMEA300 (213). These elements can be excised from the chromosome and also exist as autonomous molecules, replicating either by the RCR mechanism (e.g., pSAM2) or by a theta mechanism (e.g., pMEA300). Integration occurs by site-specific recombination mediated by a plasmid-encoded integrase via an attachment site that overlaps with a chromosomal tRNA gene (22, 157). The different plasmids integrate into different tRNA genes. Since the tRNAs are quite highly conserved, the host range of the integration system is broader than the host range for autonomous replication (133). For pSAM2, it has been demonstrated that conjugative transfer requires excision of the integrated pSAM2 molecule and its autonomous replication in the donor strain (172).
Another type of plasmid includes the large low-copy-number plasmids that replicate very stably. Only a very few low-copy-number plasmids have been characterized. The best-studied representative is SCP2, the first Streptomyces plasmid that has been physically isolated (184). SCP2 is 31,317 bp in size, replicates very stably, and accepts the cloning of large fragments encoding whole antibiotic biosynthetic pathways (129). The availability of the complete nucleotide sequence allowed the identification of two resident transposable elements, IS_1648_ and Tn_1547_ (AL645771). A derivative, SCP2*, was isolated and shown to mobilize chromosomal markers with enhanced frequency (19). The transfer features of SCP2* have been characterized by transposon mutagenesis (26).
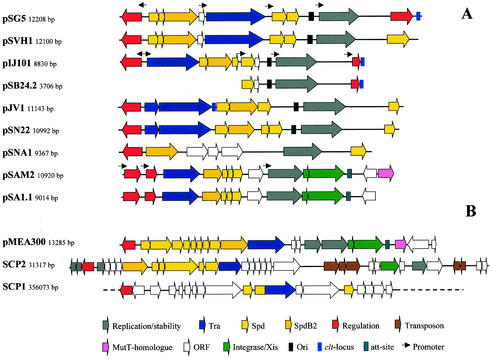
The Streptomyces plasmids also include small multicopy number plasmids. These plasmids have a molecular size of 8 to 13 kb and replicate by the rolling-circle mechanism via a single-stranded plasmid intermediate (108, 111, 147, 191, 220). All actinomycete RCR plasmids sequenced so far encode replication initiator proteins similar to pC194 RepA (146) and can be phylogenetically grouped within a single cluster of the RCR group III of the Database of Plasmid Replicons (http://www.essex.ac.uk/bs/staff/osborn/DPR/DPR_RCRIIIphylo.htm).Despite their small size, most of them are conjugative and are transferred to a plasmid-free recipient with the same efficiency as are larger plasmids (111). This class of plasmids shows a modular architecture; e.g., plasmids pSN22 and pIJ101 have a nearly identical replication region, whereas the transfer and spread genes show little to no similarity (106). The transfer and spread genes of pSN22 are very similar to those of pJV1 (191). Plasmids pSG5 and pSVH1 have a similar transfer region (60 to 70% identity), while the regulator TraR and the replication region are different (145). Very small nonconjugative plasmids such as pSB24.2 (23) and pSL33 (67) have also been isolated; they are probably deletion derivatives of larger plasmids and do not represent a typical Streptomyces plasmids.
All these different types of plasmids are conjugative, and they transferred to a plasmid-free recipient with an efficiency of nearly 100% (111). The plasmid transfer is associated with the mobilization of chromosomal markers (Cma) at a frequency ranging between 0.1 and 1% (100).
The complete nucleotide sequences of some representatives of all plasmid types have been determined. This allows the comparative analysis of the plasmid-encoded functions and the detailed characterization of the loci involved in the conjugative transfer. This comparison reveals that although the Streptomyces plasmids are not closely related to each other and have only very limited sequence similarity, most of them carry the same set of functionally homologous genes (Fig. 7).
FIG. 7.
Gene organization of actinomycete plasmids. All plasmids are drawn starting from the GntR-type regulatory gene. Identical colors indicate similar function. Red, GntR-type regulator; yellow, spread genes; orange, spdB2; blue, tra; white, Orf; grey, replication gene; light red, regulatory gene; pink, mutT homologue; green, integration/excision/recombination gene; brown, transposon. Arrows indicate regions with promoter activity. (A) Streptomyces RCR plasmids. (B) Non-RCR-type actinomycete plasmids. Only the putative transfer region of the 356-kb linear SCP1 plasmid is shown. GenBank/EMBL accession numbers: pSG5, X80774; pSVH1, Muth (unpublished); pIJ101, M21778; pSB24.2, M32513; pJV1, U23762; pSN22, D14281; pSNA1, AJ243257; pSAM2, AJ005260; pSA1.1, AB010724; pMEA300, L36679; SCP2 AL645771; and SCP1, AL590463.
Intermycelial Conjugative Transfer Mediated By a Septal DNA Translocator Protein
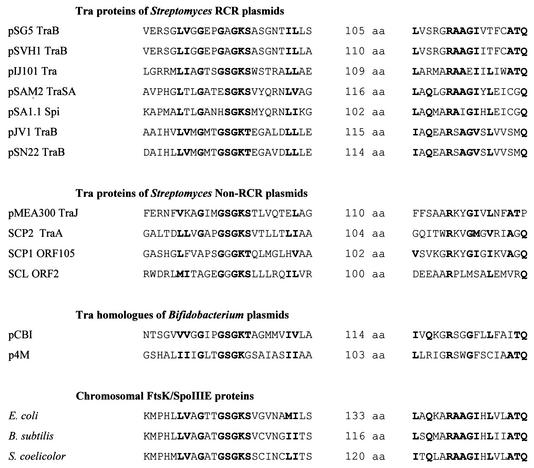
A single protein (Tra) was shown to be essential for conjugative plasmid transfer and Cma, as demonstrated for several Streptomyces plasmids by transposon and insertion mutagenesis and subcloning experiments (26, 93, 111, 130, 167, 214). The Tra proteins of pIJ101 and pSN22 have been localized in the membrane fraction of the cell (115, 168). All Tra proteins contain a nucleotide-binding site and a slightly conserved RAAGI motif at the C terminus (Fig. 8). For the pSN22 TraB protein, it was shown by site-directed mutagenesis that the NTP-binding site was essential for conjugative transfer (115). The Tra proteins of Streptomyces plasmids have only very low sequence identity to each other, but all have significant similarity to the septal DNA translocator proteins of the SpoIIIE/FtsK family (14, 130, 221). Weak similarity around the nucleotide-binding site also exists with respect to the R388 coupling protein TrwB (86). All these proteins can be grouped into the AAA superfamily (for “ATPases associated with various cellular activities”), a family of molecular motor proteins (134).
FIG. 8.
Conserved sequence motifs of the septal DNA translocator proteins. The proteins of the septal DNA translocator family share a nucleotide-binding site (Walker box A) and a conserved RAAGI motif located 100 to 133 amino acids (aa) downstream of the NTP-binding site. These motifs are also present (with wider spacing) in the hexameric ring helicase TrwB of plasmid R388 (86).
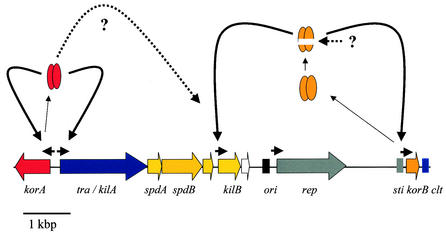
Besides the Tra protein, a _cis_-acting locus (clt) was shown to be required for the conjugative transfer of plasmids pIJ101 and pJV1. Pettis and Cohen demonstrated that if the clt locus of pIJ101 was cloned into a nontransferable plasmid, the nontransferable plasmid could be mobilized (167). However, the clt locus was not necessary for Cma. For plasmids pJV1 (192) and pSG5 (M. Elfeiturei and G. Muth, unpublished data), the clt loci were also localized (Fig. 7). They do not show any sequence similarity to the pIJ101 clt region. The pIJ101 clt locus was mapped to a 54-bp fragment (62). This fragment is most probably noncoding and contains three direct repeats and one imperfect inverted repeat. By nondenaturing polyacrylamide gel electrophoresis, the clt region was shown to be intrinsically bent or curved. Whether this DNA curvature plays a role in the conjugative transfer has not been further investigated yet.
Using an E. coli strain carrying the pIJ101 tra gene under control of the promoter Φ10 of phage T7, DNA processing at the clt site was analyzed by a sensitive primer extension analysis. However, no evidence for site-specific nicking of either strand of the clt locus was obtained (62). This is in agreement with the fact that no other DNA-relaxing enzymes beside Rep, the initiator protein of RCR, are encoded by the Streptomyces RCR plasmids.
Whether the clt locus represents the interaction site with the Tra protein was studied in gel retardation experiments. Neither Tra-containing crude extracts of E. coli or of Streptomyces lividans shifted a linear double-stranded DNA fragment containing the clt locus (62). This showed that the clt locus, at least when present on a linear DNA fragment, is not a target site for Tra or any other host protein. Therefore, the molecular function of clt in plasmid transfer remains obscure. However, the characteristics of clt indicate that clt does not represent a classical oriT region as known from other conjugative plasmids (see “Conjugative transfer in unicellular gram-positive bacteria.” above).
Temporal and Spatial Regulation of Conjugative Transfer
Conjugative transfer takes place only on solid media and in the early growth phase when Streptomyces grows as substrate mycelium. After starting morphological differentiation, conjugative transfer is abolished. The tra genes of most Streptomyces plasmids are under the transcriptional control of a GntR-type repressor (TraR/Kor). Since unregulated expression of tra is toxic, tra represents a kill function. For the S. lividans pIJ101 plasmid, it has been shown that cloning of the N-terminal half of Tra is sufficient for killing (110, 111). The mechanism of killing is unclear. However, since the N terminus contains the hydrophobic region of Tra with a predicted transmembrane helix, it can be speculated that overexpression of the membrane-binding domain of the Tra protein interferes with the integrity of the membrane. In contrast, the Tra proteins of the Streptomyces plasmids pSA1.1 from S. azureus (58) and pSG5 from S. ghanaensis (130) do not act as a kill function. Both genes can be cloned without the corresponding transcriptional repressor. Unregulated expression of these genes causes inhibition of sporulation or, in the case of pSG5, temporal retardation of the differentiation process. In most plasmids, the repressor TraR/KorA is divergently transcribed from tra and binds to repeats located in the intergenic region, repressing transcription of the tra gene (204). For the S. nigrifaciens plasmid pSN22, the TraR-binding site was determined precisely by DNase I footprinting. Purified TraR protected four 12-bp repeated “Tre”-box sequences. Protection at the Tre-box 4 region was stronger than that at the other regions. Deletion analysis and gel retardation experiments indicated that Tre-box 4, which is located between the −10 region and the start codon of tra, was critical for in vitro binding of TraR (107). Although the traR gene of pSG5 is not located upstream of traB, TraR was shown to repress transcription of the traB promoter. The promoter region of the pSG5 tra gene mediated a resistance to 600 μg of kanamycin per ml when inserted into the promoter probe plasmid pIJ487. If the DNA fragment encoding the regulatory gene traR was also inserted into the promoter probe plasmid, resistance dropped to less than 15 μg/ml (130).
The time course of Tra expression was assayed by immunologial methods. While Kosono et al. were able to detect the pSN22 Tra protein in substrate and aerial mycelium (115), Pettis and Cohen found the pIJ101 Tra protein only during the very early growth phase in the substrate mycelium (168). The highest Tra concentration was observed after 12 h. At between 18 and 21 h, the Tra concentration decreased dramatically, whereas after 30, 42, and 95 h, little or no Tra protein was detectable (168). However, transcription of the tra gene still increased after 18 h and reached its maximum level at 24 h as judged by Northern blotting (168). This suggested the existence of a further unknown factor, posttranscriptionally controlling the temporal expression of Tra. The expression of tra correlated with the efficiency of conjugative transfer of pIJ101. In “interrupted mating experiments” carried out by counterselection of the donor strain, no transfer was found up to 6 h of mating. After 12 h, a transfer rate of 0.01% was calculated, and the maximum rate of 10% in these experiments was reached after 24 h (168).
The transfer of the integrative pSAM2 plasmid was found to be differentially regulated. Conjugative transfer of pSAM2 requires excision and replication as an autonomous plasmid molecule prior to transfer (172). Therefore, the GntR repressor KorSA of pSAM2 does not directly control the expression of traSA but regulates pra, which encodes a regulator of the int-xis-rep operon (194, 195). At 7 to 8 h after mixing of donor and recipient spores, pSAM2 was excised in the donor and circular pSAM2 molecules could be detected. To that time, the transfer efficiency was 0.1%. After 22 h, the occurrence of free pSAM2 was maximal, corresponding to a transfer rate of 75%. After 48 h, conjugative plasmid transfer was completed. Then pSAM2 was integrated again into the donor and the recipient, and only a very faint band of autonomously replicating pSAM2 could be detected (172).
If a pSAM2 donor is mated with a recipient carrying a differentially marked pSAM2 derivative, no plasmid excision and no transfer is observed (172). This indicates that a kind of signaling must exist which represses the excision of the integrated pSAM2 molecule and subsequently the initiation of conjugative transfer. However, the signal or the pSAM2 region responsible for this exclusion has not been identified yet.
The KorA homologue of pMEA300 was shown to regulate the expression of the spd-tra- operon as well as the expression of the rep-int-xis operon. In gel retardation experiments, binding of KorA to a 14-bp inverted repeat upstream of korA, traA, and orfA was demonstrated (212, 214). Similar sequences have been also identified as the binding sites of ImpA, the KorA homologue of the S. coelicolor integrative plasmid SLP1 (199).
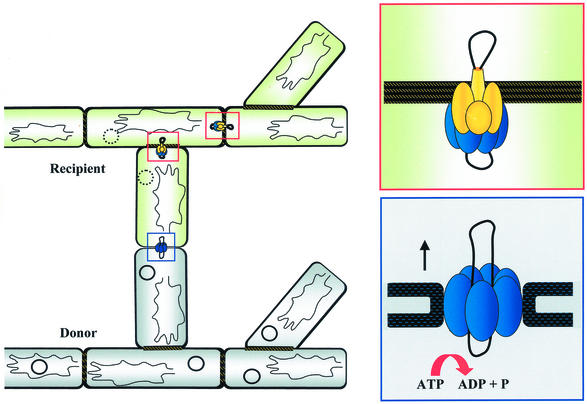
Pock Structures and Intramycelial Plasmid Spreading
As an adaptation to the mycelial growth characteristics, typical of the streptomycetes, the primary transfer event from the donor to the recipient is followed by secondary transfer events within the recipient mycelium. The incoming plasmid is subsequently distributed to the neighboring mycelial compartments, resulting in fast spreading of the plasmid within the recipient mycelium (98). On agar plates, the spreading of the plasmid is manifested by the formation of macroscopically visible growth or differentiation retardation zones (Fig. 6A and C) (20, 111). These pock structures are several millimeters across and indicate the area where the recipient mycelium has obtained a plasmid. In S. lividans this growth inhibition is accompanied by the induction of the synthesis of the red-pigmented antibiotic actinorhodin (Fig. 6B).
Whereas the primary transfer from the donor into the recipient requires only the action of the main transfer gene tra, the spreading through the cross-walls in the recipient mycelium requires three to five further genes. These spread genes are often translationally coupled and encode small hydrophobic proteins that do not show extensive sequence similarity to each other or to any other protein in databases. Inactivation of a single spd gene abolishes pock formation (106, 111).
From mutant analysis of plasmids pSN22 and pIJ101, evidence was obtained that the Tra protein is also involved in plasmid spreading (106, 169). One pIJ101 spd gene (kilB) represents a second kill function. kilB is under transcriptional control of the KorB repressor (206, 224). KorB is synthesized as a 10-kDa protein and subsequently processed to a 6-kDa peptide. The 6-kDa KorB was shown to bind to the korB promoter regions of korB and kilB. Since the binding site in the korB promoter region overlaps the minus origin for lagging-strand synthesis, sti, a role of KorB in coordinating replication and intramycelial plasmid spreading during conjugation was speculated (206). When sti was separated from the korB operator sequence, the transfer frequency decreased to 30% (207). The lethality of kilB overexpression was also suppressed by the presence of the KorA regulator (110, 111). Because KorA does not interact with the kilB promoter, the mechanism of control of KilB activity is unclear (170). A scheme of this complex regulation is shown in Fig. 9. The KilB protein of pIJ101 is present throughout the whole life cycle of Streptomyces, even in growth phases when no plasmid transfer takes place. This indicates an additional role of the KilB spread protein, independent of plasmid transfer. Of course, intramycelial plasmid spreading would also contribute to the stable maintenance of the plasmid during vegetative growth and morphological differentiation in ensuring that all mycelial compartments contain the plasmid. However, derivatives lacking the spd genes are maintained as stably as the _spd_-containing plasmids (111).
FIG. 9.
Transcriptional regulation of the transfer functions of the S. lividans plasmid pIJ101. The GntR-type repressor KorA represses the transcription of tra/kilA and korA by binding to the overlapping promoter regions (204). The KilB repressor KorB is synthesized as a 10-kDa protein and subsequently processed to a 6-kDa molecule which binds to the kilB promoter 50-fold more efficiently than to the korB promoter (206). By an unknown mechanism, KorA also overrides the lethal action of unregulated kilB expression.
Plasmids pSG5 and pSVH1 have highly homologous Spd proteins (145) with an amino acid identity of between 60 and 70%. In contrast to plasmid pSVH1, plasmid pSG5 does not cause pock formation during its conjugative transfer, probably due to missing promoter activity in front of the spdB3-orf80-spdB2 operon (130). The most intriguing spd gene encodes the multidomain membrane protein SpdB2. The SpdB2 proteins are characterized by four transmembrane helices and the presence of tandem repetitive peptide sequences, e.g., pSN22-SpdB2 (RERE, 5×), pJV1-SpdB2 (EERER, 7×), pSVH1-SpdB2 (RAEQ, 5×), and SCP2.27c (AAAEA, 12×) (bold type indicates absolutely conserved residues). Some of the SpdB2 proteins have low sequence similarity to TolA of E. coli, which also possesses tandem repetitive sequences. These repetitive sequences might enable TolA to form an amphipathic helix spanning the membrane (126). TolA has been reported to interact with other Tol proteins, TolQ and TolR, probably forming a channel through the outer and inner membrane-for the uptake of colicins and single-stranded phage DNA (47). Repetitive octapeptide units were also found in the bacteriophage T4 baseplate protein gp5. The repeats were shown to form an intertwined β-helix which acts in cooperation with three gp27 monomers as a membrane-puncturing needle for the injection of the phage DNA (105). The similarity of SpdB2 to these proteins allows speculation about the function of the Spd proteins. The Streptomyces SpdB2 protein might interact with the other Spd proteins to form a crosswall-traversing complex to support translocation of the plasmid to the neighboring mycelial compartments. Interestingly, an SpdB2 homologue (Orfgp25) was also identified on the actinophage PhiC31 (203). Orf25 belongs to the early region of PhiC31 and is expressed shortly after infection. It was suggested that the presence of spd genes might help the incoming phage to spread through the crosswalls within the Streptomyces mycelium, resulting in a more efficient infection of the total colony.
Experimental Evidence for the Transfer of a Double-Stranded Plasmid Molecule
As stated above, a DNA single strand, generated by the plasmid-encoded relaxase at the oriT, is proposed to be transferred in all conjugation systems studied in unicellular bacteria. Such an enzymatic activity does not seem to be involved in Streptomyces conjugation. The replication initiator protein for rolling-circle replication is not required for conjugative transfer of a RCR plasmid (226). Only tra and the clt locus are essential for the transfer of a Streptomyces plasmid from a donor to the recipient mycelium (167). Since Tra does not seem to possess DNA-processing activity and since no site-specific nicking at clt could be demonstrated (62), one has to suppose that conjugative transfer in Streptomyces involves unprocessed double-stranded DNA. Furthermore, the similarity of the Streptomyces Tra proteins to the septal DNA translocators SpoIIIE (221) and FtsK (14), which both translocate double-stranded chromosomal DNA, strengthens the hypothesis of double-stranded DNA transfer.
Experimental evidence for the transfer of a double-stranded plasmid molecule has been recently reported by Possoz et al. in a very elegant assay based on the differential sensitivity of single- and double-stranded DNA to _Sal_I restriction (172). Since single-stranded DNA is not a substrate for the type II restriction endonuclease _Sal_I, plasmid transfer is affected by _Sal_I only if double-stranded plasmid DNA is transferred. Mating experiments were performed with S. lividans recipient strains differing in the presence or absence of the _Sal_I restriction modification (RM) system. In a control experiment, the effect of the _Sal_I RM system on the transfer of a single-stranded molecule was studied. Since the bifunctional pSAM2 derivative pTS142 carries the RK2 oriT region, it can be mobilized from E. coli S17/1 to S. lividans as a single-stranded plasmid. In these experiments, mobilization to S. lividans occurred with the same frequency whether the _Sal_I RM system was present or not, demonstrating that a single-stranded molecule is not restricted by _Sal_I. In contrast, when the matings were performed between two different S. lividans strains, plasmid pTS142 possessing 17 _Sal_I sites could be efficiently transferred only to the recipient strain lacking the _Sal_I RM. When the recipient strain expressed the _Sal_I RM system the conjugation efficiency dropped from 1 to 10−4 (172). The sensitivity of the Streptomyces transfer process to _Sal_I restriction is convincing evidence that a double-stranded plasmid molecule may be transferred during conjugation.
Model for the Conjugative Transfer of Streptomyces Plasmids
As is evident from the comparison of the different types of plasmids and their genes involved in transfer, the mechanism of conjugative transfer in Streptomyces is clearly different from that of all other systems known so far. A model illustrating the molecular events in Streptomyces conjugation, based on experimental evidence as well as on conclusions drawn from the activities of homologous proteins, is shown in Fig. 10.
FIG. 10.
Model for conjugative plasmid transfer in Streptomyces. The hyphal tips of a plasmid-carrying donor and a recipient mycelium grow together without the need for a plasmid-encoded aggregation system. In the presence of a Tra protein, the hyphae fuse. Tra multimers form a ring-shaped structure around the double-stranded plasmid (lower inset). Depending on ATP hydrolysis, the unprocessed plasmid molecule is translocated through the Tra pore into the recipient. The newly incoming plasmid is subsequently distributed to the neighboring mycelial compartments via the hydrophobic Spd proteins, which form a pore-like structure in the septal crosswalls (upper inset). This plasmid spreading is manifested by the formation of pock-like inhibition zones (Fig. 6).
First, there is no plasmid-encoded system for mediating cell-to-cell contact between the mating partners. As a soil organism, the nonmotile Streptomyces grows on a solid substrate as a multiply branching mycelium, so that a system dedicated to the establishment of the close cell-to-cell contact (which is a prerequisite for genetic exchange between mating partners) might be dispensable. To form mating pairs, the mycelial tips could grow together without the need for special genes for pilus formation or aggregation. Only when a plasmid-encoded transfer protein is present might the tips fuse. Up to now, this fusion could not be microscopically visualized. However, the availability of sensitive reporter genes that allow differential labeling of donor and recipient mycelia should provide insight into this early stage of the Streptomyces conjugation process in the near future. The Tra protein, the only essential factor for conjugative transfer, could also be responsible for the mycelial fusion process. For the B. subtilis homologue SpoIIIE, a membrane-fusing activity has been experimentally demonstrated (197).
In all other transfer systems, conjugative transfer is initiated by a relaxase, which introduces a nick at the oriT, thus supplying the single-stranded molecule that is transported into the recipient. There is convincing evidence that plasmid transfer in Streptomyces does not require any DNA-processing activities: (i) the Streptomyces plasmids probably do not encode a conjugative relaxase (Fig. 7); (ii) the tra gene alone, when integrated into the chromosome, is sufficient to mediate conjugative gene transfer (167); (iii) the _cis_-acting locus of transfer, clt, the only region required for plasmid mobilization, is not the target site of DNA relaxation (62); and (iv) plasmid transfer between two Streptomyces strains is sensitive to _Sal_I restriction, whereas intergeneric transfer from E. coli to Streptomyces, involving a single-stranded plasmid molecule, was not affected (172).
The striking similarity of the Streptomyces Tra proteins to the DNA motor proteins SpoIIIE and FtsK, both of which were shown to translocate double-stranded circular DNA molecules, suggests that Tra has a function in translocating the plasmid into the recipient mycelium. Tra monomers aggregate in the membrane of the growing tip, giving rise to a ring-like structure around the double-stranded covalently closed circular plasmid molecule. Localization of the replication initiator protein of pSG5 within the membrane fraction (R.-M. Maas, and G. Muth, unpublished data), indicates that the Streptomyces RCR plasmids are already associated with the membrane. Recently it was demonstrated by electron microscopy that the C terminus of FtsK, which is homologous to the Streptomyces Tra proteins, aggregates to form a multimeric ring-shaped structure. The FtsK ring has a diameter of about 30 to 40 nm with a hole of about 10 nm (8), which is sufficient to accommodate two DNA double strands, as would be required for the translocation of an unprocessed double-stranded circular plasmid molecule. As is the case for FtsK during cell division or SpoIIIE during sporulation (198), the Tra protein might pump the DNA into the recipient by using the energy of ATP hydrolysis.
An important but still unsolved question is how the Tra protein recognizes and interacts with the plasmid DNA. Since the clt loci of different plasmids which are essential for plasmid transfer show no sequence similarity and are probably as diverse as the Tra proteins, the clt locus would be the first candidate for an interaction site with Tra. If clt represented a site for the association of the plasmid with the membrane, a more highly conserved sequence would be expected. However, Tra does not seem to interact directly with the clt locus. In gel retardation experiments with Tra-containing crude extracts, no binding of Tra or any other cellular protein to the clt region could be observed (62).
Intramycelial spreading of the transferred plasmid within the mycelial compartments of the recipient is accomplished by the Spd proteins. The hydrophobic Spd proteins probably form in association with Tra, a pore-like structure in the septal crosswalls of the recipient, and help distribute the plasmid to the older parts of the Streptomyces colony as well. Plasmid spreading ensures that a larger fraction of the recipient mycelium is colonized by the plasmid. Since the recipient mycelial compartments that have just acquired the incoming plasmid lack a functional TraR transcriptional repressor, tra transcription is induced. This transient overexpression of Tra results in growth inhibition and retardation of the morphological differentiation which is manifested by the characteristic pock structures.
CONJUGATIVE TRANSFER IN OTHER ACTINOBACTERIA
Conjugation has been described in a variety of other actinobacteria, including Mycobacterium, Corynebacterium, and Rhodococcus (56, 88, 128, 165), but only very few reports on the mechanism of conjugation in these organisms are available. The molecular events of the gene transfer process are not known, and the question whether conjugation follows a mechanism similar to that described for the broad-host-range plasmids (see “Conjugative transfer in unicellular gram-positive bacteria” above) or whether different models have been evolved in these organisms remains unanswered. The only evidence concerning the mechanistic aspects of transfer can be obtained by analyzing the genetic capacity of plasmids. A variety of plasmid sequences have been submitted to public databases. Recently, some large plasmids have been completely sequenced and annotated. Analysis of the predicted gene functions revealed the presence of proteins putatively involved in conjugation processes (Table 3). Putative conjugative relaxases are encoded by plasmids from Rhodococcus, Mycobacterium, Bifidobacterium, and Corynebacterium. The 80,610-bp Rhodococcus equi plasmid pREAT701 was also found to carry a trsK (pGO1) homologous gene encoding a putative coupling protein. The presence of a relaxase, the key enzyme in the initiation of conjugative transfer, indicates that conjugation in these bacteria might follow a mechanism very similar to that described for the broad-host-range plasmids, as outlined for plasmid pIP501. Therefore, conjugative plasmid transfer in these organisms probably also involves a nicking reaction at oriT and transfer of a single-stranded molecule.
TABLE 3.
Putative transfer genes encoded by plasmids from actinobacteridae
| Plasmid (accession no.) | Size (bp) | Organism | Putative transfer protein (accession no.) | Putative function |
|---|---|---|---|---|
| pREAT701 (AP001204) | 80,610 | Rhodococcus equi | TraA (Q9ETQ3) | Relaxase |
| pREAT701 (AP001204) | 80,610 | Rhodococcus equi | TrsK (Q9EU11) | Coupling protein |
| pREAT701 (AP001204) | 80,610 | Rhodococcus equi | TrbL (Q9ETB5) | Mating-pair formation |
| pREAT701 (AP001204) | 80,610 | Rhodococcus equi | TrbA (Q9EU72) | Repressor |
| pCG4 (AF164956) | 29,371 | Corynebacterium glutamicum | TraA (Q9EUN3) | Relaxase |
| pCG4 (AF164956) | 29,371 | Corynebacterium glutamicum | TraC (Q9EUN8) | Primase |
| pNG2 (AF492560) | 15,100 | Corynebacterium diphtheriae | TraA (AAM12769) | Relaxase |
| pVT2 (AY056023) | 12,868 | Mycobacterium avium | TraA (Q938A0) | Relaxase |
| pTET3 (CAD12223) | 27,856 | Corynebacterium glutamicum | TraA (Q8VVJ4) | Relaxase |
| pKJ36 (AF139129) | 3,625 | Bifidobacterium longum | MobB (Q9F152) | Relaxase |
| pKJ50 (U76614) | 4,960 | Bifidobacterium longum | MobA (Q9ZA19) | Relaxase |
| pCIBb1 (AF085719) | 5,750 | Bifidobacterium breve | FtsK homologue (Q9X3U8) | Septal DNA translocator |
| p4M (AF359574) | 4,488 | Bifidobacterium pseudocatenulatum | Tra (AAM00236) | Septal DNA translocator |
An interesting situation is found in bifidobacteria. “Classic” plasmids encoding putative relaxases, like pKJ36 and pKJ50, have been reported (164). However, there is also the 5,750-bp plasmid pCIBb1 (158) from Bifidobacterium breve, which is more closely related to the Streptomyces RCR plasmids. It encodes a replication initiator protein that aligns well with the Streptomyces RCR plasmid-encoded Rep proteins. Strikingly, plasmid pCIBb1 also encodes a septal DNA translocator protein of the FtsK/SpoIIIE family, homologous to the Streptomyces Tra proteins (e.g., 27% identity to pSG5-TraB; 253-amino-acid overlap). A further Tra homologue (28% identity to pSAM2-TraSA; 222-amino-acid overlap) is also encoded by the Bifidobacterium pseudocatenulatum plasmid p4M (AF359574).
It can therefore be speculated that two different modes of conjugative transfer might exist in bifidobacteria: (i) a classical transfer system involving a relaxase and a single-stranded plasmid intermediate and (ii) the _Streptomyces_-type transfer system involving a septal DNA translocator protein.
SUMMARY AND FUTURE PERSPECTIVES
Implications of Intergeneric Gene Transfer by Gram-Positive Transfer Systems
Conjugative transfer among gram-positive bacteria, and its implication in the transfer of resistance genes, is a process more widespread and with more clinical relevance than envisaged, especially taking into consideration that (i) actinomycetes are the most important producers of antibiotics and (ii) enterococci, streptococci, and staphylococci are the most common causes of nosocomial infections. Several examples illustrate how this process can proceed among bacteria of many different genera, especially for plasmids that have an uncommonly broad host range. Among them, mobilization of the streptococcal plasmid pMV158 by pAMβ1 from S. pneumoniae to E. faecalis has recently been documented (148). Even more strikingly, it has been shown that transfer of the streptococcal plasmid pIP501 from enterococci to multicellular gram-positive bacteria and to gram-negative bacteria was feasible. This plasmid could be transferred to E. coli and S. lividans, hosts in which the pIP501-located antibiotic resistance genes and transfer functions were functional (118a). A particular concern for public health related to transfer of antibiotic resistance genes in pathogenic gram-positive bacteria is that of resistance to vancomycin such as that encoded by conjugative transposons (79), which are quite common in the gut bacteria Enterococcus. While enterococcial infections are generally not life-threatening, the genes for vancomycin resistance may be spread to bacteria like S. aureus, which may cause fatal diseases. In 1996, the first methicillin-resistant S. aureus that had acquired resistance to vancomycin was isolated from a Japanese patient (94). However, the genetic basis for the vancomycin resistance of this isolate has not yet been elucidated. Incidences of vancomycin resistance in S. aureus have been increasing worldwide for the last 5 years. In June 2002, eight patients with confirmed clinical infections caused by vancomycin-resistant S. aureus (VRSA) have been found in the United States (201). This was the first case of infection caused by VRSA in a patient in the United States. The VRSA isolate contained the vanA vancomycin resistance gene from enterococci. The presence of vanA in this VRSA isolate suggests that the resistance determinant might have been acquired through exchange of genetic material from the vancomycin-resistant Enterococcus strain also isolated from the swab culture of the same patient, but experimental proof for the contribution of conjugative transfer to the resistance has not yet been obtained. Only under laboratory conditions could conjugative transfer of vancomycin resistance genes from enterococci to S. aureus be achieved (149). Recently, however, a natural E. faecalis conjugative plasmid (pAM368) that encoded vancomycin resistance was found to respond to the sex pheromone cAM373 secreted by S. aureus (200). A pAM373::pAD2 cointegrate plasmid could be transferred to S. aureus at a frequency of 6.5 × 10−6, demonstrating the efficiency of intergeneric transfer between E. faecalis and S. aureus (200).
Attempts to Elucidate the Role of the Type IV Components in Conjugative Plasmids from Gram-Positive Hosts
The presence of components of type IV secretion systems in gram-positive bacteria favors the hypothesis of the evolution of a common conjugative DNA/protein transport system in gram-negative and gram-positive bacteria. However, the lack of components with clear homologies to adherence and mating-channel proteins, perhaps with the exception of VirB1 in gram-positive transfer systems, suggests that these processes may differ considerably from the postulated mechanisms in gram-negative bacteria. In agreement with the presence of homologues of the NTPases, VirB4 and VirB11 in various gram-positive bacteria (Table 2) (33), the translocation energetic processes could be similar to those proposed for gram-negative bacteria. Of the 10 type IV secretory pathway protein constituents, VirB2 to VirB11, analyzed by Cao and Saier (33), only VirB11 was found ubiquitously in a wide variety of bacteria and archaea. Homologues of the TraG-TraD-TrwB-VirD4 coupling-protein family were detected in all the analyzed broad-host-range plasmids as well as the staphylococcal (pSK41 and pGO1) and lactococcal (pMRC01) plasmids from unicellular gram-positive bacteria (Table 2). These data, in combination with the high conservation of DNA relaxases across the gram-positive/gram-negative barrier, favor the existence of a relaxosome similar to that in gram-negative bacteria which could be connected to a unique gram-positive bacterial mating channel via a TraG-like coupling protein. Relationships between proteins involved in DNA-trafficking processes, such as conjugation and uptake of DNA by competent cells (as in the case of B. subtilis and several streptococcal species), await an in-depth analysis and comparative revision.
Protein interaction studies with the putative type IV components and the TraA relaxase encoded by pIP501 are currently in progress. These, together with localization experiments of the type IV homologues and NTP hydrolysis tests of the putative VirB4 and VirD4 homologues, should help confirm the proposed functions of these components in the conjugative transfer process.
Future Perspectives
There is a general lack of knowledge of many of the conjugal transfer processes in gram-positive bacteria. It has been assumed that these processes must be similar to those in gram-negative bacteria. However, some of the mechanisms observed in multicellular gram-positive bacteria, in which transfer of DNA in a double-stranded configuration seems to take place, indicate otherwise. Furthermore, the different structures of the cell envelope, and the lack of periplasm, suggest that gram-positive hosts do not need to evolve sophisticated structures, such as pili, to achieve a close cell-to-cell contact. This would lead to plasmids with a simpler genetic configuration, with regard to their tra functions, since many of the mpf functions would be superfluous in the gram-positive hosts. Mechanisms used to invade eukaryotic cells, such as adhesion functions, may also be used to transfer DNA between donor and recipient cells. Whether there is a common path between transfer, DNA uptake, and invasion functions remains to be analyzed. In addition, no structural studies of the conjugation apparatus components in gram-positive hosts have been performed, and biochemical and many biophysical studies of the proteins involved in the transfer interplay must be done before any general picture can be developed. We envisage plenty of challenges in the future for researchers working in the field of plasmid transfer among gram-positive bacteria.
Acknowledgments
Our laboratories have been funded by grant BMC2000-0550 from the Spanish Ministerio de Ciencia y Tecnología, grant 01/0956 from the Spanish Instituto de Salud Carlos III, and grant QLK2-CT-2000-01624 from the European Union, by the Program for Strategic Groups from the Comunidad Autónoma de Madrid and the Spanish Network of Infectious Pathology (M.E.), and by grants Gr1792/1-1 and Gr1792/1-2 from the Deutsche Forschungsgemeinschaft to E.G. G.M. acknowledges the financial support of the Landesstiftung Baden-Württemberg GmbH “Kompetenznetzwerk Resistenz.”
We thank K. F. Genser, B. Kurenbach, C. de Antonio, and W. Wohlleben for critical reading of the manuscript.
REFERENCES
- 1.An, F. Y., and D. B. Clewell. 1997. The origin of transfer (oriT) of the enterococcal pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid 37**:**87-94. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184**:**1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. J., and L. L. McKay. 1984. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis M3. J. Bacteriol. 158**:**954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrup, L., J. Damgaard, and K. Wassermann. 1993. Mobilization of small plasmids in Bacillus thuringiensis subsp. israelensis is accompanied by specific aggregation. J. Bacteriol. 175**:**6530-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrup, L., O. Jorgensen, A. Wilcks, L. Smidt, and G. B. Jensen. 1996. Mobilization of “nonmobilizable” plasmids by the aggregation-mediated conjugation system of Bacillus thuringiensis. Plasmid 36**:**75-85. [DOI] [PubMed] [Google Scholar]
- 6.Archer, G. L., and J. L. Johnston. 1983. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob. Agents Chemother. 24**:**70-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archer, G. L., and J. Scott. 1991. Conjugative transfer genes in staphylococcal isolates from the United States. Antimicrob. Agents Chemother. 35**:**2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aussel, L., F.-X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. J. Sherratt. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108**:**195-205. [DOI] [PubMed] [Google Scholar]
- 9.Bates, S., A. M. Cashmore, and B. M. Wilkins. 1998. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the tra2 mating system. J. Bacteriol. 180**:**6538-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15**:**1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43**:**1359-1365. [DOI] [PubMed] [Google Scholar]
- 12.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentbeiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183**:**3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker, E. C., and R. J. Meyer. 2002. MobA, the DNA strand transferase of plasmid R1162: The minimal domain required for DNA processing at the origin of transfer. J. Biol. Chem. 277**:**14575-14580. [DOI] [PubMed] [Google Scholar]
- 14.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177**:**6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180**:**4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg, T., N. Firth, and R. A. Skurray. 1997. Enterococcal pheromone-like activity derived from a lipoprotein signal peptide encoded by a Staphylococcus aureus plasmid, p. 1041-1044. In T. Horaud, M. Sicard, A. Bouve, and H. de Montelos (ed.), Streptococci and the host. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 17.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176**:**3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bey, S. J., M. F. Tsou, C. H. Huang, C. C. Yang, and C. W. Chen. 2000. The homologous terminal sequence of the Streptomyces lividans chromosome and SLP2 plasmid. Microbiology 146**:**911-922. [DOI] [PubMed] [Google Scholar]
- 19.Bibb, M. J., and D. A. Hopwood. 1981. Genetic studies of the fertility plasmid SCP2 and its SCP2* variants in Streptomyces coelicolor A3(2). J. Gen. Microbiol. 126**:**427-442. [Google Scholar]
- 20.Bibb, M. J., J. M. Ward, and D. A. Hopwood. 1978. Transformation of plasmid DNA into Streptomyces at high frequency. Nature 274**:**398-400. [DOI] [PubMed] [Google Scholar]
- 21.Bibb, M. J., J. M. Ward, T. Kieser, S. N. Cohen, and D. A. Hopwood. 1981. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol. Gen. Genet. 184**:**230-240. [DOI] [PubMed] [Google Scholar]
- 22.Boccard, F., J. Pernodet, A. Friedmann, and M. Guerineau. 1988. Site-specific integration of plasmid pSAM2 in Streptomyces lividans and S. ambofaciens. Mol. Gen. Genet. 212**:**432-439. [Google Scholar]
- 23.Bolotin, A. P., A. V. Sorokin, N. N. Aleksandrov, V. N. Danilenko, and Y. I. Kozlov. 1985. Nucleotide sequence of Actinomyces plasmid pSB24.2 DNA. Dokl. Akad. Nauk. SSSR 2834**:**1014-1017. [PubMed] [Google Scholar]
- 24.Brantl, S. 1994. The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol. Microbiol. 14**:**473-483. [DOI] [PubMed] [Google Scholar]
- 25.Brasch, M. A., and R. J. Meyer. 1987. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad-host-range plasmid R1162. J. Mol. Biol. 198**:**361-369. [DOI] [PubMed] [Google Scholar]
- 26.Brolle, D.-F., H. Pape, D. A. Hopwood, and T. Kieser. 1993. Analysis of the transfer region of the Streptomyces plasmid SCP2. Mol. Microbiol. 10**:**157-170. [DOI] [PubMed] [Google Scholar]
- 27.Bruand, C., S. D. Ehrlich, and L. Jannière. 1991. Unidirectional replication of the structurally stable Enterococcus faecalis plasmid pAMβ1. EMBO J. 10**:**2171-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruand, C., E. Le Chatelier, S. D. Ehrlich, and L. Jannière. 1993. A fourth class of theta-replicating plasmids: the pAMβ1 family from Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 90**:**11668-11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burdett, V. 1991. Purification and characterization of Tet (M), a protein that renders ribosomes resistant to tetracycline. J. Biol. Chem. 15**:**2872-2877. [PubMed] [Google Scholar]
- 30.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2**:**25-29. [DOI] [PubMed] [Google Scholar]
- 31.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25**:**1011-1022. [DOI] [PubMed] [Google Scholar]
- 32.Caillaud, F., C. Carlier, and P. Courvalin. 1987. Physical analysis of the conjugative shuttle transposon Tn_1545_. Plasmid 1**:**58-60. [DOI] [PubMed] [Google Scholar]
- 33.Cao, T. B., and M. H. Saier, Jr. 2001. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary, conclusions. Microbiology 147**:**3201-3214. [DOI] [PubMed] [Google Scholar]
- 34.Ceglowski, P., A. Boitsov, N. Karamyan, S. Chai, and J. C. Alonso. 1993. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol. Gen. Genet. 241**:**579-585. [DOI] [PubMed] [Google Scholar]
- 35.Christie, P. J. 1997. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179**:**3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8**:**354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40**:**294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clewell, D. B. 1990. Movable genetic elements and antibiotic resistance in enterococci. Eur. J. Clin. Microbiol. Infect. Dis. 9**:**90-102. [DOI] [PubMed] [Google Scholar]
- 39.Clewell, D. B. 1993. Bacterial sex pheromone-induced plasmid transfer. Cell 73**:**9-12. [DOI] [PubMed] [Google Scholar]
- 40.Clewell, D. B. 1993. Sex pheromones and the plasmid-encoded mating response in Enterococcus faecalis, p. 349-367. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 41.Clewell, D. B., F. Y. An, B. A. White, and C. Gawron-Burke. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn_918_). J. Bacteriol. 162**:**1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn_916_-Tn_1545_ family of conjugative transposons. Trends Microbiol. 3**:**229-236. [DOI] [PubMed] [Google Scholar]
- 43.Clewell, D. B. 1999. Sex pheromone systems in enterococci, p. 47-65. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 44.Clewell, D. B., and G. M. Dunny. 2002. Conjugation and genetic exchange in enterococci, p. 265-300. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 45.Clewell, D. B., F. Y. An, S. F. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35**:**246-247. [DOI] [PubMed] [Google Scholar]
- 46.Clewell, D. B., M. V. Francia, S. E. Flannagan, and F. Y. An. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48**:**193-201. [DOI] [PubMed] [Google Scholar]
- 47.Click, E. M., and R. E. Webster. 1998. The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180**:**1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Climo, M. W., V. K. Sharma, and G. L. Archer. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178**:**4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook, D. M., and S. K. Farrand. 1992. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with the T-region borders. J. Bacteriol. 174**:**6238-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Courvalin, P., and C. Carlier. 1987. Tn_1545:_ a conjugative shuttle transposon. Mol. Gen. Genet. 206**:**259-264. [DOI] [PubMed] [Google Scholar]
- 51.Covacci, A., J. L. Telford, G. del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284**:**1328-1333. [DOI] [PubMed] [Google Scholar]
- 52.Dang, T. A., X. R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol. 32**:**1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264**:**375-378. [DOI] [PubMed] [Google Scholar]
- 54.de la Cruz, F., and E. Lanka. 1998. Function of the Ti plasmid Vir proteins: T-complex formation and transfer to the plant cell, p. 281-301. In H. P. Spaink, A. Kondorosi, and P. J. Hooykaas (ed.), The Rhizobiaceae, Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 55.Derbyshire, K. M., G. Hatfull, and N. S. Willetts. 1987. Mobilization of the non-conjugative plasmid RSF1010: a genetic analysis of its origin of transfer. Mol. Gen. Genet. 206**:**154-160. [DOI] [PubMed] [Google Scholar]
- 56.Desomer, J., P. Dhaese, and M. van Montagu. 1988. Conjugative transfer of cadmium resistance plasmids in Rhodococcus fascians strains. J. Bacteriol. 170**:**2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178**:**5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doi, K., Y. Ono, E. Yokoyama, Y. Tsukagoe, and S. Ogata. 1998. Whole sequence of _spoIIIE_-like, sporulation-inhibitory, and transfer gene (spi) in a conjugative plasmid, pSA1.1, of Streptomyces azureus and detection of _spi_-like gene in the actinomycete chromosome. Biosci. Biotechnol. Biochem. 62**:**1597-1600. [DOI] [PubMed] [Google Scholar]
- 59.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol: Microbiol. 29**:**1029-1038. [DOI] [PubMed] [Google Scholar]
- 60.Drolet, M., P. Zanga, and P. C. Lau. 1990. The mobilization and origin of transfer regions of a Thiobacillus ferrooxidans plasmid: relatedness to plasmids RSF1010 and pSC101. Mol. Microbiol. 8**:**1381-1391. [DOI] [PubMed] [Google Scholar]
- 61.Dubnau, D., and M. Monod. 1986. The regulation and evolution of MLS resistance. Banbury Rep. 24**:**369-387. [Google Scholar]
- 62.Ducote, M. J., S. Prakash, and G. S. Pettis. 2000. Minimal and contributing sequence determinants of the _cis_-acting locus of transfer (clt) of streptomycete plasmid pIJ101 occur within an intrinsically curved plasmid region. J. Bacteriol. 182**:**6834-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunny, G. M., B. A. B. Leonard, and P. J. Hedberg. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: Interbacterial and host-parasite chemical communication. J. Bacteriol. 177**:**871-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dunny, G. M., and B. A. B. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51**:**527-564. [DOI] [PubMed] [Google Scholar]
- 65.Farías, M. E., and M. Espinosa. 2000. Conjugal transfer of plasmid pMV158: uncoupling of the pMV158 origin of transfer from the mobilization gene mobM, and modulation of pMV158 transfer in Escherichia coli mediated by IncP plasmids. Microbiology 146**:**2259-2265. [DOI] [PubMed] [Google Scholar]
- 66.Farías, M. E., E. Grohmann, and M. Espinosa. 1999. Expression of the mobM gene of the streptococcal plasmid pMV158 in Lactococcus lactis subsp. lactis. subsp. lactis. FEMS Microbiol. Lett. 176**:**403-410. [DOI] [PubMed] [Google Scholar]
- 67.Felsberg, J., M. Petrícek, and P. Tichy. 1993. Nucleotide sequence of the mini-plasmid pSLG33 from Streptomyces lavendulae-grasserius RIA746. Nucleic Acids Res. 21**:**3582.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Firth, N., T. Berg, and R. A. Skurray. 1999. Evolution of conjugative plasmids from Gram-positive bacteria. Mol. Microbiol. 31**:**1598-1600. [PubMed] [Google Scholar]
- 69.Firth, N., P. D. Fink, L. Johnson, and R. A. Skurray. 1994. A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone cAD1. J. Bacteriol. 176**:**5871-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhard, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 71.Firth, N., K. P. Ridgway, M. E. Byrne, P. D. Fink, L. Johnson, I. T. Paulsen, and R. A. Skurray. 1993. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene 136**:**13-25. [DOI] [PubMed] [Google Scholar]
- 72.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn_916_ from Enterococcus faecalis. Plasmid 32**:**350-354. [DOI] [PubMed] [Google Scholar]
- 73.Forbes, B. A., and D. R. Schaberg. 1983. Transfer of resistance plasmids from Staphylococcus epidermidis to Staphylococcus aureus: evidence for conjugative exchange of resistance. J. Bacteriol. 153**:**627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45**:**375-395. [DOI] [PubMed] [Google Scholar]
- 75.Francia, M. V., W. Haas, R. Wirth, E. Samberger, A. Muscholl-Silberhorn, M. S. Gilmore, Y. Ike, K. E. Weaver, F. Y. An, and D. B. Clewell. 2001. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46**:**117-127. [DOI] [PubMed] [Google Scholar]
- 76.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn_916_) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a plasmid. J. Bacteriol. 145**:**494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fürste, J. P., G. Ziegelin, W. Pansegrau, and E. Lanka. 1987. Conjugative transfer of promiscuous plasmid RP4: plasmid-specified functions essential for formation of relaxosomes, p. 553-564. In T. J. Kelly and R. McMacken (ed.), Mechanisms of DNA replication and recombination. Alan R. Liss, Inc., New York, N.Y.
- 78.Fujimoto, S., and D. B. Clewell. 1998. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc. Natl. Acad. Sci. USA 95**:**6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn_1549_, conferring VanB-type resistance in Enterococcus spp. Microbiology 146**:**1481-1489. [DOI] [PubMed] [Google Scholar]
- 80.Gasson, M. J., and F. L. Davies. 1979. Conjugal transfer of lactose genes in group N streptococci. Soc. Gen. Microbiol. Q. 6**:**87. [Google Scholar]
- 81.Gasson, M. J., J.-J. Godon, C. J. Pillidge, T. J. Eaton, K. Jury, and C. A. Shearman. 1995. Characterization and exploitation of conjugation in Lactococcus lactis. Int. Dairy J. 5**:**757-762. [Google Scholar]
- 82.Gasson, M. J., S. Swindell, S. Maeda, and H. M. Dodd. 1992. Molecular rearrangement of lactose plasmid DNA associated with high-frequency transfer and cell-aggregation in Lactococcus lactis 712. Mol. Microbiol. 6**:**3213-3223. [DOI] [PubMed] [Google Scholar]
- 83.Godon, J.-J., K. Jury, C. A. Shearman, and M. J. Gasson. 1994. The Lactococcus lactis sex-factor aggregation gene cluA. Mol. Microbiol. 12**:**655-663. [DOI] [PubMed] [Google Scholar]
- 84.Reference deleted
- 85.Gomis-Rüth, F. X., G. Moncalián, F. de la Cruz, and M. Coll. 2002. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein: detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277**:**7556-7566. [DOI] [PubMed] [Google Scholar]
- 86.Gomis-Rüth, F. X., G. Moncalián, R. Pérez-Luque, A. González, E. Cabezón, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409**:**637-641. [DOI] [PubMed] [Google Scholar]
- 87.González, J. M., Jr., and B. C. Carlton. 1984. A large transmissible plasmid is required for crystal toxin production in Bacillus thuringiensis variety israelenis. Plasmid 11**:**28-38. [DOI] [PubMed] [Google Scholar]
- 88.Gowan, B., and E. R. Dabbs. 1994. Identification of DNA involved in Rhodococcus chromosomal conjugation and self-incompatibility. FEMS Microbiol. Lett. 115**:**45-50. [DOI] [PubMed] [Google Scholar]
- 89.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182**:**1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grohmann, E., L. M. Guzmán, and M. Espinosa. 1999. Mobilisation of the streptococcal plasmid pMV158: interactions of MobM protein with its cognate oriT DNA region. Mol. Gen. Genet. 261**:**707-715. [DOI] [PubMed] [Google Scholar]
- 91.Guzmán, L., and M. Espinosa. 1997. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J. Mol. Biol. 266**:**688-702. [DOI] [PubMed] [Google Scholar]
- 92.Haase, J., R. Lurz, A. M. Grahn, D. H. Bamford, and E. Lanka. 1995. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J. Bacteriol. 177**:**4779-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hagége, J., J.-L. Pernodet, G. Sezonov, C. Gerbaud, A. Friedmann, and M. Guérineau. 1993. Transfer functions of the conjugative integrating element pSAM2 from Streptomyces ambofaciens: characterization of a kil-kor system associated with transfer. J. Bacteriol. 175**:**5529-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1**:**147-155. [DOI] [PubMed] [Google Scholar]
- 95.Hirt, H., P. M. Schlievert, and G. M. Dunny. 2002. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 70**:**716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hopwood, D. A. 1959. Linkage and the mechanism of recombination in Streptomyces coelicolor. Ann. N.Y. Acad. Sci. 81**:**887-898. [DOI] [PubMed] [Google Scholar]
- 97.Hopwood, D. A., T. Kieser, H. M. Wright, and M. J. Bibb. 1983. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J. Gen. Microbiol. 129**:**2257-2269. [DOI] [PubMed] [Google Scholar]
- 98.Hopwood, D. A., and T. Kieser. 1993. Conjugative plasmids of Streptomyces, p. 293-311. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 99.Horii, T., H. Nagasawa, and J. Nakayama. 2002. Functional analysis of TraA, the sex pheromone receptor encoded by pPD1, in a promoter region essential for the mating response in Enterococcus faecalis. J. Bacteriol. 184**:**6343-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ike, Y., K. Tanimoto, H. Tomita, K. Takeuchi, and S. Fujimoto. 1998. Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating. J. Bacteriol. 180**:**4886-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20**:**3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaffe, H. W., H. M. Sewney, R. A. Nathan, R. A. Weinstein, S. A. Kabins, and S. Cohen. 1980. Identity and interspecies transfer of gentamycin resistance plasmids in Staphylococcus aureus and Staphylococcus epidermidis. J. Infect. Dis. 141**:**738-747. [DOI] [PubMed] [Google Scholar]
- 103.Jaworski, D. D., and D. B. Clewell. 1995. A functional origin of transfer (oriT) on the conjugative transposon Tn_916_. J. Bacteriol. 177**:**6644-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jensen, G. B., A. Wilcks, S. S. Petersen, J. Damgaard, J. A. Baum, and L. Andrup. 1995. The genetic basis of the aggregation system in Bacillus thuringiensis subspecies israelenis is located on the large conjugative plasmid pXO16. J. Bacteriol. 177**:**2914-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kanamaru, S., P. G. Leiman, V. A. Kostyuchenko, P. R. Chipman, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415**:**553-557. [DOI] [PubMed] [Google Scholar]
- 106.Kataoka, M., Y. M. Kiyose, Y. Michisuji, T. Horiguchi, T. Seki, and T. Yoshida. 1994. Complete nucleotide sequence of the Streptomyces nigrifaciens plasmid, pSN22: Genetic organization and correlation with genetic properties. Plasmid 32**:**55-69. [DOI] [PubMed] [Google Scholar]
- 107.Kataoka, M., S. Kosono, T. Seki, and T. Yoshida. 1994. Regulation of the transfer genes of Streptomyces plasmid pSN22: In vivo and in vitro study of the interaction of TraR with promoter regions. J. Bacteriol. 176**:**7291-7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kataoka, M., T. Seki, and T. Yoshida. 1991. Five genes involved in self-transmission of pSN22, a Streptomyces plasmid. J. Bacteriol. 173**:**4220-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kempler, G. M., and L. L. McKay. 1979. Genetic evidence for plasmid-linked lactose metabolism in Streptococcus lactis subsp. diacetylactis. Appl. Environ. Microbiol. 37**:**1041-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kendall, K. J., and S. N. Cohen. 1987. Plasmid transfer in Streptomyces lividans: Identification of a kil-kor system associated with the transfer region of pIJ101. J. Bacteriol. 169**:**4177-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kieser, T., D. A. Hopwood, H. M. Wright, and C. J. Thompson. 1982. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol. Gen. Genet. 185**:**223-238. [DOI] [PubMed] [Google Scholar]
- 112.Kieser, H. M., T. Kieser, and D. A. Hopwood. 1992. A combined genetic and physical map of the Streptomyces coelicolor A3(2) chromosome. J. Bacteriol. 174**:**5496-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kinashi, H., M. Shimaji, and A. Sakai. 1987. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. Nature 328**:**454-456. [DOI] [PubMed] [Google Scholar]
- 114.Koonin, E. V., and T. V. Ilyina. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30**:**241-268. [DOI] [PubMed] [Google Scholar]
- 115.Kosono, S., M. Kataoka, T. Seki, and T. Yoshida. 1996. The TraB protein, which mediates the intermycelial transfer of the Streptomyces plasmid pSN22, has functional NTP-binding motifs and is localized to the cytoplasmic membrane. Mol. Microbiol. 19**:**397-405. [DOI] [PubMed] [Google Scholar]
- 116.Krause, S., M. Barcena, W. Pansegrau, R. Lurz, J. Carazo, and E. Lanka. 2000. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. USA 97**:**3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182**:**2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kurenbach, B., D. Grothe, M. E. Farías, U. Szewzyk, and E. Grohmann. 2002. The tra region of the conjugative plasmid pIP501 is organized in an operon with the first gene encoding the relaxase. J. Bacteriol. 184**:**1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118a.Kurenbach, B., C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid, in press. [DOI] [PubMed]
- 119.Lai, E. M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8**:**361-369. [DOI] [PubMed] [Google Scholar]
- 120.Lanka, E., and B. M. Wilkins. 1995. DNA processing reaction in bacterial conjugation. Annu. Rev. Biochem. 64**:**141-169. [DOI] [PubMed] [Google Scholar]
- 121.Le Blanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28**:**130-145. [DOI] [PubMed] [Google Scholar]
- 122.Leonard, B. A. B., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis binding protein, PrgZ recruits a chromosome oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93**:**260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lessl, M., D. Balzer, W. Pansegrau, and E. Lanka. 1992. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J. Biol. Chem. 267**:**20471-20480. [PubMed] [Google Scholar]
- 124.Lessl, M., D. Balzer, K. Weyrauch, and E. Lanka. 1993. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J. Bacteriol. 175**:**6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77**:**321-324. [DOI] [PubMed] [Google Scholar]
- 126.Levengood, S. K., W. F. Beyer, Jr., and R. E. Webster. 1991. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 88**:**5939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Llosa, M., J. Zupan, C. Baron, and P. Zambryski. 2000. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 182**:**3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luna, V. A., P. Coates, E. A. Eady, J. H. Cove, T. T. H. Nguyen, and M. C. Roberts. 2002. A variety of Gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44**:**19-25. [DOI] [PubMed] [Google Scholar]
- 129.Lydiate, D. J., F. Malpartida, and D. A. Hopwood. 1985. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene 35**:**223-235. [DOI] [PubMed] [Google Scholar]
- 130.Maas, R.-M., J. Götz, W. Wohlleben, and G. Muth. 1998. The conjugative plasmid pSG5 from Streptomyces ghanaensis DSM 2932 differs in its transfer functions from other Streptomyces rolling-circle-type plasmids. Microbiology 144**:**2809-2817. [DOI] [PubMed] [Google Scholar]
- 131.Machón, C., S. Rivas, A. Albert, F. M. Goni, and F. de la Cruz. 2002. TrwD, the hexameric traffic ATPase encoded by plasmid R388, induces membrane destabilization and hemifusion of lipid vesicles. J. Bacteriol. 184**:**1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Macrina, F. L., and G. L. Archer. 1993. Conjugation and broad host range plasmids in streptococci and staphylococci, p. 319-339. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 133.Martin, C., P. Mazodier, M. V. Mediola, B. Gicquel, T. Smokvina, C. J. Thompson, and J. Davies. 1991. Site specific integration of the Streptomyces plasmid pSAM2 in Mycobacterium smegmatis. Mol. Microbiol. 5**:**2499-2502. [DOI] [PubMed] [Google Scholar]
- 134.Maurizi, M. R., and C. C. Li. 2001. AAA proteins: in search of a common molecular basis. International Meeting on Cellular Functions of AAA Proteins. EMBO Rep. 2**:**980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mayford, M., and B. Weisblum. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J. 8**:**4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McDonnell, R. W., H. M. Sweeney, and S. Cohen. 1983. Conjugational transfer of gentamycin resistance plasmids intra- and interspecifically in Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 23**:**151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Merlin, C., J. Mahillon, J. Nesvera, and A. Toussaint. 2000. Gene recruiters and transporters: the modular structure of bacterial mobile elements, p. 363-409. In C. M. Thomas (ed.), The horizontal gene pool. Bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 138.Meyer, R. 2000. Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J. Bacteriol. 182**:**4875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mills, D. A., C. K. Choi, G. M. Dunny, and L. L. McKay. 1994. Genetic analysis of regions of the Lactococcus lactis subsp. lactis plasmid pRS01 involved in conjugative transfer. Appl. Environ. Microbiol. 60**:**4413-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178**:**3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mills, D. A., T. G. Phister, G. M. Dunny, and L. L. McKay. 1998. An origin of transfer (oriT) on the conjugative element pRS01 from Lactococcus lactis subsp. lactis ML3. Appl. Environ. Microbiol. 64**:**1541-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mori, M., Y. Sakagami, Y. Ishii, A. Isogai, C. Kitada, M. Fujino, J. C. Adsit, G. M. Dunny, and A. Suzuki. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263**:**14574-14578. [PubMed] [Google Scholar]
- 143.Morton, T. M., D. M. Eaton, J. L. Johnston, and G. L. Archer. 1993. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 175**:**4436-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mushegian, A. R., K. J. Fullner, E. V. Koonin, and E. W. Nester. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. USA 93**:**7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muth, G., and M. Elfeituri. 2001. Molecular characterization of the Streptomyces venezuelae plasmid pSVH1. Plasmid 45**:**152-153. [Google Scholar]
- 146.Muth, G., M. Farr, V. Hartmann, and W. Wohlleben. 1995. Streptomyces ghanaensis plasmid pSG5: nucleotide sequence analysis of the self-transmissible minimal replicon and characterization of the replication mode. Plasmid 33**:**113-126. [DOI] [PubMed] [Google Scholar]
- 147.Muth, G., W. Wohlleben, and A. Pühler. 1988. The minimal replicon of the Streptomyces ghanaensis plasmid pSG5 identified by subcloning and Tn_5_ mutagenesis. Mol. Gen. Genet. 211**:**424-429. [DOI] [PubMed] [Google Scholar]
- 148.Nieto, C., and M. Espinosa. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid, in press. [DOI] [PubMed]
- 149.Noble, W. C., Z. Virani, and R. G. A. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 93**:**195-198. [DOI] [PubMed] [Google Scholar]
- 150.Noirot-Gross, M. F., V. Bidnenko, and S. D. Ehrlich. 1994. Active site of the replication protein in the rolling circle plasmid pC194. EMBO J. 13**:**4412-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Novick, R. P., S. J. Projan, W. Rosenblum, and I. Edelman. 1984. Staphylococcal plasmid cointegrates are formed by host- and phage-mediated general rec systems that act on short regions of homology. Mol. Gen. Genet. 195**:**374-377. [DOI] [PubMed] [Google Scholar]
- 152.Núnez, B., and F. de la Cruz. 2001. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol. Microbiol. 39**:**1088-1099. [DOI] [PubMed] [Google Scholar]
- 153.Odelson, D. A., J. L. Rasmussen, C. J. Smith, and F. L. Macrina. 1987. Extrachromosomal systems and gene transmission in anaerobic bacteria. Plasmid 17**:**87-109. [DOI] [PubMed] [Google Scholar]
- 154.Okinaka, R., K. Cloud, O. Hampton, A. Hoffmaster, K. Hill, P. Keim, T. Koehler, G. Lamke, S. Kumano, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence, assembly and analysis of pXO1 and pXO2. J. Appl. Microbiol. 87**:**261-262. [DOI] [PubMed] [Google Scholar]
- 155.Okinaka, R., K. Cloud, O. Hampton, A. Hoffmaster, K. Hill, P. Keim, T. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181**:**6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Omer, C. A., and S. N. Cohen. 1984. Plasmid formation in Streptomyces: excision and integration of the SLP1 replicon at a specific chromosomal site. Mol. Gen. Genet. 196**:**429-438. [DOI] [PubMed] [Google Scholar]
- 157.Omer, C. A., and S. N. Cohen. 1986. Structural analysis of plasmid and chromosomal loci involved in site-specific excision and integration of the SLP1 element of Streptomyces coelicolor. J. Bacteriol. 166**:**999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.O'Riordan, K., and G. F. Fitzgerald. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174**:**285-294. [DOI] [PubMed] [Google Scholar]
- 159.Oskam, L., D. J. Hillenga, G. Venema, and S. Bron. 1991. The large Bacillus plasmid pTB19 contains two integrated rolling-circle plasmids carrying mobilization functions. Plasmid 26**:**30-39. [DOI] [PubMed] [Google Scholar]
- 160.Pansegrau, W., and E. Lanka. 1991. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 19**:**3455-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54**:**197-251. [DOI] [PubMed] [Google Scholar]
- 162.Pansegrau, W., and E. Lanka. 1996. Mechanisms of initiation and termination reactions in conjugative DNA processing: independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPα plasmid RP4. J. Biol. Chem. 271**:**13068-13076. [DOI] [PubMed] [Google Scholar]
- 163.Pansegrau, W., W. Schröder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269**:**2782-2789. [PubMed] [Google Scholar]
- 164.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1999. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology 145**:**585-592. [DOI] [PubMed] [Google Scholar]
- 165.Parsons, L. M., C. S. Jankowski, and K. M. Derbyshire. 1998. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol. Microbiol. 28**:**571-582. [DOI] [PubMed] [Google Scholar]
- 166.Pernodet, J. L., J. M. Simonet, and M. Guerineau. 1984. Plasmids in different strains of Streptomyces ambofaciens: free and integrated form of plasmid pSAM2. Mol. Gen. Genet. 198**:**35-41. [DOI] [PubMed] [Google Scholar]
- 167.Pettis, G. S., and S. N. Cohen. 1994. Transfer of the pIJ101 plasmid in Streptomyces lividans requires a _cis_-acting function dispensable for chromosomal gene transfer. Mol. Microbiol. 13**:**955-964. [DOI] [PubMed] [Google Scholar]
- 168.Pettis, G. S., and S. N. Cohen. 1996. Plasmid transfer and expression of the transfer (tra) gene product of plasmid pIJ101 are temporally regulated during the Streptomyces lividans life cycle. Mol. Microbiol. 19**:**1127-1135. [DOI] [PubMed] [Google Scholar]
- 169.Pettis, G. S., and S. N. Cohen. 2000. Mutational analysis of the tra locus of the broad-host-range Streptomyces plasmid pIJ101. J. Bacteriol. 182**:**4500-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pettis, G. S., N. Ward, and K. L. Schully. 2001. Expression characteristics of the transfer-related kilB gene product of Streptomyces plasmid pIJ101: implications for the plasmid spread function. J. Bacteriol. 183**:**1339-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Polzin, K. M., and M. Shimizu-Kadota. 1987. Identification of a new insertion element, similar to gram-negative IS_26_, on the lactose plasmid of Streptococcus lactis ML3. J. Bacteriol. 169**:**5481-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Possoz, C., C. Ribard, J. Gagnat, J.-L. Pernodet, and M. Guérineau. 2001. The integrative element of pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 42**:**159-166. [DOI] [PubMed] [Google Scholar]
- 173.Priebe, S., and S. A. Lacks. 1989. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J. Bacteriol. 171**:**4778-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Projan, S. J., and G. L. Archer. 1989. Mobilization of the Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J. Bacteriol. 171**:**1841-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Projan, S. J., and R. P. Novick. 1988. Comparative analysis of five related staphylococcal plasmids. Plasmid 19**:**203-221. [DOI] [PubMed] [Google Scholar]
- 176.Reniero, R., P. Cocconcelli, V. Bottazzi, and L. Morelli. 1992. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138**:**763-768. [Google Scholar]
- 177.Rice, L. B. 1998. Tn_916_ family of conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42**:**1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sakai, D., and T. Komano. 2002. Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J. Bacteriol. 184**:**444-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Salmond, G. P. C. 1994. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32**:**181-200. [Google Scholar]
- 180.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L.-Y. Li. 1995. Conjugative transposons: an unusal and diverse set of integrated gene transfer elements. Microbiol. Rev. 59**:**579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Samuels, A. L., E. Lanka, and J. E. Davies. 2000. Conjugative junctions in RP4-mediated mating of Escherichia coli. J. Bacteriol. 182**:**2709-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schaberg, D. R., and M. J. Zervos. 1986. Intergeneric and interspecies gene exchange in gram-positive cocci. Antimicrob. Agents Chemother. 30**:**817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Scherzinger, E., R. Lurz, S. Otto, and B. Dobrinski. 1992. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 20**:**41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schrempf, H., and W. Goebel. 1977. Characterization of a plasmid from Streptomyces coelicolor A3(2). J. Bacteriol. 131**:**251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H.-J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184**:**2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46**:**170-187. [DOI] [PubMed] [Google Scholar]
- 187.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn_916_: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11**:**1099-1108. [DOI] [PubMed] [Google Scholar]
- 188.Scott, J. R., and G. G. Churchward. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49**:**367-397. [DOI] [PubMed] [Google Scholar]
- 189.Senghas, E., J. M. Jones, M. Yamamoto, C. Gawron-Burke, and D. B. Clewell. 1988. Genetic organization of the bacterial conjugative transposon Tn_916_. J. Bacteriol. 170**:**245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sermonti, G., and I. Spada-Sermonti. 1955. Genetic recombination in Streptomyces. Nature 176**:**121.. [DOI] [PubMed] [Google Scholar]
- 191.Servín-González, L., A. Sampieri, J. Cabello, L. Galván, V. Juárez, and C. Castro. 1995. Sequence and functional analysis of the Streptomyces phaeochromogenes plasmid pJV1 reveals a modular organization of Streptomyces plasmids that replicate by rolling circle. Microbiology 141**:**2499-2510. [DOI] [PubMed] [Google Scholar]
- 192.Servin-González, L. 1996. Identification and properties of a novel clt locus in the Streptomyces phaeochromogenes plasmid pJV1. J. Bacteriol. 178**:**4323-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3**:**178-185. [DOI] [PubMed] [Google Scholar]
- 194.Sezonov, G., J. Hagège, J. L. Pernodet, A. Friedmann, and M. Guérineau. 1995. Characterization of pra, a gene for replication control in pSAM2, the integrating element of Streptomyces ambofaciens. Mol. Microbiol. 17**:**533-544. [DOI] [PubMed] [Google Scholar]
- 195.Sezonov, G., C. Possoz, A. Friedmann, J. L. Pernodet, and M. Guerineau. 2000. KorSA from the Streptomyces integrative element pSAM2 is a central transcriptional repressor: target genes and binding sites. J. Bacteriol. 182**:**1243-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sharma, V. K., J. L. Johnston, T. M. Morton, and G. L. Archer. 1994. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pGO1. J. Bacteriol. 176**:**3445-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96**:**14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295**:**137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shiffman, D., and S. N. Cohen. 1993. Role of the imp operon of the Streptomyces coelicolor genetic element SLP1: Two _imp_-encoded proteins interact to autoregulate imp expression and control plasmid maintenance. J. Bacteriol. 175**:**6767-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Showsh, S. A., E. H. De Boever, and D. B. Clewell. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob. Agents Chemother. 45**:**2177-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sievert, D. M., M. L. Boulton, G. Stoltman, D. Johnson, M. G. Stobierski, F. P. Downes, P. A. Somsel, J. T. Rudrik, W. Brown, W. Hafeez, T. Lundstrom, E. Flanagan, R. Johnson, J Mitchell, and S. Chang. 2002. Staphylococcus aureus resistant to vancomycin-United States, 2002. Morb. Mortal. Wkly. Rep. 51**:**565-567. [PubMed] [Google Scholar]
- 202.Smith, C. J., and A. C. Parker. 1998. The transfer origin for Bacteroides mobilizable transposon Tn_4555_ is related to a plasmid family from gram-positive bacteria. J. Bacteriol. 180**:**435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Smith, M. C., R. N. Burns, S. E. Wilson, and M. A. Gregory. 1999. The complete genome sequence of the Streptomyces temperate phage straight phiC31: evolutionary relationships to other viruses. Nucleic Acids Res. 27**:**2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Stein, D. S., K. J. Kendall, and S. N. Cohen. 1989. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J. Bacteriol. 171**:**5768-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Szpirer, C. Y., M. Faelen, and M. Couturier. 2001. Mobilization functions of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 183**:**2101-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tai, J. T. N., and S. N. Cohen. 1993. The active form of the KorB protein encoded by the Streptomyces plasmid pIJ101 is a processed product that binds differentially to the two promoters it regulates. J. Bacteriol. 175**:**6996-7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tai, J. T. N., and S. N. Cohen. 1994. Mutations that affect regulation of the korB gene of Streptomyces lividans plasmid pIJ101 alter plasmid transmission. Mol. Microbiol. 12**:**31-39. [DOI] [PubMed] [Google Scholar]
- 208.Townsend, D. E., N. Ashdown, D. I. Annear, and W. B. Grubb. 1985. A conjugative plasmid encoding production of a diffusible pigment and resistance to aminoglycosides and macrolides in Staphylococcus aureus. Aust. J. Exp. Biol. Med. 63**:**573-586. [DOI] [PubMed] [Google Scholar]
- 209.van der Lelie, D., S. Bron, G. Venema, and L. Oskam. 1989. Similarity of minus origins of replication and flanking open reading frames of plasmids pUB110, pTB913 and pMV158. Nucleic Acids Res. 17**:**7283-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.van der Lelie, D., F. Chavarri, G. Venema, and M. J. Gasson. 1991. Identification of a new genetic determinant for cell aggregation associated with lactose plasmid transfer in Lactococcus lactis. Appl. Environ. Microbiol. 57**:**201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vivian, A. 1971. Genetic control of fertility in Streptomyces coelicolor A3(2): plasmid involvement in the interconversion of UF and IF strains. J. Gen. Microbiol. 69**:**353-364. [DOI] [PubMed] [Google Scholar]
- 212.Vrijbloed, J. W., M. Jelínková, G. I. Hessels, and L. Dijkhuizen. 1995. Identification of the minimal replicon of plasmid pMEA300 of the methylotrophic actinomycete Amycolatopsis methanolica. Mol. Microbiol. 18**:**21-31. [DOI] [PubMed] [Google Scholar]
- 213.Vrijbloed, J. W., J. Madon, and L. Dijkhuizen. 1994. A plasmid from the methylotrophic actinomycete Amycolatopsis methanolica capable of site-specific integration. J. Bacteriol. 176**:**7087-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vrijbloed, J. W., N. M. J. Van der Put, and L. Dijkhuizen. 1995. Identification and functional analysis of the transfer region of plasmid pMEA300 of the methylotrophic actinomycete Amycolatopsis methanolica. J. Bacteriol. 177**:**6499-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang, A., and F. L. Macrina. 1995. Characterization of six linked open reading frames necessary for pIP501-mediated conjugation. Plasmid 34**:**206-210. [DOI] [PubMed] [Google Scholar]
- 216.Waters, V. L. 1999. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 4**:**416-439. [DOI] [PubMed] [Google Scholar]
- 217.Waters, C. M., and G. M. Dunny. 2001. Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance. J. Bacteriol. 183**:**5659-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Waters, V. L., and D. G. Guiney. 1993. Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol. Microbiol. 9**:**1123-1130. [DOI] [PubMed] [Google Scholar]
- 219.Weisblum, B. 1985. Inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression. J. Antimicrob. Chemother. 16**:**63-90. [DOI] [PubMed] [Google Scholar]
- 220.Wohlleben, W., and G. Muth. 1993. Streptomyces plasmid vectors, p. 147-175. In K. G. Hardy (ed.), Plasmids: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 221.Wu, L. J., P. J. Lewis, R. Allmannsberger, P. M. Hauser, and J. Errington. 1995. A conjugation-like mechanism for prespore chromosome-partitioning during sporulation in Bacillus subtilis. Genes. Dev. 9**:**1316-1326. [DOI] [PubMed] [Google Scholar]
- 222.Yang, C.-C., C.-H. Huang, C.-Y. Li, Y.-G. Tsay, S.-C. Lee, and C. W. Chen. 2002. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol. Microbiol. 43**:**297-305. [PubMed] [Google Scholar]
- 223.Yeo, H.-J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6**:**1461-1472. [DOI] [PubMed] [Google Scholar]
- 224.Zaman, S., H. Richards, and J. Ward. 1992. Expression and characterisation of the korB gene product from the Streptomyces lividans plasmid pIJ101 in Escherichia coli and determination of its binding site on the korB and kilB promoters. Nucleic Acids Res. 20**:**3693-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zatyka, M., and C. M. Thomas. 1998. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev. 21**:**291-319. [DOI] [PubMed] [Google Scholar]
- 226.Zechner, E. L., F, de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 227.Zhou, L., D. A. Manias, and G. M. Dunny. 2000. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 37**:**639-651. [DOI] [PubMed] [Google Scholar]
- 228.Zupan, J., and P. Zambryski. 1997. The Agrobacterium DNA transfer complex. Crit. Rev. Plant Sci. 16**:**279-295. [Google Scholar]