No Accelerated Rate of Protein Evolution in Male-Biased Drosophila pseudoobscura Genes (original) (raw)
Abstract
Sexually dimorphic traits are often subject to diversifying selection. Genes with a male-biased gene expression also are probably affected by sexual selection and have a high rate of protein evolution. We used SAGE to measure sex-biased gene expression in Drosophila pseudoobscura. Consistent with previous results from D. melanogaster, a larger number of genes were male biased (402 genes) than female biased (138 genes). About 34% of the genes changed the sex-related expression pattern between D. melanogaster and D. pseudoobscura. Combining gene expression with protein divergence between both species, we observed a striking difference in the rate of evolution for genes with a male-biased gene expression in one species only. Contrary to expectations, D. pseudoobscura genes in this category showed no accelerated rate of protein evolution, while D. melanogaster genes did. If sexual selection is driving molecular evolution of male-biased genes, our data imply a radically different selection regime in D. pseudoobscura.
FEMALES and males often exhibit substantial differences in behavior, physiology, and morphology. Although only a small number of genes contribute to the first steps of sexual differentiation, a significant proportion of the genome is involved in the establishment and maintenance of sexual dimorphism (Hughes 2001). It is well established that such sexually dimorphic traits are often subject to directional selection (Luck and Joly 2005; Snook et al. 2005). While initial studies focused mainly on morphology, behavior, and physiology (Hoy 1990; Dickens et al. 1998; Holland and Rice 1999), more recently attention has turned to the pattern of molecular evolution (Meiklejohn et al. 2003; Ranz et al. 2003). Several proteins involved in male reproductive function are shown to exhibit an accelerated rate of molecular evolution, presumably due to positive selection driving amino acid replacements (Coulthart and Singh 1988; Tsaur and Wu 1997; Begun and Lindfors 2005; Wagstaff and Begun 2005). However, some of these genes may also evolve rapidly due to reasons other than positive selection (Rooney et al. 2000; Torgerson and Singh 2004; Civetta et al. 2006). Interestingly, recent studies in Drosophila and mice showed that this pattern could be generalized to genes that are more highly expressed in the male than in the female (male-biased genes) (Swanson et al. 2001; Zhang et al. 2004; Good and Nachman 2005). In mice, however, the accelerated rate of evolution seems to be restricted to genes with a male-biased gene expression during late spermatogenesis (Good and Nachman 2005).
An important assumption that has been implicitly made for all studies determining the rate of molecular evolution is the maintenance of function/gene expression across species. Interestingly, a comparative analysis of gene expression in Drosophila melanogaster and D. simulans, which diverged <3 million years ago, indicated that for a significant number of genes (∼20%) the sex bias in gene expression has been lost or even reversed between these two species (Ranz et al. 2003). Thus, the interpretation of the observed high rate of sequence evolution of genes with a male-biased gene expression in one species is less clear. It was proposed that the accelerated rate of protein evolution of male-biased genes is caused by sexual selection driven by male–male competition, male–female interaction, or both (Swanson and Vacquier 2002). Nevertheless, to rule out other evolutionary forces, such as a rapid rate of protein evolution associated with a shift in function/gene expression, it is important to determine rates of sequence evolution in combination with patterns of gene expression in more than one species.
We performed an analysis of sex-biased gene expression in D. pseudoobscura, the second Drosophila species for which a completely sequenced genome is available (Richards et al. 2005). Combining sequence and expression data, we provide evidence that the accelerated rate of protein evolution is restricted to D. melanogaster, while D. pseudoobscura male-biased genes evolve at lower rates, comparable to nonbiased genes.
MATERIALS AND METHODS
Serial analysis of gene expression library:
Total RNA was extracted with Trizol from 220-mg males and 370-mg females of D. pseudoobscura (obtained from the M. Akam Lab). Only virgin individuals from both sexes between 3 and 7 days old were used. The serial analysis of gene expression (SAGE) library construction followed published protocols (Velculescu et al. 1995) at the EMBL core facility. In brief, the mRNA was isolated using a biotin-labeled T_n_ oligo and Dynal (Great Neck, NY) beads. Double-stranded cDNA was generated with Superscript II reverse transcriptase and DNA polymerase I. The resulting cDNA library was digested with _Nla_III (anchoring enzyme). 3′-Restriction fragments were recovered with Dynal beads and split into two populations. Each aliquot was ligated to one of the two annealed linker pairs. After extensive washing tags were created by digestion with _Bsm_FI (tagging enzyme). Tags were blunted with T4 polymerase and ligated to generate ditags. Ditags were PCR amplified and digested with _Nla_III. Restriction fragments were separated on a 12% polyacrylamide gel and the band containing the ditags was isolated from the gel. The purified inserts were concatemerized, ligated into pZero, and transformed into EDH5α electromax cells. Similar to other SAGE experiments in Drosophila (Jasper et al. 2001, 2002), our design was aiming for ∼20,000 tags/library.
D. melanogaster/D. simulans expression data:
The D. melanogaster genes in our analyses were classified as male, female, or unbiased on the basis of gene expression data for adult males and females reported in Parisi et al. (2003). We selected this data set, as it represents the largest set of genes and the adult flies covered an age range similar to our study (5–7 days). Furthermore, the expression data from Ranz et al. (2003) and Arbeitman et al. (2002) were underrepresented for male-biased genes. Expression intensity and sex bias were determined by averaging replicate experiments. D. simulans expression intensity and sex bias used in this study were obtained from Ranz et al. (2003) as provided in the online supplement (http://www.sciencemag.org/cgi/content/full/300/5626/1742/DC1). If not otherwise noted, we used a twofold expression difference between males and females to define a sex-biased gene expression. To classify genes according to expression level (i.e., to identify highly expressed genes) in D. melanogaster, we considered the absolute intensity values (after background subtraction) in the two channels from microarray experiments of Parisi et al. (2003). As multiple experiments in adult flies were performed and some genes were represented more than once on each array, we averaged the absolute intensity values of each gene across experiments and duplicate spots. On the basis of this average we classified genes according to their expression level. We note that the intensity on a two-color array is not an optimal estimator of expression level (Townsend 2003); nevertheless, the misestimates introduced may result only in greater noise. Hence, we do not expect that our procedure produced a strong bias in our results.
Data analysis:
SAGE tag extraction:
Vector sequence was removed using an in-house perl script. Duplicated clones were identified and removed with the xmatchdt.pl software (Dinel et al. 2005). Fourteen base-pair SAGE tags (10 bp plus CATG, the _Nla_III recognition sequence) were extracted with the SAGEparser software (Dinel et al. 2005). This software also screens for overrepresented ditags and corrects for this PCR artifact. Data were submitted to Gene Expression Omnibus (GEO) database (GSM60839, GSM60840) of NCBI.
Comparison of male and female SAGE libraries:
Sequencing errors could inflate the number of unique tags. Therefore, we used only tags that were identified more than once in the full data set containing male and female libraries. This strategy should not make a difference in the results, as unique tags would not be considered a significant difference between both libraries. The tags from the male and female libraries were compared using a two-tailed Fisher's exact test as implemented by SAGE analysis tools (http://www.mbgproject.org/mbgp_tools.html). Consistent with previous reports (Ruijter et al. 2002), we obtained almost identical results when we used the Audic-Claverie statistics (Audic and Claverie 1997) as implemented in Discoveryspace 3.2.3 (http://www.bcgsc.ca/bioinfo/software/discoveryspace/); thus we report only results based on Fisher's exact test. We expressed the sex bias by the log2 ratio of male-to-female tags. In cases for which tags were detected in one sex only, we set the tag count to one for the other sex to avoid problems in calculating the log2 ratio.
Tag-to-gene mapping:
We used the complete transcriptome from the D. pseudoobscura annotation 1.0 to build a database consisting of the sequence between the 3′-end of the transcript and the most 3′ _Nla_III recognition sequence. Transcripts without the _Nla_III recognition site were not considered. This database was used to match the SAGE tags. Only perfect and unique matches were considered.
Gene conservation:
We used a precompiled list of D. melanogaster genes that are conserved in D. pseudoobscura, which is available from FlyBase upon request, to link sex-biased gene expression with gene conservation.
Functional enrichment of SAGE libraries:
To test whether male/female-biased tags in our SAGE library are enriched for genes with sex-specific function, we used GeneMerge software, which relies on gene ontology (GO) terms for a functional classification of genes (Castillo-Davis and Hartl 2003).
Substitution rates, codon usage, and Grantham's distance:
D. melanogaster transcripts from release 4.0 and D. pseudoobscura transcripts from release 1.0 were aligned using the gene prediction software GeneWise (Birney and Durbin 2000) and protein sequences from D. melanogaster (release 4.0). We used an in-house perl script to parse the GeneWise output and to generate aligned fasta files. The rates of nonsynonymous substitutions (_d_N) were calculated from pairwise alignments of D. melanogaster_–_D. pseudoobscura genes using the Yang and Nielsen method (Yang and Nielsen 2000) as implemented in the program yn00 in PAML 3.14 (Yang 1997). We used _d_N rather than the ratio of nonsynonymous-to-synonymous substitutions (_d_N/_d_S), as synonymous substitutions are saturated due to the high divergence of the two species (Richards et al. 2005). Given that the time of divergence for all genes is the same (within the limitations of lineage sorting) and that Drosophila shows no male-biased mutation rates (Bauer and Aquadro 1997), nonsynonymous substitution rates are sufficient to compare rates of protein evolution (Cusack and Wolfe 2005). We note, however, that genes with a male bias in D. melanogaster show a trend toward a slightly higher _d_S than other genes (Zhang et al. 2004; Jagadeeshan and Singh 2005). The magnitude of the effect in our analysis is, however, too large to explained by a variation in _d_S. Furthermore, the elevated _d_S may be a result of recurrent selective sweeps (Betancourt and Presgraves 2002); hence, the _d_N/_d_S ratio may be overly conservative. The level of codon usage bias was measured as the frequency of optimal codons (_F_op) (Ikemura 1981) and was calculated using the program codonW (Peden 1999). Rather than comparing the codon usage for each of the species separately, we focused our analysis on the difference in codon usage, as this conveniently combines the data from both species into a single statistic. Grantham distance was used as a measure to calculate the amino acid similarity and was calculated by using in-house perl scripts, implementing the Grantham matrix (Grantham 1974).
RESULTS
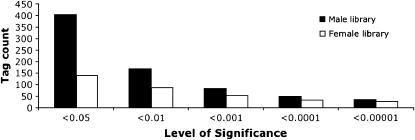
We generated two SAGE libraries for _D. pseudoobscura_—one for males and one for females. Each library contained at least 23,400 tags. Conservatively, we restricted our analyses to tags that were identified more than once in the two libraries, resulting in 3618 and 3481 unique tags (genes) for the male and female libraries, respectively. The total number of unique tags from both libraries was 4450, which corresponds to 42.3% of the annotated genes in D. pseudoobscura. Consistent with the gene expression pattern in D. melanogaster, we detected more male-biased genes (402) than female-biased genes (138) and this difference was robust to changes in the significance thresholds (Figures 1 and 2). The variance in gene expression among female-biased genes was significantly higher than in male-biased genes (P < 0.001, Levene's test). This contrasts with D. melanogaster, for which a higher variance was observed for male-biased genes expressed in adults (Parisi et al. 2003).
Figure 1.—
Number of male- and female-biased tags based on different levels of significance (Fisher's exact test).
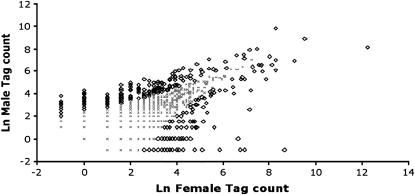
Figure 2.—
Gene expression of D. pseudoobscura males and females measured by SAGE tag counts. The open diamonds indicate SAGE tags that differ significantly (P ≤ 0.05, Fisher's exact test) between both sexes. The diamonds above the diagonal are male biased and below the diagonal are female biased. Shaded crosses indicate unbiased tags. A value of −1 was assigned to tags with no counts for one of the sexes.
We used the release 1.0 of the D. pseudoobscura genome annotation (Richards et al. 2005) to map the SAGE tags of both libraries to the corresponding genes (see materials and methods for more details). Consistent with previous SAGE experiments (Pleasance et al. 2003; Lee et al. 2005), a significant proportion of the SAGE tags (e.g., 45–66% in D. melanogaster; Jasper et al. 2001, 2002; Lee et al. 2005) could not be unambiguously mapped to a gene. On average, we mapped 1183 (27%) of the SAGE tags to a single D. pseudoobscura gene. Interestingly, the proportion of genes that could be mapped differed among male-biased, female-biased, and unbiased genes. Only 19 (14%) of the female-biased tags could be mapped, while 32 and 27% were mapped for male-biased and unbiased tags, respectively. The difference between male- and female-biased tags that could be mapped is highly significant (P < 0.001, Fisher's exact test).
After mapping the SAGE tags to D. pseudoobscura genes, we identified genes for which the available information was consistent with a sex-biased expression pattern (supplemental Table 1 at http://www.genetics.org/supplemental/). The female SAGE library contained several genes with a typical female-specific function. We used GeneMerge for a formal test of significant overrepresentation and found the GO terms oogenesis, insect chorion formation, vitelline membrane formation, and protein biosynthesis to be overrepresented (e < 0.05). The male library contained several ubiquitin- and microtubule-associated genes. Nevertheless, GeneMerge did not find a significant overrepresentation of these classes.
We further corroborated our tag to gene mapping using the method called _g_eneration of _l_onger cDNA fragments from serial analysis of gene expression tags for _g_ene _i_dentification (GLGI) to generate longer cDNA fragments on the basis of the poly(A) tail and the tag sequence (Chen et al. 2000). Of 21 tags haphazardly selected from different classes of sex bias, 20 identified the correct gene. One tag resulted in a cDNA sequence that could not be identified in the entire D. pseudoobscura genome. Hence, we estimate the fraction of inaccurately mapped tags to fall into the interval of 0.001 to 0.237 (95% confidence interval, binomial distribution). Overall, these results indicate a reasonable accuracy of our tag to gene mapping.
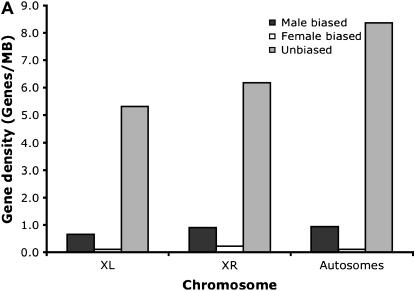
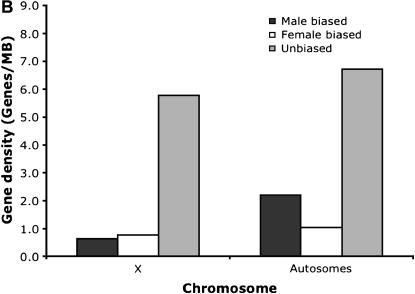
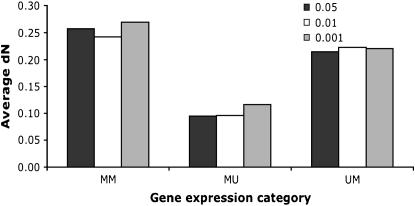
Contrary to the case in D. melanogaster, the ratio of male-biased genes to unbiased genes did not differ between X chromosomes and autosomes (P = 0.393, Fisher's exact test) in D. pseudoobscura. Furthermore, no pronounced difference could be detected between chromosomes XL and XR (which corresponds to 3L in D. melanogaster; see Figure 3, A and B). In total, 1102 genes with expression data for males and females in both species, D. melanogaster and D. pseudoobscura, were available. About 34% of the genes changed the sex bias of the expression pattern between both species. Eight classes of gene expression could be distinguished (Table 1). We measured the rate of evolution by the proportion of nonsynonymous substitutions. Consistent with an accelerated rate of evolution of male-biased genes, we found the highest rate of nonsynonymous substitutions for genes with a male-biased gene expression in both species. Genes with a male bias in D. melanogaster and no sex bias in D. pseudoobscura also showed a high rate of sequence evolution. The complementary group of genes with male-biased gene expression in D. pseudoobscura and no sex bias in D. melanogaster, however, did not have a high rate of nonsynonymous changes. The difference between these two groups was statistically significant (P < 0.001, Mann–Whitney _U_-test). Overall, female-biased genes showed a trend toward a lower rate of protein evolution, but the sample sizes were too small to provide robust estimates. Interestingly, a similar pattern emerged when we used the _d_N/_d_S ratio rather than _d_N. The difference between D. melanogaster and D. pseudoobscura for genes with a male expression pattern only in one species was also highly significant on the basis of the _d_N/_d_S ratio (P < 0.001, Mann–Whitney _U_-test). We also note that the differences in the rate of evolution remain when more stringent criteria were used to define sex-biased genes (Figure 4).
Figure 3.—
Density of male-biased, female-biased, and unbiased genes on (A) chromosomes XL and XR and the autosomes in D. pseudoobscura and on (B) the X chromosome and autosomes in D. melanogaster.
TABLE 1.
Rate of evolution, Grantham distance, and codon usage for genes in different categories of sex-biased expression pattern
| Expression bias | No. of genes | Grantham distance | _F_op | |||||
|---|---|---|---|---|---|---|---|---|
| D. pseudoobscura | D. melanogaster | _d_N | _d_S | _d_N/_d_S | D. pseudoobscura | D. melanogaster | ||
| Male biased | Male biased | 34 | 0.257 | 2.795 | 0.109 | 67.990 | 0.601 | 0.504 |
| Male biased | Unbiased | 77 | 0.095 | 2.061 | 0.049 | 61.880 | 0.592 | 0.582 |
| Male biased | Female biased | 6 | 0.059 | 2.425 | 0.026 | 52.860 | 0.712 | 0.693 |
| Unbiased | Male biased | 187 | 0.216 | 2.698 | 0.087 | 64.803 | 0.594 | 0.536 |
| Unbiased | Unbiased | 681 | 0.106 | 2.295 | 0.054 | 61.091 | 0.560 | 0.568 |
| Unbiased | Female biased | 99 | 0.078 | 1.910 | 0.043 | 56.041 | 0.599 | 0.635 |
| Female biased | Unbiased | 7 | 0.041 | 1.383 | 0.027 | 46.572 | 0.608 | 0.596 |
| Female biased | Female biased | 11 | 0.096 | 1.517 | 0.056 | 61.140 | 0.704 | 0.699 |
Figure 4.—
Average _d_N values between D. melanogaster and D. pseudoobscura at different significance level cutoffs. MM, genes with male-biased expression in both species; MU, genes with male-biased expression in D. pseudoobscura and unbiased expression in D.melanogaster; UM, genes with unbiased expression in D. pseudoobscura and male-biased expression in D. melanogaster.
We further substantiated the different evolutionary behavior of genes with a male-biased expression pattern in D. melanogaster by calculating Grantham's distance (Grantham 1974) between D. melanogaster and D. pseudoobscura genes (Table 1). Grantham's distance considers the carbon composition, polarity, and volume of amino acids to determine their exchangeability. Thus, a higher Grantham distance indicates more radical amino acid changes. Our results of the Grantham distances nicely parallel the pattern of nonsynonymous substitutions. The highest Grantham's distances were found for genes with a male-biased gene expression in both species. Again, genes with a male-biased gene expression in D. pseudoobscura and no sex bias in D. melanogaster had a significantly lower Grantham distance (P = 0.019, Mann–Whitney _U_-test) than the complementary group of genes with no sex bias in D. pseudoobscura and male bias in D. melanogaster.
We also determined the influence of gene expression on codon usage by comparing the frequency of the optimal codon (_F_op) between the two species. Consistent with the previously reported similarity in codon usage between D. melanogaster and D. pseudoobscura (Moriyama and Powell 1997), unbiased genes have a very similar optimal codon frequency (Table 1). Pronounced differences, however, were found for genes with a male-biased gene expression pattern in D. melanogaster (Table 1). Comparing the difference in codon usage [|_F_op (pseudoobscura) − _F_op (melanogaster)|] between the two categories of genes with male-biased gene expression in one species, but no sex bias in the other species, we detected a significant difference (P < 0.001, Mann–Whitney _U_-test) with only D. melanogaster male-biased genes showing a lower codon usage bias.
We calculated Cohen's d (Cohen 1988) to determine whether the effect size is sufficiently large that the statistical significances reported are biologically relevant. A moderate effect size for Grantham's distance (d = 0.3) and a high effect size for codon bias (d = 0.47) were observed. These results suggest that ∼21% of the observations of Grantham's distance and 33% of the observations of codon bias are not overlapping (Cohen 1988).
Rates of evolution were based on all genes to which we could unambiguously map a SAGE tag. As these genes may not be completely conserved between D. melanogaster and D. pseudoobscura, we repeated our analyses using only those genes for which we could align the complete protein between both species, but the same trends were observed. Furthermore, to exclude that a differential coverage of lowly expressed genes in the SAGE and microarray experiments affected our interpretation of the results, we repeated the analysis using only the 20 and 40% most highly expressed genes from the microarray experiments in D. melanogaster. However, the same trend was observed (data are available from the authors upon request).
We also compared D. melanogaster data from Ranz et al. (2003) with D. pseudoobscura expression data to see whether a similar pattern could be observed. About 50% of the genes that are included in the analysis by Parisi et al. (2003) are missing in this comparison. Comparing genes with a male-biased expression in D. pseudoobscura and an unbiased expression in D. melanogaster (12 genes rather than 77 genes in the Parisi et al. 2003 data) and the complementary class (161 genes rather than 187 genes in the Parisi et al. 2003 data), the later class manifests higher nonsynonymous substitution rates (0.13 against 0.07). However, this difference lacks sufficient statistical power, which may be due to smaller sample sizes.
The XR chromosome of D. pseudoobscura is originated by translocation from an autosome (3L) in D. melanogaster. Hence, we were interested if we could detect evidence for different rates of evolution for genes located on chromosomes XL and XR and on autosomes. Nevertheless, within the limitations of a moderate number of genes, we observed similar rates of evolution within each class of sex-biased genes across the chromosomes (data are available from authors upon request).
DISCUSSION
In this study, we compared both sequence and expression data for males and females in two different Drosophila species. We measured gene expression in D. pseudoobscura using SAGE and compared it to microarray-based expression data in D. melanogaster. While the comparison of gene expression data generated by different experimental systems could potentially lead to some bias, we note that SAGE and microarray data were found to be highly correlated (Ishii et al. 2000). Furthermore, we do not compare the expression data directly, but we compare the gene expression differences between males and females. Thus, the relative gene expression is determined for each species separately using a consistent experimental design, which further minimizes the risk of a possible bias (Draghici et al. 2006). Assuming that both SAGE and microarray experiments measure the expression level accurately, the only possible bias could arise from genes with a low expression level that are not included in the SAGE data. We therefore repeated our analysis using only those genes that are highly expressed in D. melanogaster, but obtained the same results. A different significance level for identifying sex-biased genes in D. pseudoobscura also did not change the overall picture (Figure 1). Hence we are confident that the comparison of SAGE and microarray data did not bias our results.
Impact of tag-to-gene mapping on the comparison of male and female gene expression:
SAGE allows the analysis of sex-biased gene expression independent of an annotated genome. SAGE provides a set of tags, which are overrepresented in males or females. A genome annotation, however, is required when these tags should be associated with the corresponding gene (tag-to-gene mapping). Thus, the proportion of SAGE tags mapped depends on the accuracy of genome annotation. Three major factors could affect the efficiency of our tag-to-gene mapping: (1) as the annotation of D. pseudoobscura relied heavily on genes known in D. melanogaster, the sequence conservation of D. melanogaster genes in D. pseudoobscura is a major determinant of the tag-to-gene mapping efficiency; (2) many SAGE tags are located in the 3′-UTR (Pleasance et al. 2003), but this region is very poorly annotated in D. pseudoobscura; (3) genes specific to D. pseudoobscura and not yet discovered genes in D. melanogaster.
Our analyses indicated that a lower proportion of female-biased tags than unbiased or male-biased tags could be mapped to D. pseudoobscura genes. In the following paragraphs, we discuss to what extent this result could be an artifact of the incomplete genome annotation available for D. pseudoobscura.
In principle, the low tag-to-gene mapping efficiency of female-biased genes could be the result of a lower sequence conservation of female-biased genes. We tested this hypothesis by determining the proportion of D. melanogaster genes that are conserved in D. pseudoobscura. We found that 83% of the genes with a female-biased gene expression in D. melanogaster were conserved in D. pseudoobscura. Seventy-nine percent of the unbiased and only 63% of the male-biased genes were conserved. Hence, on the basis of D. melanogaster gene expression data, a lower proportion of male-biased genes are expected, but not a lower proportion of female-biased genes.
The gene expression data from D. melanogaster and D. pseudoobscura are based on different methods. In contrast to the microarray data for D. melanogaster, the dynamic range of a SAGE analysis depends on the number of tags sequenced. As the depth of our SAGE sequencing was not extensive, we probably covered only the more highly expressed genes. To exclude that the differential coverage of lowly expressed genes affected our results, we repeated our analyses restricting the microarray data to the 20/40% most highly expressed genes in D. melanogaster. Nevertheless, we still found approximately the same degree of conservation. Hence, we conclude that the lower efficiency of tag-to-gene mapping for female-biased tags in D. pseudoobscura is not an artifact of a different dynamic range of SAGE and microarray data.
Alternative splicing could result in transcripts with different SAGE tags. If genes with a female-biased gene expression in D. pseudoobscura are more frequently alternatively spliced, this could result in a lower tag to gene mapping efficiency. We explored this possibility and determined the proportion of genes that would result in different SAGE tags due to alternative splicing. However, only a very small proportion of alternatively spliced genes in D. melanogaster (4%) yielded alternative SAGE tags. No significant difference was detected between male- and female-biased genes. Unless this pattern has dramatically changed in D. pseudoobscura, we can discount this explanation for the low efficiency of tag-to-gene mapping for female-biased genes in D. pseudoobscura.
Tag-to-gene mapping is also affected by polymorphism (Ng et al. 2005). As we used a different D. pseudoobscura strain than the sequenced one, it is possible that sequence polymorphism reduces the tag-to-gene mapping efficiency. The lower efficiency to map female-biased tags to D. pseudoobscura genes could be explained if female-biased genes have more sequence polymorphism than male-biased ones. Currently, too few polymorphism data are available in D. pseudoobscura to address this question.
Tag-to-gene mapping may be affected by the incomplete 3′-UTR annotation in D. pseudoobscura. If female-biased genes have a longer 3′-UTR than male-biased genes, this could lead to an underrepresentation of female-biased genes. We compared the length of 3′-UTRs in D. melanogaster and found that male-biased genes on average have shorter 3′ UTRs (293 bp) than female-biased genes (333 bp). The longest 3′-UTRs, however, were observed for the unbiased genes with 433 bp on average. As we mapped a higher proportion of unbiased tags than female biased tags, we conclude that UTR length is unlikely to be the sole reason for the low efficiency of tag-to-gene mapping of female-biased SAGE tags.
Finally, a high proportion of hitherto unrecognized genes could explain the low tag-to-gene mapping efficiency for female-biased genes. Either the genes have not yet been annotated in D. melanogaster (e.g., Lee et al. 2005) or they are present in D. pseudoobscura only. In-depth sequencing of cDNA clones obtained from female D. pseudoobscura will be highly instrumental in addressing this question further.
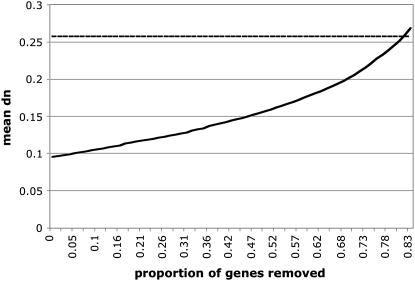
We further tried to estimate the error rate in tag-to-gene mapping. Given the limitations of the current genome annotation of D. pseudoobscura, it is possible that a certain fraction of the tags were mapped to the wrong gene. If male-biased genes in D. pseudoobscura evolve at a high rate, similar to those in D. melanogaster, the lower _d_N value in our data could be caused by an erroneous inclusion of non-male-biased genes. We estimated the proportion of falsely assigned genes (with a lower _d_N value) by iteratively removing the gene with the lowest _d_N value from the genes that were male biased in D. melanogaster, but unbiased in D. pseudoobscura. Figure 5 shows that ∼80% of the genes would need to be wrongly assigned to obtain a similar _d_N value for genes with male-biased gene expression in both species.
Figure 5.—
Mean _d_N value of genes with unbiased gene expression in D. pseudoobscura and male-biased gene expression in D. melanogaster (solid line). The _x_-axis gives the proportion of genes with low _d_N value that were deleted from the full data set. Note that the genes were sorted according to _d_N value and iteratively the gene with the lowest _d_N value was removed. The dashed line provides the mean _d_N value for genes with a male-biased gene expression in both species (no genes were deleted).
Finally, the best support for the accuracy of our tag-to-gene mapping procedure is provided by the experimental validation using the GLGI method: only 1 of 21 tags analyzed did not confirm our tag-to-gene mapping. Hence, we conclude that errors in tag-to-gene mapping could not explain our results.
Stability of male-biased gene expression:
We found that ∼34% of the genes changed the sex-related expression pattern between D. melanogaster and D. pseudoobscura. A recent comparison between D. melanogaster and D. simulans found a very similar proportion of genes with a change in expression pattern between males and females despite a much closer phylogenetic relationship (Ranz et al. 2003). Hence, it may be possible that the sex-related expression of several genes changed multiple times. As male-biased genes were particularly unstable (Ranz et al. 2003), we were interested in whether genes with a male-biased gene expression in D. melanogaster and D. pseudoobscura maintained their sex bias during evolution. We used the limited set of genes for which expression data are available in three species—D. melanogaster, D. simulans, and D. pseudoobscura. Interestingly, all 7 genes with a male-biased gene expression in D. melanogaster and D. pseudoobscura were also male biased in D. simulans (Table 2A). Nevertheless, it should be noted that we used a different criterion to define male bias (twofold difference in expression) than Ranz et al. (2003) used. Using the significance threshold of Ranz et al. (2003), we found that 1 of 21 genes with a male-biased gene expression in D. melanogaster and D. pseudoobscura was not male biased in D. simulans (Table 2B). Given that ∼20% of the male-biased genes in D. melanogaster changed their expression pattern in D. simulans, this high concordance may suggest that only a subset of the male-biased genes changes its expression pattern, while others remain male biased. The analysis of sex-biased gene expression in other species will be instrumental in determining their evolutionary stability.
TABLE 2.
Comparison of genes with male-biased expression in D. pseudoobscura and D. melanogaster with D. simulans
| D. pseudoobscura | D. melanogaster | D. simulans: | ||
|---|---|---|---|---|
| Gene | Log2 expression ratio (male/female) | Gene | Log2 expression ratio (male/female)a | Log2 expression ratio (male/female)a |
| A. Based on fold difference | ||||
| GA10438-RA | 2.32 | CG10616 | 1.21 | 1.13 |
| GA13209-RA | 2.00 | CG14735 | 5.34 | 5.07 |
| GA15417-RA | 1.67 | CG2668 | 5.90 | 6.10 |
| GA15920-RA | 3.32 | CG3092 | 5.01 | 4.38 |
| GA18342-RA | 2.81 | CG4669 | 4.76 | 4.49 |
| GA18974-RA | 3.32 | CG5565 | 4.46 | 3.49 |
| GA20544-RA | 2.00 | CG7722 | 2.88 | 2.52 |
| B. Based on level of significance | ||||
| GA10438-RA | 2.32 | CG10616 | 1.21 | 1.13 |
| GA10736-RA | 2.32 | CG11064 | 0.73 | 1.50 |
| GA10887-RA | 2.32 | CG11280 | 0.31 | 0.38 |
| GA11127-RA | 0.76 | CG11661 | 0.77 | 0.68 |
| GA12802-RA | 2.32 | CG1417 | 0.47 | 0.74 |
| GA13209-RA | 2.00 | CG14735 | 5.34 | 5.07 |
| GA14194-RA | 1.26 | CG16884 | 0.71 | 0.49 |
| GA14224-RA | 2.32 | CG16932 | 0.28 | 0.18 |
| GA14923-RA | 2.00 | CG18408 | 0.64 | 0.44 |
| GA15417-RA | 1.67 | CG2668 | 5.90 | 6.10 |
| GA15456-RA | 1.74 | CG2765 | 0.68 | 0.70 |
| GA15920-RA | 3.32 | CG3092 | 5.01 | 4.38 |
| GA18342-RA | 2.81 | CG4669 | 4.76 | 4.49 |
| GA18974-RA | 3.32 | CG5565 | 4.46 | 3.49 |
| GA19329-RA | 0.72 | CG6058 | 0.81 | 0.71 |
| GA20243-RA | 2.32 | CG7292 | 0.84 | 0.83 |
| GA20345-RA | 2.00 | CG7430 | 0.51 | 0.50 |
| GA20544-RA | 2.00 | CG7722 | 2.88 | 2.52 |
| GA20881-RA | 1.00 | CG8189 | 0.76 | −0.07b |
| GA20964-RA | 2.58 | CG8295 | 0.76 | 0.59 |
| GA22030-RA | 1.74 | CG9779 | 0.39 | 0.42 |
No increase in _d_N by change in sex-biased gene expression:
An analysis of sex-biased gene expression in D. melanogaster and D. simulans indicated a rapid change in expression pattern, in particular for male-biased genes (Ranz et al. 2003). Assuming that such changes in expression pattern reflect functional shifts, it is interesting to determine if a change in gene expression is associated with an increased rate of protein evolution. Two different evolutionary scenarios would predict an accelerated rated of evolution: (1) a change in gene expression relaxes the functional constraint leading to more amino acid replacements and (2) the change in gene expression is associated with an expansion/modulation of the functional repertoire. Thus, directional selection is expected to drive the required changes in the protein.
Of the possible changes in expression pattern, three categories contained >50 genes: two categories with male-biased–unbiased genes and one category with a female-biased gene expression in D. melanogaster but unbiased in D. pseudoobscura. Contrary to expectations, none of the categories with a change in gene expression had a higher _d_N than genes with a male-biased gene expression in both species. In fact, genes with a female-biased gene expression in D. melanogaster and no sex bias in D. pseudoobscura had a _d_N lower than all categories with no change in gene expression pattern. This indicates that a change in sex-biased gene expression pattern does not necessarily increase the nonsynonymous substitution rate.
Male-biased genes in D. pseudoobscura do not evolve fast:
It is well documented that genes with a male-biased gene expression have an accelerated rate of protein evolution (Meiklejohn et al. 2003). Nevertheless, most of the inference is based on the expression pattern in one of the species only. Consistent with the previous results, we also found that genes in D. melanogaster and D. pseudoobscura with a male-biased gene expression evolved at the highest rate in our data set. Nevertheless, the comparison to genes that changed their expression pattern is not consistent with an unconditionally higher rate of protein evolution for male-biased genes. If male-biased genes evolved faster, genes with a change in expression should have an intermediate rate of evolution. Moreover, this pattern should be independent of the species that shows the male bias. Genes with a male-biased gene expression only in D. melanogaster evolve fast and with a rate very similar to that of genes that are male biased in both species. However, contrary to expectation, genes with a male-biased gene expression only in D. pseudoobscura do not evolve fast and have a rate of protein evolution similar to that of unbiased genes. This pattern is better explained by a change in the evolution rate of male-biased genes between D. pseudoobscura and D. melanogaster.
It was previously suggested that the accelerated rate of evolution of male-biased genes is driven by positive selection (Swanson and Vacquier 2002). Consistent with this hypothesis, we also found a higher Grantham distance and a lower codon usage for genes with a male-biased gene expression pattern in both species. Interestingly, genes with a male-biased gene expression only in D. pseudoobscura did not show this effect, further emphasizing the difference of male-biased genes between D. melanogaster and D. pseudoobscura. Nevertheless, we note that relaxed constraint, rather than directional selection, results in a similar pattern for the male-biased genes in D. melanogaster.
Our analyses were based on genes that are conserved between D. melanogaster and D. pseudoobscura. Thus, rapidly evolving genes are missed in our analysis. We note that it is not possible to estimate the proportion of rapidly evolving genes that are still functional in D. pseudoobscura and whether or not they have changed their sex bias. For this reason, we focused our attention on those genes that are male biased in one species only. Our analysis was expected to be unbiased unless we preferentially missed fast-evolving genes with a male bias in D. pseudoobscura. However, we do not think that this applies to our data. Assuming that genes with a male-biased gene expression evolve so fast that they are difficult to detect in D. pseudoobscura, this would result in a downward bias in mutation rate affecting all genes with a male expression bias. Hence, the most dramatic effect would be seen for genes with a male-biased gene expression in both species. Genes with a male bias in only one of the species are expected to be affected to a lesser extent and should affect both species to the same extent (assuming that the change of male-biased gene expression is randomly distributed over the divergence between D. melanogaster and D. pseudoobscura). Contrary to these predictions, the highest rate of evolution is observed for genes with a male bias in both species. Genes with a male bias only in D. melanogaster evolve at slightly lower rate, while genes with a male bias only in D. pseudoobscura show no evidence of a high rate of evolution.
Assuming that the accelerated rate of protein evolution, low codon usage, and high Grantham distance of male-biased genes in D. melanogaster are driven by positive selection, probably associated with sexual selection, the important question arises as to how D. pseudoobscura differs from D. melanogaster. Is there any evidence that sexual selection is reduced in D. pseudoobscura males?
In the following, we discuss some differences between D. melanogaster and D. pseudoobscura that could potentially be related to sexual selection. Nevertheless, it is not clear if these differences could explain the pattern observed in this report. Both species, D. melanogaster (Griffiths et al. 1982; Harshman and Clark 1998; Imhof et al. 1998) and D. pseudoobscura (Cobbs 1977; Anderson et al. 1987), remate frequently, but the variation within a given species across studies is too large to identify possible differences between species. Nevertheless, D. melanogaster and D. pseudoobscura differ in their tendency to remate. While D. melanogaster has a refractory period of 2 hr (Bundgaard and Christiansen 1972), D. pseudoobscura starts remating after 12 hr (Beckenbach 1981). Furthermore, D. pseudoobscura females approach males more often than D. melanogaster females do (Gowaty et al. 2003), suggesting that D. pseudoobscura females are choosier than D. melanogaster females. While it is conceivable that a longer refractory period and choosier females could reduce some aspects of sexual selection in male D. pseudoobscura, it is not clear if this is sufficient to explain the observed differences in molecular evolution. Further work is required to test whether these differences between D. melanogaster and D. pseudoobscura could explain the contrasting evolution of male-biased genes.
Acknowledgments
We thank D. Ibberson and V. Benes for the construction of the SAGE library and for many helpful discussions. Many thanks to the M. Akam lab for providing the D. pseudoobscura strain. M. Schäfer and C. Vogl provided helpful comments. V. Nolte assisted in GLGI experiments. This work is funded through Fonds zur Förderung der wissenschaftlichen Forschung grants to C.S. M.M. is supported by a Doktoratsstipendium of the Veterinärmedizinischen Universität Wien.
References
- Anderson, P. R., W. R. Knibb and J. G. Oakeshott, 1987. Observations on the extent and temporal stability of latitudinal clines for alcohol dehydrogenase allozymes and four chromosome inversions in Drosophila melanogaster. Genetica 75**:** 81–88. [DOI] [PubMed] [Google Scholar]
- Arbeitman, M. N., E. E. Furlong, F. Imam, E. Johnson, B. H. Null et al., 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297**:** 2270–2275. [DOI] [PubMed] [Google Scholar]
- Audic, S., and J. M. Claverie, 1997. The significance of digital gene expression profiles. Genome Res. 7**:** 986–995. [DOI] [PubMed] [Google Scholar]
- Bauer, V. L., and C. F. Aquadro, 1997. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol. Biol. Evol. 14**:** 1252–1257. [DOI] [PubMed] [Google Scholar]
- Beckenbach, A., 1981. Multiple mating and the “sex-ratio” trait in Drosophila pseudoobscura. Evolution 35**:** 275–281. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and H. A. Lindfors, 2005. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol. Biol. Evol. 22**:** 2010–2021.15987879 [Google Scholar]
- Betancourt, A. J., and D. C. Presgraves, 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99**:** 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., and R. Durbin, 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10**:** 547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundgaard, J., and F. B. Christiansen, 1972. Dynamics of polymorphisms. I. Selection components in an experimental population of Drosophila melanogaster. Genetics 71**:** 439–460. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C. I., and D. L. Hartl, 2003. GeneMerge: post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19**:** 891–892. [DOI] [PubMed] [Google Scholar]
- Chen, J. J., J. D. Rowley and S. M. Wang, 2000. Generation of longer cDNA fragments from serial analysis of gene expression tags for gene identification. Proc. Natl. Acad. Sci. USA 97**:** 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., S. A. Rajakumar, B. Brouwers and J. P. Bacik, 2006. Rapid evolution and gene-specific patterns of selection for three genes of spermatogenesis in Drosophila. Mol. Biol. Evol. 23**:** 655–662. [DOI] [PubMed] [Google Scholar]
- Cobbs, G., 1977. Multiple insemination and male sexual selection in natural populations of Drosophila pseudoobscura. Am. Nat. 111**:** 641–656. [Google Scholar]
- Cohen, J., 1988. Statistical Power Analysis for the Behavioral Sciences. Lawrence Earlbaum Associates, Hillsdale, NJ.
- Coulthart, M. B., and R. S. Singh, 1988. High level of divergence of male-reproductive-tract proteins, between Drosophila melanogaster and its sibling species, D. simulans. Mol. Biol. Evol. 5**:** 182–191. [DOI] [PubMed] [Google Scholar]
- Cusack, B. P., and K. H. Wolfe, 2005. Changes in alternative splicing of human and mouse genes are accompanied by faster evolution of constitutive exons. Mol. Biol. Evol. 22**:** 2198–2208. [DOI] [PubMed] [Google Scholar]
- Dickens, J. C., F. E. Callahan, W. P. Wergin, C. A. Murphy and R. G. Vogt, 1998. Odorant-binding proteins of true bugs. Generic specificity, sexual dimorphism, and association with subsets of chemosensory sensilla. Ann. NY Acad. Sci. 855**:** 306–310. [DOI] [PubMed] [Google Scholar]
- Dinel, S., C. Bolduc, P. Belleau, A. Boivin, M. Yoshioka et al., 2005. Reproducibility, bioinformatic analysis and power of the SAGE method to evaluate changes in transcriptome. Nucleic Acids Res. 33**:** e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici, S., P. Khatri, A. C. Eklund and Z. Szallasi, 2006. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 22**:** 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, J. M., and M. W. Nachman, 2005. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol. Biol. Evol. 22**:** 1044–1052. [DOI] [PubMed] [Google Scholar]
- Gowaty, P. A., R. Steinichen and W. W. Anderson, 2003. Indiscriminate females and choosy males: within- and between-species variation in Drosophila. Evolution 57**:** 2037–2045. [DOI] [PubMed] [Google Scholar]
- Grantham, R., 1974. Amino acid difference formula to help explain protein evolution. Science 85**:** 862–864. [DOI] [PubMed] [Google Scholar]
- Griffiths, R. C., S. W. McKenchnie and J. A. McKenzie, 1982. Multiple mating and sperm displacement in a natural population of Drosophila melanogaster. Theor. Appl. Genet. 62**:** 89–96. [DOI] [PubMed] [Google Scholar]
- Harshman, L. G., and A. G. Clark, 1998. Inference of sperm competition from broods of field-caught Drosophila. Evolution 52**:** 1334–1341. [DOI] [PubMed] [Google Scholar]
- Holland, B., and W. R. Rice, 1999. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc. Natl. Acad. Sci. USA 96**:** 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy, R. R., 1990. Evolutionary innovation in behavior and speciation: opportunities for behavioral neuroethology. Brain Behav. Evol. 36**:** 141–153. [DOI] [PubMed] [Google Scholar]
- Hughes, I. A., 2001. Minireview: sex differentiation. Endocrinology 142**:** 3281–3287. [DOI] [PubMed] [Google Scholar]
- Ikemura, T., 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151**:** 389–409. [DOI] [PubMed] [Google Scholar]
- Imhof, M., B. Harr, G. Brem and C. Schlötterer, 1998. Multiple mating in wild Drosophila melanogaster revisited by microsatellite analysis. Mol. Ecol. 7**:** 915–917. [DOI] [PubMed] [Google Scholar]
- Ishii, M., S. Hashimoto, S. Tsutsumi, Y. Wada, K. Matsushima et al., 2000. Direct comparison of GeneChip and SAGE on the quantitative accuracy in transcript profiling analysis. Genomics 68**:** 136–143. [DOI] [PubMed] [Google Scholar]
- Jagadeeshan, S., and R. S. Singh, 2005. Rapidly evolving genes of Drosophila: differing levels of selective pressure in testis, ovary, and head tissues between sibling species. Mol. Biol. Evol. 22**:** 1793–1801. [DOI] [PubMed] [Google Scholar]
- Jasper, H., V. Benes, C. Schwager, S. Sauer, S. Clauder-Munster et al., 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell 1**:** 579–586. [DOI] [PubMed] [Google Scholar]
- Jasper, H., V. Benes, A. Atzberger, S. Sauer, W. Ansorge et al., 2002. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev. Cell 3**:** 511–521. [DOI] [PubMed] [Google Scholar]
- Lee, S., J. Bao, G. Zhou, J. Shapiro, J. Xu et al., 2005. Detecting novel low-abundant transcripts in Drosophila. RNA 11**:** 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, N., and D. Joly, 2005. Sexual selection and mating advantages in the giant sperm species, Drosophila bifurca. J. Insect Sci. 5**:** 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100**:** 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1997. Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 45**:** 514–523. [DOI] [PubMed] [Google Scholar]
- Ng, P., C. L. Wei, W. K. Sung, K. P. Chiu, L. Lipovich et al., 2005. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods 2**:** 105–111. [DOI] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299**:** 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, J. F., 1999. Analysis of codon usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK.
- Pleasance, E. D., M. A. Marra and S. J. Jones, 2003. Assessment of SAGE in transcript identification. Genome Res. 13**:** 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300**:** 1742–1745. [DOI] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15**:** 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, A. P., J. Zhang and M. Nei, 2000. An unusual form of purifying selection in a sperm protein. Mol. Biol. Evol. 17**:** 278–283. [DOI] [PubMed] [Google Scholar]
- Ruijter, J. M., A. H. Van Kampen and F. Baas, 2002. Statistical evaluation of SAGE libraries: consequences for experimental design. Physiol. Genomics 11**:** 37–44. [DOI] [PubMed] [Google Scholar]
- Snook, R. R., A. Robertson, H. S. Crudgington and M. G. Ritchie, 2005. Experimental manipulation of sexual selection and the evolution of courtship song in Drosophila pseudoobscura. Behav. Genet. 35**:** 245–255. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3**:** 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98**:** 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson, D. G., and R. S. Singh, 2004. Rapid evolution through gene duplication and subfunctionalization of the testes-specific α4 proteasome subunits in Drosophila. Genetics 168**:** 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J. P., 2003. Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics 4**:** 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur, S. C., and C.-I Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14**:** 544–549. [DOI] [PubMed] [Google Scholar]
- Velculescu, V. E., L. Zhang, B. Vogelstein and K. W. Kinzler, 1995. Serial analysis of gene expression. Science 270**:** 484–487. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura. Mol. Biol. Evol. 22**:** 818–832. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13**:** 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17**:** 32–43. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., T. M. Hambuch and J. Parsch, 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21**:** 2130–2139. [DOI] [PubMed] [Google Scholar]