Opacity-Associated Adhesin Repertoire in Hyperinvasive Neisseria meningitidis (original) (raw)
Abstract
The opacity (Opa) proteins mediate a variety of interactions between the bacterium Neisseria meningitidis and its human host. These interactions are thought to be of central importance in both the asymptomatic colonization of the nasopharynx and the sporadic occurrence of meningococcal disease. The receptor specificities of a limited number of Opa protein variants have been explored, but the high level of amino acid sequence diversity among variants has complicated the assignment of specific roles to individual Opa variants or combinations of variants. In addition, the distribution of Opa protein variants among diverse meningococci, information that is potentially informative for studies of Opa function, is poorly understood. A systematic survey of the genetic diversity in the four opa gene loci in each of 77 meningococcal isolates was undertaken. These isolates were representative of the seven hyperinvasive meningococcal clonal complexes that caused the majority of meningococcal disease over the last 50 years. Consistent with previous studies, a high level of sequence diversity was observed among the opa genes and the proteins that they encoded; however, particular sets of Opa protein variants were consistently associated with each of the clonal complexes over time periods often spanning decades and during global spread. These observations were consistent with the postulate that particular combinations of Opa proteins confer fitness advantages to individual clonal complexes and have implications for studies of Opa function and the inclusion of Opa proteins in novel meningococcal vaccines.
The genus Neisseria comprises bacteria that live in close association with the mucosal surfaces of animals. The colony opacity-associated (Opa) outer membrane proteins of these gram-negative organisms are adhesins that play a major role in interactions with mucosal epithelia and endothelia. Opa proteins have been implicated in the virulence of the pathogenic species Neisseria meningitidis, the meningococcus, and Neisseria gonorrhoeae, the gonococcus (21). The Opa proteins may also act as immunomodulators, enabling interactions with macrophages, neutrophils, and CD4+ T cells (2, 6, 18, 32, 52). These effects are mediated by interactions with members of the carcinoembryonic antigen cell adhesion molecule (CEACAM) family of proteins (9, 18, 52, 53) and host cell surface polysaccharides (10, 36, 49, 50).
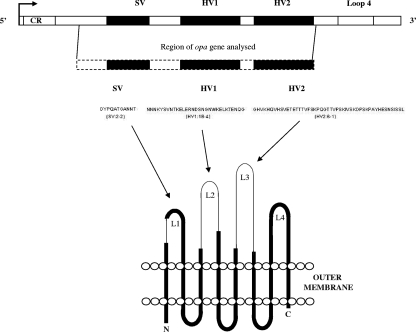
Meningococcal Opa proteins are highly diverse, with different variants exhibiting specific interactions with human cells. The outcomes of in vitro infections of human cells by meningococci expressing different Opa proteins range from no adhesion to adhesion only or adhesion followed by invasion. These differences are determined by combinations of amino acid sequence variations present at each of three variable regions of Opa protein sequences (4, 5, 14, 17, 19, 35, 36, 38, 51). The Opa proteins are thought to exhibit the β-barrel topology typical of a number of outer membrane proteins of gram-negative bacteria, with the variable sequences corresponding to three of four putative extramembranous loops that extend from the surface of the bacterium and are consequently available for interaction with host molecules (Fig. 1) (13, 33).
FIG. 1.
Predicted secondary structural topology of Opa proteins. SV, semivariable region located in putative loop 1; HV1 and HV2, two hypervariable regions, located in putative loops 2 and 3, respectively. The regions of opa genes sequenced in this study are indicated.
Most meningococcal isolates express up to four Opa protein variants, encoded at four loci (opaA, opaB, opaD, and opaJ) dispersed throughout the genome (39, 45). These genes are constitutively transcribed, with expression of a full-length protein controlled at the translational level by a phase-variable pentameric repeat region within the opa gene open reading frame (29, 43). Nucleotide sequence analysis of a number of opa genes revealed that the high level of diversity of Opa proteins is generated by mutation within and genetic recombination between opa loci, both among loci in the same genome and among loci in different meningococci (23, 24).
Horizontal genetic exchange plays a major role in the generation of genetically and antigenically diverse populations of N. meningitidis, which comprise large numbers of diverse genotypes (27). These genotypes can be identified using a number of methods, but multilocus sequence typing (MLST) (31), which characterizes the sequence variation in seven housekeeping genes, has become the standard technique for the investigation of meningococcal population structure (47, 56). Genotypes are identified by MLST as sequence types (STs), which are combinations of alleles at each of the seven loci. Groups of related STs are referred to as clonal complexes, which are equivalent to the clusters, complexes, clones, subgroups, and lineages previously described by studies employing multilocus enzyme electrophoresis (8).
Comparisons of the genotypes of meningococcal isolates obtained from patients with invasive meningococcal disease with those collected from asymptomatic carriers have shown that whereas carried isolates are genetically and antigenically highly diverse (27), most meningococcal disease is caused by a limited number of clonal complexes known as the hyperinvasive lineages (7, 56). Individual clonal complexes are often associated with particular combinations of surface antigens, including capsular polysaccharides (which are used to assign the meningococcal serogroup) and outer membrane proteins (which define the serotype and subtype). These associations, however, are not absolute (44). Meningococcal clonal complexes are valuable in epidemiological analysis as well as representing evolutionary units (28, 48, 56).
In the present study, the distribution of Opa protein variants within and among clonal complexes representing the major hyperinvasive lineages of N. meningitidis was determined by nucleotide sequencing of all opa loci in each isolate. Although the level of diversity of the opa genes and Opa proteins was high, the distribution of variants among meningococci was analogous to that of a number of other meningococcal antigens, with particular combinations of variants consistently associated with the same clonal complexes over decades of global epidemic spread. These data have important implications for studies of the functional roles of individual Opa proteins and their combinations in mediating adhesion to and invasion of host cells by meningococci.
MATERIALS AND METHODS
Meningococcal isolates, propagation, and DNA isolation.
A total of 77 meningococcal isolates were examined, representing the seven hyperinvasive meningococcal lineages responsible for the majority of invasive meningococcal disease worldwide during the second half of the 20th century, i.e., the ST-1, ST-4, ST-5, ST-8, ST-11, ST-32, and ST-41/44 complexes. The isolates were collected between 1937 and 1996 from diverse geographic locations and were chosen to include diverse members of each meningococcal hyperinvasive lineage. These isolates have been characterized extensively (31, 48), and a full description of them, including year and location of isolation and MLST and antigen gene sequencing data, is available at http://pubmlst.org/neisseria/links.shtml.
Genomic DNA was prepared by culturing isolates as previously described (27) before genomic DNA extraction using a DNA Mini kit (QIAGEN, Crawley, United Kingdom) according to the manufacturer's instructions.
PCR amplification and nucleotide sequence determination of opa genes.
Meningococcal opa loci were amplified in separate PCRs with locus-specific oligonucleotide primer sets (Eurogentec, Southampton, United Kingdom). The opaA locus was amplified with primers O3510 (TAC GCT GCA GAA AAT GAA TCC AGC CCC C) (37) and tyrA (ACA TCG GAA ATC CAA GTG TGT TCC) at a final concentration of 1 μM (each), the opaB locus was amplified with primers O3510 and O464 (AAG GCG AGG TAG GAT TGC) (37) at a final concentration of 1 μM (each), the opaD locus was amplified with primers O87 (GCG CAC GCC CAA TGA GAC TTC GTG GG) (37) and ppx (TTT GAA CGA ATC GAT AAC TTT CAA CTG TCC) at a final concentration of 0.4 μM (each), and the opaJ locus was amplified with primers O87 and pipP2 (CTC AAC CGC CTG AAC CAA CG) (31) at a final concentration of 0.05 μM (each). Individual amplification reaction mixtures (50 μl) contained premixed deoxynucleoside triphosphates (dA, dC, dT, and dG; Applied Biosystems, Warrington, United Kingdom) at 0.2 mM (each), Taq DNA polymerase (2.5 units), and reaction buffer (QIAGEN, Crawley, United Kingdom). The volume of the reaction mix was adjusted to 48 μl using MilliQ water before the addition of approximately 100 ng of meningococcal genomic DNA in a volume of 2 μl.
The standard PCR cycling conditions employed were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 68°C for 1 min 30 seconds. The reaction mixtures were subsequently cooled to 4°C for storage. Reaction conditions were optimized for the amplification of each locus by varying the annealing temperature and extension time, as follows: for the opaA and opaJ loci, the annealing temperature was 60°C and the extension time was 10 min; for the opaB locus, the annealing temperature was 55°C and the extension time was 5 min; and for the opaD locus, the annealing temperature was 53°C and the extension time was 8 min.
Following PCR amplification, the nucleotide sequences, including the regions of opa genes encoding the three hypervariable loops of Opa proteins (Fig. 1), were determined for both DNA strands by using the oligonucleotide primers MH4 (CGT GAT GTC GAA ACC GAC ACC G) (24) and fwdseq (GCA GGC GGC AAG TGA AGA CG), which were targeted to conserved sequences present in all known opa gene sequences. Extension products were subsequently separated using either an ABI377 or ABI3700 DNA analyzer (Applied Biosystems, Warrington, United Kingdom). Nucleotide and deduced amino acid sequence data are available at http://neisseria.org/nm/typing/opa/.
Manipulation of sequence data.
The nucleotide sequence at each locus was determined at least once for each DNA strand. Sequence data from forward- and reverse-strand chromatograms were assembled into single contiguous sequences using the Staden software package (42), and the resultant nucleotide sequences were trimmed to the first nucleotide of the codon encoding the tyrosine residue of the sequence YAAERITH, located in the putative first transmembrane strand, and the nucleotide preceding the translated amino acid sequence GLG in the putative sixth transmembrane strand. This region defined the opa alleles employed in this work, encompassing the three variable putative extramembranous loops.
Amino acid sequence homology groups for the variable regions were identified by multiple sequence alignments constructed manually in the SEQLAB program, which is part of the GCG10 sequence analysis package (55). Alignments were based on amino acid sequence similarity, and codon integrity was maintained. The SV region was defined as spanning the amino acid sequence between, but not including, RITH and STVS; the HV1 region was defined as spanning the amino acid sequence between, but not including, YRKW (at the end of the third putative β-strand) and NGSF (at the start of the fourth β-strand); and the HV2 region was defined as spanning the amino acid sequence between, but not including, RVAY (at the end of the fifth β-strand) and GLGV (at the start of the sixth β-strand).
Phylogenetic analysis.
Genetic p distances were calculated using the MEGA v3.0 software package (30), with compensation for sequence alignment gaps by using a pairwise deletion method. Phylogenetic trees were constructed using the maximum likelihood (ML) method of the PAUP* package (D. L. Swofford, Sinauer Associates, Sunderland, Mass., 2003). The general time reversible model of nucleotide substitution was used, with values for the nucleotide substitution matrix, the proportion of invariable sites, and the shape parameter (α) of rate variation among sites (with four categories) estimated during tree reconstruction. ML phylogenies were reconstructed by analyzing concatenated aligned sequences of the opa loci from each isolate, giving a single sequence with a total length of 2,211 bp. The ML method was employed to group similar isolates and was not a reliable indicator of deeper phylogenetic structure in meningococci due to the high level of recombination in this species (15, 16, 28). Other tree-drawing methods were also applied and gave results consistent with those obtained with the ML method (data not shown). Phylogenies of concatenated opa sequences were compared to those reconstructed for concatenated MLST alleles and concatenated antigen gene sequences (porA, porB, and fetA) from the same collection of isolates (48).
Nomenclature.
The nomenclature system proposed for opa loci and Opa proteins (22) combines an allele number with the locus designation; for example, opaA1 designates allele 1 at the opaA locus and opaB1 designates an identical sequence at the opaB locus. Identical Opa protein sequences encoded by different nucleotide sequences are assigned the number of the allele with which they were first associated; for example, the alleles opaA1 and opaA2 may both encode an identical protein designated Opa1. This system has the advantage that the highest possible level of discrimination among opa alleles is obtained by the assignment of unique numbers to individual alleles that may differ by as little as a single nucleotide change. A major disadvantage is that no information on the degree of similarity among Opa proteins is included, as proteins differing by only a single amino acid are assigned different allele numbers. Such closely related sequences may be more likely to possess similar functional abilities than two Opa proteins sharing little sequence similarity in their variable regions. A simple, comprehensive method of describing this diversity and the relationships among variants would therefore greatly facilitate comparisons of functional studies of Opa proteins.
In the present study, a modified nomenclature for Opa proteins and opa genes is employed that accommodates the diversity in opa genes and Opa proteins and the relationships among diverse variants or different loci. In this scheme, the locus origin of the gene is included after the allele number; for example, _opaA132_L93/4286 and _opaD132_L93/4286 represent the same allele found in the opaA and opaD loci of isolate L93/4286. For the amino acid diversity of Opa proteins, a scheme similar to that employed for the meningococcal PorA serosubtyping antigen was adopted (40). Unique identifying numbers were assigned to individual sequence homology families and their members according to the following convention: variable region designation, followed by amino acid sequence homology family number, followed by the individual amino acid sequence variant identifying number. For example, opa1, SV:2-1, HV1:1B-4, HV2:6-1 indicates the unique nucleotide allele opa1, which encodes an Opa protein with the first amino acid variant of the second homology family in the SV region, the fourth variant of the 1B family (the alphabetical designation indicates a subgroup of the first homology family) in the HV1 region, and the first variant of the sixth family in the HV2 region. The major advantage of this system is that the regions which are likely to define receptor specificity can be characterized and compared among different Opa proteins, within and among isolates, and also among Neisseria species, as the opa genes from different members of this genus are homologous (46, 54). Allele and variable region sequences are available at http://neisseria.org/nm/typing/opa/. The correlation of the nomenclature scheme described herein with that previously published is given in the supplemental material, which provides a comprehensive list of the previously published nomenclature (33; M. Achtman, personal communication) along with the relevant GenBank accession numbers and the new nomenclature proposed.
Nucleotide sequence accession numbers. The GenBank accession numbers for the opa alleles sequenced in this study are DQ777637 to DQ777726.
RESULTS
Diversity of opa genes and Opa proteins.
In the 308 loci examined, corresponding to 4 loci in each of the 77 isolates, 90 opa alleles were observed, encoding 83 different polypeptides. The average nucleotide p distance among alleles was 13.4%. For the SV region, nine peptide variants which grouped into four sequence homology families were observed, with an average nucleotide p distance of 8.3% for the corresponding part of the gene. The SV:2 amino acid sequence homology family was most common in this data set, being present in 217/308 (70.5%) loci examined, and the SV:4 family accounted for a further 62/308 (20.1%) loci. For the HV1 region, 45 peptide variants were observed, which were grouped into 16 homology families (average corresponding nucleotide p distance, 24.2%), with the HV1:1 family being the most common, as it was present at 49/308 (15.9%) loci. For the HV2 region, there were 49 variants assigned to 19 homology families (average corresponding nucleotide p distance, 25.7%), and the HV2:1 sequence family was most common, being present at 83/308 (26.9%) loci.
There were 40 combinations of HV1/HV2 sequence homology families and 65 combinations of HV1/HV2 sequence homology family variants among the 90 alleles identified. The most prevalent HV1/HV2 family combination was HV1:11, HV2:1, which was present at 34/308 (11%) loci examined. This combination was found in 12 ST-1 complex isolates (with all variations present at the opaJ locus), 11 ST-11 complex isolates (9 opaD and 2 opaA variants), and 3 ST-41/44 complex isolates (all opaJ variants) and at the opaD loci of all 8 ST-8 complex isolates. The hypervariable region combination HV1:19, HV2:14 was found at 31 loci, with 1 locus in an ST-1 complex isolate (opaA), 8 loci in ST-11 complex isolates (all opaJ), 20 loci in ST-32 complex isolates (10 opaA and 10 opaJ), and 2 loci in an ST-8 complex isolate, at the opaA and opaJ loci of isolate B6116/77. The other most common combinations were HV1:3, HV2:1, which was present at 24 loci; the combinations HV1:1, HV2:8 and HV1:2, HV2:1, which were present at 22 loci each; and HV1:17, HV2:17, which was present at 21 loci.
Distribution of opa gene diversity within and among hyperinvasive clonal complexes.
In each of the seven clonal complexes examined, particular opa alleles were consistently observed at each locus (Table 1). The most common alleles at each locus in the ST-11 complex were opaA244 (8/10 isolates), opaD132 (9/10 isolates), and opaJ317 (6/10 isolates). Two further isolates possessed genes encoding related Opa proteins, which varied in their HV1 regions from HV1:19-1 to HV1:19-11. At the opaB locus, the most common allele was opaB34 (3/10 isolates), with another combination (SV:4-3, HV1:5-5, HV2:8C-1, which was encoded by two alleles, opa244 and opa339) present in three other isolates. The most common alleles in the ST-32 complex were opaA96 (9/10 isolates), opaB185 (6/10 isolates), opaD147 (8/10 isolates), and opaJ218 (9/10 isolates). The opaA and opaJ loci of all ST-32 complex isolates were similar, with identical HV1 regions and similar HV2 regions.
TABLE 1.
opa gene and Opa protein repertoires of 77 hyperinvasive meningococci
| Group and isolate | Year, country of isolation | Phenotype (serogroup: serotype: serosubtype)a | ST | opaA locusb | opaB locusb | opaD locusb | opaJ locusb | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | SV | HV1 | HV2 | Allele | SV | HV1 | HV2 | Allele | SV | HV1 | HV2 | Allele | SV | HV1 | HV2 | ||||
| ST-1 complex/ subgroup I/II | |||||||||||||||||||
| 6748 | 1971, Canada | A:4,21:P1.3,6 | 1 | 243 | 2-1 | 1-1 | 9-3 | 326 | 2-2 | 18-7 | 14-1 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| 20 | 1963, Niger | A:4,21:P1.10 | 1 | 252 | 4-2 | 13-1 | 17-7 | 340 | 2-2 | 1A-1 | 8A-1 | 145 | 4-2 | 12-2 | 21-1 | 345 | 2-1 | 11-1 | 1-10 |
| 254 | 1966, Djibouti | A:4,21:P1.10 | 1 | 252 | 4-2 | 13-1 | 17-7 | 333 | 4-2 | 13-1 | 1-10 | 145 | 4-2 | 12-2 | 21-1 | 349 | 4-2 | 1A-1 | 8A-8 |
| 129 | 1964, West Germany | A:4,21:P1.10 | 1 | 250 | 2-1 | 1-4 | 9A-1 | 304 | 2-1 | 1-4 | 9-7 | 311 | 2-2 | 3-8 | 9-4 | 325 | 2-1 | 11-1 | 1-4 |
| 371 | 1980, India | A:4,21:P1.10 | 1 | 243 | 2-1 | 1-1 | 9-3 | 326 | 2-2 | 18-7 | 14-1 | 344 | 2-2 | 18-7 | 14-1 | 346 | 2-2 | 11-1 | 1-4 |
| 139M | 1968, Philippines | A:— | 1 | 96 | 2-1 | 19-10 | 14-3 | 326 | 2-2 | 18-7 | 14-1 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| 120M | 1967, Pakistan | A:4,21:P1.10 | 1 | 249 | 4-2 | 1-1 | 9-3 | 326 | 2-2 | 18-7 | 14-1 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| S5611 | 1977, Australia | A:— | 1 | 34 | 4-3 | 18-3 | 14-1 | 298 | 4-2 | 3-2 | 19-2 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| 106 | 1967, Morocco | A:4,21:P1.10 | 1 | 243 | 2-1 | 1-1 | 9-3 | 326 | 2-2 | 18-7 | 14-1 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| 393 | 1968, Greece | A:— | 1 | 243 | 2-1 | 1-1 | 9-3 | 326 | 2-2 | 18-7 | 14-1 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| 322/85 | 1985, East Germany | A:4,21:P1.10 | 2 | 255 | 4-1 | 1A-1 | 8A-6 | 14 | 2-2 | 18-1 | 15-2 | 243 | 2-1 | 1-1 | 9-3 | 208 | 2-2 | 3-2 | 8-A3 |
| 79128 | 1979, China | A:— | 3 | 248 | 4-3 | 16-2 | 5-13 | 327 | 2-2 | 5-8 | 9-6 | 248 | 4-3 | 16-2 | 5-13 | 346 | 2-2 | 11-1 | 1-4 |
| BZ133 | 1977, The Netherlands | B:NT:— | 1 | 243 | 2-1 | 1-1 | 9-3 | 326 | 2-2 | 18-7 | 14-1 | 77 | 2-2 | 1B-4 | 6-1 | 325 | 2-1 | 11-1 | 1-4 |
| 79126 | 1979, China | A:4:P1.7 | 3 | 248 | 4-3 | 16-2 | 5-13 | 341 | 2-1 | 1B-4 | 6-3 | 248 | 4-3 | 16-2 | 5-13 | 346 | 2-2 | 11-1 | 1-4 |
| ST-4 complex/ subgroup IV | |||||||||||||||||||
| A4/M1027 | 1937, United States | A:4,21:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 95 | 2-1 | 1-1 | 9-3 | 310 | 2-1 | 17-3 | 17-8 | 242 | 2-1 | 2-3 | 1-8 |
| 26 | 1963, Niger | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 297 | 2-2 | 11-2 | 9B-1 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| 243 | 1966, Cameroon | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| 2059001 | 1990, Mali | A:4,21:P1.7 | 4 | 242 | 2-1 | 2-3 | 1-8 | 278 | 2-1 | 2-3 | 9B-1 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| 10 | 1963, Burkina Faso | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| 255 | 1966, Burkina Faso | A:4,21:P1.7 | 4 | 242 | 2-1 | 2-3 | 1-8 | 297 | 2-2 | 11-2 | 9B-1 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| S3131 | 1973, Ghana | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 297 | 2-2 | 11-2 | 9B-1 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| 690 | 1980, India | A:4,21:P1.7 | 4 | 254 | 2-1 | 17-3 | 1-8 | 278 | 2-1 | 2-3 | 9B-1 | 315 | 2-1 | 17-3 | 9B-1 | NP | NP | NP | NP |
| C751 | 1983, Gambia | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 278 | 2-1 | 2-3 | 9B-1 | 315 | 2-1 | 17-3 | 9B-1 | NP | NP | NP | NP |
| 1014 | 1985, Sudan | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 328 | 2-2 | 11-2 | 9B-1 | 296 | 2-1 | 17-3 | 17-8 | NP | NP | NP | NP |
| D8 | 1990, Mali | A:— | 4 | 242 | 2-1 | 2-3 | 1-8 | 278 | 2-1 | 2-3 | 9B-1 | 309 | 2-1 | 17-3 | 9B-1 | NP | NP | NP | NP |
| ST-5 complex/ subgroup III | |||||||||||||||||||
| IAL2229 | 1976, Brazil | A:— | 5 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 153 | 1966, China | A:4,21:P1.9 | 5 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 301 | 4-3 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 154 | 1966, China | A:4,21:P1.9 | 6 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 14/1455 | 1970, USSR | A:4,21:P1.20,9 | 5 | 242 | 2-1 | 2-3 | 1-8 | 127 | 4-3 | 3-6 | 1-1 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| S4355 | 1974, Denmark | A:— | 5 | 253 | 2-10 | 1-3 | 8A-5 | 253 | 2-10 | 1-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 7891 | 1975, Finland | A:4,21:P1.9 | 5 | 242 | 2-1 | 2-3 | 1-8 | 296 | 2-1 | 17-3 | 17-8 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| F4698 | 1987, Saudi Arabia | A:— | 5 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 299 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| H1964 | 1987, United Kingdom | A:— | 5 | 242 | 2-1 | 2-3 | 1-8 | 281 | 2-1 | 6-1 | 9-2 | 299 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| F6124 | 1988, Chad | A:— | 5 | 242 | 2-1 | 2-3 | 1-8 | 281 | 2-1 | 6-1 | 9-2 | 299 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 92001 | 1992, China | A:— | 7 | 242 | 2-1 | 2-3 | 1-8 | 253 | 2-10 | 1-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 11-004 | 1984, China | A:— | 5 | 242 | 2-1 | 2-3 | 1-8 | 329 | 2-1 | 2-3 | 8A-5 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| 80049 | 1963, China | A:4:P1.10 | 5 | 242 | 2-1 | 2-3 | 1-8 | 342 | 2-2 | 5-1 | 8C-2 | 296 | 2-1 | 17-3 | 17-8 | 127 | 4-3 | 3-6 | 1-1 |
| ST-8 complex/ cluster A4 | |||||||||||||||||||
| BZ 10 | 1967, The Netherlands | B:2b:P1.2 | 8 | 246 | 2-2 | 15-3 | 12-2 | 277 | 2-2 | 15-3 | 12-2 | 161 | 2-2 | 11-1 | 1-4 | 257 | 4-2 | 15-3 | 12-2 |
| B6116/77 | 1977, Iceland | B:— | 10 | 258 | 4-3 | 19-4 | 14-2 | 277 | 2-2 | 15-3 | 12-2 | 161 | 2-2 | 11-1 | 1-4 | 258 | 4-3 | 19-4 | 14-2 |
| BZ 163 | 1979, The Netherlands | B:2b:P1.16 | 9 | 256 | 2-2 | 15-3 | 12-2 | 343 | 2-2 | 3-2 | 8A-8 | 161 | 2-2 | 11-1 | 1-4 | 256 | 2-2 | 15-3 | 12-2 |
| G2136 | 1986, England | B:— | 8 | 257 | 4-2 | 15-3 | 12-2 | 285 | 2-1 | 19-3 | 2-1 | 161 | 2-2 | 11-1 | 1-4 | 257 | 4-2 | 15-3 | 12-2 |
| SB25 | 1990, South Africa | C:— | 8 | 246 | 2-2 | 15-3 | 12-2 | 246 | 2-2 | 15-3 | 12-2 | 161 | 2-2 | 11-1 | 1-4 | 246 | 2-2 | 15-3 | 12-2 |
| AK22 | 1992, Greece | B:— | 8 | 260 | 2-2 | 3-2 | 6-2 | 294 | 2-2 | 3-2 | 12-2 | 161 | 2-2 | 11-1 | 1-4 | 323 | 4-2 | 3-2 | 12-2 |
| 94/155 | 1994, New Zealand | C:— | 66 | 246 | 2-2 | 15-3 | 12-2 | 292 | 2-1 | 5-2 | 18-1 | 161 | 2-2 | 11-1 | 1-4 | 246 | 2-2 | 15-3 | 12-2 |
| 312 901 | 1996, England | C:— | 8 | 259 | 2-2 | 5-2 | 18-1 | 292 | 2-1 | 5-2 | 18-1 | 161 | 2-2 | 11-1 | 1-4 | 292 | 2-1 | 5-2 | 18-1 |
| ST-11 complex/ ET-37 complex | |||||||||||||||||||
| 38VI | 1964, United States | B:—:P1.5,2 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 34 | 4-3 | 18-3 | 14-1 | 132 | 2-2 | 11-2 | 1-6 | 317 | 2-2 | 19-1 | 14-1 |
| NG P20 | 1969, Norway | B:2a:P1.2 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 279 | 2-2 | 1A-5 | 9B-2 | 132 | 2-2 | 11-2 | 1-6 | 317 | 2-2 | 19-1 | 14-1 |
| F1576 | 1984, Ghana | C:2a:P1.5,2 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 305 | 4-3 | 5-10 | 9B-2 | 132 | 2-2 | 11-2 | 1-6 | 317 | 2-2 | 19-1 | 14-1 |
| 500 | 1984, Italy | C:2a | 11 | 244 | 4-3 | 5-5 | 8C-1 | 300 | 2-2 | 5-9 | 9B-2 | 132 | 2-2 | 11-2 | 1-6 | 317 | 2-2 | 19-1 | 14-1 |
| MA-5756 | 1985, Spain | C:2a:P1.5 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 339 | 4-3 | 5-5 | 8C-1 | 132 | 2-2 | 11-2 | 1-6 | 317 | 2-2 | 19-1 | 14-1 |
| M597 | 1988, Israel | C:2a:P1.5 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 339 | 4-3 | 5-5 | 8C-1 | 132 | 2-2 | 11-2 | 1-6 | 347 | 2-2 | 19-11 | 14-1 |
| D1 | 1989, Mali | C:2a:P1.5 | 11 | 132 | 2-2 | 11-2 | 1-6 | 34 | 4-3 | 18-3 | 14-1 | 132 | 2-2 | 11-2 | 1-6 | 317 | 2-2 | 19-1 | 14-1 |
| 90/18311 | 1990, Scotland | C:NT:P1.5 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 244 | 4-3 | 5-5 | 8C-1 | 132 | 2-2 | 11-2 | 1-6 | 347 | 2-2 | 19-11 | 14-1 |
| L93/4286 | 1993, England | C:— | 11 | 132 | 2-2 | 11-2 | 1-6 | 295 | 4-3 | 10-3 | 10-5 | 132 | 2-2 | 11-2 | 1-6 | 34 | 4-3 | 18-3 | 14-1 |
| BRAZ10 | 1976, Brazil | C:2a:P1.5 | 11 | 244 | 4-3 | 5-5 | 8C-1 | 34 | 4-3 | 18-3 | 14-1 | 316 | 4-2 | 12B-1 | 1-9 | 348 | 2-2 | 3-6 | 1-1 |
| ST-32 complex/ ET-5 complex | |||||||||||||||||||
| 8680 | 1987, Chile | B:15:P1.3 | 32 | 96 | 2-1 | 19-10 | 14-3 | 185 | 5-1 | 10-5 | 3-1 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| BZ 83 | 1984, The Netherlands | B:NT | 34 | 96 | 2-1 | 19-10 | 14-3 | 335 | 2-1 | 10-5 | 3-1 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| 204/92 | 1992, Cuba | B:— | 33 | 261 | 2-2 | 19-7 | 14-3 | 185 | 5-1 | 10-5 | 3-1 | 147 | 2-2 | 1A-2 | 8-1 | 322 | 2-2 | 19-7 | 14-5 |
| EG 329 | 1985, East Germany | B:15:P1.16 | 32 | 96 | 2-1 | 19-10 | 14-3 | 185 | 5-1 | 10-5 | 3-1 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| NG 80 | 1981, Norway | B:15:P1.16 | 32 | 96 | 2-1 | 19-10 | 14-3 | 288 | 5-1 | 1A-2 | 8-1 | 71 | 4-2 | 19-7 | 4-1 | 218 | 2-1 | 19-10 | 14-5 |
| BZ 169 | 1985, The Netherlands | B:NT:P1.16 | 32 | 96 | 2-1 | 19-10 | 14-3 | 185 | 5-1 | 10-5 | 3-1 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| 44/76 | 1976, Norway | B:— | 32 | 96 | 2-1 | 19-10 | 14-3 | 288 | 5-1 | 1A-2 | 8-1 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| NG144/82 | 1982, Norway | B:15:P1.16 | 32 | 96 | 2-1 | 19-10 | 14-3 | 313 | 4-2 | 7-1 | 5-11 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| 196/87 | 1987, Norway | C:15 | 32 | 96 | 2-1 | 19-10 | 14-3 | 185 | 5-1 | 10-5 | 3-1 | 147 | 2-2 | 1A-2 | 8-1 | 218 | 2-1 | 19-10 | 14-5 |
| NG PB24 | 1985, Norway | B:NT:P1.16 | 32 | 96 | 2-1 | 19-10 | 14-3 | 185 | 5-1 | 10-5 | 3-1 | 185 | 5-1 | 10-5 | 3-1 | 218 | 2-1 | 19-10 | 14-5 |
| ST-41/44 complex/ lineage 3 | |||||||||||||||||||
| 931905 | 1993, The Netherlands | B:— | 41 | 265 | 3-2 | 7-1 | 5-9 | 265 | 3-2 | 7-1 | 5-9 | 265 | 3-2 | 7-1 | 5-9 | 213 | 2-1 | 3-6 | 1-1 |
| 50/94 | 1994, Norway | B:— | 45 | 245 | 2-1 | 1-5 | 9A-2 | 293 | 2-1 | 4-2 | 7-3 | 245 | 2-1 | 1-5 | 9A-2 | 213 | 2-1 | 3-6 | 1-1 |
| 88/03415 | 1988, Scotland | B:— | 46 | 245 | 2-1 | 1-5 | 9A-2 | 201 | 2-2 | 4-1 | 7-3 | 265 | 3-2 | 7-1 | 5-9 | 320 | 4-2 | 11-1 | 1-4 |
| 91/40 | 1991, New Zealand | B:4:P1.4 | 42 | 201 | 2-2 | 4-1 | 7-3 | 201 | 2-2 | 4-1 | 7-3 | 308 | 3-2 | 7-1 | 5-9 | 213 | 2-1 | 3-6 | 1-1 |
| AK50 | 1992, Greece | B:— | 41 | 262 | 2-1 | 7-1 | 5-9 | 201 | 2-2 | 4-1 | 7-3 | 303 | 3-2 | 1B-7 | 5-9 | 213 | 2-1 | 3-6 | 1-1 |
| BZ198 | 1986, The Netherlands | B:NT | 41 | 245 | 2-1 | 1-5 | 9A-2 | 264 | 2-1 | 1B-4 | 6-1 | 265 | 3-2 | 7-1 | 5-9 | 213 | 2-1 | 3-6 | 1-1 |
| M-101/93 | 1993, Iceland | B:— | 41 | 263 | 2-2 | 12-1 | 4B-5 | 265 | 3-2 | 7-1 | 5-9 | 213 | 2-1 | 3-6 | 1-1 | 213 | 2-1 | 3-6 | 1-1 |
| M40/94 | 1994, Chile | B:— | 41 | 264 | 2-1 | 1B-4 | 6-1 | 265 | 3-2 | 7-1 | 5-9 | 265 | 3-2 | 7-1 | 5-9 | 132 | 2-2 | 11-2 | 1-6 |
| 400 | 1991, Austria | B:— | 40 | 245 | 2-1 | 1-5 | 9A-2 | 201 | 2-2 | 4-1 | 7-3 | 201 | 2-2 | 4-1 | 7-3 | 213 | 2-1 | 3-6 | 1-1 |
| NG E30 | 1988, Norway | B:4:P1.16 | 44 | 24 | 4-3 | 19-10 | 11-2 | 332 | 2-1 | 3-9 | 8-4 | 24 | 4-3 | 19-10 | 11-2 | 213 | 2-1 | 3-6 | 1-1 |
| NG H15 | 1988, Norway | B:8:P1.15 | 43 | 471 | 2-1 | 4-2 | 7-3 | 142 | 4-6 | 12A-1 | 20-1 | 248 | 4-3 | 16-2 | 5-13 | 346 | 2-2 | 11-1 | 1-4 |
| NG H36 | 1988, Norway | B:8:P1.2 | 47 | 24 | 4-3 | 19-10 | 11-2 | 313 | 4-2 | 7-1 | 5-11 | 313 | 4-2 | 7-1 | 5-11 | 213 | 2-1 | 3-6 | 1-1 |
In the ST-4 complex, the predominant alleles were opaA242 (10/11 isolates) and opaD296 (7/11 isolates). In this clonal complex, the opaD310 allele, which is related to opaD296, was present in one other isolate, and the combination SV:2-1, HV1:17-3, HV2:9B-1, encoded by two alleles, opa309 and opa315, was present at this locus in the remaining three isolates. The opaB278 allele was present at the opaB locus in 4 of 11 isolates, and the gene at the opaJ locus was absent in all isolates except for A4/M1027, in which the opaJ242 allele (identical to opaA242 in this clonal complex) was observed. The opaA and opaD loci of the ST-5 complex were similar to those of the ST-4 complex, with opaA242 observed in 11 of 12 isolates and opaD296 observed in 8 of 12 isolates, and all but one isolate (isolate 153; SV:4-3) had the combination SV:2-1, HV1:17-3, HV2:17-8 at the opaD locus. The opaB253 allele was present in 6 of 12 ST-5 complex isolates, and the opaJ127 allele was present in all isolates.
In the ST-8 complex, the HV1:15-3, HV2:12-2 combination predominated at 13 of the 32 loci examined and was present at three of the four opa loci, including opaA (five of eight isolates), opaB (three of eight isolates), and opaJ (five of eight isolates). This combination was encoded by four alleles (opa246, opa256, opa257, and opa277). For three of these alleles, the SV:2-2 variant was present, accounting for 10 of the 13 loci, whereas three other isolates had the SV:4-2 variant encoded by opa257. At the opaD locus, opaD161 was observed in all isolates.
The predominant allele at each locus for the ST-1 complex was opaA243 (5 of 14 isolates), with another isolate, 120 M, sharing an identical HV1/HV2 combination. The opaB326 (7 of 14 isolates), opaD77 (7 of 14 isolates), and opaJ325 (8 of 14 isolates) alleles were also common. Four other isolates shared similar opaJ variable regions with opaJ325: isolate 20 shared an SV/HV1 combination, and isolates 79128, 79126, and 371 shared HV1 and HV2 combinations.
In the ST-41/44 complex, opaA245 (4 of 12 isolates) and opaB201 (4 of 12 isolates) were the most common alleles, with another isolate, 50/94, sharing an identical HV2 region to that encoded by opaB201. The opa265 allele was present at the opaB locus in another 3 isolates, whereas this allele was also observed at the opaD locus in 4 of 12 isolates, with another isolate (91/40) sharing identical variable region variants. In contrast to the diversity at the other three loci, the opaJ213 allele was present in the majority (9/12) of ST-41/44 complex isolates.
Recombinational reassortment of opa gene sequences.
Evidence for recombinational reassortment of opa sequences, including gene mosaic structures and whole gene conversion events, was present throughout the data set. Examples where common descent could not be differentiated from recombinational replacement as an explanation for the possession of identical sequences included the similarity of the opaA and opaD loci in isolates belonging to the ST-4 and ST-5 complexes; the identical HV1:3-6, HV2:1-1 combination at the opaJ loci of most ST-5 and ST-41/44 complex isolates; the presence of the HV2:14-1 variant at the opaB locus in the majority of ST-1 and ST-11 complex isolates; and the identical hypervariable region variant combination (HV1:11-1, HV2:1-4) at the opaD locus in all ST-8 complex isolates and at the opaJ locus in most ST-1 complex isolates.
Further support for the effect of recombination in shaping the Opa repertoire was provided by isolates belonging to the ST-8 and ST-41/44 complexes, in which high allelic diversity was observed at each locus but identical alleles, candidate whole gene conversion events, or identical hypervariable regions were present at multiple loci. Evidence of frequent genetic exchange was observed among the opaA, opaB, and opaJ loci of members of the ST-8 complex and among the opaA and opaJ loci of members of the ST-32 complex. For the ST-41/44 complex, the effect of recombination was also observed with the exchange, possibly by whole gene conversion, of the opa201 and opa265 alleles among the opaA, opaB, and opaD loci. A possible exchange of identical sequences among clonal complexes was also observed, for example, in the presence of the opa132 allele at the opaD loci of the majority of ST-11 complex isolates and also in the opaJ locus of isolate M40/94 of the ST-41/44 complex. Further examples include the shared identical HV1:3-6/HV2:1-1 combination belonging to the ST-41/44 and ST-5 complexes, which was also found in isolate BRAZ10, belonging to the ST-11 complex, and the opaB253 allele, which is present in members of the ST-4 and ST-5 complexes.
Temporal and geographic distribution of Opa protein variants and combinations of variants.
Both temporal and geographic structuring of the Opa protein repertoire was apparent. For example, the variable region combination SV:2-1, HV1:2-3, HV2:1-8 was encoded by the opaA gene in most (21/23) isolates belonging to the ST-4 and ST-5 complexes, while the combination SV:2-1, HV1:17-3, HV2:17-8 was present at the opaD locus in 19 of 23 isolates. These combinations persisted for at least 55 years during spread over five continents. The opaJ127 allele in the ST-5 complex also persisted for 29 years over four continents. For the ST-11 complex, the combination of the opaA244 and opaD132 alleles persisted over 4 decades and four continents. The opa317 allele at the opaJ locus and the opa34 allele at the opaB locus persisted from 1964 to 1989. Similar structuring was apparent at the opaA (opa96), opaD (opaD147), and opaJ (opa218) loci in ST-32 complex isolates, and this combination of alleles persisted for 11 years after this complex first caused an outbreak in 1976 which spread between Europe and South America. The opaD161 allele was associated with the ST-8 complex for 29 years over three continents, despite the allelic variability at other loci in this clonal complex, at which the HV1:15-3, HV2:12-2 combination had also persisted for 24 years across the same three continents. For the ST-1 complex, temporal structuring was apparent for all loci, but the duration of persistence of alleles was shorter than in the cases described above. For example, the opaA243, opaB326, and opaJ325 alleles were observed in isolates spanning periods of at least 13 years and four or five continents each, whereas opaD77 was present in isolates collected over a period of 10 years from five continents. The earliest isolate belonging to the ST-41/44 complex in the data set was collected in 1986, and between then and 1994, the opaA245, opaD265, and opaJ213 alleles persisted on multiple continents.
Phylogenetic analysis of hyperinvasive meningococcal Opa protein repertoires.
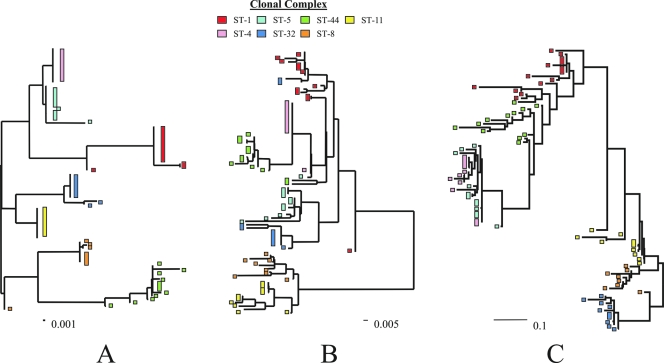
The sequences corresponding to the opa repertoire of each isolate (all opa alleles present) were concatenated, translated, and aligned manually (nucleotide p distance, 13.1%); this alignment was then analyzed to construct an ML tree (Fig. 2). The ST-4 and ST-5 complexes formed a single cluster on the tree, consistent with their similar Opa repertoires, which varied only in the opaB allele and the opaJ locus (the gene was absent from ST-4 complex subgroup IV-1 isolate A4/M1027). The presence of the shared opaB253 allele caused the ST-4 complex isolates 10 and 243 to cluster among ST-5 complex isolates. Isolate S4355, belonging to the ST-5 complex, appeared to cluster away from related isolates due to the presence of the opaA253 allele, potentially due to a recombination event involving the identical allele at the opaB locus. Isolates belonging to the ST-11 and ST-32 complexes and all but one isolate belonging to the ST-8 complex formed individual clusters, as did most (9/12) isolates belonging to the ST-41/44 complex. Isolate B6116/77 differed from other ST-8 complex isolates at the opaA and opaJ loci, with a single, more divergent allele present at both loci. Isolates NG E30 and NG H36 differed from other members of the ST-41/44 complex by their possession of a common opaA24 allele. Isolate NG H15 appeared among a group of ST-1 complex isolates due to their common opaD and opaJ loci, which are distinct from those of other members of each complex. The remaining (8/14) ST-1 complex isolates formed a single cluster. The phylogeny of concatenated opa sequences was consistent with the ML phylogenies reconstructed for concatenated MLST alleles and concatenated antigens (porA, porB, and fetA) (48).
FIG. 2.
Maximum likelihood phylogenies of concatenated MLST alleles (A), porA, porB, and fetA alleles (B), and opa sequences (C). Based on concatenated antigen allele sequences alone (phylogenies B and C), isolates clustered into genetically related groups, consistent with a phylogeny reconstructed from concatenated MLST alleles (phylogeny A).
DISCUSSION
The diversity of meningococcal Opa proteins has hampered the elucidation of the roles of individual variants and combinations of variants in both asymptomatic colonization and the development of disease. In the present study, a total of 90 opa alleles encoding 83 Opa protein variants were identified in 308 loci from 77 meningococcal isolates examined, representing seven hyperinvasive lineages. In addition to confirming (33) and defining the great diversity in these proteins, the data provided evidence for extensive structuring of this diversity within meningococcal populations.
Comparison of the ML phylogenies for concatenated opa genes with equivalent phylogenies generated from either concatenated MLST genes or three concatenated variable outer membrane protein genes (porA, porB, and fetA) showed that the opa gene repertoire reflected both clonal complex structure and structuring in the repertoires of other antigens. In addition, in isolates belonging to the same clonal complex, particular alleles were consistently found at each of the four opa loci. In most clonal complexes, these variants were consistently observed at the same locus during global spread, often spanning decades, indicating that particular meningococcal genotypes encode characteristic Opa repertoires. These observations imply the existence of a mechanism that operates to stabilize Opa repertoires in meningococci.
Several mechanisms can explain the genetic structuring in bacterial population data. The simplest of these is descent, with the asexual nature of bacterial reproduction inevitably leading to a clonal population structure characterized by linkage disequilibrium and a tree-like phylogeny. This mechanism operates in the absence of frequent horizontal genetic exchange and therefore is unlikely to be the explanation for the structuring of the Opa repertoire in the meningococcus, which exhibits high levels of horizontal genetic exchange and a nonclonal population structure (15, 16, 28). Furthermore, there was extensive evidence of horizontal genetic exchange in the opa data, including the distribution of identical alleles among diverse meningococci. This, combined with the strong diversifying selection that acts on these loci, would tend to randomize opa repertoires. The epidemic clone model postulates that a degree of clonal structure may occur in recombining populations as a consequence of the short-term expansion of particular genotypes, such that individual clones arise from time to time and come to dominate in the population (34); however, this is not a likely explanation for Opa repertoire structuring, as the isolates investigated here were collected over several decades and from widely separated geographic areas (31).
The association of particular Opa repertoires with individual clonal complexes over decades of global spread could be maintained if the particular combinations of Opa proteins confer fitness advantages for transmission from host to host (58). Such a model could also accommodate minor variations: such variants could be a consequence of sequence variation that arises in individual hosts, conferring a fitness advantage within that host as a consequence of immune evasion but resulting in less fitness for transmission among hosts.
The biology of the Opa proteins suggests two mechanisms that might stabilize repertoires by introducing fitness differences among meningococci. These mechanisms could act separately or in combination. First, Opa proteins are important surface antigens that are exposed to the immune system, and it has been shown that host immune responses can shape antigenic repertoires at the population level, even in recombining populations. Simulations have shown that the host immune response can act at the population level to structure pathogen populations into nonoverlapping antigen islands, where pathogen variants are diverse and do not share individual variants at different loci. Such structures have been observed for other meningococcal antigens, in addition to that reported here for the Opa proteins (20).
The second mechanism stems from the role of Opa proteins as mediators of bacterium-host cell interactions in that sequence variation among opa variants is likely to lead to functional variation among Opa proteins, resulting in differences in, for example, adhesion to host receptors (Table 2). In experiments performed in vitro, most Opa proteins tested bind to CEACAM1 (38, 52, 53), and sequence differences in their three variable regions determine their specificities for other receptors (4, 5, 14, 17, 19, 35, 36, 38, 51). Conservation of the Opa repertoire may therefore be a consequence of conservation of functionally important combinations of Opa proteins, with particular combinations conferring different effects on transmission fitness through alternative affinities and avidities for different receptors.
TABLE 2.
Nomenclature of meningococcal Opa protein variants with known receptor specificitiesa
| Protein | Isolate | Reference | GenBank accession no. | Opa variable region | Interaction with CEACAM familyd | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| SV | HV1 | HV2 | CEACAM1 | CEACAM3 | CEACAM5 | CEACAM6 | ||||
| OpaA127 | H44/76 | 14 | AF016286 | 2-1 | 19-10 | 14-3 | Y | N | N | N |
| OpaB128 | H44/76 | 14 | AF016289 | 5-1 | 1A-2 | 8-1 | Y | N | Y | N |
| OpaD126b | H44/76 | 14 | NAc | 2-2 | 1A-2 | 8-1 | Y | N | Y | N |
| OpaJ129 | H44/76 | 14 | AF016285 | 2-1 | 19-10 | 14-5 | Y | N | N | N |
| OpaA132 | 00170/F6124 | 38 | AF001180 | 2-1 | 2-3 | 1-8 | Y | N | Y | Y |
| OpaB94 | F6124 | 38 | AF001201 | 2-1 | 6-1 | 9-2 | Y | N | Y | N |
| OpaD100 | 00170/F6124 | 38 | AF001195 | 2-1 | 17-3 | 17-8 | Y | N | N | N |
| OpaJ101 | 00170/F6124 | 38 | AF001179 | 4-3 | 3-6 | 1-1 | N | N | N | N |
| OpaB92 | 00170 | 38 | AF001199 | 2-10 | 1-3 | 8A-5 | Y | N | Y | N |
| C1938-1 | C1938 | 38 | NA | NA | NA | NA | N | N | Y | N |
| 41 | C1938 | 38 | X06445 | 4-3 | 19-4 | 14-2 | Y | N | N | N |
| 40 | C1938 | 38 | X06446 | 2-1 | 11-2 | NA | Y | N | Y | N |
| C1938-4 | C1938 | 38 | NA | NA | NA | NA | N | N | N | N |
| OpaA | C751 | 51-53 | U03405 | 2-1 | 2-3 | 1-8 | Ye | N | Y | Y |
| OpaB | C751 | 51-53 | U03406 | 2-1 | 2-3 | 9B-1 | Ye | Y | Y | Y |
| OpaD | C751 | 51-53 | U03404 | 2-1 | 17-3 | 9B-1 | Ye | N | Y | N |
The evolution of the Opa repertoire in Neisseria species may be an example of protein promiscuity (1, 25), in which a genetic duplication event is followed by functional specialization of the duplicated gene product with an alternative substrate while maintaining an affinity for the original substrate. In the case of Opa proteins, an evolutionary requirement for multiple receptor specificities could explain the differences in receptor specificities of individual variants but the maintenance of CEACAM1 binding. Furthermore, this concept might explain the presence of multiple opa loci in the genomes of meningococci and other Neisseria species (3, 43, 46, 54).
Multiple mechanisms could act in concert, reconciling the need for nonoverlapping antigenicity with functional promiscuity, with both of these requirements enhanced by the phase variability of opa genes. The extent to which the Opa repertoire exerts effects on the fitness of individual clonal complexes has yet to be determined, however, and will require elucidation of the functions of individual variants and of clonal complex-associated combinations.
Whatever the mechanism leading to the structuring of the Opa repertoire, the observed repertoire structure has a number of implications for investigations of Opa function. First, notwithstanding the great diversity in Opa proteins, the number of Opa variants requiring investigation in a given clonal complex is limited. Second, a quarter of the total number of loci examined contained alleles encoding the HV2:1 sequence homology family (the HV1:11 and HV2:1 family combination was the most predominant). Alleles encoding this HV2 family were widespread and predominated in at least one locus in all clonal complexes except the ST-32 complex. These data may reflect conservation of an advantageous functional ability.
Although the genetic traits defining the differing pathogenicities among meningococcal clonal complexes remain to be completely elucidated, differences in their Opa repertoires are potentially important. A number of examples of associations between individual genetic traits and particular clonal complexes have been reported, including differently distributed restriction-modification systems, plasmids (11, 12), a disease-associated prophage, and the OpcA adhesin (41, 57). As with the Opa proteins, particular variant combinations of the porin proteins PorA and PorB and the iron transport protein FetA are associated with given hyperinvasive lineages during global epidemic spread over periods measured in decades. Such structuring has been proposed as the basis for future meningococcal vaccines (48). The conserved antigenicity of individual clonal complexes is also being exploited in the development of “tailor-made” outer membrane vesicle vaccines. Together with these studies, the investigation of the Opa repertoire described here further demonstrates the value of population-based analyses in the generation of novel prophylactic strategies for antigenically diverse pathogens.
Supplementary Material
[Supplemental material]
Acknowledgments
We gratefully acknowledge Ian Feavers for the provision of meningococcal isolates, the University of Oxford Department of Zoology sequencing service for separation of cycle sequencing reaction products, Rachel Urwin and Edward Holmes for assistance with phylogenetic analysis, and Man-Suen Chan for sequence analysis scripts. This study made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley et al. (26) and sited at the University of Oxford.
The development of the Neisseria Multi Locus Sequence Typing website was funded by the Wellcome Trust and the European Union. M.C.J.M. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences. This study was funded by a Wellcome Prize Ph.D. studentship awarded to M.C.J.M. for M.J.C.
Footnotes
REFERENCES
- 1.Aharoni, A., L. Gaidukov, O. Khersonsky, S. McQ Gould, C. Roodveldt, and D. S. Tawfik. 2005. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37**:**73-76. [DOI] [PubMed] [Google Scholar]
- 2.Belland, R. J., T. Chen, J. Swanson, and S. H. Fischer. 1992. Human neutrophil response to recombinant neisserial Opa proteins. Mol. Microbiol. 6**:**1729-1737. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, K. S., C. P. Gibbs, O. Barrera, S. G. Morrison, F. Jahnig, A. Stern, E. M. Kupsch, T. F. Meyer, and J. Swanson. 1991. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 5**:**1889-1901. [DOI] [PubMed] [Google Scholar]
- 4.Bos, M. P., F. Grunert, and R. J. Belland. 1997. Differential recognition of members of the carcinoembryonic antigen family by Opa variants of Neisseria gonorrhoeae. Infect. Immun. 65**:**2353-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos, M. P., D. Kao, D. M. Hogan, C. C. Grant, and R. J. Belland. 2002. Carcinoembryonic antigen family receptor recognition by gonococcal Opa proteins requires distinct combinations of hypervariable Opa protein domains. Infect. Immun. 70**:**1715-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulton, I. C., and S. D. Gray-Owen. 2002. Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nat. Immunol. 3**:**229-236. [DOI] [PubMed] [Google Scholar]
- 7.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106**:**505-525. [PubMed] [Google Scholar]
- 8.Caugant, D. A., K. Bovre, P. Gaustad, K. Bryn, E. Holten, E. A. Høiby, and L. O. Frøholm. 1986. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J. Gen. Microbiol. 132**:**641-652. [DOI] [PubMed] [Google Scholar]
- 9.Chen, T., F. Grunert, A. Medina-Marino, and E. C. Gotschlich. 1997. Several carcinoembryonic antigens (CD66) serve as receptors for gonococcal opacity proteins. J. Exp. Med. 185**:**1557-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, T., J. Swanson, J. Wilson, and R. J. Belland. 1995. Heparin protects Opa+ Neisseria gonorrhoeae from the bactericidal action of normal human serum. Infect. Immun. 63**:**1790-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claus, H., A. Friedrich, M. Frosch, and U. Vogel. 2000. Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J. Bacteriol. 182**:**1296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claus, H., J. Stoevesandt, M. Frosch, and U. Vogel. 2001. Genetic isolation of meningococci of the electrophoretic type 37 complex. J. Bacteriol. 183**:**2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jonge, M. I., M. P. Bos, H. J. Hamstra, W. Jiskoot, P. van Ulsen, J. Tommassen, L. van Alphen, and P. van der Ley. 2002. Conformational analysis of opacity proteins from Neisseria meningitidis. Eur. J. Biochem. 269**:**5215-5223. [DOI] [PubMed] [Google Scholar]
- 14.de Jonge, M. I., H. J. Hamstra, L. van Alphen, J. Dankert, and P. van der Ley. 2003. Mapping the binding domains on meningococcal Opa proteins for CEACAM1 and CEA receptors. Mol. Microbiol. 50**:**1005-1015. [DOI] [PubMed] [Google Scholar]
- 15.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98**:**182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feil, E. J., M. C. J. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16**:**1496-1502. [DOI] [PubMed] [Google Scholar]
- 17.Grant, C. C., M. P. Bos, and R. J. Belland. 1999. Proteoglycan receptor binding by Neisseria gonorrhoeae MS11 is determined by the HV-1 region of OpaA. Mol. Microbiol. 32**:**233-242. [DOI] [PubMed] [Google Scholar]
- 18.Gray-Owen, S. D., C. Dehio, A. Haude, F. Grunert, and T. F. Meyer. 1997. CD66 carcinoembryonic antigens mediate interactions between Opa-expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J. 16**:**3435-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray-Owen, S. D., D. R. Lorenzen, A. Haude, T. F. Meyer, and C. Dehio. 1997. Differential Opa specificities for CD66 receptors influence tissue interactions and cellular responses to Neisseria gonorrhoeae. Mol. Microbiol. 26**:**971-980. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, S., M. C. J. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2**:**437-442. [DOI] [PubMed] [Google Scholar]
- 21.Hauck, C. R., and T. F. Meyer. 2003. ‘Small’ talk: Opa proteins as mediators of Neisseria-host-cell communication. Curr. Opin. Microbiol. 6**:**43-49. [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock, P. J. 1989. Unified nomenclature for pathogenic Neisseria species. Clin. Microbiol. Rev. 2**:**S64-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs, M. M., B. Malorny, P. Prasad, G. Morelli, B. Kusecek, J. E. Heckels, J. G. Cannon, and M. Achtman. 1998. Recombinational reassortment among opa genes from ET-37 complex Neisseria meningitidis isolates of diverse geographical origins. Microbiology 144**:**157-166. [DOI] [PubMed] [Google Scholar]
- 24.Hobbs, M. M., A. Seiler, M. Achtman, and J. G. Cannon. 1994. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol. Microbiol. 12**:**171-180. [DOI] [PubMed] [Google Scholar]
- 25.James, L. C., and D. S. Tawfik. 2003. Conformational diversity and protein evolution—a 60-year-old hypothesis revisited. Trends Biochem. Sci. 28**:**361-368. [DOI] [PubMed] [Google Scholar]
- 26.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5**:**86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. J. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38**:**4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolley, K. A., D. J. Wilson, P. Kriz, G. McVean, and M. C. Maiden. 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22**:**562-569. [DOI] [PubMed] [Google Scholar]
- 29.Kawula, T. H., E. L. Aho, D. S. Barritt, D. G. Klapper, and J. G. Cannon. 1988. Reversible phase variation of expression of Neisseria meningitidis class 5 outer membrane proteins and their relationship to gonococcal proteins II. Infect. Immun. 56**:**380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5**:**150-163. [DOI] [PubMed] [Google Scholar]
- 31.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95**:**3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makepeace, B. L., P. J. Watt, J. E. Heckels, and M. Christodoulides. 2001. Interactions of Neisseria gonorrhoeae with mature human macrophage opacity proteins influence production of proinflammatory cytokines. Infect. Immun. 69**:**1909-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malorny, B., G. Morelli, B. Kusecek, J. Kolberg, and M. Achtman. 1998. Sequence diversity, predicted two-dimensional protein structure, and epitope mapping of neisserial Opa proteins. J. Bacteriol. 180**:**1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90**:**4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCaw, S. E., E. H. Liao, and S. D. Gray-Owen. 2004. Engulfment of Neisseria gonorrhoeae: revealing distinct processes of bacterial entry by individual carcinoembryonic antigen-related cellular adhesion molecule family receptors. Infect. Immun. 72**:**2742-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, J., S. E. Bailey, Z. Benmechernene, C. Tzitzilonis, N. J. Griffiths, M. Virji, and J. P. Derrick. 2005. Recognition of saccharides by the OpcA, OpaD, and OpaB outer membrane proteins from Neisseria meningitidis. J. Biol. Chem. 280**:**31489-31497. [DOI] [PubMed] [Google Scholar]
- 37.Morelli, G., B. Malorny, K. Muller, A. Seiler, J. F. Wang, J. del Valle, and M. Achtman. 1997. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol. Microbiol. 25**:**1047-1064. [DOI] [PubMed] [Google Scholar]
- 38.Muenzner, P., C. Dehio, T. Fujiwara, M. Achtman, T. F. Meyer, and S. D. Gray Owen. 2000. Carcinoembryonic antigen family receptor specificity of Neisseria meningitidis Opa variants influences adherence to and invasion of proinflammatory cytokine-activated endothelial cells. Infect. Immun. 68**:**3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404**:**502-506. [DOI] [PubMed] [Google Scholar]
- 40.Russell, J. E., K. A. Jolley, I. M. Feavers, M. C. J. Maiden, and J. Suker. 2004. PorA variable regions of Neisseria meningitidis. Emerg. Infect. Dis. 10**:**674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seiler, A., R. Reinhardt, J. Sarkari, D. A. Caugant, and M. Achtman. 1996. Allelic polymorphism and site-specific recombination in the opc locus of Neisseria meningitidis. Mol. Microbiol. 19**:**841-856. [DOI] [PubMed] [Google Scholar]
- 42.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5**:**233-241. [DOI] [PubMed] [Google Scholar]
- 43.Stern, A., and T. F. Meyer. 1987. Common mechanism controlling phase and antigenic variation in pathogenic neisseriae. Mol. Microbiol. 1**:**5-12. [DOI] [PubMed] [Google Scholar]
- 44.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94**:**271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287**:**1809-1815. [DOI] [PubMed] [Google Scholar]
- 46.Toleman, M., E. Aho, and M. Virji. 2001. Expression of pathogen-like Opa adhesins in commensal Neisseria: genetic and functional analysis. Cell. Microbiol. 3**:**33-44. [DOI] [PubMed] [Google Scholar]
- 47.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11**:**479-487. [DOI] [PubMed] [Google Scholar]
- 48.Urwin, R., J. E. Russell, E. A. Thompson, E. C. Holmes, I. M. Feavers, and M. C. Maiden. 2004. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect. Immun. 72**:**5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Putten, J. P., and S. M. Paul. 1995. Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 14**:**2144-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Putten, J. P. M., T. D. Duensing, and R. L. Cole. 1998. Entry of OpaA+ gonococci into HEp-2 cells requires concerted action of glycosaminoglycans, fibronectin and integrin receptors. Mol. Microbiol. 29**:**369-379. [DOI] [PubMed] [Google Scholar]
- 51.Virji, M., D. Evans, A. Hadfield, F. Grunert, A. M. Teixeira, and S. M. Watt. 1999. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34**:**538-551. [DOI] [PubMed] [Google Scholar]
- 52.Virji, M., K. Makepeace, D. J. Ferguson, and S. M. Watt. 1996. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22**:**941-950. [DOI] [PubMed] [Google Scholar]
- 53.Virji, M., S. M. Watt, S. Barker, K. Makepeace, and R. Doyonnas. 1996. The N-domain of the human CD66a adhesion molecule is a target for Opa proteins of Neisseria meningitidis and Neisseria gonorrhoeae. Mol. Microbiol. 22**:**929-939. [DOI] [PubMed] [Google Scholar]
- 54.Wolff, K., and A. Stern. 1995. Identification and characterization of specific sequences encoding pathogenicity associated proteins in the genome of commensal Neisseria species. FEMS Microbiol. Lett. 125**:**255-263. [DOI] [PubMed] [Google Scholar]
- 55.Womble, D. D. 2000. GCG: the Wisconsin package of sequence analysis programs. Methods Mol. Biol. 132**:**3-22. [DOI] [PubMed] [Google Scholar]
- 56.Yazdankhah, S. P., P. Kriz, G. Tzanakaki, J. Kremastinou, J. Kalmusova, M. Musilek, T. Alvestad, K. A. Jolley, D. J. Wilson, N. D. McCarthy, D. A. Caugant, and M. C. Maiden. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42**:**5146-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, P., G. Morelli, and M. Achtman. 1999. The opcA and (psi)opcB regions in Neisseria: genes, pseudogenes, deletions, insertion elements and DNA islands. Mol. Microbiol. 33**:**635-650. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, P., A. van der Ende, D. Falush, N. Brieske, G. Morelli, B. Linz, T. Popovic, I. G. Schuurman, R. A. Adegbola, K. Zurth, S. Gagneux, A. E. Platonov, J. Y. Riou, D. A. Caugant, P. Nicolas, and M. Achtman. 2001. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl. Acad. Sci. USA 98**:**5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]