Differences in vertebrate microRNA expression (original) (raw)
Abstract
MicroRNAs (miRNAs) attenuate gene expression by means of translational inhibition and mRNA degradation. They are abundant, highly conserved, and predicted to regulate a large number of transcripts. Several hundred miRNA classes are known, and many are associated with cell proliferation and differentiation. Many exhibit tissue-specific expression, which aids in evaluating their functions, and it has been assumed that their high level of sequence conservation implies a high level of expression conservation. A limited amount of data supports this, although discrepancies do exist. By comparing the expression of ≈100 miRNAs in medaka and chicken with existing data for zebrafish and mouse, we conclude that the timing and location of miRNA expression is not strictly conserved. In some instances, differences in expression are associated with changes in miRNA copy number, genomic context, or both between species. Variation in miRNA expression is more pronounced the greater the differences in physiology, and it is enticing to speculate that changes in miRNA expression may play a role in shaping the physiological differences produced during animal development.
Keywords: chick, evolution, medaka, miRNA, zebrafish
MicroRNAs (miRNAs) represent an abundant class of regulatory elements controlling a range of physiological processes, including developmental timing, differentiation, and cell proliferation (reviewed in refs. 1–3). Many are highly conserved, and their misexpression is associated with cancer (4–9). MiRNAs influence gene expression through subtle combinatorial effects (10). Natural selection could act on these subtle changes to gene expression induced by individual miRNAs to drive evolution (3).
Most miRNAs are expressed in a tissue-specific manner (11–15). Array as well as in situ expression data for many zebrafish, a few mouse, and several Drosophila miRNAs demonstrate that overlap in expression exists (11, 13, 15–17). Based on these data and the high degree of miRNA sequence conservation, it has been assumed that there is a high degree of expression conservation. To evaluate the overall conservation of miRNA expression, we set out to compare expression in two animals, medaka and chicken, with existing data from zebrafish and mouse. One hundred and one miRNAs were compared between medaka and zebrafish, 71 between chicken, and 11 between mice. These results reveal that the timing and location of miRNA expression is not strictly conserved, with variation in miRNA expression being more pronounced the greater the differences in physiology. Comparison of miRNA copy number and genomic context is also examined, and, together, these data serve as a base for examining the role of miRNA expression in the evolution of animal physiology.
Results and Discussion
Comparison of miRNA Expression Between Medaka and Zebrafish.
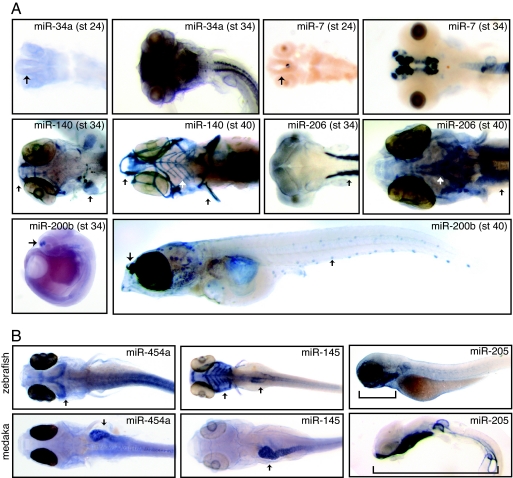
To compare conservation of miRNA expression, two fish species, medaka and zebrafish, were examined. Medaka (Oryzias latipes) is a small fresh water fish separated by ≈150–200 million years from zebrafish (18). This distance is substantial, considering that the separation between mouse and human is estimated to be 75 million years and between human and chimpanzee 6 million years (19, 20). In general, medaka develops more slowly than zebrafish, although the timing for specific tissues varies (18, 21). The majority of miRNAs begin to be expressed between 1–2 days postfertilization (dpf) in zebrafish. For many miRNAs, comparable expression is observed between stages 30 and 34 in medaka (3.5–5.5 dpf). However, based on miRNA expression, the brain, eyes, and body wall develop more quickly compared with the head, jaw, and sensory epithelia in medaka relative to zebrafish. Representative examples of this delay in miRNA expression for several tissues involved in sensation and feeding are shown in Fig. 1A. In each case, expression is delayed in the head and lateral line relative to comparable expression in other tissues. This result is found for five lateral line-specific miRNAs, which specifically stain the neuromasts or small sensory cells along the body (miR-96, miR-182, miR-183, miR-200a, and miR-200b). MiR-200b is shown in Fig. 1A. Although pro-neuromasts are present much earlier in medaka, by stage 32 (4.5 dpf), the timing of observed miRNA expression within the lateral line coincides with the period of neuromast differentiation, stage 39 (10 dpf) (22). Both miRNA expression and lateral line differentiation are observed much earlier in zebrafish, 2 dpf (11, 23), consistent with earlier hatching. Our observed differences in the timing of miRNA expression between medaka and zebrafish, therefore, fit well with the observed differences in the timing of lateral line maturation between these two species. Interestingly, neuromast location along the body of medaka and zebrafish is markedly different. Migration is coupled to differentiation for neuromast development (22), and miRNA expression affects differentiation (24). Thus, the delay in miRNA expression could be linked to the changes in location of neuromasts along the body of each fish. In zebrafish, simultaneous overexpression of three lateral line-specific miRNAs (miR-96, 200a, and 200b) impairs neuromast migration with partial penetrance adding support to the association of miRNA expression to neuromast location (Fig. 3, which is published as supporting information on the PNAS web site).
Fig. 1.
MiRNA expression in medaka. (A) MiRNAs are expressed late in medaka development. Representative examples are shown. Brain tissue is clearly visible by stage 24 (see miR-34a and miR-7, arrows), yet peak expression occurs much later, stage 34. MiRNA expression is further delayed in tissues associated with feeding such as the jaw and head muscles, as well as sense organs such as the lateral line and taste buds. Representative examples of cartilage (miR-140), muscle (miR-206), and sensory epithelia (miR 200b) specific expression are shown. In each case, peak expression occurs later in these tissues (stage 40, arrows) compared with expression of the same miRNA in related tissues (stage 34, arrows). (B) Several miRNAs exhibit spatial expression differences between medaka and zebrafish. Representative examples are shown. MiR-454a is expressed in the gut (arrow) in medaka and in the dorsal midbrain (arrow) in zebrafish. MiR-145 is observed in zebrafish pharyngeal arches (arrow) and fins with weaker expression observed in the gut and gall bladder (arrow). In medaka, expression is most pronounced in the heart and gut (arrow), with weak expression observed late in the pharyngeal arches and head skeleton. MiR-205 is expressed in the epidermis in both medaka and zebrafish. Expression extends through the entire length of the lateral surface (bracket) in medaka (stage 34) but strong expression is limited to the epidermis surrounding the pharyngeal arches in zebrafish (bracket). During later developmental stages, strong expression in medaka is limited to the pharyngeal arches, mimicking observed expression in zebrafish (data not shown).
This pattern of delayed expression is also observed for many miRNAs associated with the head skeleton and cartilage. In total, 9 miRNAs associated with the pharyngeal arches were probed (miR-23a, miR-27a, miR-27b, miR-140, miR-140*, miR-145, miR-146, miR-199a, miR-199a*, and miR-214). Seven exhibit delayed expression (miR-27a, miR-27b, miR-140, miR-140*, miR-199a and miR-199a*, and miR-214), whereas two exhibit weak to no expression in these structures (miR-145 and miR-146). MiR-140 is shown in Fig. 1A. In medaka, pharyngeal arch formation begins at stage 34, but maturation is not complete until hatching, stage 39 (25). This pattern of delayed expression is also observed in the head for three muscle-specific miRNAs (miR-1, miR-133a, and miR-206) relative to their expression in body wall muscle, mimicking the pattern observed for pharyngeal arch development. MiR-206 is shown in Fig. 1A. Both the head and jaw are morphologically different between medaka and zebrafish, and changes in animal physiology are coupled to changes in the timing of differentiation (26). Given that miRNA expression is associated with the regulation of differentiation, changes in miRNA expression could potentially influence the timing of differentiation in these tissues.
Several miRNAs exhibit clear spatial expression differences between medaka and zebrafish. The most dramatic differences result from changes in expression in the gut and head. MiR-454a and miR-145 are shown in Fig. 1B. For miR-145, strong expression is observed in the pharyngeal arches and gut in zebrafish (Fig. 1B, arrows), whereas gut expression is visible in medaka (Fig. 1B, arrow) but expression in the pharyngeal arches is significantly weaker. MiR-454a is also a gut-specific miRNA in medaka (Fig. 1B, arrow). In zebrafish, miR-454a exhibits brain-specific expression (Fig. 1B, arrow). Such dramatic differences in miRNA expression could be due to changes in their genomic location or their copy number, as well as their promoter or enhancer sequences. For the complete list of differences in miRNA expression, see Tables 1 and 2, which are published as supporting information on the PNAS web site.
The largest class of miRNA patterns analyzed, 42, is neuronal tissue-specific. A representative example of the variety of patterns in medaka brain relative to zebrafish is shown in Fig. 4_A_, which is published as supporting information on the PNAS web site. MiRNA expression in the brain changes during development, and the majority of the observed expression differences are due to differences in the timing of brain development (Fig. 4_B_). One notable exception is miR-187, which exhibits brain-specific expression in medaka, but only background expression is observed in zebrafish.
Comparison of miRNA Expression Among Medaka, Zebrafish, Chicken, and Mouse.
Chicken embryos, Hamburger and Hamilton (1951) (HH) stages 3–26, were compared for 71 miRNAs where in situ expression data were available for either medaka or zebrafish, and 11 expression patterns in mouse were compared with each. In general, when expression differences are observed, miRNA expression in chicken and mouse appear more similar to each other than either appears to either fish. This is also observed for medaka and zebrafish, where their expression appears more similar to each other when compared with mouse or chicken, suggesting that variation in miRNA expression is more pronounced the greater the differences in physiology. Comparisons of miRNA expression among chicken, mouse, and fish are highlighted for several. For the complete list of comparisons, see Tables 1 and 2.
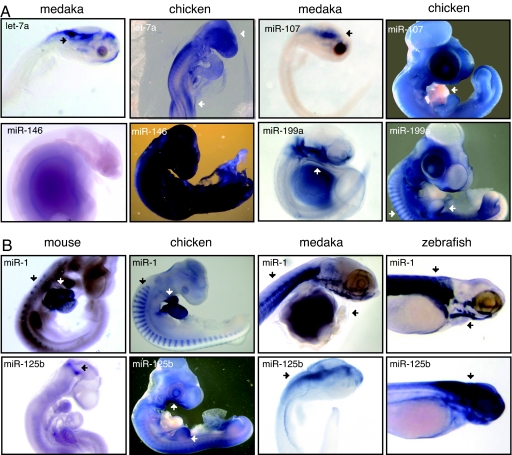
In fish, in situ expression of the let-7 family is limited to neuronal tissues during embryonic development. In medaka, the expression of let-7 members can be grouped into two categories, predominately ventricular brain expression (let-7a, -b, -c, and -f) and ubiquitous brain expression leading to no expression late in development (let-7g and -i). This pattern contrasts observed expression in mouse and chicken embryos where members of the let-7 family exhibit expression in developing limbs (let-7a, -b, and -j) and the epiblast (let-7b) (36). Let-7a is shown in Fig. 2A (arrow) (14). In mouse, another let-7 family member, let-7c, is implicated in limb development (14, 27). It remains possible that members of the let-7 family are general regulators of tissue proliferation. Let-7a, -b, -c, and -f are found in the proliferating regions of the brain in medaka, and misexpression of members of this family is associated with a variety of cancers (28, 29).
Fig. 2.
Comparison of miRNA expression. (A) Regional differences in miRNA expression are observed between chicken and medaka. For let-7a, brain-specific expression is seen for both medaka and chicken (arrows). In chicken, additional expression is also observed in the developing limb buds (arrow). MiR-199a is expressed in the epithelia surrounding the pharyngeal arches in medaka (arrows), whereas expression is observed in head, limb, and body mesoderm in chicken (arrows). In chicken, miR-107 is expressed in all tissues, except the heart (arrow). In medaka, miR-107 expression is brain-specific (arrow). MiR-146 is specifically expressed in the gut late in medaka development (stage 40), exhibiting no expression in earlier stages (shown). In chicken, miR-146 is expressed in the epidermis early in development, leading to ubiquitous expression in later developmental stages. (B) Comparison of miRNA expression among mouse, chicken, medaka, and zebrafish. Representative examples of differences in expression are shown. MiR-1 is expressed in skeletal muscle in both medaka and zebrafish (arrows). Similar expression is observed in the progenitor to muscle, or somites, in both chicken and mouse (arrows) and is additionally expressed in the heart (arrows). MiR-125b is expressed in the brain and spinal cord in both medaka and zebrafish (arrows). It is ubiquitously expressed in chicken, with stronger expression in the pharyngeal arches and limb buds (arrows). In mouse, expression is observed at the midbrain–hindbrain boundary (arrow).
Several miRNAs are known to be actively expressed during tissue proliferation, and most are found within a genomic cluster up-regulated in cancer (4, 9). Several (miR-17-3p, miR-15a, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, miR-20b, miR-92, miR-107, miR-125b, miR-130a, miR-130b, and miR-363) are ubiquitously expressed, excluding the heart, in chicken embryos (36). Interestingly, many of these miRNAs are ubiquitously expressed early in both medaka and zebrafish, with expression for most becoming attenuated and generally restricted to proliferative tissues late in development (miR-17-5p, miR-19b, miR-20a, and miR-92). MiR-92 is shown in Fig. 4_B_). Of note, miR-107 and miR-130a show clear spatial expression differences, exhibiting brain-specific expression in medaka and zebrafish and ubiquitous, no heart expression in chicken. MiR-107 is shown in Fig. 2A. Additionally, miR-125b exhibits a higher level of tissue-specific expression in the brain, motor horns, gonads, pharyngeal arches (Fig. 2B, arrow), and limb buds (Fig. 2B, arrow) in chicken embryos, whereas expression in mouse is limited further to the mid/hindbrain boundary (Fig. 2B, arrow). MiR-125b is a brain and spinal chord-specific miRNA in medaka and zebrafish (Fig. 2B, arrows).
Several muscle-specific miRNAs may serve as some of the clearest examples of the changes in tissue-specific miRNA expression among fish, chicken, and mouse. MiR-1, for example, is expressed in the skeletal muscle in developing medaka and zebrafish embryos (Fig. 2B, arrows). In both mouse and chicken, miR-1 is expressed in the somites (Fig. 2B, arrows), the progenitor to skeletal muscle, and is additionally expressed early in the developing heart (Fig. 2B, arrows). This same pattern is also observed for miR-133a (data not shown). Interestingly, heart physiology is notably different between medaka and zebrafish when compared with chicken and mouse. Differences in miRNA expression may therefore reflect physiological differences in this tissue.
Together, these examples demonstrate that miRNA conservation does not necessarily imply expression conservation. For many miRNAs, expression differences are seen with absolute conservation of the miRNA sequence between species. For others, expression differences are marked by minor changes to the mature miRNA. Most often, these differences are due to the addition of one or two nucleotides to the end of the miRNA in one species compared with another. For miRNAs exhibiting similar expression patterns, minor changes to the miRNA sequence are also seen, suggesting that there is no strong correlation between minor changes in the miRNA sequence and expression differences. For the complete listing of any changes made to the miRNA probes, see Table 2.
MiRNA copy number was compared across vertebrate genomes, and sequences were mapped to their respective locations (Table 3 and Data Set 1, which are published as supporting information on the PNAS web site). In this analysis, fish (zebrafish, medaka, fugu, and tetraodon) exhibit a higher copy number for several miRNAs, reflected in the sum of the miRNAs for each species. When expression conservation is compared with conservation in genomic location, examples are found for miRNAs exhibiting similar expression and comparable genomic locations (miR-10 and miR-196) as well as examples of conserved expression (miR-204 and miR-153) and expression differences (miR-125 and miR-34) with partial overlap in their genomic locations. In the case of miR-125, three copies are found in both zebrafish and mouse, but in different genomic contexts (exclusively intergenic vs. intronic and intergenic), whereas five copies are found in medaka, compared with only one in chicken. It should be noted that the sequence of the chicken miRNA precursor was not identified using our filters and was alternatively mapped by means of its miRBase annotation.
Numerous studies link miRNA expression to cell proliferation and differentiation. As such, differences in miRNA expression between animals likely reflect changes in growth and differentiation. Heterochrony, or a change in the timing of developmental growth, is known to alter the overall shape of developing tissues, leading to differences in physiology (26). The vast majority of observed expression differences between medaka and zebrafish likely reflect changes in the timing of miRNA expression rather than changes in the type of tissue regulated by miRNAs. Medaka and zebrafish exhibit relatively subtle differences in physiology, which is in line with the high number of conserved miRNA expression patterns. The most striking physiological differences are seen in the overall pattern of the lateral line as well as the structure of the head and jaw. It is therefore notable that the most dramatic changes to the timing of miRNA expression involve changes in miRNAs associated with the head and sensory epithelia such as the lateral line.
Differences in miRNA expression between either fish species when compared with chicken or mouse are more pronounced, and, together, these data fit well with the expectation that miRNA expression may be linked to differences in animal physiology. Changes to gene regulation between species have been proposed to be a driving force behind evolutionary changes to body structure (30), and transcriptional regulation has recently been implicated as one mechanism involved in evolution (31). Here, we speculate that changes to miRNA expression may act in a similar manner.
To conclude, many miRNAs exhibit a high degree of conservation between species, with greater variation in miRNA expression seen, the larger the differences in physiology. Differences between species most often result from heterochronic changes in the timing of miRNA expression and to a lesser extent from spatial expression differences. In some instances, such as lateral line-specific expression, heterochronic differences in miRNA expression track differences in the timing of differentiation. In the case of spatial expression differences, differences in some instances are associated with changes in miRNA copy number, genomic context, or both between species.
Materials and Methods
Animals and Reagents.
Chicken embryo care, collection, and analysis were performed as described in ref. 36. Both medaka and zebrafish were kept and cared for using standard laboratory conditions (32). Medaka (wild-type and albino mutant) and zebrafish (albino mutant) embryos were used for in situ hybridizations. Collected embryos were staged according to refs. 21 and 33. Because miRNA expression is largely tissue-specific and animal development is heterochronic, comparison of miRNA expression between species was based on the stage of development for individual tissues (18, 21, 33, 34). The miRNA expression patterns were analyzed by using locked nucleic acid (LNA) probes as described (11). LNA probes were digoxigenin (DIG) labeled by using 3′-end DIG labeling kits (Roche, Basel, Switzerland). DIG-labeled LNA probes were purified by using sephadex G-25 microspin columns (Amersham, Buckinghamshire, U.K.) before use.
Whole-Mount in Situ Hybridizations.
Chicken and mouse in situs were performed as described in refs. 36 and 16, respectively. Medaka in situ hybridizations were performed as described in ref. 16. Briefly, collected embryos were fixed overnight in 4% paraformaldehyde (PFA) and washed with PBST (PBS, 0.1% Tween 20) before dechorionation. Dechorionated embryos were transferred to methanol (MetOH) and stored at −20°C for one day or up to several months. All hybridizations were performed 20°C-25°C below the predicted melting temperature (_T_m) of the LNA probe. Best results were observed at ≈25°C below this _T_m. Embryos were treated with proteinase K (10 μg/ml) at 37°C for 30 min (stage 34 and younger) or 60 min (older than stage 34). After proteinase K treatment, embryos were refixed in 4% PFA. Embryos were incubated in hybridization buffer for 1–3 h before the addition of the LNA probe (10 nM, final). Embryos were incubated with the LNA probe for 2–18 h. Embryos were incubated in blocking buffer (PBST, 2% sheep serum, 2 μg/ml BSA) for 1–3 h. Antibody hybridization was performed overnight at 4°C in blocking buffer containing 1:2,000 dilution of anti-DIG-AP FAB fragments (Roche). Embryos were transferred to APT (100 mM Tris·HCl, pH 9.5/50 mM MgCl2/100 mM NaCl/0.1% Tween 20). NBT (nitro blue tetrazolium; Roche) and BCIP (5-bromo-4-chloro-3-indoyl phosphate; Roche) were added (3.5 μl/ml APT) to stain the embryos. Embryos were stained overnight at 4°C. Zeiss (Oberkochen, Germany) Axioplan and Leica (Wetzlar, Germany) MZFLIII microscopes mounted with digital cameras were used to photograph the embryos and sections. Adobe (San Jose, CA) Photoshop software was used to globally adjust the brightness, contrast, color, and saturation of the images.
Microinjections.
Annealed miRNA duplexes were injected into one-cell stage zebrafish embryos. One nanoliter of injection solution containing 10 μM miRNA duplex(es) was injected per embryo. The lateral line was visualized by staining live (3–4 dpf) embryos by using a solution of 200 μM 2-Di-4-Asp (D-3418; Sigma, St. Louis, MO) in system water for 3–5 min. Embryos were washed and anesthetized by using MS-222. Fluorescent staining was visualized by using the GFP settings on a fluorescence microscope (Zeiss Axioscope II).
MiRNA Mapping.
MiRNA sequence identification was performed as described in ref. 35 and in Data Set 1.
Acknowledgments
Chicken in situs were performed by Simran Kaur and Stacey Stansilaw. We thank Rene Ketting, Titia Sijen, and Dana Jongejan-Zivkovic for critically reading the manuscript and Karen Lyle for helpful discussions. The medaka genome data have been provided freely by the National Institute of Genetics and the University of Tokyo for use in this publication only. B.A. is funded by a National Research Service Award postdoctoral fellowship. Chicken miRNA expression and analysis were funded by National Institutes of Health Grant R01HD044767 (to P.B.A.).
Abbreviations
miRNA
microRNA
dpf
days postfertilization
LNA
locked nucleic acid.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Garcia I, Miska EA. Development (Cambridge, UK) 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 3.Plasterk RH. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 4.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AM, Byrom MW, Shelton J, Ford LP. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 10.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 11.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 12.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 13.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler G, Ntonuia-Fousara S, Granda B, Rathjen T, Dalmay T. FEBS Lett. 2006;580:2195–2200. doi: 10.1016/j.febslet.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 17.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furutani-Seiki M, Wittbrodt J. Mech Dev. 2004;121:629–637. doi: 10.1016/j.mod.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Gregory SG, Sekhon M, Schein J, Zhao S, Osoegawa K, Scott CE, Evans RS, Burridge PW, Cox TV, Fox CA, et al. Nature. 2002;418:743–750. doi: 10.1038/nature00957. [DOI] [PubMed] [Google Scholar]
- 20.The Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 21.Iwamatsu T. Mech Dev. 2004;121:605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Sapede D, Gompel N, Dambly-Chaudiere C, Ghysen A. Development (Cambridge, UK) 2002;129:605–615. doi: 10.1242/dev.129.3.605. [DOI] [PubMed] [Google Scholar]
- 23.Gompel N, Cubedo N, Thisse C, Thisse B, Dambly-Chaudiere C, Ghysen A. Mech Dev. 2001;105:69–77. doi: 10.1016/s0925-4773(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 24.Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 25.Langille RM, Hall BK. J Morphol. 1987;193:135–158. doi: 10.1002/jmor.1051930203. [DOI] [PubMed] [Google Scholar]
- 26.Huxley JS. Problems of Relative Growth. London: Methuen; 1932. [Google Scholar]
- 27.Schulman BR, Esquela-Kerscher A, Slack FJ. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 29.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 30.King M, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 31.Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 32.Westerfield M. The Zebrafish Book. Eugene: Univ Oregon Press; 1993. [Google Scholar]
- 33.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 34.Bellairs R, Osmond M. The Atlas of Chick Development. New York: Academic; 1998. [Google Scholar]
- 35.Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. Genome Res. 2006 doi: 10.1101/gr.5159906. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darnell DK, Stanislaw SK, Konieczka JK, Yatskievych TA, Antin PB. Dev Dyn. 2006 doi: 10.1002/dvdy.20956. in press. [DOI] [PubMed] [Google Scholar]