Gene Expression Analysis of Immune-Mediated Arrest of Tumorigenesis in a Transgenic Mouse Model of HER-2/neu-Positive Basal-Like Mammary Carcinoma (original) (raw)
Abstract
We previously showed that a vaccine combining interleukin 12 and allogeneic p185neu-positive mammary carcinoma cells completely prevented multifocal mammary carcinogenesis in HER-2/neu transgenic mice. To identify the molecular events responsible for effective tumor prevention and to define the tumor gene expression signature, we used microarrays to analyze the expression profile of mammary tissue of untreated transgenic mice and of vaccine-treated, tumor-free mice at different time points. Mammary tissue from vaccinated mice displayed a gene expression profile different from that of untreated, tumor-bearing mice but similar to that of normal/hyperplastic mammary gland. Comparison of treated and untreated mice at 15 weeks of age revealed up-regulation of genes encoding antibodies, chemokines, γ-interferon-induced genes and inflammatory molecules, and down-regulation of early genes induced by tumor development. The gene expression signature of HER-2/neu-transformed tumor cells showed modulation of genes promoting proliferation, angiogenesis, migration, invasion, and metastasis and inhibiting apoptosis and immune response. Meta-analysis of microarray data on human breast cancer showed that the signature of tumors arising in murine HER-2/neu transgenic model correctly classified human HER-2/neu-expressing tumors and normal breast tissue. Moreover murine and human HER-2/neu-positive tumors share the signature of basal-like breast cancers. This gene expression analysis reveals the immune events associated with prevention of tumor development and shows that HER-2/neu transgenic mice represent a good model of a poor-prognosis group of human breast tumors.
Immunological prevention of tumors is a feasible possibility especially in light of both the encouraging results obtained in preclinical models of neoplasia and of the recent advances in detection of healthy individuals at high risk of developing cancer.1,2 Transgenic mice expressing the HER-2/neu oncogene under tissue-specific transcriptional control of the mouse mammary tumor virus promoter (MMTV-LTR) represent a suitable model of mammary carcinogenesis. HER-2/neu is overexpressed in 25 to 30% of human breast cancers, influencing biological features and prognosis of the tumor, and the natural history of HER-2/neu transgenic mammary tumors closely resembles that of human breast carcinoma, from atypical hyperplasia to carcinoma in situ and invasive tumor.3 Among HER-2/neu transgenic mice, BALB-NeuT mice harboring a mutated version of the rat neu oncogene represent a very aggressive model of mammary carcinogenesis because they develop multifocal mammary carcinomas with a short latency (4 to 5 months of age).4 Mammary tumors arising in this strain of transgenic mice could mimic human lobular carcinoma of alveolar type. The HER-2/neu transgene is expressed in the epithelium of lobular ducts and lobules. Consecutive stages of tumor progression are associated with a high epithelial proliferation rate and with the activation of the angiogenic program.3 Among the various immunopreventive approaches performed on HER-2/neu models,4–12 vaccination with allogeneic mammary carcinoma cells expressing HER-2/neu combined with interleukin (IL)-12 is able to reach a striking percentage of long-term protection from mammary carcinoma development, maintaining 90 to 100% of mice free from tumor up to at least 1 year of age.13,14 Molecular analysis of the immune response in the mammary gland environment and of changes in the tumor genetic program induced by vaccination would improve the design of new immunopreventive approaches by revealing the gene expression signature of both the achievement and the failure of the immune protection from tumor development.
Human breast cancer arises as a consequence of multiple genetic lesions that accumulate in a specific tissue environment influencing the response to different treatment approaches. Gene expression profiling studies on breast carcinomas have pointed out that molecular subtypes of breast cancer exist, and can be correlated to prognosis and other clinically relevant parameters.15–20 Some other studies have focused on the genetic profile changes associated with the expression of a definite oncogene in cell lines,21–23 that only partially approximate the in vivo behavior of the tumor, or in transgenic mouse models but only in late-stage tumors.24 This study analyzes a time-course gene expression profile in the mammary gland of untreated and vaccinated, tumor-free HER-2/neu transgenic mice, thus monitoring the tumor’s transcriptome changes and associating the genetic profile with the efficacy of immune treatments. We show here that mammary glands of vaccinated mice display a gene expression profile that closely resembles that of normal/hyperplastic mammary gland. This analysis also provides a genetic signature of HER-2/neu-induced mammary carcinogenesis and shows that HER-2/neu transgenic mice develop tumors that resemble at the gene expression level human breast cancers of the basal-like subtype.
Materials and Methods
Transgenic Mice
Female BALB-NeuT transgenic mice (H-2d),4 that overexpress the mutated rat HER-2/neu oncogene under control of the MMTV promoter were bred as reported.14 Animal experiments were authorized by the local animal use and care committee. Mammary glands were inspected weekly: masses whose mean diameter exceeded 3 mm were regarded as tumors. Mice were sacrificed for humane reasons when all of the 10 mammary glands were tumor bearing or when a mass exceeded a mean diameter of 1.5 cm. All of the five mammary glands of each side were collected and immediately frozen in liquid nitrogen.
Cell Lines
Cell lines were derived from mammary carcinomas of FVB-NeuN #202 mice (H-2q), transgenic for the rat HER-2/neu proto-oncogene under control of the MMTV promoter.13,25 Two HER-2/neu-positive cell clones (N202.1A and TT12.E2, hereafter referred to as Neu/A and Neu/B) and one HER-2/neu-negative cell clone (N202.1E, a brother clone of N202.1A, hereafter referred to as Neuneg/A) were used. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal bovine serum (Invitrogen, Milan, Italy).
Production of IL12-Engineered Cell Vaccines
Cell lines Neu/A and Neu/B were transfected in vitro, respectively, with 10 μg of pIL12-IREShygro or pIL12-IRES1neo polycistronic expression plasmids by FuGene transfection reagent (Roche, Milan, Italy), selected, and cloned by limiting dilution. The two clones used as cell vaccines produced 50 ± 12 and 285 ± 87 ng/ml/106 cells/72 hours of IL-12, respectively, as determined by enzyme-linked immunosorbent assay (R&D Systems Inc., Minneapolis, MN). Transfectants were routinely cultured in the presence of the selective agent.
Vaccination Protocols
Starting at the 6th week of age, BALB-NeuT mice were subjected to intraperitoneal vaccination with 2 × 106 IL12-engineered Neu/A or Neu/B cells twice a week for 2 weeks, followed by 2 weeks of rest. This 4-week cycle was repeated lifelong. In vaccination experiments cells were pretreated with 40 μg/ml of mitomycin C (Sigma, Milan, Italy), to block cell proliferation. Mice vaccinated with IL12-transfected Neu/A or Neu/B cells will hereafter be referred to as Vax/A and Vax/B.
Morphological Analysis
Histological analysis and immunohistochemistry was performed as described;3 the antibodies used were anti-p185neu (C-18; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and anti-proliferating cell nuclear antigen (PCNA; Ylem, Rome, Italy). Whole mounts of mammary glands were performed as described at http://ccm.ucdavis.edu/tgmouse/HistoLab/wholmt1.htm. Briefly, skinned pelts were fixed in buffered formalin, then mammary glands were defatted in acetone, rehydrated, and stained with ferric hematoxylin, dehydrated again, and cleared with histo-lemon. Storage and image acquisition of mammary glands was performed in methyl salicylate.
RNA Extraction and Microarray Analysis
Total RNA was extracted from mammary gland areas including lymph node, unless otherwise specified, and from cell lines with TriZol reagent (Invitrogen). Total RNA was extracted from individual mice and each individual RNA was labeled and hybridized to an independent array. RNA quality was checked on ethidium bromide-stained gel. Fragmented biotin-labeled cRNA for chip hybridization was prepared according to the Affymetrix Expression Analysis Sample Prep protocol (Affymetrix, Santa Clara, CA). Briefly, double-stranded cDNA was synthesized from 16 μg of total RNA with the Superscript Choice system for cDNA synthesis (Invitrogen) and an oligo-dT carrying a T7 RNA polymerase promoter. Biotin-labeled cRNA was synthesized from cDNA with the Bioarray High Yield RNA transcript labeling kit (Affymetrix), purified with RNeasy spin columns (Qiagen, Milan, Italy) and fragmented to an average size of 50 to 200 bp. Hybridization to MU74Av2 chips (Affymetrix) was performed with 20 μg of fragmented biotin-labeled cRNA at the Biopolo consortium (University of Milano-Bicocca, Milan, Italy). Hybridized arrays were washed, stained, and scanned, and the digitized images were processed with Microarray Suite 5.0 gene expression analysis software (MAS 5.0, Affymetrix) to obtain cell intensity files.
Data Filtering, Normalization, and Clustering
Probe set intensities were background-corrected, normalized, and summarized by the Robust Multi-Array analysis (RMA) method26 implemented in the Affy package of Bioconductor (www.bioconductor.org). Gene expression ratios were calculated with respect to the mean intensity of 6-week samples. Standard correlation among the different time points and treatments was calculated on the entire dataset of 12,489 probes. Principal component analysis was calculated on a filtered gene list containing only probes that displayed at least a fourfold difference in the expression level in at least one comparison between two single samples (1125 probes). The principal components were calculated using TIGR MultiExperiment Viewer 3.027 and the first two, that together accounted for 83% of the total variance, were visualized graphically.
Genes induced or repressed by vaccination at 15 weeks were selected as having a 1.5-fold change difference in the expression level and statistically significant (Wilcoxon test) at the 0.05 cutoff P value; Benjamini and Hochberg correction for multiple testing was applied thus retrieving a list of 155 probes that was called “vaccination” gene list. Among vaccination-induced genes we selected a subgroup of probes that were down-regulated by at least twofold in lymph node-deprived with respect to lymph node-containing mammary glands of vaccinated mice. This list is provided as Supplemental Table B on http://www.amjpathol.org.
Genes that changed along with tumor growth in untreated mice were selected as having at least a twofold difference in the expression level between at least two time points and statistically significant at the 0.01 cutoff P value (Kruskal-Wallis test) and applying Benjamini and Hochberg correction for multiple testing (1069 probes). This list was called “tumor” gene list. Vaccination and tumor gene lists were clustered by means of self organizing maps28 on the three time points of the vaccination time course (6 × 1 topology) and on the four time points of the tumor growth time course (3 × 3 topology), respectively. Complete linkage hierarchical clustering was performed on normalized log2 ratios calculating Euclidean distance as a metrics of similarity. TIGR MultiExperiment Viewer 3.027 was used to perform both types of clustering. Gene ontology classification was performed with the DAVID/EASE annotation tool (http://david.niaid.nih.gov/david/ease.htm).
The subset of genes specifically expressed by the epithelial neu-positive cells was extracted from the tumor gene list by selecting the genes that displayed also a concordant twofold difference in the expression between Neu/A and Neuneg/A cells, and this list was called “HER-2/neu” gene list. Two independent biological replicates of the two clones were performed and used for microarray analysis. Raw data (log2 probe intensities) of vaccination, tumor, and HER-2/neu gene lists are shown on http://www.amjpathol.org in Supplemental Table A, C and D, respectively.
Meta-Analysis of Human Breast Cancer Data
Mouse orthologs of publicly available gene lists coming from Sotiriou and colleague’s19 study (706 cDNA clones) and Sørlie and colleague’s16 study (intrinsic gene list, 456 cDNA clones corresponding to 427 unique genes) and conversely human orthologs of mouse probes were found using MatchMiner29 and GeneSpring software (Silicon Genetics, Redwood City, CA). Lists of mouse orthologs of the intrinsic gene list and the list from the Sotiriou and colleague’s19 study are supplied as Supplemental Tables E and F on http://www.amjpathol.org.
Human orthologs of the tumor gene list that were also present in the array used in the Sørlie and colleagues16 publication were used to cluster human HER-2/neu-expressing tumors (11 samples) and normal breast tissues and benign fibroadenomas (7 samples) taken from the Sørlie and colleagues16 dataset. Mouse orthologs of the intrinsic gene list from the Sørlie and colleagues16 study that were also present in the murine MU74Av2 array were used to build a prediction classifier on human basal-like (14 samples) and luminal-like tumors (47 samples) taken from their study. A threshold was selected to minimize the number of classifier genes and keep the misclassification error rate on the training set at 0%, thus identifying an optimal 66-probe classifier. This classifier was used to predict the basal or luminal features of our murine samples. Prediction Analysis of Microarrays30 software was used.
Real-Time Polymerase Chain Reaction
RNA extracted from tumor specimens and mammary glands was retrotranscribed as described previously.13 TaqMan primers and probes for rat-specific HER-2/neu (dir, 5′-GCAACTTGGAGCTTACCTACG-3′; reverse, 5′-CGATGAGCATGTAACCCTGA-3′; MGB-probe 5′-6-FAM-CCAGCCTCTCATTCC-minor groove binder/nonfluorescent quencher-3′), were chosen by Primer Express 2.0 and custom synthesized by Applera, Milan, Italy. The murine housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase was used as the reference gene and amplified with commercially available TaqMan reagents (Applera). Quantitative real-time polymerase chain reaction was performed with a 5700 Applied Biosystems (Applera) apparatus.
Results
Prevention of Mammary Carcinoma in HER-2/neu Transgenic Mice
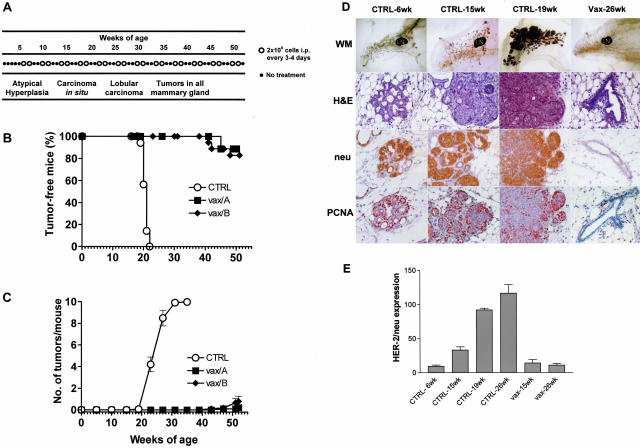
Mammary carcinoma onset in the BALB-NeuT model can be completely prevented with a vaccination consisting of FVB-NeuN-derived mammary carcinoma cells (that express HER-2/neu oncogene and allogeneic MHC), combined with the administration of systemic IL-12.13 The same extent of protection was achieved with IL-12-transduced mammary carcinoma cells (see Figure 1A for vaccination schedule and mammary carcinoma progression steps in untreated controls): at 1 year of age 83 to 90% of vaccinated mice were tumor-free, whereas all untreated mice developed palpable mammary carcinomas within 22 weeks (Figure 1B). The low percentage of vaccinated mice developing tumors showed a highly prolonged latency and a very low tumor multiplicity (Figure 1C). In this study we compared the gene expression profiles in the mammary glands of mice that will succumb to mammary carcinoma with those of vaccinated mice that will survive indefinitely. Because our vaccine was administered intraperitoneally, the effects on the mammary gland transcriptome should be induced by systemic immune responses. We have previously shown that the lack of any single component (HER-2/neu, allogeneic MHC, or IL-12) caused a dramatic fall in vaccine efficacy.13,14 As an example, without IL-12 all vaccinated mice eventually succumb to mammary carcinoma.13 Therefore a vaccine without IL-12 is feasible, but it is ineffective. For this reason we directly compared the three-component vaccine with the mock vaccination.
Figure 1.
Inhibition of mammary tumorigenesis in BALB-NeuT transgenic mice receiving immunopreventive vaccine treatments. A: Tumor progression stages in untreated mice and vaccination schedule. B: Tumor-free survival curve. Open symbol, no vaccination; closed symbols, HER-2/neu-expressing, IL12-transduced, allogeneic cell vaccines. Vax/A, IL12-transduced Neu/A cells; Vax/B, IL12-transduced Neu/B cells. Groups of 11 to 18 mice received the indicated treatments. Both vaccinations are statistically significant (P < 0.01, at least) with respect to control mice. C: Tumor multiplicity per mouse (mean ± SEM). Significance: any vaccine versus control; P < 0.01 at least from week 21 onwards. D: Morphological and immunohistochemical analysis of mammary glands from untreated (CTRL) or vaccinated (Vax) mice at different time points. WM, whole mount analysis; H&E, hematoxylin and eosin morphological analysis; neu, p185neu staining; PCNA, proliferating cell nuclear antigen. The fourth mammary gland is shown. LN indicates the lymph node. Untreated 6-week-old BALB-NeuT mice mammary glands show rare nests of cells displaying atypical hyperplasia that begin to express p185neu and PCNA. At 15 weeks of age mammary glands show diffuse hyperplasia and some foci of carcinoma in situ that invariably express p185 and PCNA. At 19 weeks of age mammary glands display lobular invasive carcinomas that are merging together and that are neu-positive and hyperproliferating. On the contrary, 26-week-old vaccinated mice show mammary glands with no sign of carcinoma and only some hyperplastic buds; ductal branches are formed by a single layer of epithelial cells that do not express HER-2/neu and have a low proliferative activity. E: Expression of rat-HER-2/neu measured by TaqMan real-time polymerase chain reaction in mammary glands of untreated (CTRL) and vaccinated mice (Vax) at different time points. The mean ± SEM of rat-HER-2/neu expression calculated versus housekeeping gene GAPDH is shown.
Morphological Analysis
Previous pathological studies allowed us to identify key stages in the natural history of mammary carcinoma in HER-2/neu transgenic mice.3 Samples for microarray studies were therefore taken at different time points to investigate tumor progression and the preventive effects of vaccination. Figure 1D illustrates the successive stages of tumor progression in this system. At the 6th week of age mammary glands began to show atypical hyperplasia, with some small nests of p185neu-positive proliferating cells. At 15 weeks of age mammary glands already showed several in situ carcinomas in which all of the cells were p185-positive and PCNA-positive. At 19 weeks mammary glands of untreated mice revealed the presence of multiple lobular carcinomas that were merging together to form the palpable tumor mass that was actively proliferating and expressing p185neu. Vaccinated mammary glands from 26-week-old mice were completely free from carcinoma, and only showed some hyperplastic foci in the terminal end buds; histologically the gland resembled a normal mammary gland with lobular structures, that displayed no HER-2/neu expression and low PCNA positivity. A conspicuous macrophage, neutrophil, and CD8-lymphocyte infiltrate was present in the hyperplastic foci of mice treated with the vaccine. Vaccination also increased the number of dendritic and natural killer cells. This recruitment of reactive cells was accompanied by overexpression of endothelial adhesion molecules in the small vessels. The expression of proinflammatory cytokines was also induced in mammary glands from vaccinated mice.13,14,31 The expression profile was analyzed also in 26-week-old untreated mice mammary glands that displayed palpable invasive lobular carcinomas, and in 15-week-old vaccinated mice mammary glands. The progressive growth of p185neu-positive cells at different time points in untreated mice was mirrored by the increased mRNA expression of the rat HER-2/neu oncogene, whereas mammary tissue from vaccinated mice expressed HER-2/neu at levels comparable to that of 6-week-old mice (Figure 1E).
Global Gene Expression Analysis
We analyzed two to three individual mice for each time point and treatment, because correlation between the expression profile of the replicas was satisfactory (mean r2 ∼ 0.98). In the case of 6-week-old mice we had to analyze four mice to reach the same correlation. The wider variability between replicates was seen in untreated mice at preneoplastic stages suggesting that both tumor growth and vaccination impose a distinctive and reproducible shift in the mammary gland’s gene expression profile.
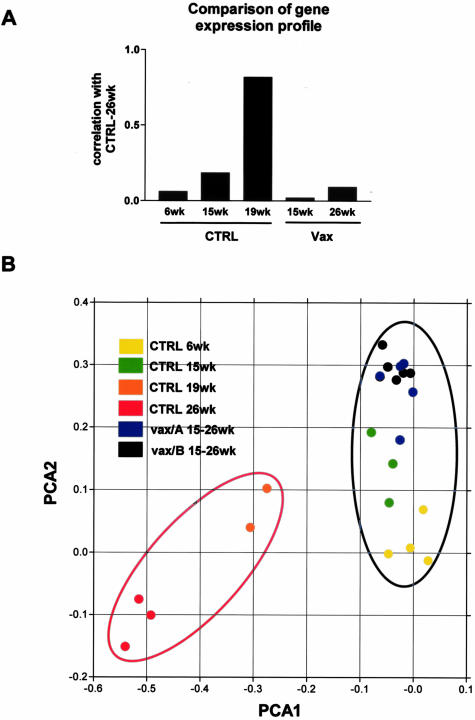
The whole transcriptome changed profoundly with tumor growth; the tumor transcriptome correlation index was very low both compared to normal mammary gland from 6-week-old mice and to mammary glands from vaccinated mice at any time point (Figure 2A). Moreover, principal component analysis (Figure 2B) of the gene expression differences clearly separated tumor-bearing (red ellipse) from tumor-free (blue ellipse) mammary glands on the first component (PCA1, horizontal axis in Figure 2B). The shift on first component between 6-week-old mice that displayed only hyperplastic foci and 15-week-old mice that already showed in situ carcinomas, is minimal; however this result should be expected because the increase in tumor cell proportion from 6 to 15 weeks is small if compared to later stages, and consequently the weight of this change on the whole mammary gland transcriptome is comparably small. Almost all mammary glands from vaccinated mice could be separated along the second principal component from those of untreated mice at 6 and 15 weeks of age, thus showing that vaccination was able to induce gene expression changes in the mammary tissue. These data taken together show that this vaccination is an efficient strategy to block HER-2/neu-induced hyperproliferation and tumor development. Thus this system represents a good model to study both HER-2/neu oncogenic pathway in vivo and the immune events elicited by vaccination locally in the mammary gland.
Figure 2.
Global analysis of gene expression changes. A: Correlation coefficient calculated on all of the probe sets versus mammary glands from 26-week-old untreated mice. Tumor-bearing mice (26 weeks old) display a mammary gland transcriptome that shows a very low correlation with normal/hyperplastic mammary glands from 6-week-old mice or vaccinated mice mammary glands at any time point. B: Two-dimensional representation of the first two components of principal components analysis (PCA) calculated on genes that showed at least a fourfold difference in at least one sample-to-sample comparison. CTRL, untreated; Vax/A or Vax/B, vaccinated; red ellipse, tumor-bearing samples; blue ellipse, tumor-free samples. During mammary carcinoma growth samples acquire more negative values of PCA1 component, and samples from vaccinated mice with either vaccination and at any time point are positioned at the same PCA1 level of early stage untreated samples at 6 and 15 weeks of age.
Vaccination-Induced Changes in the Gene Expression Profile
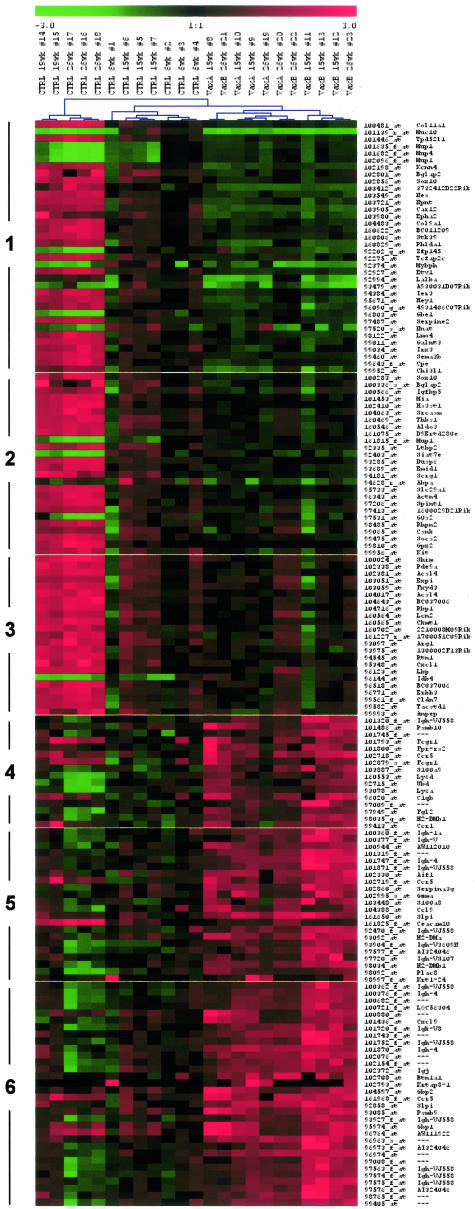
Previous analysis on the complex immune responses elicited by vaccination in HER-2/neu transgenic mice indicated that key actors of long-term tumor prevention were T-cell-derived cytokines, such as interferon (IFN)-γ, and antibodies, whereas cytotoxic T lymphocyte-mediated lysis of tumor cells apparently played a minor role.13,31 To study early changes in gene expression induced by vaccination we compared the expression patterns of mammary tissues from untreated and vaccinated 15-week-old mice. Figure 3 shows the 155 probes differentially expressed between untreated and vaccinated transgenic mice at 15 weeks of age (vaccination gene list). Raw data of these probes in all samples are provided as Supplemental Table A. Genes were clustered with self-organizing maps on the three time points of the vaccination time course, yielding six clusters of genes with similar expression dynamics (Supplemental Figure S1 on http://www.amjpathol.org). The first three clusters contained the probes down-regulated in vaccinated mice with respect to untreated at 15 weeks of age. These genes could be useful to elucidate the mechanisms of carcinoma progression and to detect new possible targets for prevention. Some outliers related to the mammary gland stroma and not to tumor cells were included in these clusters just because these genes are down-regulated in vaccinated mice with respect to untreated at 6 weeks of age. The first two clusters contained mainly tumor-related genes whose level of expression is differently affected by vaccination. Cluster 1 grouped mainly tumor-specific genes that were brought by vaccination at levels even lower than those at 6 weeks of age; these genes control cell proliferation (Stk39, Ier3, Hey1, Etv1), invasion, and metastatic spread (serine proteinase inhibitor, clade E, member 2, serpine 2; semaphorin 3B, Sema3b). This result can indeed be explained by the fact that at 6 weeks of age in many strains of mice the mammary gland is undergoing a stage of normal proliferation as the ducts extend to fill the fat pad during puberty. Cluster 2 contained all of the genes specific of tumor cells whose expression was blocked by vaccination at levels identical to that at 6 weeks of age. Cluster 3 included some genes already associated with HER-2/neu activation or overexpression in different rodent models,32–34 such as chitinase 3-like 1 (Chi3l1), lipocalin 2 (Lcn2), FXYD domain-containing ion transport regulator 3 (Fxyd3), and extracellular proteinase inhibitor (Expi1). This cluster likely consists of genes highly expressed in carcinoma cells that could be sensitive markers of small tumor burdens. More experimental evidence is needed to validate it. The expression of these genes was efficiently blocked by vaccination only at 15 weeks of age, whereas at 26 weeks the expression started to increase (Figure 3).
Figure 3.
Genes differentially expressed at 15 weeks between untreated and vaccinated mice. Samples were clustered by complete linkage hierarchical clustering. Legend on top of the hierarchical clustering reports the treatment of individual mice (CTRL, untreated; VaxA or VaxB, vaccinated), their age, and individual mouse identification number. Genes were clustered by self-organizing maps on the three time points of the vaccination time course. Affymetrix IDs and gene symbols of the differentially expressed genes are indicated on the right of the heat map, whereas cluster membership is shown on the left. Red indicates genes more expressed with respect to the mean of 6-week-old samples and green indicates genes less expressed with respect to the mean of 6-week-old samples. Raw data from all of the samples of the full vaccination gene list are provided as Supplemental Table A.
Two distinctive features of the tumor transcriptome found with this analysis and not reported previously were related to retinoid metabolism (retinol-binding protein 1, Rbp1; lipocalin 2, Lcn2; N10 gene for a nuclear hormonal binding receptor, nur77) and immune suppression (suppressor of cytokine signaling 2, Socs2; arginase 1, Arg1). Correlation to human breast tumorigenesis was evident because several genes are overexpressed also in human breast carcinomas, as tumor-associated calcium signal transducer 1 (Tacstd1 or Ep-CAM), tumor protein D52-like 1 (Tpd52l1), AP-2 gamma (AP-2γ), LIM domain only 4 (Lmo4).35–38
Genes more expressed in vaccinated samples (clusters 4, 5, and 6) were almost exclusively immune response genes, as gene ontology functional categories immunoglobulin and immunity protein were overrepresented with a P value ≪ 0.01. Genes highly induced in vaccinated samples with respect to untreated were grouped into cluster 6 and are almost exclusively immunoglobulin chains and γ-interferon-induced genes, such as Cxcl9 or MIG (monokine induced by IFN-γ), guanylate nucleotide-binding protein 1 (Gbp1), and proteasome subunit, β type 9 (Psmb9 or LMP2). Cluster 5 grouped other antibody chains and inflammation markers, such as S100 calcium-binding protein A9 (S100A9), placenta-specific 8 (Plac8), and allograft inflammatory factor 1 (Aif1). This suggests that the predominant immunological response is based on B cells and γ-interferon-secreting T cells. Other immune cell populations such as neutrophils and dendritic cells play a minor role, as exemplified by the lower induction of genes specific of these cell types (cluster 4). The same marginal role is played by cytotoxic T cells, because microarray data show only a mild induction of granzyme A (Gzma).
We also dissected mammary glands from untreated and vaccinated mice at 15 weeks of age to exclude the lymph node of the fourth gland (Figure 1D). Vaccine up-regulated genes could be divided into two clusters: one containing immunoglobulin-coding genes, the other one grouping all of the IFN-mediated and inflammatory response genes. Immunoglobulin genes did not show any difference in the lymph node-deprived tissues from vaccinated and untreated mice. The second group of genes was still up-regulated in the mammary glands of vaccinated mice versus untreated even when these were evaluated without the lymph node. A list of the probes significantly down-regulated in lymph node-deprived versus lymph node-containing mammary glands from vaccinated mice is provided as Supplemental Table B; this list almost exclusively contains immunoglobulin-coding genes whereas the expression of IFN-induced or inflammation-related genes was not modified by lymph node removal. No gene was up-regulated in lymph node-deprived with respect to lymph node-containing mammary glands. Thus we can infer that the B-cell response resides mainly in the lymph node, whereas all of the other arms of the immune response to the vaccine take place in the mammary tissue.
Tumor-Specific HER-2/neu-Induced Gene Expression Signature
The identification of genes expressed by HER-2/neu-positive tumors provides new targets for specific therapies. In an animal model we can better define the tumor-specific signature, because we can follow the progressive enrichment of tumor cells and the acquisition of more malignant features by comparing the expression profile of the mammary tissue at consecutive points in time. Thus we selected the genes that displayed a progressive change in untreated samples from the 6th week up to the 26th week. A high proportion of genes displayed at least a twofold difference in the expression level and statistical significance at the 1% P value cutoff (tumor gene list, 1069 probes; provided as Supplemental Table C). To analyze the different expression dynamics of these genes on the four time points of tumor growth we clustered the tumor gene list with self-organizing maps, associating with each cluster the most represented gene ontology annotations (Supplemental Figure S2 on http://www.amjpathol.org). Genes heavily down-regulated in the tumor reflect mainly the progressive loss of the normal stromal compartment of the mammary gland, exemplified by the GO categories muscle development, fatty acid metabolism, defense response. On the contrary, up-regulated genes belonged both to the tumor cell-specific transcriptome and to the tumor-induced changes in the microenvironment, reflecting both the enrichment in tumor cell population and the acquisition of a more malignant phenotype during tumor progression. In fact among the up-regulated genes we found the categories cell cycle, proteolysis, and phosphorylation.
The architecture of the mammary gland in the early stages of tumor development, the co-existence of multiple distinct cell types,39 and the heterogeneous level of HER-2/neu expression during early steps of tumorigenesis3 do not allow a meaningful use of HER-2/neu-positive tissue microdissection in this system. To extract the signature of HER-2/neu-positive tumor cells from the transcriptome of the whole tumor tissue we compared the expression profile of neu-positive versus neu-negative mammary carcinoma cell clones derived from HER-2/neu transgenic tumors.25 Neu/A cell clone and Neuneg/A, that lost the expression of HER-2/neu in vitro and was derived from the same tumor, were used for microarray analysis. Genes up-regulated or down-regulated on HER-2/neu-driven tumor development were selected by crossing the list of 1069 genes that changed during tumor growth in vivo with the list of twofold different genes between Neu/A cell clone and Neuneg/A. The resulting 103 differential genes represent the signature of HER-2/neu-driven tumorigenesis both in vivo and in vitro (HER-2/neu gene list; raw data are provided in Supplemental Table D). Functional classification of a selection of the signature genes is shown in Table 1. These results show that on HER-2/neu expression mammary carcinoma cells up-regulated or down-regulated genes that collectively induce an increase in proliferation and inhibition of apoptosis, and acquired a phenotype that promotes angiogenesis, invasion, metastasis, and branching morphogenesis and represses local immune responses. Several genes related to mammary gland differentiation were also induced. Interestingly samples taken from vaccinated mice expressed these genes at levels comparable to neu-negative samples at early points in time.
Table 1.
Functional Role and Differential Expression of Genes Belonging to the HER-2/neu Signature
| Gene name | Symbol | In vivo fold change | In vitro fold change | |
|---|---|---|---|---|
| Angiogenesis | Chitinase 3-like 1 | Chi3l1 | 10.6 | 9.1 |
| CEA-related cell adhesion molecule 1 | Ceacam1 | 4.2 | 7.8 | |
| Coagulation factor III | F3 | 4.3 | 2.6 | |
| Ras homolog gene family, member C (RhoC) | Arhc | 2.1 | 2.0 | |
| Invasion-metastasis | Serine (or cysteine) proteinase inhibitor, clade E, member 2 | Serpine2 | 15.5 | 6.3 |
| Ets variant gene 1 | Etv1 | 3.4 | 8.9 | |
| Extracellular proteinase inhibitor | Expi | 6.2 | 6.5 | |
| GalNAc alpha-2,6-sialyltransferase | St6gal1 | 2.4 | 2.3 | |
| Cytidine monophospho-_N_-acetylneuraminic acid synthetase | Cmas | 2.8 | 4.5 | |
| Ras homolog gene family, member C (RhoC) | Arhc | 2.1 | 2.0 | |
| E74-like factor 5 | Elf5 | 2.6 | 3.9 | |
| Matrix metalloproteinase 15 | Mmp15 | 2.0 | 4.8 | |
| Cathepsin H | Ctsh | 2.1 | 2.3 | |
| Caveolin, caveolae protein | Cav1 | −10.1 | −6.5 | |
| Proliferation | Transcription factor AP-2, gamma | Tcfap2c | 3.6 | 8.2 |
| LIM domain only 4 | Lmo4 | 2.9 | 3.2 | |
| Ras homolog gene family, member C (RhoC) | Arhc | 2.1 | 2.0 | |
| Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | Kcnn4 | 3.1 | 7.6 | |
| RAS p21 protein activator 3 | Rasa3 | −4.4 | −4.7 | |
| Kruppel-like factor 4 | Klf4 | −3.0 | −4.2 | |
| Caveolin, caveolae protein | Cav1 | −10.1 | −6.5 | |
| Migration-branching | Downstream of tyrosine kinase 1 | Dok1 | 2.0 | 3.3 |
| Development and differentiation enhancing | Ddef1 | −2.0 | −2.6 | |
| Ets variant gene 1 | Etv1 | 3.4 | 8.9 | |
| LIM domain only 4 | Lmo4 | 2.9 | 3.2 | |
| Apoptosis inhibition | Glutathione peroxidase 2 | Gpx2-ps1 | 3.4 | 26.5 |
| Pleckstrin homology-like domain, family A, member 1 | Phlda1 | 2.5 | 7.5 | |
| Nuclear receptor subfamily 4, group A, member 1 | Nr4a1 | 2.0 | 4.0 | |
| Immune suppression | 24p3 gene (Lipocalin 2) | Lcn2 | 5.6 | 13.5 |
| Suppressor of cytokine signaling 2 | Socs2 | 2.0 | 10.7 | |
| MRC-OX2 antigen homolog gene (CD200) | Mox2 | 2.5 | 3.1 | |
| Lectin, galactose binding, soluble 1 (Galectin 1) | Lgals1 | −11.6 | −5.1 | |
| Mammary gland differentiation | CCAAT/enhancer binding protein (C/EBP), delta | Cebpd | 5.4 | 2.6 |
| FXYD domain-containing ion transport regulator 3 | Fxyd3 | 4.9 | 9.1 | |
| 24p3 gene (Lipocalin 2) | Lcn2 | 5.6 | 13.5 | |
| Extracellular proteinase inhibitor | Expi | 6.2 | 6.5 | |
| Lectin, galactose binding, soluble 1 (Galectin 1) | Lgals1 | −11.6 | −5.1 |
Tumors from HER-2/neu Transgenic Mice as a Model of Human HER-2/neu-Positive, Basal-Like Tumors
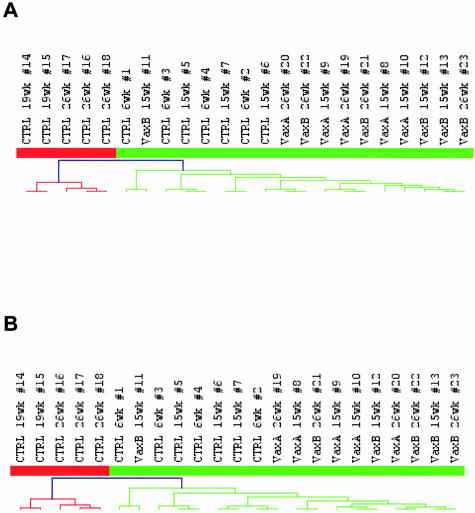
From a morphological point of view, HER-2/neu transgenic mammary carcinogenesis mimics human breast tumor progression,3 and early microarray studies of mouse tumors suggested a parallel with human breast tumorigenesis also at the gene expression level.34 We collected data from two independent studies on the classification of human breast tumors to analyze the homology between the transcriptome of HER-2/neu transgenic murine mammary carcinomas and of human breast tumors. Sørlie and colleagues16 identified an intrinsic gene list that describes inner biological features of human breast cancers; more recently Sotiriou and colleagues19 described a 706-probe element gene list that also identifies natural subclasses of breast tumors. In both cases when clustering our samples with mouse orthologs of the two lists obtained from clinically and biologically relevant human data we always ended up with a two-arm dendrogram, in which on one side there were the two branches of 26- and 19-week-old untreated samples, that correspond to overt tumors, whereas on the other side there were tumor-free samples from 6-week-old and 15-week-old untreated mice and all of the mammary glands from vaccinated mice (Figure 4, A and B). A list of the orthologs used for classification is provided in Supplemental Tables E and F. In the branch of tumor-free samples, 6- and 15-week-old mice were mixed up, mainly because the weight of the increase in tumor cell population between these two stages is small if compared to the whole mammary gland transcriptome profile.
Figure 4.
Homologies between the HER-2/neu transgenic tumor model and human breast cancer. Gene lists taken from the Sørlie and colleague’s16 publication (A) and from the Sotiriou and colleague’s19 publication (B), both describing inner biological features of human breast carcinomas, were used to cluster samples from untreated and vaccinated HER-2/neu transgenic mice. Legend on top of the hierarchical clustering reports the treatment of individual mice (CTRL, untreated; VaxA or VaxB, vaccinated), their age, and individual mouse identification number. Red tree branches, tumor-bearing samples from 19-week and 26-week untreated mice and green tree branches, tumor-free samples (6- and 15-week untreated samples and all vaccinated ones).
In the HER-2/neu transgenic model we identified a tumor gene list that is specifically associated with HER-2/neu-positive tumors and that clearly separated tumor-bearing tissues from the normal mammary gland. To determine whether these genes were also related to human HER-2/neu-induced breast tumor formation we used human orthologs of the tumor gene list to cluster human HER-2/neu-positive tumor samples and normal breast tissues taken from the Sørlie and colleagues16 dataset. Figure 5A shows that our mouse-derived gene list separates human HER-2/neu-expressing tumors from normal mammary gland tissue and from nonneoplastic fibroadenomas.
Figure 5.
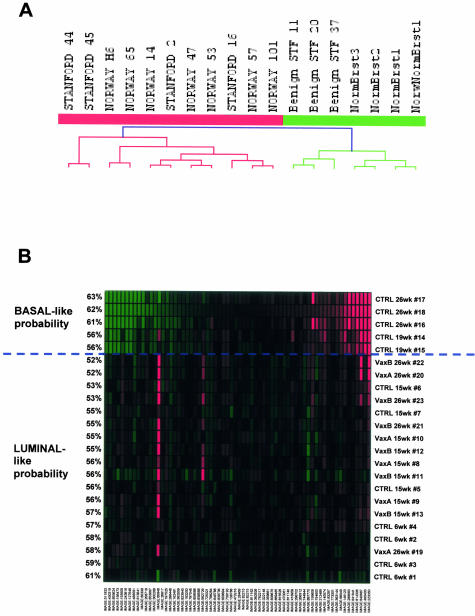
A: Hierarchical clustering of 11 human HER-2/neu-expressing tumor samples (Stanford and Norway) and 7 normal mammary tissue (four normal breast and three benign fibroadenomas) taken from the Sørlie and colleague’s16 dataset with human orthologs of our tumor gene list that differentiates transgenic HER-2/neu-positive tumors from normal mammary glands in the transgenic model. Our gene list separates tumor samples (red tree branches) from normal tissue (green tree branches). B: Classification of HER-2/neu transgenic murine samples regarding basal-like and luminal-like signatures. The prediction classifier was trained on the 14 basal-like and 47 luminal-like human tumor samples from the Sørlie and colleague’s16 dataset on those probes in the intrinsic gene list whose mouse orthologs were present in the murine MU74Av2 array. The probes selected to best classify the two groups on the human dataset are identified with human IMAGE clone number, and the probability of every sample to belong to one of the two groups is shown on the left. Above the dashed line there are the samples classified as basal-like (tumors from 26- and 19-week-old mice) while below the dashed line there are all of the other samples that are classified as luminal-like.
Most studies on human breast tumor profile-based classifications showed that at least two subgroups of breast tumors exist, the luminal-like and the basal-like, and that HER-2/neu-positive tumors tend to cluster together with basal-like tumors and to share with this group a poor prognosis. No information is at the moment available on how much different transgenic tumor models mimic one or the other subgroup of human breast tumors. To answer this question we used mouse orthologs of the intrinsic gene list from Sørlie and colleagues16 to build a predictor that correctly classifies human basal- and luminal-like tumors from their study. An optimal separation with the minimal number of genes was reached with a list of 66 IMAGE clones corresponding to the top scoring genes. The predictor generated on human samples was used to classify our mouse samples, showing that tumors from HER-2/neu transgenic mice were classified as basal-like, whereas samples from mammary glands that do not show overt signs of tumorigenesis (6- and 15-week-old untreated, and all of the vaccinated ones) were classified as luminal-like (Figure 5B). Samples from 19-week-old untreated mice were classified as basal-like, even if with a probability inferior to the one of 26-week-old untreated mice, because at 19 weeks of age the tumors co-exist with tissues from residual normal mammary glands.
Discussion
In this work we analyzed the gene expression changes associated with HER-2/neu-driven tumorigenesis and with immune prevention of transgenic mammary carcinoma onset. Vaccination consisting of allogeneic HER-2/neu-positive mammary carcinoma cells engineered to secrete IL-12 was able to maintain almost all mice tumor-free up to at least 1 year of age.14 Our results showed that there is a high concordance between molecular profiling and morphological appearance of the mammary gland exemplified both by whole mount and immunohistochemical analysis. In fact samples with a similar morphology and immunohistochemical pattern, such as 6-week-old and vaccinated mice mammary glands, display almost superimposable gene expression profiles. These results demonstrate that our immune prevention strategy effectively blocks carcinoma onset in the mammary gland even at the genetic level and fits in well with recent findings on the inhibition afforded by a different vaccination modality.40 Finding such a concordance between traditional histological classification and molecular profiles suggests that morphological similarity could imply parallel similarity at the molecular and genetic level.40
Despite the interest in immune prevention strategies, particularly in the HER-2/neu transgenic model, information on the molecular events associated with prevention of tumor development is still scarce. By comparing the genetic profile of untreated and vaccinated mice at 15 weeks we chose to focus our attention mainly on the early events associated with immune prevention, when the mammary glands of untreated mice show only some subclinical foci of carcinoma in situ and when the immune response in vaccinated mice is mounting up, as shown by the increase in serum anti-vaccine antibody titers.13,14 The vaccine-elicited immune response combines a lymph node-specific, antibody response, and other immune events that take place in the mammary gland, such as the T-helper-mediated, IFN-γ-dependent response, shown by the increase in the expression of the chemokine Cxcl9 (MIG), the proteasome subunit, β type 9 (Psmb9 or LMP2), the macrophage-expressed allograft inflammatory factor 1 (AIF1), and several other genes. IFN-γ acts both on cells of the immune compartment inducing the isotype switch on B cells toward the production of Th1-immunoglobulins and promoting the recruitment of a reactive leukocyte infiltrate, but also directly on tumor cells modifying their phenotype. In fact the induction of the anti-angiogenic chemokine MIG represses the proangiogenic phenotype of tumor cells, whereas the increase in the LMP2 subunit of the proteasome restores the expression of class I MHC. Loss of expression of MHC molecules is an event that commonly occurs not only in the HER-2/neu transgenic tumor model,41 but also in human tumors and is known to impair the recognition of tumor cells by the host immune system.
The role of antibody production and IFN-γ-mediated responses was shown by crossing BALB-NeuT mice with IFN-γ knockout mice or with antibody-deficient μMT mice: in both cases vaccination is almost completely ineffective.13,31 In this system antibodies mainly acted as inductors of antibody-dependent cell cytotoxicity and complement-mediated cytotoxicity, and complement subunits were indeed overexpressed in mammary tissue from vaccinated mice. Cytotoxic T lymphocyte response played a marginal role because only a slight overexpression of granzyme A was found in the mammary glands of vaccinated mice, whereas no other sign of cytotoxic T lymphocyte activation was evident.
Our vaccination induced a massive recruitment of cells of the immune system in the mammary gland; from the molecular point of view neutrophil migration could be directed by calgranulin overexpression (S100A9),42 whereas the contemporary induction in mammary glands of vaccinated mice of CCR1 by nonepithelial cells and of its ligand CCL9 (MIP-1γ) by epithelial cells could explain the recruitment of activated dendritic cells and T cells. This microarray analysis also revealed that vaccination represses the expression of genes related to immune suppression promoted by tumor development, such as suppressor of cytokine signaling-2 (Socs2)43 and arginase-1.44
The first hints at the homology between HER-2/neu transgenic model of mammary carcinogenesis and human breast tumors are suggested by the fact that several genes overexpressed during tumor growth in this model (Fxyd3, Tacstd1, Lcn2, Chi3l1, AP-2-γ) are important in human breast tumorigenesis being either breast tumor markers or indicators of poor prognosis. This homology is not obvious, because HER-2/neu transgenic mice express the mutated oncogene, whereas in humans activation of this oncogene occur by amplification. This fact hampered an a priori extrapolation of data from the mouse model to the human situation. We show here for the first time that human mammary tumorigenesis is correctly reproduced in the HER-2/neu transgenic model at the global gene expression level. In fact, genes that describe human breast cancer inner biology and predict clinical outcome16,19 identify tumor-bearing and tumor-free murine mammary glands as separate groups. Moreover, the genes that are different between the tumor and the normal mammary gland in the HER-2/neu transgenic model separate correctly also human HER-2/neu-expressing tumor samples from normal mammary tissue. These results eventually demonstrate at the molecular level that the HER-2/neu transgenic model is a good model of human breast tumorigenesis.
Many independent studies on microarray-based classification of human breast cancers have shown that at least two main subgroups of tumors exist, the luminal-like and the basal-like; the first one prevalently includes estrogen receptor-positive tumors that express genes relatively highly expressed by breast luminal epithelial cells, whereas the second is characterized by estrogen receptor-negative tumors that tend to express markers of basal epithelial cells. Human HER-2/neu-positive tumors tend to cluster with basal-like tumors and share with them a negative prognosis.16,17,19 We show here for the first time that tumors developing in the murine HER-2/neu transgenic model have a basal-like signature, in agreement with human data. The fact that normal mammary glands tend to be classified as luminal-like probably stems from the fact that the epithelial compartment of the normal mammary gland is prevalently composed of cells that show luminal-like features.45 This result further suggests that this model closely mimics HER-2/neu-induced tumorigenesis in humans even in its similarity to tumors of the basal-like subtype. Microarray data that extensively monitor gene expression in human tumors and in their murine models counterpart will offer an unprecedented opportunity to identify new tumor antigens1 that can be targeted by specific immunopreventive approaches, whose efficacy and safety can easily be evaluated in the murine model. Therefore, data-driven microarray experiments can turn out into hypothesis-driven applications that would probably be more effectively translated into clinical applications. Hopefully these and other results might guide the application of the effective preventive or therapeutic approaches developed in this and other similar models to subgroups of human tumors with the proper signature characterized by a negative prognosis and by a low responsiveness to commonly used chemotherapeutic and chemopreventive agents.
Supplementary Material
Supplemental Material
Footnotes
Address reprint requests to Pier-Luigi Lollini, Cancer Research Section, Department of Experimental Pathology, viale Filopanti 22, I-40126 Bologna, Italy. E-mail: pierluigi.lollini@unibo.it.
Supported by the Italian Association for Cancer Research, Italian Foundation for Cancer Research, Italian Ministry for Education, and University and Research, University of Bologna.
A.A. and S.C. are recipients of fellowships from the Italian Foundation for Cancer Research.
Supplemental material for this article can be found at http://www.amjpathol.org.
References
- Lollini PL, Forni G. Cancer immunoprevention: tracking down persistent tumor antigens. Trends Immunol. 2003;24:62–66. doi: 10.1016/s1471-4906(02)00030-3. [DOI] [PubMed] [Google Scholar]
- Finn OJ. Cancer vaccines: between the idea and the reality. Nat Rev Immunol. 2003;3:630–641. doi: 10.1038/nri1150. [DOI] [PubMed] [Google Scholar]
- Di Carlo E, Diodoro MG, Boggio K, Modesti A, Modesti M, Nanni P, Forni G, Musiani P. Analysis of mammary carcinoma onset and progression in HER-2/neu oncogene transgenic mice reveals a lobular origin. Lab Invest. 1999;79:1261–1269. [PubMed] [Google Scholar]
- Boggio K, Nicoletti G, Di Carlo E, Cavallo F, Landuzzi L, Melani C, Giovarelli M, Rossi I, Nanni P, De Giovanni C, Bouchard P, Wolf S, Modesti A, Musiani P, Lollini PL, Colombo MP, Forni G. Interleukin 12-mediated prevention of spontaneous mammary adenocarcinomas in two lines of Her-2/neu transgenic mice. J Exp Med. 1998;188:589–596. doi: 10.1084/jem.188.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amici A, Venanzi FM, Concetti A. Genetic immunization against neu/erbB2 transgenic breast cancer. Cancer Immunol Immunother. 1998;47:183–190. doi: 10.1007/s002620050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cefai D, Morrison BW, Sckell A, Favre L, Balli M, Leunig M, Gimmi CD. Targeting HER-2/neu for active-specific immunotherapy in a mouse model of spontaneous breast cancer. Int J Cancer. 1999;83:393–400. doi: 10.1002/(sici)1097-0215(19991029)83:3<393::aid-ijc16>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Dakappagari NK, Douglas DB, Triozzi PL, Stevens VC, Kaumaya PT. Prevention of mammary tumors with a chimeric HER-2 B-cell epitope peptide vaccine. Cancer Res. 2000;60:3782–3789. [PubMed] [Google Scholar]
- Dela Cruz JS, Lau SY, Ramirez EM, De Giovanni C, Forni G, Morrison SL, Penichet ML. Protein vaccination with the HER2/neu extracellular domain plus anti-HER2/neu antibody-cytokine fusion proteins induces a protective anti-HER2/neu immune response in mice. Vaccine. 2003;21:1317–1326. doi: 10.1016/s0264-410x(02)00741-7. [DOI] [PubMed] [Google Scholar]
- Esserman LJ, Lopez T, Montes R, Bald LN, Fendly BM, Campbell MJ. Vaccination with the extracellular domain of p185neu prevents mammary tumor development in neu transgenic mice. Cancer Immunol Immunother. 1999;47:337–342. doi: 10.1007/s002620050539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata M, Okudaira T, Samanta A, Clark DP, Drebin JA, Jolicoeur P, Greene MI. Prevention of breast tumour development in vivo by downregulation of the p185neu receptor. Nat Med. 1995;1:644–648. doi: 10.1038/nm0795-644. [DOI] [PubMed] [Google Scholar]
- Piechocki MP, Pilon SA, Wei WZ. Complementary antitumor immunity induced by plasmid DNA encoding secreted and cytoplasmic human ErbB-2. J Immunol. 2001;167:3367–3374. doi: 10.4049/jimmunol.167.6.3367. [DOI] [PubMed] [Google Scholar]
- Pupa SM, Invernizzi AM, Forti S, Di Carlo E, Musiani P, Nanni P, Lollini P-L, Meazza R, Ferrini S, Menard S. Prevention of spontaneous neu-expressing mammary tumor development in mice transgenic for rat proto-neu by DNA vaccination. Gene Ther. 2001;8:75–79. doi: 10.1038/sj.gt.3301360. [DOI] [PubMed] [Google Scholar]
- Nanni P, Nicoletti G, De Giovanni C, Landuzzi L, Di Carlo E, Cavallo F, Pupa SM, Rossi I, Colombo MP, Ricci C, Astolfi A, Musiani P, Forni G, Lollini PL. Combined allogeneic tumor cell vaccination and systemic interleukin 12 prevents mammary carcinogenesis in HER-2/neu transgenic mice. J Exp Med. 2001;194:1195–1205. doi: 10.1084/jem.194.9.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giovanni C, Nicoletti G, Landuzzi L, Astolfi A, Croci S, Comes A, Ferrini S, Meazza R, Iezzi M, Di Carlo E, Musiani P, Cavallo F, Nanni P, Lollini PL. Immunoprevention of HER-2/neu transgenic mammary carcinoma through an interleukin 12-engineered allogeneic cell vaccine. Cancer Res. 2004;64:4001–4009. doi: 10.1158/0008-5472.CAN-03-2984. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de RM, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein LP, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der KK, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Sinha C, Ignatoski KW, Lippman ME, Ethier SP, Chinnaiyan AM. Transcriptome analysis of HER2 reveals a molecular connection to fatty acid synthesis. Cancer Res. 2003;63:132–139. [PubMed] [Google Scholar]
- Wilson KS, Roberts H, Leek R, Harris AL, Geradts J. Differential gene expression patterns in HER2/neu-positive and -negative breast cancer cell lines and tissues. Am J Pathol. 2002;161:1171–1185. doi: 10.1016/S0002-9440(10)64394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay A, Jones C, Dexter T, Silva RL, Bulmer K, Jones A, Simpson P, Harris RA, Jat PS, Neville AM, Reis LF, Lakhani SR, O’Hare MJ. cDNA microarray analysis of genes associated with ERBB2 (HER2/neu) overexpression in human mammary luminal epithelial cells. Oncogene. 2003;22:2680–2688. doi: 10.1038/sj.onc.1206349. [DOI] [PubMed] [Google Scholar]
- Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, Simon R, Liu ET, Green JE. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni P, Pupa SM, Nicoletti G, De Giovanni C, Landuzzi L, Rossi I, Astolfi A, Ricci C, De Vecchi R, Invernizzi AM, Di Carlo E, Musiani P, Forni G, Menard S, Lollini PL. p185(neu) protein is required for tumor and anchorage-independent growth, not for cell proliferation of transgenic mammary carcinoma. Int J Cancer. 2000;87:186–194. [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey KJ, Kane D, Sunshine M, Narasimhan S, Nishizuka S, Reinhold WC, Zeeberg B, Ajay W, Weinstein JN. MatchMiner: a tool for batch navigation among gene and gene product identifiers. Genome Biol. 2003;4:R27. doi: 10.1186/gb-2003-4-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni P, Landuzzi L, Nicoletti G, De Giovanni C, Rossi I, Croci S, Astolfi A, Iezzi M, Di Carlo E, Musiani P, Forni G, Lollini PL. Immunoprevention of mammary carcinoma in HER-2/neu transgenic mice is IFN-gamma and B cell dependent. J Immunol. 2004;173:2288–2296. doi: 10.4049/jimmunol.173.4.2288. [DOI] [PubMed] [Google Scholar]
- Stoesz SP, Gould MN. Overexpression of neu-related lipocalin (NRL) in neu-initiated but not ras or chemically initiated rat mammary carcinomas. Oncogene. 1995;11:2233–2241. [PubMed] [Google Scholar]
- Morrison BW, Leder P. Neu and ras initiate murine mammary tumors that share genetic markers generally absent in c-myc and int-2-initiated tumors. Oncogene. 1994;9:3417–3426. [PubMed] [Google Scholar]
- Andrechek ER, Laing MA, Girgis-Gabardo AA, Siegel PM, Cardiff RD, Muller WJ. Gene expression profiling of neu-induced mammary tumors from transgenic mice reveals genetic and morphological similarities to ErbB2-expressing human breast cancers. Cancer Res. 2003;63:4920–4926. [PubMed] [Google Scholar]
- Gastl G, Spizzo G, Obrist P, Dunser M, Mikuz G. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet. 2000;356:1981–1982. doi: 10.1016/S0140-6736(00)03312-2. [DOI] [PubMed] [Google Scholar]
- Turner BC, Zhang J, Gumbs AA, Maher MG, Kaplan L, Carter D, Glazer PM, Hurst HC, Haffty BG, Williams T. Expression of AP-2 transcription factors in human breast cancer correlates with the regulation of multiple growth factor signalling pathways. Cancer Res. 1998;58:5466–5472. [PubMed] [Google Scholar]
- Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O’Reilly L, White D, Williams R, Armes J, Lindeman GJ. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Mattei MG, Basset P. Definition of the tumor protein D52 (TPD52) gene family through cloning of D52 homologues in human (hD53) and mouse (mD52). Genomics. 1996;35:523–532. doi: 10.1006/geno.1996.0393. [DOI] [PubMed] [Google Scholar]
- Smalley M, Ashworth A. Stem cells and breast cancer: a field in transit. Nat Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- Quaglino E, Rolla S, Iezzi M, Spadaro M, Musiani P, De Giovanni C, Lollini PL, Lanzardo S, Forni G, Sanges R, Crispi S, De Luca P, Calogero R, Cavallo F. Concordant morphologic and gene expression data show that a vaccine halts HER-2/neu preneoplastic lesions. J Clin Invest. 2004;113:709–717. doi: 10.1172/JCI19850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lollini PL, Nicoletti G, Landuzzi L, De Giovanni C, Rossi I, Di Carlo E, Musiani P, Muller WJ, Nanni P. Down regulation of major histocompatibility complex class I expression in mammary carcinoma of HER-2/neu transgenic mice. Int J Cancer. 1998;77:937–941. doi: 10.1002/(sici)1097-0215(19980911)77:6<937::aid-ijc24>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- Krebs DL, Hilton DJ. SOCS: physiological suppressors of cytokine signaling. J Cell Sci. 2000;113:2813–2819. doi: 10.1242/jcs.113.16.2813. [DOI] [PubMed] [Google Scholar]
- Bronte V, Serafini P, De Santo C, Marigo I, Tosello V, Mazzoni A, Segal DM, Staib C, Lowel M, Sutter G, Colombo MP, Zanovello P. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J Immunol. 2003;170:270–278. doi: 10.4049/jimmunol.170.1.270. [DOI] [PubMed] [Google Scholar]
- Smith GH, Chepko G. Mammary epithelial stem cells. Microsc Res Tech. 2001;52:190–203. doi: 10.1002/1097-0029(20010115)52:2<190::AID-JEMT1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material