Canonical notch signaling functions as a commitment switch in the epidermal lineage (original) (raw)
Abstract
Mammalian epidermis consists of a basal layer of proliferative progenitors that gives rise to multiple differentiating layers to provide a waterproof envelope covering the skin surface. To accomplish this, progenitor cells must detach from the basal layer, move upward, and execute a terminal differentiation program consisting of three distinct stages: spinous, granular layer, and stratum corneum. Notch signaling has been implicated in late stages of differentiation, but the commitment switch remains unknown. Here we show with loss and gain-of-function studies that active Notch intracellular domain (NICD) and its obligate canonical signaling partner RBP-J act at the basal/suprabasal juncture to induce spinous and down-regulate basal fate. Spinous layers are absent in RBP-J conditional null epidermis and expanded when Notch1 signaling is elevated transgenically in epidermis. We show that RBP-J is essential for mediating both spinous gene activation and basal gene repression. In contrast, the NICD/RBP-J target gene Hes1 is expressed in spinous layers and mediates spinous gene induction but not basal gene repression. These data uncover an early role for RBP-J and Notch in commitment of epidermal cells to terminally differentiate and reveal that spinous gene induction is mediated by a Hes1-dependent mechanism, while basal gene repression occurs independently of Hes1.
Keywords: RBP-J, Hes1, Notch, epidermis, stem cell fate
The skin epidermis is a stratified epithelium, in which only the innermost (basal) layer is mitotically active. Epidermal stratification is thought to rely on two mechanisms: delamination, where basal cells lose their attachment to the basement membrane (BM) and exit the basal layer (Watt and Green 1982), and asymmetrical cell division, in which the plane of cell division is asymmetric relative to the BM, generating a committed suprabasal daughter cell and a proliferative basal cell (Lechler and Fuchs 2005). In both cases, commitment to terminally differentiate is marked by withdrawal from the cell cycle concomitant with suppressed expression of basal integrins, extracellular matrix (ECM) proteins, p63, and keratins K5/K14 (Fuchs and Raghavan 2002).
Once an epidermal cell exits the basal layer and enters the first suprabasal (spinous) layer, it induces expression of keratins K1/K10 as well as transcription factors involved in the sequential program of terminal differentiation (for review, see Dai and Segre 2004). As cells transit to the granular layers, they express the keratin-bundling protein filaggrin, cornified envelope proteins such as involucrin and loricrin, and a variety of hydrophobic molecules (Segre 2006). Finally, as cells reach the outermost stratum corneum layers, they enter an apoptotic-like destructive phase, becoming metabolically inert as they extrude lipid bilayers and lose cytoplasmic organelles, including the nucleus. Terminally differentiated squames are sloughed from the skin surface, continually replaced by inner cells moving outward.
Little is known about the signaling pathways that orchestrate this differentiation program. One candidate is Notch, which functions broadly in specifying cell fates during differentiation and morphogenesis. Upon signaling, Notch is cleaved, releasing its intracellular domain (NICD). Most of NICD's effects have been attributed to its ability to bind the transcriptional repressor RBP-J, enabling it to activate target genes—e.g., Hes and Hey— that are normally suppressed in the absence of Notch activity (Artavanis-Tsakonas et al. 1999; Iso et al. 2003; Lai 2004). Depending on tissue and context, Notch can either restrict or promote cell fate determination.
In epidermal development, Notch receptors 1–3 and their ligand Jagged 1 are expressed suprabasally, whereas Jagged 2 is expressed basally (Powell et al. 1998; Pan et al. 2004). Postnatally, _K5-CreER_-mediated conditional ablation of Notch1 results in a hyperproliferative epidermis with expansion of proliferating basal-like cells, leading to the view that Notch functions as a tumor suppressor (Rangarajan et al. 2001; Nicolas et al. 2003). Late-stage differentiation is impaired in Notch1 conditional knockout (cKO) mice, with an increased expression of granular markers (loricrin and filaggrin). Conversely, using Notch1 gain of function by transgenically expressing active NICD1 under the control of the involucrin promoter, researchers have noted that Notch1 can promote granular differentiation (Nickoloff et al. 2002; Uyttendaele et al. 2004). A role for Notch in granular layer differentiation has also been supported by in vitro studies (Nickoloff et al. 2002).
In addition, Notch/RBP-J signaling also functions in hair follicle maintenance and terminal differentiation (Yamamoto et al. 2003; Pan et al. 2004; Vauclair et al. 2005). To date, the only evidence for a role for Notch signaling at the commitment step of epidermal differentiation comes from in vitro studies, where NICD1 was found to induce K1 expression. However, this mechanism appeared to be independent of RBP-J, suggesting a noncanonical role for Notch in cultured keratinocyte differentiation (Rangarajan et al. 2001). In contrast, no defects in spinous layer morphology or biochemistry have been noted in the Notch1 cKO mice (Rangarajan et al. 2001). Thus, it remains unclear whether Notch functions physiologically at an early step in epidermal differentiation, and if so, whether this is through canonical and/or noncanonical mechanisms.
In this report, we provide in vivo molecular and functional evidence to show that in addition to acting as a regulator of epidermal differentiation, Notch also operates at the commitment step to govern the balance between proliferative basal progenitors and terminally differentiating suprabasal progeny. We show that Notch exerts its effects at the basal–suprabasal juncture to specify spinous layers and repress basal epidermal genes by a canonical mechanism dependent on RBP-J. Moreover, we show that NICD/RBP-J induces spinous fate through a Hes1-dependent mechanism while repressing basal fate through a Hes1-independent mechanism.
Results
Evidence for canonical Notch/RBP-J-dependent activity in the epidermis
Using semiquantitative RT–PCR and immunofluorescence, we first examined which Notch pathway members might be responsible for mediating Notch function in the epidermis (Fig. 1; Supplementary Fig. S1). As reported previously, anti-RBP-J displayed nuclear staining throughout all of the epidermal layers (Supplementary Fig. S1C; Yamamoto et al. 2003). In contrast, Notch1 and Notch 3 were restricted to suprabasal layers, where they decorated the cell borders of most, if not all cells (Fig. 1A; Supplementary Fig. S1B). Anti-NICD1 antibodies (Abs) also labeled the nuclei of suprabasal cells (Fig. 1A; Okuyama et al. 2004; Pan et al. 2004).
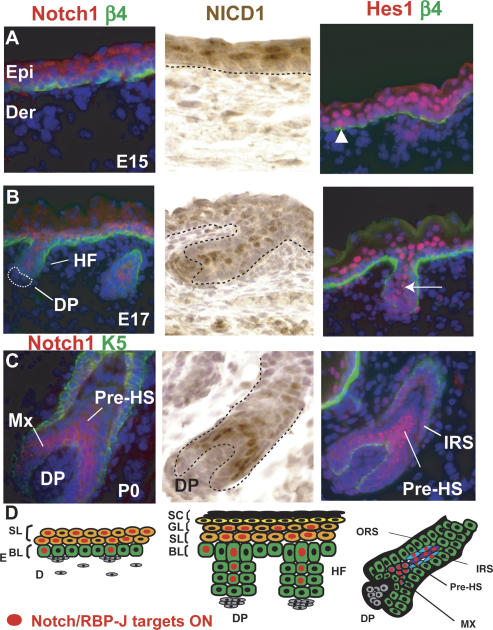
Figure 1.
Notch/RBP-J-dependent signaling during epidermal development. In response to Notch signaling, NICDs are generated that associate with DNA-binding protein RBP-J to activate Notch target gene Hes1 and/or Hey1, encoding bHLH transcription factors (see also Supplementary Fig. S1). (A–C) Immunofluorescence microscopy and immunohistochemistry to monitor Notch signaling during skin embryogenesis. During the early stage of stratification (E15), Hes1 is expressed in the first suprabasal cell layer, although occasional basal cells are also positive (arrowhead). At E17, Hes1-positive cells are restricted to the spinous cell layer and in the inner core of the hair follicle (arrow). As hair follicles mature (postnatal day 0, P0), Hes1 is expressed in IRS and HS cells. Abs are color-coded according to fluorescently tagged secondary Abs. (DAPI) Blue. (D) Summary of Notch signaling patterns. (NICD1) Notch1 intracellular domain; (Hes1) Hairy Enhancer of Split 1; (Epi) epidermis; (Der) dermis; (β4) β4 integrin; (HF) hair follicle; (Mx) matrix; (pre-HS) differentiating hair shaft precursors; (ORS) outer root sheath; (IRS) inner root sheath; (SL) spinous layer; (BL) basal layer; (GL) granular layer; (SC) stratum corneum; (DP) dermal papilla, the mesenchymal component of the hair follicle.
To determine whether canonical Notch signaling occurs in the epidermis, we conducted microarray analyses and semiquantitative RT–PCR to test for expression of the 10 hairy enhancer of split family members (Hes/ Hey), some of which have been identified as direct downstream target genes of NICD1/RBP-J (Iso et al. 2003). In skin of embryonic day 17.5 (E17.5) embryos, only Hes1, and to a lesser extent Hey1, were expressed and up-regulated upon Notch/RBP-J signaling in skin (Supplementary Table S1>). We conducted immunofluorescence microscopy with antibodies against these two transcription factors (Fig. 1; Supplementary Fig. S1). Of the two, Hes1 was detected in the epidermis, where it appeared concomitant with stratification. As development proceeded, Hes1 became concentrated in the nuclei of the first suprabasal (spinous) layer, with only an occasional positive basal cell (Fig. 1A,B, arrowhead). Taken together, these data demonstrated the existence of canonical Notch/ RBP-J activity in the epidermis, and suggested a particular prominence at the basal–suprabasal juncture.
Hes1, NICD,1 and Notch1 were also detected in the suprabasal core of cells emerging within embryonic hair germs (Fig. 1B, arrow). The timing and location at E17 coincided with the appearance of the prospective hair channel (inner root sheath, IRS) of the developing follicle, and was consistent with the established role of Notch in the follicle IRS (Kopan and Weintraub 1993; Pan et al. 2004). As the precursor cells of the hair shaft (Pre-HS) began to appear internally to the IRS, Hes1 expression extended to these cells (Fig. 1C). In mature follicles, Hey1 was detected in cortical and cuticle precursor cells of the hair shaft, while Hes 1 was most prominent in the IRS (Supplementary Fig. S1D,E). Thus, in the hair follicle, canonical Notch/RBP-J was reflected by Hes1 and Hey1, which appeared to be activated as matrix cells commit to terminally differentiate to form the hair shaft and its channel (Fig. 1D).
An essential role for RBP-J and canonical Notch signaling at an early step in epidermal differentiation
To avoid possible redundant functions of Notch receptors in the epidermis, and to directly determine the role of canonical Notch/RBP-J signaling during epidermal development, we conditionally ablated (cKO) the RBP-J gene by crossing RBP-J flox/flox mice (Tanigaki et al. 2002) with RBP-J flox/+ mice expressing K14-CRE (Vasioukhin et al. 1999). The K14 promoter is activated at E14.5 in embryonic basal progenitor cells, resulting in production of sufficient Cre recombinase to inactivate both floxed RBP-J alleles by the end of epidermal development. At birth, mice lacking both RBP-J alleles showed a complete loss of RBP-J in skin epithelium (Fig. 2A). Correspondingly, Hes1 was no longer detected in epidermis, consistent with a loss of canonical RBP-J/Notch signaling in the skin (Fig. 2B).
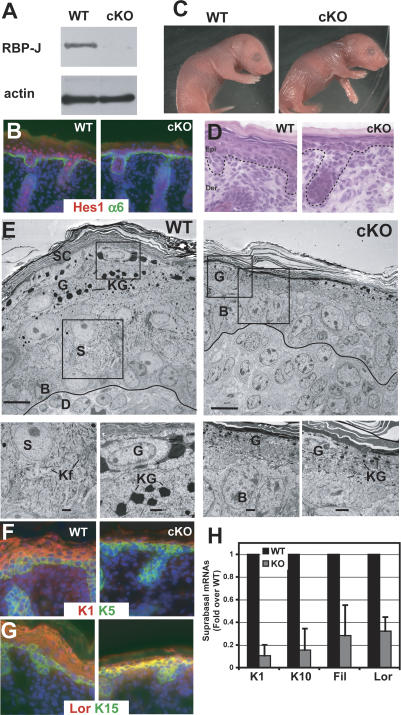
Figure 2.
Conditional loss of RBP-J suppresses the commitment of epidermal cells to terminally differentiate. (A) Quantitative loss of expression of the Notch target Hes1 in RBP-J cKO skin. (B) Notch/RBP-J target gene Hes1 expression is greatly diminished in the absence of RBP-J. (C) K14-Cre/RBP-J cKO newborn animals display a wrinkled and translucent appearance. (D) Histology reveals a thin cKO skin epidermis. (E) Transmission electron microscopy of ultrathin sections of newborn wild-type (WT) and cKO epidermis. Boxed areas are magnified in images along bottom. Electron-dense keratohyalin granules (KG) mark the granular layer (G) cells, and are diminished in RBP-J cKO epidermis. cKO epidermis also shows an absence of spinous layer (S) cells, typified by dense keratin filament (Kf) bundles. (F–H) Immunofluorescence of spinous (F) and granular (G) markers and RT–PCR (H) revealed a marked reduction in differentiation markers in RBP-J cKO epidermis.
RBP-J conditional null (cKO) mice were born with an expected Mendelian ratio but died from severe dehydration within a day. At birth, RBP-J cKO skin was wrinkled and translucent (Fig. 2C). Histological and ultrastructural analyses showed that RBP-J cKO epidermis was much thinner than wild-type epidermis (Fig. 2D,E). Suprabasal cells lacked the dense network of keratin filament bundles that are typical of spinous layer cells. Additionally, KO epidermis displayed fewer granular layer cells, which were also distinguished by a reduction in keratohyalin granules.
To further pinpoint the defect in RBP-J cKO epidermis, we examined biochemical markers of epidermal differentiation and proliferation. Consistent with the morphological defects, RBP-J cKO skin displayed a dramatic reduction in spinous and granular layer markers, both at the level of protein and mRNA (Fig. 2F,G). Taken together, these data reveal that the loss of RBP-J and canonical RBP-J/Notch signaling results in a defect in the commitment switch between the basal to spinous cell fate, leading to a severely impaired differentiation program. This contrasts strongly with Notch1 cKO epidermis, which is typified by an increase rather than decrease of granular marker expression and no noticeable defect in spinous layers (Rangarajan et al. 2001). This could be attributable to noncanonical roles for Notch1, as previously suggested (Rangarajan et al. 2001), functional redundancy among Notch receptors, and/or postnatal indirect effects due to a defective epidermal barrier.
In contrast to the repression of spinous and granular markers, basal markers appeared relatively normal in RBP-J cKO epidermis (Fig. 3). This included p63, integrins, and keratins K5 and K14 (Fig. 3A–D). Only slight increases in the overall intensity of signals were detected, and in most regions only a single layer of basal cells was seen (Figs. 2E, 3). This too contrasted strikingly with Notch1 cKO epidermis, where hyperproliferation and an expansion in basal layers were observed (Rangarajan et al. 2001; Nicolas et al. 2003).
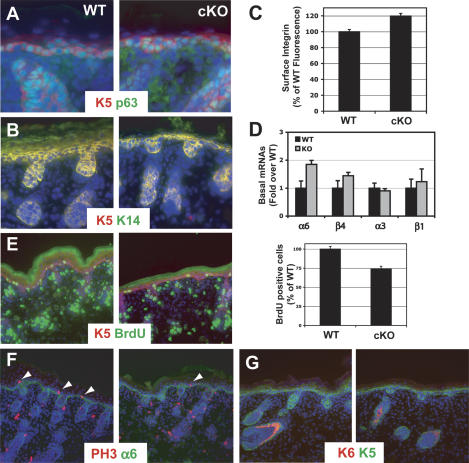
Figure 3.
Conditional loss of RBP-J does not disrupt basal gene expression but does decrease proliferation. (A–D) Basal markers Keratin 5, Keratin 14, p63, and integrins are not altered in RBP-J KO epidermis. Immunostaining (A,B), FACS analysis (C), and RT–PCR (D) for basal markers shows that these markers are maintained if not slightly increased in the absence of RBP-J KO. (E,F) Staining and quantification of BrdU incorporation (E) and phospho-Histone H3 (F) demonstrates diminished proliferation in RBP-J KO epidermis. (G) Comparable immunofluorescence between wild-type (WT) and RBP-J cKO newborn skin stained with Abs against Keratin 6 (K6), a marker of hyperproliferative disorders in the epidermis.
To more rigorously test the proliferative status of RBP-J cKO skin, we performed 5-Bromo-2′-deoxyuridine (BrdU)-labeling experiments and conducted immunofluorescence microscopy with biochemical markers of proliferation and apoptosis. Surprisingly, RBP-J cKO epidermis actually exhibited a reduction in proliferation as demonstrated by reductions in the number of cells positive for BrdU incorporation (Fig. 3E), for mitotic H3 histone phosphorylation (Fig. 3F), and proliferating nuclear antigen, Ki67 (data not shown). Moreover, _RBP-J_-null skin displayed only the normal hair follicle pattern of antibody staining for K6, which in epidermis is a reliable suprabasal marker of hyperproliferation (Fig. 3G). Finally, anti-active capase 3 immunostaining was comparable between wild-type and cKO cells, revealing no apparent difference in apoptosis (data not shown).
Postnatally, the epidermis is required to keep microbes out and essential body fluids in. When the epidermal barrier is compromised, hyperproliferation is frequently observed as a secondary response (for review, see Segre 2006). To determine whether this might happen in postnatal RBP-J-deficient skin, we grafted newborn RBP-J cKO and wild-type littermate skins onto the backs of Nude mice, and examined the grafts 24 d later. As judged by immunofluorescence microscopy with antibodies against Ki67 and K6, the RBP-J cKO epidermis displayed signs of elevated proliferation (Supplementary Fig. S2). Taken together, these findings suggest that loss of RBP-J/Notch signaling directly impairs epidermal differentiation, eliciting an indirect proliferative reaction in postnatal skin. Whether this explanation also accounts for the hyperproliferation observed in postnatal Notch1 cKO skin cannot be automatically inferred, as noncanonical Notch effects could also be involved.
Canonical Notch/RBP-J pathway is required for hair follicle terminal differentiation
The first follicles to develop, the guard hairs, are thought to initiate prior to K14 promoter activity, while additional waves of follicle morphogenesis occur throughout embryonic development (Vasioukhin et al. 1999). This said, overall numbers of hair follicles appeared to be comparable in newborn RBP-J cKO and wild-type animals, suggesting that the initiation step occurs even in the absence of RBP-J. Guard follicles still displayed markers of hair follicle maturation: companion layer (K6+), IRS (AE15+), and hair shaft precursors (AE13+) (Fig. 3G; Supplementary Fig. S3A–C). However, these follicles were runted and exhibited signs of impaired differentiation. In particular, the IRS marker Gata3 (Kaufman et al. 2003) was expressed at a lower level and in fewer cells, suggesting that Notch/RBP-J is already required during the earliest stages of IRS differentiation (Supplementary Fig. S3B).
To determine whether Notch/RBP-J signaling is involved in later stages of hair follicle differentiation, we grafted the skins of wild-type and RBP-J cKO neonatal mice onto the backs of Nude mice deficient in follicle morphogenesis. Within 2 wk after grafting, wild-type but not RBP-J cKO skin presented visible hairs (Supplementary Figs. S2, S3D). Microscopic examinations revealed gross defects in hair follicle maturation in the absence of RBP-J (Supplementary Fig. S3E–J). Those hair channels that formed were filled with keratinized material, but lacked IRS and hair shaft structures and displayed a paucity of hair differentiation markers. Oil-Red-O staining was largely negative in the RBP-J cKO skin, indicating that sebaceous gland differentiation is also impaired in the absence of canonical Notch/RBP-J signaling (Supplementary Fig. S3F). Ultrastructural analyses further revealed follicles as empty shells consisting of a dermal papilla, matrix cells, and an outer root sheath (ORS), encasing highly keratinized cells at the core. RBP-J-deficient hair follicles eventually degenerated into cystic structures, which could further compromise the epidermal barrier and contribute to indirect hyperproliferative defects in postnatal, but not newborn skin epidermis (Supplementary Fig. S3).
Overall, these results were consistent with prior hair follicle studies using Nestin-Cre RBP-J cKO mice, where ablation occurred in a mosaic manner in the hair follicle epithelium (Yamamoto et al. 2003). Our findings were also in good agreement with those of Pan et al. (2004), who conducted an _Msx2-Cre_-mediated matrix cell gene ablation of γ-secretase, required for production of NICD (Pan et al. 2004). Our results thus confirm and support the view that canonical Notch/RBP-J signaling does not affect hair follicle initiation but acts downstream to impair differentiation of matrix to IRS and/or hair shaft.
Notch signaling in the basal layer represses basal fate, promotes spinous fate, and results in skin blistering
We next asked whether the absence of spinous layers in RBP-J cKO epidermis occurs as a consequence of defective cell fate specification or aberrant terminal differentiation. To answer this question, we constitutively activated Notch signaling in skin epithelium by selectively targeting bicistronic expression of GFP and the intracellular domain of Notch1 (NICD1) (Lox-stop–Lox-RosaN-ICD-ires-GFP X K14-Cre) (Vasioukhin et al. 1999; Murtaugh et al. 2003). By E15, NICD-ires-GFP mRNA was detected in the epidermis, and both NICD and GFP proteins were detected shortly thereafter (Fig. 4A; Supplementary Fig. S4A,B). The NICD protein was active, as judged by the parallel increase in expression of three NICD/RBP-J target genes including Hes1 (Fig. 4B; Supplementary Fig. S4C; Supplementary Table S1; Krebs et al. 2001; Lamar et al. 2001; Iso et al. 2003).
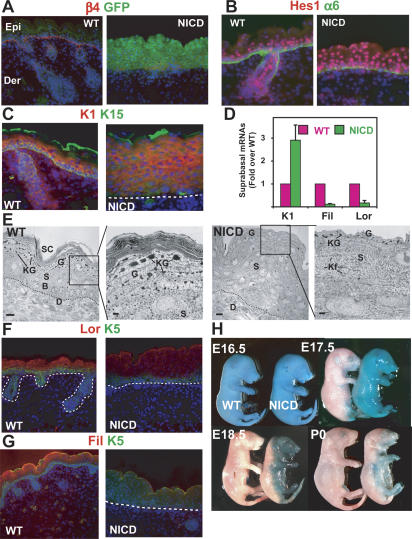
Figure 4.
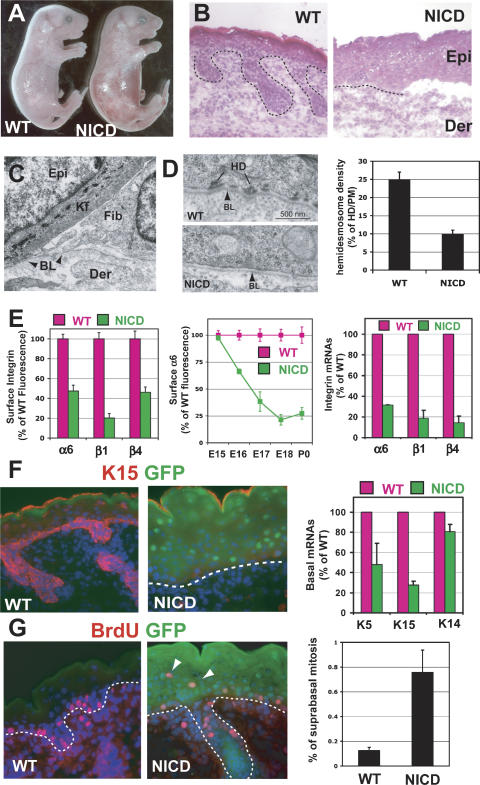
Increased Notch signaling through NICD promotes activation of the target gene Hes1 and the first step of lineage commitment in the epidermis. (A) Skin-specific expression of the NICD-ires-GFP bicistronic transgene during embryogenesis, as measured by GFP epifluorescence. (B) NICD transgene expression results in NICD/RBP-J target gene Hes1 induction in the epidermis. (C) NICD transgenic animals display expansion of spinous layers as marked by Keratin 1 (K1) staining. (D)RT–PCR for spinous (K1) and granular markers (Filaggrin and Loricrin) shows altered differentiation in NICD1-expressing epidermis. (E) Ultrastructural analyses show that keratohyalin granules (G) are reduced and dense keratin bundles typical of spinous layers (S) are increased in NICD1 epidermis. (Kf) Keratin filaments. (F,G) Immunofluorescence microscopy reveals that both Loricrin and Filaggrin, the principal components of granular cells, are decreased in NICD transgenic epidermis. (H) Barrier assay, as determined by penetration of blue dye. The barrier is normally acquired and dye excluded by E17.5, but is defective in NICD transgenic animals, even after birth.
Consistent with an inductive role for Notch signaling in spinous fate determination, immunostaining, real-time RT–PCR, and immunofluorescence microscopy all revealed a massive expansion of the spinous layers in NICD transgenic epidermis (Fig. 4C–E). In contrast, granular cell differentiation was severely impaired, as demonstrated morphologically by the reduction in the number and the size of keratohyalin granules (Fig. 4E) and biochemically by the reduction in the expression of filaggrin and loricrin, two major components of granular cells (Fig. 4F,G). The functional outcome of these defects in terminal differentiation was a defective epidermal barrier (Fig. 4H).
Newborn NICD1 transgenic mice also exhibited severe skin blistering, which by histological analyses, was attributable to epidermal detachment from the underlying dermis (Fig. 5A,B). Ultrastructurally, the separation appeared to occur between the basal epidermis and the underlying basal lamina (BL), and in nonblistered regions, a marked reduction in hemidesmosomes was observed (Fig. 5C,D). This type of blistering is known in humans as junctional epidermolysis bullosa, and it can be caused by mutations in the hemidesmosomal integrins (α6β4) or in the laminin 5 chains that compose their ECM ligand (Fine et al. 2000).
Figure 5.
Increased Notch signaling results in inhibition of basal character and blistering disorder. (A) NICD1 transgenic animals display skin blistering at birth. (B) Histology reveals skin blistering at birth in NICD1 animals. (C) While the BL remains intact, there is a marked decrease in hemidesmosomes in NICD1 epidermis as quantified in D. (E) Temporal reduction in surface integrins and their mRNAs correlates with NICD induction in transgenic basal cells, analyzed by FACS (dispase and trypsin-treated epidermis, GFP+ gated cells). (F) Immunofluorescence microscopy and RT–PCR to monitor NICD-mediated changes in expression of basal markers. (G) Presence of atypical suprabasal S-phase epidermal cells in E18 transgenic mice pulsed 4 h with BrdU. (Graph) Quantification of suprabasal mitotic cells is determined by FACS analysis of BrdU-positive cells in α6-negative cells.
Fluorescence-activated cell sorting (FACS) revealed a marked decline in α6, β4, and β1 surface levels, with similar kinetics to the rise in NICD1 expression (Fig. 5E). These changes were further reflected at the level of mRNA (data shown). Based on these data, we attribute the skin blistering and loss of hemidesmosomes to loss of integrin expression that occurred concomitant with NICD1 induction. Hair follicle downgrowths were also diminished in NICD transgenic skin, although we did not pursue it further as this is a common feature associated with the loss of basal integrins (Dowling et al. 1996; Raghavan et al. 2000). In addition to the repression of basal integrins, other basal markers were also markedly down-regulated in NICD1 transgenic epidermis (Fig. 5F).
Although caspase-3 is not thought to be activated or involved in epidermal differentiation (Fischer et al. 2005), mice deficient for caspase 3 in the epidermis display a decrease in granular gene expression, markers that are up-regulated in Notch1 cKO epidermis (Okuyama et al. 2004). Based on this parallel and on additional in vitro studies, it has been proposed that Notch1 induces epidermal differentiation by increasing caspase3 activity and caspase 3 gene expression (Okuyama et al. 2004), which is also a hallmark of apoptosis.
Given this report, we therefore examined caspase 3 activity in the NICD transgenic skin. In regions where the skin epidermis had blistered, caspase 3 activity and other signs of apoptosis were detected (Supplementary Fig. S4D). This was not surprising, since cell survival is thought to be dependent on attachment to an underlying BM. However, localization of Hes1 indicated that Notch activity was strong throughout the epidermis of NICD transgenic skin (Fig. 4B), and yet we did not observe signs of enhanced caspase 3 activity, nor did we detect morphological changes that would suggest an increase in apoptosis in the nonblistered areas (data not shown). We also did not detect signs of altered caspase-3 activity or apoptosis in our RBP-J loss-of-function mice. Thus, we have not been successful in finding support for canonical Notch/RBP-J signaling in controlling caspase 3 activity in vivo.
NICD1 transgenic epidermis did exhibit a mild hyper-proliferative response, and Ki67 and BrdU labeling revealed an increase in cycling cells, which atypically extended into the suprabasal layers (Fig. 5G; Supplementary Fig. S4E). FACS analyses confirmed the presence of both α6-positive and α6-negative mitotically active cells within the NICD1 epidermal population, whereas wild-type mitotic cells were located largely, if not exclusively, in the α6-positive cells. Although cell division is not typically associated with spinous fate, proliferation normally occurs suprabasally in wild-type embryonic epidermis during the early stages of epidermal stratification (Lechler and Fuchs 2005). Notch signaling is active at this stage (Supplementary Fig. S1A), suggesting the possibility that these findings may be physiologically relevant in early development. Another possibility is that Notch signaling only promotes certain aspects of spinous differentiation and regulation of proliferation and spinous fate determination are disconnected in NICD1 transgenic epidermis.
Spinous cell gene expression is dependent on canonical Notch/RBP-J signaling
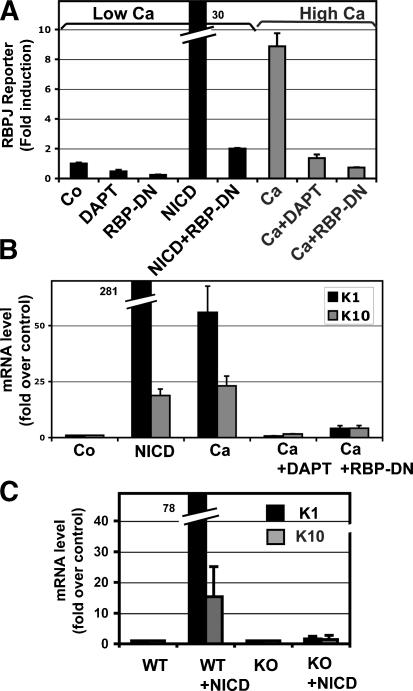
To elucidate the molecular mechanism by which Notch promotes the basal to suprabasal transition and the acquisition of spinous fate, we exploited the ability to culture primary murine keratinocytes (MK) and induce them to differentiate upon increased calcium exposure (Hennings et al. 1980). For these experiments, we infected MK at high efficiency with GFP-tagged retroviral expression vectors and FACS purified the cells for analyses (Supplementary Fig. S5). We first showed that an RBP-J reporter gene could be selectively activated when MK were calcium switched to induce terminal differentiation (Fig. 6A). This activation was dependent on Notch signaling since (1) NICD expression alleviated the requirement for calcium; (2) the γ-secretase inhibitor DAPT, which prevents NICD production (Fortini 2002; Geling et al. 2002), also inhibited RBP-J reporter activity induced by the calcium switch; and (3) the activity appeared to be dependent on NICD's association with RBPJ, since dominant-negative RBP-J (RBP-DN) blocked reporter activity (Fig. 6A).
Figure 6.
Calcium induces spinous differentiation through canonical Notch/RBP-J signaling. (A–C) Calcium induces RBP-J reporter gene activity (A), and endogenous K1/K10 gene expression (B) by a Notch/RBP-J-dependent mechanism, as this effect is blocked by expression of a dominant-negative RBP-J. (C) Stimulation of spinous markers by NICD1 is completely blocked in the absence of RBP-J. RBP-J reporter assays and quantifications of K1 and K10 were performed on FACS-isolated MK cultured in low-calcium (differentiation-restricted) or high-calcium (differentiation-promoting) medium, as indicated. Where noted, cells were also transduced with IRES-GFP retroviral expression vectors encoding NICD, dominant-negative RBP-J (RBP-DN), or empty vector (Co, control). Where indicated, cells were also treated with DAPT (_N_-[_N_-{3,5-difluorophenacetyl}-lalanyl]-_S_-phenylglycine _t_-butyl ester) to inhibit Notch processing and NICD production (Geling et al. 2002).
Similarly, calcium induced the endogenous expression of the two major spinous layer genes, K1 and K10 (Hennings et al. 1980). K1/K10 mRNA levels were dramatically elevated 25- to 75-fold in MK exposed to calcium or expressing NICD1 (Fig. 6B). Again, this effect could be quantitatively repressed by DAPT or RBP-J-DN. Spinous gene induction by NICD1 was dependent on RBP-J, as these genes could not be induced in RBP-J KO keratinocytes (Fig. 6C). Taken together, these findings firmly establish that spinous gene expression is dependent on both NICD and RBP-J, and that canonical Notch signaling is responsible for these effects.
Induction of spinous cell differentiation is dependent on Hes1
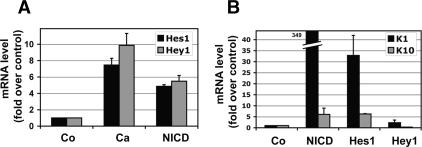
To evaluate whether spinous layer differentiation is dependent on the Hes/Hey downstream targets of RBP-J/ NICD, we first examined mRNA levels for Hes/Hey family members in MK. MK appeared to differ somewhat from epidermis in its expression of these proteins. As judged by microarray analyses, Hes1 and Hey1 mRNAs were the main family members expressed in MK (Supplementary Table S1). In vitro, both Hes1 and Hey1 mRNAs were elevated by approximately sixfold to 10-fold in MK exposed to calcium (Fig. 7A). NICD expression had a similar effect, in this case, with approximately five-to sixfold enhancement of Hes1 and Hey1 mRNAs (Fig. 7A). Unexpectedly, endogenous K1/K10 mRNA levels were enhanced by as much as 40-fold in MK retrovirally expressing Hes1, but not Hey1, in low-calcium conditions (Fig. 7B). Although Hes1 is conventionally considered a transcriptional repressor, it can also act as transcriptional activator, as shown during the differentiation of neuronal stem cells (Ju et al. 2004). In preadipocytes, Hes1 overexpression can both induce and repress genes (Ross et al. 2006). Whether Hes1 induces spinous genes directly or indirectly is an interesting question, but it is beyond the scope of the present study.
Figure 7.
The Notch target gene Hes1 induces spinous gene expression. (A) Endogenous Hes1 and Hey1 can both be induced by either calcium or NICD1 expression. (B) Hes1 but not Hey1 can induce expression of spinous markers K1 and K10. MK were cultured in low calcium and infected with IRES-GFP retroviral expression vectors encoding NICD, Hes1, Hey1, or empty vector (Co, control). Proper expression was confirmed by immunoblot (see Supplementary Fig. S5B).
Basal gene repression is dependent on canonical Notch/RBP-J signaling, but does not require Hes1
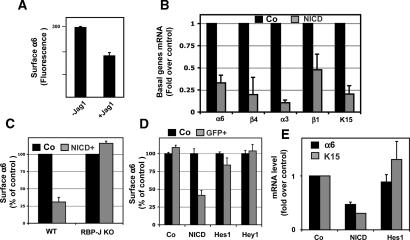
NICD exerted a potent effect in vitro as it did in vivo in repressing basal gene expression (Fig. 8A,B). When MK were exposed to either the Notch ligand Jagged-1 (Jag1) or retrovirally infected with a NICD expression vector, all five basal genes tested exhibited reduced levels of mRNA expression. Repression of basal genes was also dependent on RBP-J. In _RBP-J_-null MK expressing NICD1, basal gene expression was comparable with that seen in control wild-type MK (Fig. 8C). Taken together, these findings suggest that Notch-mediated repression of basal genes is dependent on canonical Notch/RBP-J signaling.
Figure 8.
Basal markers are reduced by active Notch signaling. (A) Coculturing K14-GFP MK with cells expressing the Notch ligand Jagged1 (Jag1) results in a reduction in surface α6 integrin levels. MK were cocultured and then subjected to FACS for quantification. (B) MK were infected as in Figure 7 and then assayed for basal layer genes as monitored by RT–PCR of GFP-FACS-isolated keratinocyte mRNAs. (C) The absence of down-regulation of α6 integrin expression by NICD1 in RBP-J cKO cells demonstrated that Notch repression of basal integrin is RBP-J dependent. (D,E) MK were infected with NICD, Hes1, and Hey1, but assayed by FACS (D) and RT–PCR (E) for basal marker expression, and neither Hes1 nor Hey1 repressed basal markers.
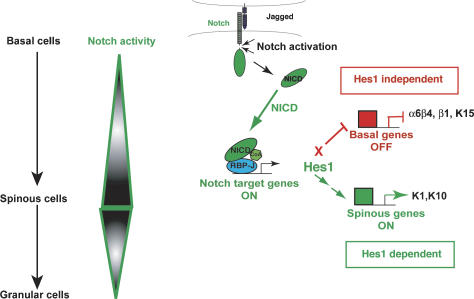
While both spinous gene activation and basal gene repression were dependent on canonical Notch signaling, they differed in their response to Hes1. In contrast to spinous genes, the basal markers tested displayed no repression by either Hes1 or Hey1 (Fig. 8D,E). The model in Figure 9 summarizes these findings.
Figure 9.
A model depicting the roles of Notch signaling pathways in governing the transition from proliferation to differentiation in epidermal progenitors. Upon ligand engagement, Notch is activated, releasing NICD. NICD plays two roles in driving the transition from basal to spinous cells. NICD interacts with RBP-J to drive expression of Hes1, a canonical target gene. This leads to the downstream induction of spinous layer genes encoding differentiation-specific proteins. NICD represses basal genes including integrins, allowing basal cells to detach from the BL during stratification by a mechanism independent of Hes1.
Discussion
Canonical Notch signaling as a molecular switch in commitment of basal cells to a spinous cell fate
The Notch pathway is highly conserved and is known to function by cell–cell interactions in a wide variety of tissues where such diverse roles as cell fate specification, stem cell renewal, maintenance, proliferation, apoptosis, and differentiation have been described (Artavanis-Tsakonas et al. 1999). In response to ligand engagement, Notch undergoes proteolysis to release NICD, which enters the nucleus and acts as a transcription cofactor. In the canonical Notch signaling pathway, NICD associates with the DNA-binding protein RBP-J to stimulate transcription of downstream target genes, most notably Hes and Hey of the basic helix–loop–helix (bHLH) family (for review, see Iso et al. 2003).
It has previously been reported that in epidermis lacking Notch1, spinous cell layers appear normal, and yet there is a moderate increase in expression of granular markers, suggestive of a defect late in terminal differentiation (Rangarajan et al. 2001). In vitro studies have further suggested that NICD1 overexpression promotes keratinocyte differentiation, including some spinous layer genes such as K1; however, based on those studies, the mechanism appeared to be independent of RBP-J, suggestive of a noncanonical mechanism of action (Rangarajan et al. 2001). More recent studies in vitro have indicated that NICD1 also exerts a negative control on expression of p63 (Nguyen et al. 2006), which is a key basal transcription factor implicated in stratification and stem cell maintenance (Mills et al. 1999; Yang et al. 1999). Prior to our ablation of RBP-J in the current study, there was no in vivo evidence to clarify whether the effects of Notch are dependent on RBP-J or a noncanonical pathway.
By targeting RBP-J for ablation in the skin epidermis, we uncovered a hitherto unappreciated role for canonical Notch signaling in the epidermis. Moreover, our studies further unveiled a physiological role for Notch/RBP-J signaling at the earliest step in the commitment of a proliferative basal cell to embark on the differentiation program of the epidermal lineage. By fully ablating canonical Notch signaling in the epidermis, we have defined RBP-J/Notch function as a governor of the basal to spinous cell fate. Our gain-of-function studies using NICD activation are in good agreement with this positioning and further underscore the importance of RBP-J/Notch in spinous cell fate specification at the basal–suprabasal juncture. The preferential expression of Hes1 in the spinous layer and the defects in granular differentiation observed in NICD1 transgenic mice further suggest that Notch/RBP-J signaling is most prevalent in the spinous layer. That said, additional roles in later stages of the differentiation cannot be ruled out.
Since the Notch ligand Jagged2 is expressed in basal cell progenitors, while Jagged1 is suprabasal (Powell et al. 1998), the molecular event that allows canonical Notch signaling to function as a master switch might be the regulation of Notch itself whether at the transcriptional, translational, and/or post-translational levels. The differential expression in the skin epidermis of the Fringe-related glycosyl-transferase known to regulate Notch activity might be relevant to this process (Thelu et al. 1998). The presence of occasional Hes1-positive basal cells could be a sign that the differentiation switch induced by Notch/RBP-J may even occur before cells leave the basal compartment. An early role for Notch could explain why K1/K10-positive cells have sometimes been detected within the basal layer (Schweizer et al. 1984). In this regard, it is also tempting to speculate that Notch might be activated in suprabasal daughter cells arising from asymmetrical cell divisions, as occurs during Drosophila neurogenesis (Bardin et al. 2004; see also Lechler and Fuchs 2005).
The induction of spinous fate by Notch/RBP-J in the epidermis contrasts with the restrictive function of Notch signaling in cell fate determination observed in the central nervous system, intestine, and pancreas (Apelqvist et al. 1999; Jensen et al. 2000; Yang et al. 2001; Murtaugh et al. 2003; Fre et al. 2005; van Es et al. 2005; Yoon and Gaiano 2005; Fujikura et al. 2006). In those tissues, Notch regulates cell fate determination by up-regulation of Hes/Hey family members, which repress the expression of atonal and achaete–scute complex family members (Kageyama et al. 2005). The downstream effectors of Hes/Hey functions in the epidermis remain to be determined.
Based on in vitro studies utilizing a dominant-negative form of RBP-J, the ability of NICD to induce spinous gene expression was suggested to be independent of RBP-J (Rangarajan et al. 2001). However, in our genetic studies in vivo, we observed a loss of spinous genes in _RBP-J_-null epidermis and no induction of spinous genes by NICD in keratinocytes lacking RBP-J. These data provide compelling evidence that in vivo, the spinous fate induced by Notch is dependent on the canonical—i.e., RBP-J-dependent—pathway.
Canonical Notch signaling and cell proliferation
Elevated Notch signaling has been implicated in many human cancers, including T-cell leukemias where activating mutations of the Notch pathway were first identified (for review, see (Maillard and Pear 2003; Wilson and Radtke 2006) In the intestine, for example, canonical Notch signaling seems to function as a governor of the crypt progenitor cells: Postnatal inactivation of RBP-J results in a loss of proliferating cells, while NICD overexpression inhibits differentiation of the crypt progenitors (Fre et al. 2005; van Es et al. 2005). In skin, however, Notch has been viewed as a tumor suppressor (Rangarajan et al. 2001; Nicolas et al. 2003). This view has been largely predicated on the basis that Notch1 targeted epidermis displays an increase in basal cell proliferation and a hyperthickening of the skin epidermis.
Based on the striking hyperproliferation seen in the absence of Notch1 (Rangarajan et al. 2001; Nicolas et al. 2003), we were surprised to find that rather than promoting proliferation, quantitative loss of canonical Notch signaling resulted in a marked thinning of embryonic epidermis, and this was not accompanied by an overt increase in the status of proliferation within the RBP-J-deficient epidermis. How do we reconcile these seemingly disparate results?
A priori, it could be that Notch1 functions as a tumor suppressor through a noncanonical pathway (Rangarajan et al. 2001; Nicolas et al. 2003). While this is formerly possible, our studies suggest that perturbations in canonical Notch signaling compromise epidermal barrier function, which in turn leads to postnatal hyperproliferation as an indirect secondary reaction. Our interpretation is supported by recent studies showing that squamous cell carcinomas develop in transgenic mice over-expressing a dominant-negative form of mastermind, which is a necessary coactivator of canonical Notch signaling (Proweller et al. 2006).
It has been speculated that the epidermal basal layer attempts to compensate for a defective barrier by proliferating to generate a thickened epidermis. In this scenario, mutations in Notch1 still contribute to tumorigenesis, but do so in an indirect fashion. In addition, the loss of follicle stem cells and hair degeneration following Notch1 ablation could contribute to the hyperproliferative state of the epidermis (Vauclair et al. 2005). Finally, we cannot rule out the possibility that simply removing one member of the Notch family might have pleiotropic effects not found upon the removal of the entire family or that RBP-J can function independently of Notch signaling (Barolo et al. 2000; Beres et al. 2006). Future studies will be necessary to sift through these possibilities, which need not be mutually exclusive.
Hes1-dependent and Hes1-independent pathways in canonical Notch signaling in vivo
In normal epidermis, Notch1, Notch3, and Hes1 are all expressed suprabasally, where basal genes are down-regulated. Hence, the simplest physiological explanation for these observations is that canonical Notch signaling suppresses basal gene expression after a cell commits to a spinous cell fate. In agreement with this notion, basal markers are markedly down-regulated when Notch signaling is artificially activated in the basal layer of the epidermis. However, in our vitro studies, although NICD1’s suppressive effects on basal gene expression required RBP-J, it was not dependent on the activation of Hes1. In this regard, the ability of Notch signaling to regulate integrin expression appeared to be distinct from that of p63, where a role for Hes1 has been proposed (Nguyen et al. 2006).
Although we did not observe a role for Hes1 in regulating basal integrin and keratin gene expression, Hes1 did appear to function in promoting spinous gene expression in MK. Consistent with this notion, the NICD1 transgenic mice displayed a dramatic expansion of spinous gene expression including Hes1, and conversely, Hes1 and expression of other spinous genes were obliterated in _RBP-J_-null epidermis. Taken together, our studies support a role for Hes1 in spinous gene expression, but suggest that NICD/RBP-J-mediated repression of at least some basal markers is independent of the classical bHLH Notch target genes (Fig. 9; see also Supplementary Fig. S6). In the future, genome-wide chromatin immuno-precipitation analyses of RBP-J might provide new candidates of the downstream effectors of Notch signaling that mediate basal gene repression in the skin epidermis.
Summary
In closing, we have defined a novel role for Notch/RBP-J signaling as a molecular switch at the basal–spinous cell juncture during epidermal development. Our studies further suggest a role for the Notch signaling pathway in the detachment of progenitor cells from the BM as well as their commitment to activate early genes in the terminal differentiation program that culminates in epidermal barrier formation. In orchestrating this key transition in the epidermal lineage, cells utilize the canonical downstream target Hes1 for some, but not other steps in the pathway. Overall, these studies provide new and important findings in the global understanding of the complex mechanisms by which Notch governs its diverse processes in the epidermis and in other tissues of the body.
Materials and methods
Mice
Lox-stop–Lox-RosaNICD-ires-GFP mice (Murtaugh et al. 2003), a generous gift from D. Melton (Harvard University, Boston, MA), were crossed with K14-Cre mice (Vasioukhin et al. 1999). RBP-J floxed mice, a generous gift from T. Honjo (Kyoto University, Kyoto, Japan) (Tanigaki et al. 2002), were crossed with K14-Cre mice. BrdU (Sigma-Aldrich) was administered to pregnant mice by injection (50 μg/g BrdU).
Immunostaining and blots
Histology and immunofluorescence were performed as described (Blanpain et al. 2004). For immunohistochemistry, tissues were fixed in formaldehyde and then dehydrated and embedded in paraffin. Antigen unmasking was performed in Retreiver 2100 for 20 min (Pick Cell Laboratories BV). Abs and dilutions used were α6-integrin, β4-integrin (rat, 1:100; BDPharmingen), Notch1 full-length (rabbit, 1:200; Santa Cruz Bio-technology), Notch3 (Goat, 1:50; R&D Systems), cleaved NICD1 (rabbit, 1:100; Cell Signaling), K5 (chicken, 1:500; Fuchs Laboratory), AE13 and AE15 (mouse, 1:50; T.T. Sun, New York University, New York), c-Kit (rat, 1:100; BD-Pharmingen), K15 (rabbit, 1:500; Fuchs Laboratory), BrdU (rat, 1:100; Abcam), Hey1 (rabbit, 1:500; L. Kedes, University of Southern California, Los Angeles, CA), Hes1 (rabbit, 1:250; N. Brown, Cincinnati Children's Hospital Medical Center, Cincinnati, Ohio), K1 (rabbit, 1:250; Fuchs Laboratory), K6 (rabbit, 1:1000; Fuchs Laboratory), Filaggrin (rabbit, 1:100; Fuchs Laboratory), Involucrin (rabbit, 1:400; Fuchs Laboratory), Loricrin (rabbit, 1:400; Fuchs Laboratory), Ki67 (rabbit, 1:100; Castro-nova), and active capase-3 (rabbit, 1:500; R&D Systems). Secondary Abs coupled to FITC,
Alexa488, or Texas-Red were from Jackson Laboratories. Nuclei were labeled by 4′v6′-diamidino-2-phenylindole (DAPI) for immunofluoresence.
For immunoblots, total cell lysates for both protein and RNA isolation were prepared in lysis buffer (Absolutely RNA, Stratagene). For protein isolation, samples were precipitated in acetone, washed in ETOH, and resuspended in Laemli protein buffer. Samples were run on 4%–12% gradient gels, transferred to nitrocellulose, and blotted overnight in the indicated antibodies.
Electron microscopy
Tissues were fixed for >1 h in 2% glutaraldehyde, 4% formaldehyde, and 2 mM CaCl2 in 0.05 M sodium cacodylate buffer, and then processed for Epon embedding. Samples were visualized with a Tecnai 12-G2 transmission electron microscope. For hemidesmosome quantification, 100 digital images (49,000×) were randomly taken at sites of the dermal–epidermal boundary for each sample. Total continuous membrane length and individual hemidesmosome lengths along the plasma membrane were measured using ImageJ (NIH). Results were expressed as micrometer of hemidesmosome length per micrometer of plasma membrane.
FACS analyses
FACS analyses of keratinocyes from embryonic mouse back-skin were performed as described (Blanpain et al. 2004). Cell cycle analyses were performed as described (Blanpain et al. 2004) using the BrdU Flow Kit (Pharmingen). FACS purification of transduced keratinocytes was performed on a FACSVantage SE system equipped with FACS DiVa software (BD Biosciences). Cells were gated for single events and viability, then sorted according to their GFP expression.
Real-time PCR
RNAs were isolated from cells. Following DNase treatment (Absolutely RNA, Stratagene), RNA quality and concentration were measured by Lab on a Chip (Agilent). Equal RNA amounts were added to reverse-transcriptase reaction mix (Invitrogen) with oligo-dT(12) as primer. All PCR test primers flanked exon–intron boundaries to avoid misinterpretation from genomic contamination. RT–PCRs of RNA (i.e., not reverse-transcribed) were used as negative controls. Real-time PCR was conducted with a LightCycler system (Roche Diagnostics). Reactions were performed using the indicated primers and template mixed with the LightCycle DNA master SYBR Green kit and run for 45 cycles. Specificity of the reactions was determined by subsequent melting curve analysis. LightCycle analysis software was used to remove background fluorescence (noise band). The number of cycles needed to reach the crossing point for each sample was used to calculate the amount of each product using the 2−ΔΔCP method. Levels of PCR product were expressed as a function of GAPDH and/or HPRT.
Cell culture and reporter assays
MK were plated in E medium (15% serum, 0.05 mM CaCl) (Blanpain et al. 2004), infected with retrovirus or transfected using fugene 6 (Roche), and cultures were analyzed for gene expression 3 d later. L cells stably expressing Jagged1 were a gift of Geraldine Weimaster (University of California at Los Angeles, Los Angeles, CA). Switches in calcium concentration were performed by adding calcium chloride at 1.2 mM final concentration. For all of the experiments using the γ-secretase inhibitor, cells were cultured with _N_-(_N_-[3,5-difluorophenacetyl]-L-alanyl)-_S_-phenylglycine _t_-butyl ester (DAPT) in 0.1% DMSO at 1 μM final concentration or in 0.1% DMSO for the control cells (Geling et al. 2002). The medium was refreshed every day to maintain the inhibition by DAPT.
For reporter assays, we used the dual-glo luciferase assay kit to monitor both the firefly luciferase activity of the construct indicated and Renilla luciferase under control of a constitutive promoter to control for transfection efficiency. Results were expressed as a ratio of firefly to Renilla luciferase and set to a baseline as indicated.
Plasmids/constructs
Retroviral (MSCV) constructs encoding a Flag version of Hey1 was performed by inserting Hey cDNA obtained from M. Gessler (Wurzburg University, Wurzburg, Germany) into FlagX3 vector (Sigma) and subcloning the N-terminal Flag-tagged Hey1 into MSCV. Retroviral constructs expressing various cDNAs containing an IRES-GFP element to monitor expression were kindly provided by W. Pear, University of Pennsylvania, Philadelphia, PA (Hes1); T. Kadesh, University of Pennsylvania, Philadelphia, PA (NICD). RBP-J, RBP-J-DN, and RBP-J-DA cDNAs in mammalian expression vectors (RDB1801, RDB3022, and RDB3023), were a gift of the Riken BioResource Center DNA Bank with permission from Dr. T. Honjo (Riken, Kyoto, Japan). The RBP-J reporter construct was a gift from Dr. S.D. Hayward (Johns Hopkins University, Baltimore, MD).
Acknowledgments
In the Materials and Methods, we cite our many colleagues for their generous contribution of mice and reagents. We acknowledge colleagues in the Fuchs laboratory for constructive discussions and criticisms. We thank M. Nikolova, J. de la Cruz-Racelis, L. Polak, and N. Stokes for technical assistance. A special thank you goes to H. Rhee and R. Yi for their valuable comments and their willingness to help in the preparation of this manuscript. C.B. was supported by an HFSP fellowship, and W.L. was supported by an NRSA fellowship. E.F. is an HHMI investigator. This work was supported in part by R01 AR27883 (E.F.).
Footnotes
References
- Apelqvist, A., Li, H., Sommer, L., Beatus, P., Anderson, D.J., Honjo, T., Hrabe de Angelis, M., Lendahl, U., Edlund, H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., Rand, M.D., Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bardin, A.J., Le Borgne, R., Schweisguth, F. Asymmetric localization and function of cell-fate determinants: A fly's view. Curr. Opin. Neurobiol. 2004;14:6–14. doi: 10.1016/j.conb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Barolo, S., Walker, R.G., Polyanovsky, A.D., Freschi, G., Keil, T., Posakony, J.W. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Beres, T.M., Masui, T., Swift, G.H., Shi, L., Henke, R.M., MacDonald, R.J. PTF1 is an organ-specific and Notch-independent basic helix–loop–helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain, C., Lowry, W.E., Geoghegan, A., Polak, L., Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Dai, X., Segre, J.A. Transcriptional control of epidermal specification and differentiation. Curr. Opin. Genet. Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, J., Yu, Q.C., Fuchs, E. β4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine, J.D., Eady, R.A., Bauer, E.A., Briggaman, R.A., Bruckner-Tuderman, L., Christiano, A., Heagerty, A., Hintner, H., Jonkman, M.F., McGrath, J., et al. Revised classification system for inherited epidermolysis bullosa: Report of the Second International Consensus Meeting on diagnosis and classification of epidermolysis bullosa. J. Am. Acad. Dermatol. 2000;42:1051–1066. [PubMed] [Google Scholar]
- Fischer, H., Rossiter, H., Ghannadan, M., Jaeger, K., Barresi, C., Declercq, W., Tschachler, E., Eckhart, L. Caspase-14 but not caspase-3 is processed during the deveopment of fetal mouse epidermis. Differentiation. 2005;73:406–413. doi: 10.1111/j.1432-0436.2005.00046.x. [DOI] [PubMed] [Google Scholar]
- Fortini, M.E. γ-Secretase-mediated proteolysis in cell-sur-face-receptor signalling. Nat. Rev. Mol. Cell Biol. 2002;3:673–684. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- Fre, S., Huyghe, M., Mourikis, P., Robine, S., Louvard, D., Artavanis-Tsakonas, S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., Raghavan, S. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- Fujikura, J., Hosoda, K., Iwakura, H., Tomita, T., Noguchi, M., Masuzaki, H., Tanigaki, K., Yabe, D., Honjo, T., Nakao, K. Notch/Rbp-j signaling prevents premature endocrine and ductal cell differentiation in the pancreas. Cell Metab. 2006;3:59–65. doi: 10.1016/j.cmet.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Geling, A., Steiner, H., Willem, M., Bally-Cuif, L., Haass, C. A γ-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings, H., Michael, D., Cheng, C., Steinert, P., Holbrook, K., Yuspa, S.H. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980;19:245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- Iso, T., Kedes, L., Hamamori, Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jensen, J., Pedersen, E.E., Galante, P., Hald, J., Heller, R.S., Ishibashi, M., Kageyama, R., Guillemot, F., Serup, P., Madsen, O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Ju, B.G., Solum, D., Song, E.J., Lee, K.J., Rose, D.W., Glass, C.K., Rosenfeld, M.G. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kageyama, R., Ohtsuka, T., Hatakeyama, J., Ohsawa, R. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Kaufman, C.K., Zhou, P., Pasolli, H.A., Rendl, M., Bolotin, D., Lim, K.C., Dai, X., Alegre, M.L., Fuchs, E. GATA-3: An unexpected regulator of cell lineage determination in skin. Genes & Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan, R., Weintraub, H. Mouse notch: Expression in hair follicles correlates with cell fate determination. J. Cell Biol. 1993;121:631–641. doi: 10.1083/jcb.121.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, L.T., Deftos, M.L., Bevan, M.J., Gridley, T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 2001;238:110–119. doi: 10.1006/dbio.2001.0408. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. Notch signaling: Control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lamar, E., Deblandre, G., Wettstein, D., Gawantka, V., Pollet, N., Niehrs, C., Kintner, C. Nrarp is a novel intra-cellular component of the Notch signaling pathway. Genes & Dev. 2001;15:1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler, T., Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard, I., Pear, W.S. Notch and cancer: Best to avoid the ups and downs. Cancer Cell. 2003;3:203–205. doi: 10.1016/s1535-6108(03)00052-7. [DOI] [PubMed] [Google Scholar]
- Mills, A.A., Zheng, B., Wang, X.J., Vogel, H., Roop, D.R., Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Murtaugh, L.C., Stanger, B.Z., Kwan, K.M., Melton, D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, B.C., Lefort, K., Mandinova, A., Antonini, D., Devgan, V., Della Gatta, G., Koster, M.I., Zhang, Z., Wang, J., di Vignano, A.T., et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes & Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff, B.J., Qin, J.Z., Chaturvedi, V., Denning, M.F., Bonish, B., Miele, L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-κB and PPARγ. Cell Death Differ. 2002;9:842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- Nicolas, M., Wolfer, A., Raj, K., Kummer, J.A., Mill, P., van Noort, M., Hui, C.C., Clevers, H., Dotto, G.P., Radtke, F. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Okuyama, R., Nguyen, B.C., Talora, C., Ogawa, E., Tommasi di Vignano, A., Lioumi, M., Chiorino, G., Tagami, H., Woo, M., Dotto, G.P. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1–caspase 3 regulatory mechanism. Dev. Cell. 2004;6:551–562. doi: 10.1016/s1534-5807(04)00098-x. [DOI] [PubMed] [Google Scholar]
- Pan, Y., Lin, M.H., Tian, X., Cheng, H.T., Gridley, T., Shen, J., Kopan, R. γ-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Powell, B.C., Passmore, E.A., Nesci, A., Dunn, S.M. The Notch signalling pathway in hair growth. Mech. Dev. 1998;78:189–192. doi: 10.1016/s0925-4773(98)00177-4. [DOI] [PubMed] [Google Scholar]
- Proweller, A., Tu, L., Lepore, J.J., Cheng, L., Lu, M.M., Seykora, J., Millar, S.E., Pear, W.S., Parmacek, M.S. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- Raghavan, S., Bauer, C., Mundschau, G., Li, Q., Fuchs, E. Conditional ablation of β1 integrin in skin. Severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J. Cell Biol. 2000;150:1149–1160. doi: 10.1083/jcb.150.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan, A., Talora, C., Okuyama, R., Nicolas, M., Mammucari, C., Oh, H., Aster, J.C., Krishna, S., Metzger, D., Chambon, P., et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, D.A., Hannenhalli, S., Tobias, J.W., Cooch, N., Shiekhattar, R., Kadesch, T. Functional analysis of hes-1 in preadipocytes. Mol. Endocrinol. 2006;20:698–705. doi: 10.1210/me.2005-0325. [DOI] [PubMed] [Google Scholar]
- Schweizer, J., Kinjo, M., Furstenberger, G., Winter, H. Sequential expression of mRNA-encoded keratin sets in neonatal mouse epidermis: Basal cells with properties of terminally differentiating cells. Cell. 1984;37:159–170. doi: 10.1016/0092-8674(84)90311-8. [DOI] [PubMed] [Google Scholar]
- Segre, J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki, K., Han, H., Yamamoto, N., Tashiro, K., Ikegawa, M., Kuroda, K., Suzuki, A., Nakano, T., Honjo, T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- Thelu, J., Viallet, J.P., Dhouailly, D. Differential expression pattern of the three Fringe genes is associated with epidermal differentiation. J. Invest. Dermatol. 1998;111:903–906. doi: 10.1046/j.1523-1747.1998.00372.x. [DOI] [PubMed] [Google Scholar]
- Uyttendaele, H., Panteleyev, A.A., de Berker, D., Tobin, D.T., Christiano, A.M. Activation of Notch1 in the hair follicle leads to cell-fate switch and Mohawk alopecia. Differentiation. 2004;72:396–409. doi: 10.1111/j.1432-0436.2004.07208006.x. [DOI] [PubMed] [Google Scholar]
- van Es, J.H., van Gijn, M.E., Riccio, O., van den Born, M., Vooijs, M., Begthel, H., Cozijnsen, M., Robine, S., Winton, D.J., Radtke, F., et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vasioukhin, V., Degenstein, L., Wise, B., Fuchs, E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclair, S., Nicolas, M., Barrandon, Y., Radtke, F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev. Biol. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Watt, F.M., Green, H. Stratification and terminal differentiation of cultured epidermal cells. Nature. 1982;295:434–436. doi: 10.1038/295434a0. [DOI] [PubMed] [Google Scholar]
- Wilson, A., Radtke, F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 2006;580:2860–2868. doi: 10.1016/j.febslet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Yamamoto, N., Tanigaki, K., Han, H., Hiai, H., Honjo, T. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 2003;13:333–338. doi: 10.1016/s0960-9822(03)00081-2. [DOI] [PubMed] [Google Scholar]
- Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R.T., Tabin, C., Sharpe, A., Caput, D., Crum, C., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Bermingham, N.A., Finegold, M.J., Zoghbi, H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- Yoon, K., Gaiano, N. Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat. Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]