Sequence-specific binding of single-stranded RNA: is there a code for recognition? (original) (raw)
Abstract
A code predicting the RNA sequence that will be bound by a certain protein based on its amino acid sequence or its structure would provide a useful tool for the design of RNA binders with desired sequence-specificity. Such de novo designed RNA binders could be of extraordinary use in both medical and basic research applications. Furthermore, a code could help to predict the cellular functions of RNA-binding proteins that have not yet been extensively studied. A comparative analysis of Pumilio homology domains, zinc-containing RNA binders, hnRNP K homology domains and RNA recognition motifs is performed in this review. Based on this, a set of binding rules is proposed that hints towards a code for RNA recognition by these domains. Furthermore, we discuss the intermolecular interactions that are important for RNA binding and summarize their importance in providing affinity and specificity.
INTRODUCTION
One of the prime motivations for studying the structures of protein–RNA complexes is to gain a better understanding of the patterns that determine specific RNA binding and help to predict the sequences that are recognized by a protein based on the amino acid sequence. Such predictions are a prerequisite for engineering RNA-binding domains for medical or basic research applications as was done for DNA-binding proteins (1). Furthermore, accurate predictions could lead to a better understanding of the cellular functions of RNA-binding proteins.
Many different types of single-stranded RNA (ssRNA)-binding domains have been identified to date and a very instructive review on their structures has been published recently (2). Although some of these domains are very abundant, i.e. found in hundreds of proteins within one species and present across all kingdoms of life, such as the RNA recognition motif domain (RBD/RRM/RNP domain) and the hnRNP K homology (KH) domain, others are quite unique, either because the domain is confined to a single species or a specific function (i.e. viral or cap-binding proteins).
Here, we present a comparative structural analysis of the RNA recognition modes of four different types of RNA-binding units, namely PUF repeats, zinc-binding domains, KH domains and RRM domains. All of these RNA-binding entities consist of small protein domains or repeats of 35–90 amino acids in size that bind sequence-specifically to ssRNA and are often found in multiple copies within a single protein. Furthermore, recent complex structures have extended the knowledge of the modes of RNA recognition employed by these domains. We summarize the nature and origin of the intermolecular interactions that drive ssRNA binding by proteins and discuss their contribution to affinity and sequence-specificity. Finally, based on these analyses, we propose a set of binding rules that could be useful for rational design of de novo sequence-specific RNA binders.
SMALL PROTEIN DOMAINS THAT BIND ssRNA SEQUENCE-SPECIFICALLY
The Pumilio homology domain
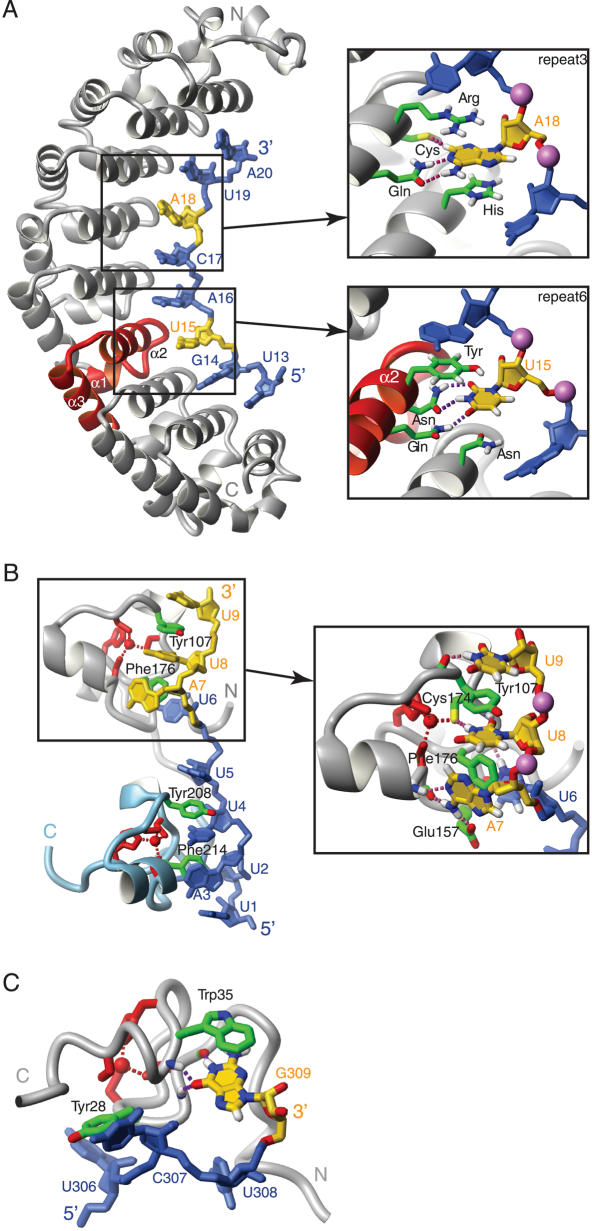
Members of the PUF protein family (named based on the initially identified members Drosophila Pumilio and Caenorhabditis elegans FBF) play an important role in the regulation of development in a wide variety of species. PUF proteins influence mRNA stability and translation by sequence-specifically binding to 3′-untranslated regions (3,4). PUF proteins contain a C-terminal RNA-binding domain known as Pumilio homology domain (PUM-HD). The PUM-HD of human Pumilio1 is composed of eight 37 amino acid PUF repeats flanked by an N- and a C-terminal PUF related sequence. The structure of human Pumilio1 in complex with a 10 nt ssRNA has been determined by X-ray crystallography (5). The PUF repeats, which consist of three α-helices each, pack together in a curved structure that resembles about half of a donut with a diameter of ∼80 Å (6). The RNA is bound as an extended strand to the inner surface with each nucleotide contacting two consecutive repeats (5). All the phosphates are solvent exposed, while the bases make the contacts to the protein side-chains (Figure 1A). The second helix (α2) of each repeat participates in RNA binding. For each nucleotide, the side-chain of the fourth amino acid in helix 2 stacks on top of the base while the side-chains of the third and seventh amino acid of the helix are hydrogen-bonded to its Watson–Crick edge. In addition, the fourth amino acid side-chain of the following repeat is stacked underneath the base (Figure 1A). Thus, there is a continuous alternate stacking between RNA bases and protein side-chains. Intermolecular stacking is mediated by aromatic, positive and neutral side-chains (5).
Figure 1.
Pumilio and zinc-binding domains. (A) Human Pumilio1 in complex with RNA (PDB code: 1M8Y). (B) Complex structure of Tis11d (PDB code: 1RGO). (C) Zinc knuckle of the MMLV nucleocapsid protein in complex with RNA (PDB code: 1U6P). The proteins are shown as grey ribbons; individual protein side-chains are shown in green. Repeat 6 of Pumilio is represented by a red ribbon, the C-terminal zinc finger of Tis11d is represented as a light blue ribbon and the zinc coordinating side-chains in (B and C) are in red. The RNA molecules are in blue and yellow, individual phosphate atoms are shown as purple spheres. Intermolecular hydrogen-bonds are depicted as purple dashed lines. Figures were generated with MOLMOL (88).
Zinc-binding domains
The structures of two proteins containing small zinc-binding domains [namely Tis11d (7) and MMLV nucleocapsid (8,9)] in complex with ssRNA have been determined recently by NMR spectroscopy. Tis11d is a protein implicated in the regulation of mRNA stability that contains two 35 amino acid tandem zinc finger domains of the type CX8CX5CX3H. Each domain binds sequence-specifically to one UAUU stretch within the single-stranded class II AU-rich element (ARE) RNA 5′-UUAUUUAUU-3′ (7). The RNA backbone points away from the protein surface while each of the four bases fits into a specific binding pocket created mostly by the protein main-chain and two aromatic side-chains (Figure 1B). U6, A7 and U8 wrap around a conserved phenylalanine which is part of the loop between the third cysteine and the histidine of the zinc finger. U6 and A7 stack on both sides of the phenylalanine and U8 interacts with one edge of the ring. Furthermore, U8 and U9 sandwich a conserved tyrosine of the loop between the second and the third cysteine of the domain. Sequence-specific recognition is primarily achieved by the fold of the domain as almost all the hydrogen bonds involving the base-specific groups of the RNA are mediated by the main-chain of the protein or by cysteine side-chains coordinated to the zinc atom (Figure 1B) with only one exception (see Glu157 in Figure 1B).
The nucleocapsid protein of MMLV contains a 28 amino acid zinc knuckle (Arg16-Pro43) of the type CX2CX3HX4C. Several structures of this protein in complex with various ssRNA sequences have been determined (8,9). Although the zinc knuckle binds with highest affinity to a CUCG sequence, binding to other 4 nt sequences occurs as long as they contain a guanine at the 3′ end (9). As for Tis11d, two aromatic residues of the zinc knuckle are involved in RNA binding. Tyrosine 28 (between the first and second cysteine) stacks with U306 and contacts C307 and tryptophan 35 (between the histidine and the third cysteine) stacks with G309 (Figure 1C). Base-specific contacts to U306, C307 and U308 are mediated by several protein side-chains, while specific recognition of G309 is achieved by three hydrogen bonds involving the protein main-chain (Figure 1C) (8,9). Hence, the fold of this CCCH zinc knuckle appears to be specific for an NNNG ssRNA tetranucleotide, while side-chains decide on the preferred identity of the three 5′ nucleotides. Interestingly, a G-specific binding pocket is found in other CCCH zinc-knuckles as well, even though the domain fold in these cases is different and a smaller number of nucleotides is bound (10–12).
The KH domain
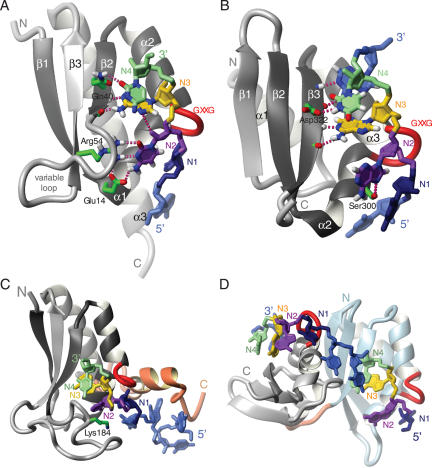
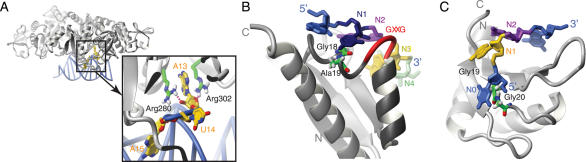
The KH domain is highly abundant and found in various proteins that mediate regulation of gene expression. The KH domain is ∼70 amino acid residues in size and characterized by a (I/L/V)-I-G-X-X-G-X-X-(I/L/V) motif in the middle of the domain (13,14). All KH domains whose structures have been solved to date share the same fold, which is composed of a three-stranded β-sheet packed against three α-helices. However, the domain family can be subdivided into two distinct types (13): type I KH domains fold in a βααββα topology with an antiparallel β-sheet that features β3 as the central strand [e.g. see Nova in Figure 2A (15)], while type II domains have a αββααβ topology and a β-sheet in which β2 is the central strand that is parallel to β3 and antiparallel to β1 [see NusA in Figure 2B (16)]. The two consecutive α-helices are connected by the so-called ‘GXXG loop’, which is part of the conserved sequence motif.
Figure 2.
KH domains. (A) Type I KH domain of Nova (PDB code: 1EC6). (B) Type II KH domain of NusA (PDB code: 2ATW). (C) KH and QUA2 domains of SF1 (PDB code: 1K1G). (D) Tandem KH domains of NusA (2ATW). The proteins are depicted as grey ribbons, the GXXG loop is shown in red and RNA contacting side-chains are represented by green sticks. The RNA nucleotides N1, N2, N3 and N4 are shown in dark blue, purple, yellow and green, respectively. Other nucleotides are in light blue. Individual intermolecular hydrogen bonds are shown as purple dashed lines. The QUA2 domain of SF1 and the N-terminal KH domain of NusA are shown as red and light blue ribbons. Figures were generated with MOLMOL (88).
Two structures of type I KH domains (15,17) and a structure of two type II KH domains (16), both in complex with ssRNA, have been determined. In addition, five type I KH domain structures in complex with ssDNA have been solved (18–21). In all these structures, the ssRNA or ssDNA is bound in a cleft formed by the GXXG loop, the two consecutive helices, the following β-strand (β2 for type I and β3 for type II) and the so-called ‘variable loop’ (the β2β3 loop in type I and the β3α2 loop in type II) (Figure 2). Each KH domain binds at least 4 nt (referred to as N1 to N4 in Figure 2 and Table 1). The first 3 nt N1, N2 and N3 are spread on the surface of the domain. The base of N1 is stacked onto a peptide bond within α1 (α2 in type II) between a conserved glycine and the following residue, while N2 and N3 lie on a hydrophobic surface made up of two side-chains, one from α1 and one from β2 (α2 and β3 in type II) that act as a wedge between the 2 nt (not shown in Figure 2) (15–18,21). The backbone carbonyl and amide oxygen of the same conserved hydrophobic residue in β2 are also hydrogen-bonded to the N3 base (Figure 2A and B). These two hydrogen bonds favour an adenine or a cytosine in the N3 position (Table 1). The conformation is further maintained by contacts between the sugar-phosphate backbone of N1 and N2 and the highly conserved GXXG loop, which run almost parallel to one another (Figure 2). In particular, the phosphate group between N1 and N2 is hydrogen bonded to the backbone amide of the third residue of the GXXG loop (not shown in Figure 2). Finally, N4 stacks over N3 and interacts with side-chains of β2 (β3 in type II) (Figure 2A and B).
Table 1.
Register of the RNA or DNA sequences in complex structures of KH domain containing proteins
| Position on the KH domain | N1 | N2 | N3 | N4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein and sequences bound | ||||||||||
| Nova1 (15) | A | G | A | U | C | A | C | C | ||
| SF1 (17) | A | U | A | C | U | A | A | C | A | A |
| NusA KH1 (16) | A | G | A | A | ||||||
| NusA KH2 (16) | C | U | C | A | A | U | A | |||
| hnRNPK KH3 (18) | C | C | C | C | ||||||
| hnRNPK KH3 (18) | T | C | C | C | ||||||
| PCBP2 KH1 (21) | A | C | C | C | ||||||
| Number of bases in each position | ||||||||||
| A | 2 | 2 | 4 | 1 | ||||||
| C | 2 | 4 | 3 | 5 | ||||||
| U/T | 3 | 0 | 0 | 1 | ||||||
| G | 0 | 1 | 0 | 0 |
Outside the canonical binding of these 4 nt, binding of additional nucleotides is mediated either by the variable loops [e.g. Nova-1 (15) and SF1 (17)] or by an extension of the domain (e.g. the long helix 3 in Nova-1, Figure 2A). In SF1, the presence of an additional small domain (QUA2 domain) C-terminal to the KH domain allows the binding of three additional nucleotides (Figure 2C) (17). Finally, in NusA, the juxtaposition of two type II KH domains leads to binding of two additional nucleotides (Figure 2D) (16).
The RRM/RNP/RBD domain
The RRM/RNP/RBD domain has a typical size of ∼90 amino acids and is the most abundant RNA-binding domain in higher vertebrates. Furthermore, it is the most extensively studied RNA-binding domain, both in terms of structure and biochemistry (22). The structures of 11 different RRM proteins in complex with RNA (23–35) or DNA (36,37) have been determined to date by either X-ray crystallography (25,26,28,30,31,33,34,36) or NMR spectroscopy (23,24,27,29,32,35,37,38). Since several of these proteins contain more than one RRM, the structures of a total of 20 RRM–nucleic acid complexes are currently available.
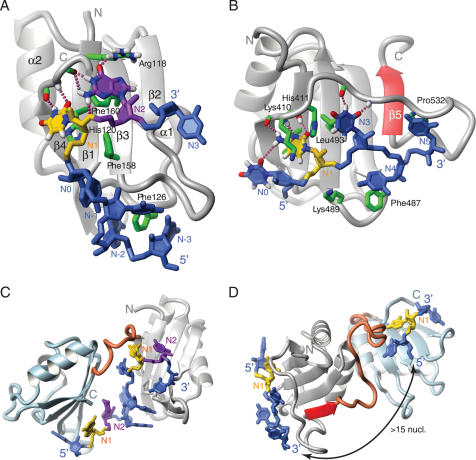
In terms of primary sequence, the RRM is characterized by two conserved sequence stretches referred to as RNP1 (consensus K/R-G-F/Y-G/A-F/Y-V/I/L-X-F/Y) and RNP2 (V/I/L-F/Y-V/I/L-X-N/L). Structurally, RRMs consist of a four-stranded antiparallel β-sheet which is backed by two α-helices in a βαββαβ topology (39). Each RRM binds a variable number of nucleotides, ranging from a minimum of two in the cases of CBP20 (28,34) and Nucleolin RRM2 (27,35) to a maximum of eight for U2B′ (31). The 4-stranded β-sheet is the primary RNA-binding surface. It typically contains three conserved aromatic side-chains in the two central β-strands (β1 and β3) that accommodate two RNA nucleotides as follows: the 5′ nucleotide (N1 in Figure 3A) and the 3′ nucleotide (N2 in Figure 3A) stack on aromatic rings located on β1 (position 2 of the RNP2 sequence) and on β3 (position 5 of RNP1), respectively. The third aromatic ring, which is usually located on β3 (position 3 of RNP1), is often inserted between the two sugar rings of the dinucleotide. However, deviations from this basic mode of binding are found. For example, in the RRM of CBP20 (28,34) and in all four RRMs of PTB (29), no binding on the β3 strand is observed (i.e. there is no base equivalent to the canonical N2, Figure 3B).
Figure 3.
RRM domains. (A) The RRM of Fox-1 (PDB code: 2ERR). (B) RRM3 of PTB (PDB code: 2ADC). (C) The tandem RRMs of Sex-lethal (PDB code: 1B7F). (D) RRMs 3 and 4 of PTB (PDB code: 2ADC). The proteins are depicted as grey ribbons, except for the C-terminal RRMs of Sex-lethal and PTB, which are in light blue, and the fifth β-strand of PTB RRM3 and the interdomain linkers, which are in red. Individual side-chains that contact the RNA are represented by green sticks. The RNA nucleotides N1 and N2 are shown in yellow and purple, respectively. Other nucleotides are in blue. Individual hydrogen bonds are shown as purple dashed lines. Figures were generated with MOLMOL (88).
In most RRM complexes, 1 or 2 nt are bound in addition to this dinucleotide. N0, the nucleotide 5′ to N1, is either bound to β4 (8 RRMs, see PTB RRM3 in Figure 3B) or resides in a binding pocket formed by the β1α1 and β2β3 loops (6 RRMs, see Fox-1 RRM in Figure 3A). N3, the nucleotide 3′ to N2, is frequently found in contact with the RRM but can be bound in several different locations. For example, in 5 RRMs, N3 stacks with N2 and is recognized by the protein region C-terminal of the RRM, while in another 4 RRMs, N3 is residing on the β2 strand (see Fox1 RRM in Figure 3A). Hence, like the KH domain and the zinc-binding domains, a typical RRM contains 4 nt binding sites (Table 2).
Table 2.
Register of the RNA or DNA sequences in complex structures of RRM domain containing proteins
| Position on the RRM domain | N−3 | N−2 | N−1 | N0 | N1 | N2 | N3 | |
|---|---|---|---|---|---|---|---|---|
| Protein and sequence bound | ||||||||
| U1A (24,30) | A | U | U | G | C | A | C | |
| Sex-lethal RRM1 (26) | U | U | U | U | U | U | U | |
| Sex-lethal RRM2 (26) | U | G | U | |||||
| PABP RRM1 (25) | A | A | A | A | ||||
| PABP RRM2 (25) | A | A | A | A | ||||
| U2B″ (31) | A | U | U | G | C | A | G | U |
| hnRNPA1 RRM1 (36) | T | A | G | G | ||||
| hnRNPA1 RRM2 (36) | T | T | A | G | G | |||
| Nucleolin RRM1 (27,35) | C | G | A | |||||
| Nucleolin RRM2 (27,35) | U | C | C | |||||
| HuD RRM1 (33) | U | U | A | U | U | U | ||
| HuD RRM2 (33) | U | U | ||||||
| HuD RRM2 (33) | U | A | U | |||||
| CBP20 RRM (28,34) | G | N | ||||||
| PTB RRM1 (29) | U | C | U | |||||
| PTB RRM2 (29) | C | U | N | |||||
| PTB RRM3 (29) | U | C | U | N | N | |||
| PTB RRM4 (29) | U | C | N | |||||
| Fox-1 RRM (23) | U | G | C | A | U | G | U | |
| hnRNPD RRM (37) | T | A | G | G | ||||
| Number of bases in each position | ||||||||
| A | 3 | 6 (1 syn) | 4 | 3 | ||||
| C | 0 | 7 | 1 | 2 | ||||
| U/T | 11 | 4 | 5 | 5 | ||||
| G | 2 | 2 | 5 (all syn) | 4 (1 syn) |
In addition to this canonical RNA binding surface, binding sites for another three nucleotides 5′ to N0 are found in the RRMs of U1A (30), U2B′′ (31), Sex-lethal RRM1 (26), HuD RRM1 (33) and Fox-1 (23) (Table 2). In all these complexes, RNA binding of these nucleotides is mediated by loops β1α1, β2β3 and α2β4. Nevertheless, the structures adopted by these nucleotides reveal three different topologies. In U1A and U2B′′, N−2 stacks over N−3 and N0 stacks over N−1 with almost a 90° angle between the two stacks, while in Sex-lethal and HuD only N−1 and N−2 stack (Figure 3C), and finally, in Fox-1, no intra-RNA stacking is found but a base pair between N−2 and N0 is formed (Figure 3A). In Sex-lethal and HuD, a tyrosine in the first position of the β1α1 loop stacks with N−3, and in Fox-1, a phenylalanine in the third position of the β1α1 loop stacks with both N−3 and N−1 (Figure 3A), whereas in U1A and U2B′′, no aromatic rings are found in this loop. Thus, it appears that like on the surface of the β-sheet, aromatic rings in the β1α1 loop can shape the structure of the RNA. Interestingly, in the case of Fox-1, binding mediated by the β-sheet and by the loops is independent, since phenylalanine to alanine mutations in either the loop or the β-sheet abolish binding to one site, but not the other (23).
Binding of additional nucleotides 3′ to N3 is much less common and has so far only been observed for U2B′′ (31) and PTB RRM2 and RRM3 (29) (Figure 3B) (Table 2). The additional nucleotides (two for U2B′′ and PTB RRM3 and one for PTB RRM2) are bound beyond the β2 strand. In U2B′′, binding is mediated by the β2β3 loop and the N-terminus of helix 1; in PTB RRM2 and RRM3, it is achieved by the β2β3 loop and the loop between β4 and an additional β5 strand unique to these two RRMs (Figure 3B). The origin of these additional RNA-binding sites originates from extensions of the RRM: an additional β-strand for PTB RRM2 and RRM3 and an elongated α-helix 1 for U2B′′.
Several structures of two tandem RRMs bound to RNA have been determined. In most cases (25–27,33,35), both RRMs are separated by a small linker and bind two adjacent stretches within the same RNA molecule (Figure 3C). This topology provides a large RNA-binding surface. However, there are exceptions to this rule, like, for example, RRMs 3 and 4 of PTB (29,40). In this protein, the two RRMs interact in such a way that their RNA-binding surfaces point away from each other (Figure 3D). This topology prevents the two domains from binding immediately adjacent pyrimidine tracts but instead favours RNA looping if the two pyrimidine tracts are separated by at least 15 nt (29).
Sequence-specific versus non-sequence-specific ssRNA-binding proteins
Examination of these sequence-specific ssRNA-binding domains reveals a few common structural features. The binding surface of the protein is primarily hydrophobic in order to maximize intermolecular contact with the bases of the RNA. The RNA bases are usually spread on the surface of the protein domains while the RNA phosphates point away toward the solvent. Only a few intramolecular RNA stacking interactions are observed, while many intermolecular stacking interactions, often mediated by aromatic amino acids, are observed (with the notable exception of the KH domain). This mode of binding contrasts with how non-sequence-specific RNA binding proteins recognize ssRNA. For example, in the structures of RNA polymerases bound with DNA–RNA hydrids (41,42) and in the recently determined structures of the DEAD-box protein Vasa (43) and of two viral nucleoproteins (44,45) bound with ssRNA, RNA binding is mostly mediated by positively charged side-chains that contact the sugar-phosphate backbone of the RNA (Figure 4). As a consequence, the RNA bases are exposed to the solvent and are stacked with neighbouring RNA bases rather than with protein side-chains.
Figure 4.
(A) Structures of the DEAD-box protein Vasa (43) and (B) of the rabies virus nucleoprotein (44), two recent non-sequence-specific ssRNA binding proteins in complex with RNA (PDB code: 2DB3 and 2GTT). The protein ribbon is shown as a grey ribbon and the RNA is in dark blue or in color (yellow, green and red) with the phosphate atoms shown as purple spheres. The ATP analogue AMPPNP is shown in orange.
THE INTERMOLECULAR INTERACTIONS RESPONSIBLE FOR ssRNA BINDING
Aromatic interactions of the RNA bases
π−π Interactions
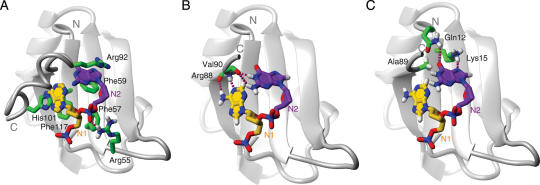
A common feature of complexes of proteins with ssRNA is the so-called ‘stacking’ of aromatic moieties. In such a stack, the planes of the aromatic rings are in parallel orientation with an average distance of ∼3.3 Å in between the planes (46). At protein–RNA interfaces, stacks can be either intermolecular, i.e. formed by rings of the nucleic acid bases with the aromatic side-chains of phenylalanine, tyrosine, tryptophane and histidine, or within the RNA, involving two or more bases. In the zinc-binding domains mentioned above, for example, only intermolecular stacking is observed (Figure 1B and C). In RRMs, on the other hand, both intra-RNA and intermolecular stacking is frequently encountered, e.g. N2 often stacks simultaneously on an aromatic protein side-chain and with N3 [see U1A Phe56, A11 and C12 (30) in Figure 5C]. Finally, in KH domains, only intra-RNA stacking has so far been observed (see N3 and N4 in Figure 2).
Figure 5.
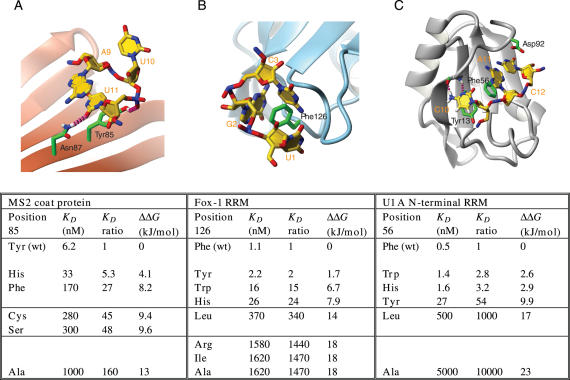
The energies associated with intermolecular stacking interactions. (A) Stacking of U11 and A9 on top of Tyr85 in the MS2 coat protein complex and the effect of Tyr85 mutants on affinity and binding free energy. (B) Contacts between Phe126 and U1, G2 and C3 in the Fox-1 complex and the changes in affinity and binding free energy upon mutating Phe126. (C) Stacking contacts at the U1A RNA binding interface and energetic effects of mutating Phe56. RNA bases are shown in yellow, protein side-chains in green and intermolecular hydrogen bonds as red dashed lines. The table shows dissociation constants (_K_Ds), ratios of K_Ds and corresponding differences in binding free energy (ΔΔ_G). Data are taken from (23,50,51). PDB accession codes are 1ZDI, 2ERR and 1URN. Figures were generated with MOLMOL (88).
Experiments with isolated nucleosides and single-stranded polynucleotides show that each nucleotide has distinct stacking properties with purines being better stacking partners than pyrimidines [reviewed in chapters 2 and 8 of (47)]. Furthermore, studies on various benzene compounds indicate that the strength of a stacking interaction depends on the ring substituents (48). This might suggest that stacking interactions take part in sequence-specific recognition. However, examinations of the different RRM–RNA complexes reveal examples of stacking between each of the four bases with a phenylalanine or a tyrosine aromatic ring of the RNP1 or RNP2 motives (22). Furthermore, a more general statistical analysis of protein–RNA complexes confirms that all four bases are found involved in a stacking interaction more or less equally often and all four bases stack most often with phenylalanine (49). Hence, it seems that stacking interactions do not provide much sequence-specificity in protein–RNA complexes. However, the number of known protein–RNA complexes is still limited which hampers statistical analyses.
Instead, do stacking interactions in protein–RNA complexes provide binding affinity? Isolated nucleosides in solution form stacks rather than base pairs, indicating that the stacking interaction provides some favourable energy in aqueous solution. In the case of isolated nucleosides, these energies are quite small, however (47). Interestingly, they are associated with unfavourable entropy and favourable enthalpy, ruling out hydrophobic interactions as the dominant driving force, as hydrophobic interactions originate from the ‘liberation’ of ordered water molecules and hence increasing entropy. Since there has been no evidence so far for a specific π−π interaction, it therefore seems that van der Waals bonding is dominating the stacking attraction (46). In contrast to experiments on isolated nucleosides or ssRNA (47), stacking interactions at the protein–RNA interface seem to be associated with substantial free energies. Mutation of the three stacking aromatic side-chains of the Fox-1 complex, a phenylalanine in a loop, as well as a histidine and a phenylalanine on the β-sheet of the RRM, to alanine, leads to a 1500-, 160- and ∼30 000-fold loss in affinity, respectively (23). Similar results have been obtained for the N-terminal RRM of U1A and for the MS2 bacteriophage coat protein. In U1A, replacement by alanine of a conserved phenylalanine in the β-sheet of the RRM leads to ∼10 000-fold loss of binding affinity and in MS2 coat protein, substitution of a stacking tyrosine by alanine leads to a 160-fold increase of the dissociation constant _K_D (Figure 5) (50,51). Similar results have also been obtained in other studies (52,53).
Additionally, in the cases of Fox-1, U1A and MS2 coat protein, mutant proteins have been studied in which the stacking amino acid was replaced by either another aromatic residue or various other side-chains (Figure 5). A general trend is apparent from these measurements. Replacement by another aromatic side-chain generally leads to a fairly small loss in binding affinity. However, this small loss of affinity is always present in these complexes, indicating that the binding pockets have been optimized evolutionarily for a particular aromatic side-chain such that, for example, the hydroxyl group of tyrosine might be required in one case (MS2 coat protein, where it makes a hydrogen bond to a phosphate group in the RNA) and might be sterically disfavoured in another case (Fox-1) (Figure 5A and B). However, an aromatic side-chain always provides higher affinity than replacement by non-aromatic side-chains. Leucine seems to play an intermediate role, being an amino acid with a fairly large van der Waals interaction surface and being sterically similar to the aromatic side-chains. Cysteine and serine mutants also have intermediate binding affinities in the MS2 coat protein, which might be due to the fact that they can hydrogen bond with the RNA (Figure 5A). The largest loss in affinity occurs when the entire side-chain is removed, i.e. in the alanine mutants (23,50,51).
In these mutation experiments, it might be argued that removal of the aromatic side-chain disrupts more than just the stacking interaction, e.g. by affecting the hydrogen-bond network of the stacking RNA base or by leading to larger conformational rearrangements, such that the energetic effect of stacking cannot be separated from other effects. To address this problem, a F56L mutant of the N-terminal RRM of U1A was used together with modified RNA bases in which individual hydrogen-bonding groups had been removed (51). Disruption of one hydrogen bond leads to a similar loss of binding free energy of ∼4–7 kJ/mol in the wild-type and mutant proteins, indicating that the hydrogen-bond network is intact despite the removal of the stacking partner (Table 3) (51). However, these results were obtained for a leucine mutant, which still provides a considerable binding interface for van der Waals attractive forces. For MS2 coat protein, photocrosslinking experiments showed that there were no large structural rearrangements in case of the Tyr, Phe, His and Cys mutants (50). Hence, the general trends found in these experiments are consistent with a powerful role for stacking interactions at the protein–RNA interface in providing binding affinity of ∼13–23 kJ/mol and base.
Table 3.
Number of hydrogen-bonds lost and corresponding differences in binding free energy (ΔΔ_G_) for adenine mutants of the RNA binding to U1A (wild-type and F56L) and Fox-1
| U1A N-terminal RRM | Fox-1 RRM | |||||
|---|---|---|---|---|---|---|
| RNA mutation | Number of H-bonds lost | ΔΔ_G_ (kJ/mol) wt | ΔΔ_G_ (kJ/mol) F56L | RNA mutation | Number of H-bonds lost | ΔΔ_G_ (kJ/mol) |
| A6 to Tubercidin | 1 | 4.6 | 4.2 | U1 to A | 1 | 4.0 |
| A6 to 1-Deazaadenosine | 1 | 9.6 | 5.9 | U1 to C | 1 | 4.0 |
| A6 to Purine | 1 | 10.5 | 6.7 | C3 to U | 2 | 14 |
| A4 to Purine | 1 | 5.2 | ||||
| A4 to Inosine | 2 | 13 | ||||
| U5 to C | 1 | 3.9 | ||||
| G6 to A | 4 | 19 |
In the interaction of aromatic rings, two possible orientations are found. The parallel orientation described above, as well as a perpendicular orientation, which is sometimes called a ‘T-stack’. These two orientations represent energy minima and can be observed at protein–RNA interfaces. In the structure of Tis11d, for example, both types of interactions are found (7) (Figure 1B). In the case of the π−π edge-to-face interaction, electrostatic attraction seems to dominate the interaction: the electron-rich central core of the aromatic ring makes a favourable interaction with the partially positive ring protons of the other aromatic moiety [(46,48) and references therein].
Cation–π interactions
Another protein side-chain that can be found to make stacking interactions with RNA bases is the guanidino group of arginine residues. The guanidinium moiety is protonated at physiological pH, which leads to a planar, positively charged, resonance-stabilized structure capable of engaging in stacking interactions. Interestingly, statistical analyses hint at a sequence preference for arginine stacking with the order of preference being U, A, C > G (49,54). Energetically, in the case of the positively charged guanidinium group, electrostatic interactions play an important role in the attractive forces (55). Consequently, a larger spectrum of angles between the planes is observed as compared to the stacking of neutral species. In fact, in analyses of protein structures and ATP-binding proteins, almost all possible angles between the planes of arginine and aromatic side-chains or adenine bases could be found (55,56). Nevertheless, the parallel and the T-shaped orientation seem to represent energy minima (55). Hence, van der Waals forces as well as electrostatic forces between the electron-negative center of the aromatic ring and the positively charged side-chain (cation–π interactions) play a role in arginine-base interactions. The parallel conformation, however, can have the additional energetic advantage of a better hydrogen-bond network with the surroundings. Other cation–π interactions at the protein–RNA interface involve interactions between the RNA bases and lysine and even histidine residues as histidine can be either neutral or positively charged at physiological pH, depending on its chemical environment within the complex. For lysine, the interaction is dominated by electrostatic forces, whereas van der Waals terms play a negligible role (57).
Cation–π interactions are a very common feature of nucleic acid recognition. In statistical analyses of protein–DNA complexes and ATP-binding proteins, cation–π interactions are seen in more than half of the known structures (56,58). This also true for protein–RNA complexes; the most striking example being the recently determined structure of a splicing endonuclease where a bulge adenine near the cleavage site is found sandwiched between two arginines (Figure 6A) (59). In the ssRNA-binding domains described above, interactions between arginine side-chains and RNA bases can be seen, for example, in Pumilio repeat 3 (Figure 1A) and in all RRMs of PTB in complex with pyrimidine tracts (5,29). Furthermore, a lysine–adenine interaction has been shown to be important for RNA binding by SF1 (17) (see its interaction with N2 in Figure 2C), a lysine stacking on top of a base was found in many RRMs including PTB (Figure 3B), and histidines are commonly found as stacking partners on RNA-binding proteins (Figures 1 and 3).
Figure 6.
Arginine and peptide bond stacking. (A) General view and close-up view of the splicing endonuclease in complex with RNA (PDB code: 2GJW) At the splicing endonucleoase active-site, A13 is sandwiched between two arginine side-chains. (B) In the Nova KH domain, N1 stacks on a peptide bond within α1. (C) The N0 nucleotide stacks on a peptide bond that lies at the end of β1 of the RRM of hnRNP A1. The colour scheme is as in Figures 2 and 3. PDB accession codes are 1EC6 (Nova) and 2UP1 (hnRNPA1). Figures were generated with MOLMOL (88).
Other π interactions
The amino groups of asparagine and glutamine are also frequently found to be in contact with aromatic moieties. Again, there are two possible interaction modes. Either the amino group is oriented perpendicularly to the aromatic ring, pointing a δ+ hydrogen atom towards the electron-rich aromatic ring, forming what is in essence a hydrogen bond. Or the planar sp2 nitrogen stacks on top of the aromatic ring due to favourable van der Waals energies, as it is seen, e.g. in Pumilio repeat 6 (Figure 1A) or for the RRMs of U1A, U2B″ and PTB RRM 1 and 4 (5,29–31). Calculations suggest that the energies of the unusual hydrogen bonds are rather weak as compared to conventional hydrogen bonds and an analysis of amino–π interactions in protein structures, as well as in structures of adenine binding proteins, shows that the parallel conformation is generally preferred (57,60,61). Again, this could be due to the fact that the parallel conformation allows the amino bearing side-chains to engage in a larger number of conventional, energetically more favourable hydrogen bonds.
Aspartate and glutamate bear planar, resonance-stabilized formamide groups which can be found as stacking partners at protein–RNA interfaces. For example Asp92 stacks on C12 at the RNA-binding interface of U1A (Figure 5C). A computational study confirmed the importance of Asp92 for stabilizing the quadruple stack F56–A11–C12–D92 (62).
Finally, even peptide bond planes can serve as stacking platforms. In KH domains, the N1 residue stacks on the peptide bond between a conserved glycine and the following residue within an α-helix (Figure 6B), whereas in several RRMs, the N0 nucleotide stacks on a peptide bond between a glycine and the following residue within a β-strand (26,33,36,37) (Figure 6C).
Electrostatic interactions
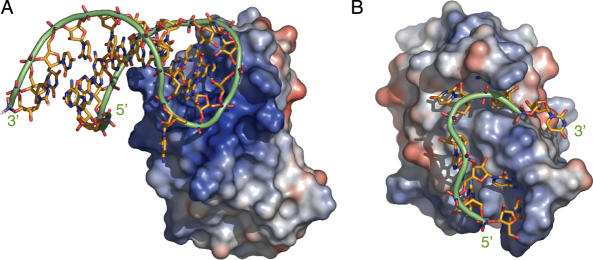
Electrostatic attraction, the attractive force between two particles of opposite charge, plays a crucial role in protein–nucleic acid interactions, as nucleic acids are highly negatively charged molecules. For many proteins that bind to double-stranded DNA or RNA molecules, there are extensive positively charged patches on the protein surface so that it is often fairly easy to predict where the nucleic acid will bind from the protein structure alone (Figure 7A). Furthermore, in the recognition of RNA molecules with a characteristic tertiary structure, electrostatic interactions can play a role in specific recognition of their shape (63,64). Sequence-specific protein contacts to single-stranded nucleotides, on the other hand, commonly occur via the accessible nucleic acid bases, while the phosphate moieties point towards the bulk solution. Hence, the protein surface that contacts the nucleotide is often not extensively positively charged but rather hydrophobic and direct contacts to the nucleic acid backbone can be rare (Figure 7B). Nevertheless, some studies have shown that even in these cases, electrostatic interactions play a highly important role in binding of the RNA (23,65,66). However, since the distribution of charges on an ssRNA is independent of its sequence, they are not important in providing sequence-specificity (53).
Figure 7.
Surface potential of RNA binding proteins. Blue areas indicate a positive potential, red areas a negative potential. (A) Vts1, a protein that recognizes a structured RNA loop. The RNA binding surface of the protein is a highly positive patch. (B) Fox-1 RRM, which binds ssRNA. Positive and negative potentials surround the RNA and the area where most contacts are made is primarily apolar. Figures were generated with PyMOL (http://www.pymol.org) and the surface potential was calculated according to (89). PDB accession codes are 2ESE and 2ERR.
Two methods are typically employed to test the contribution of electrostatic interactions to a biomolecular binding process. Either charged groups are removed from the binding partners (usually by site-directed mutagenesis of charged amino acids or by varying the number of phosphate groups in an oligonucleotide) or the salt dependence of the dissociation constant is measured. If the binding is favoured by electrostatic attraction, increasing the salt concentration of the buffer will reduce affinity. The first approach has revealed, for example, that at 10 mM NaCl, the nucleocapsid zinc knuckle of MMLV shows ∼250 times higher affinity for an UCUG sequence if it carries a phosphate group at the 5′ end and prefers UAUCUG-P over UAUCUG by a factor of ∼2.5 (9). Furthermore, lysine to alanine mutations of residues that are close but not in hydrogen-bond contact to the RNA backbone in U1A reduce the affinity for U1hpII ∼15- to 40-fold at 150 mM NaCl (66). Finally, increasing the number of phosphate groups of cap analogues increases their affinity for eukaryotic translation initiation factor 4E (eIF4E) by ∼6-fold per phosphate group, or even more when comparing m7GMP to m7GDP (67). The second approach shows that in the case of the Fox-1 complex, binding at 150 and 75 mM NaCl is ∼70 and 500 times stronger, respectively, than at 600 mM (23) (Table 4). Similarly, a ∼80-fold decrease of affinity was determined for the U1A U1hpII interaction when the salt concentration was increased from 150 to 500 mM NaCl (65) (Table 4). A particularly thorough way of testing the contribution of individual positive amino acids is a combination of the two methods: the charged amino acid side-chain is mutated and the difference in salt dependence of the affinity of mutant and wild-type are compared (65–69). Studies of this kind can provide information about the exact electrostatic contributions of individual charged residues to RNA binding. In conclusion, all the above measurements show that even for ssRNA-binding proteins, electrostatic interactions strongly contribute to the overall affinity. However, the exact contribution of a particular charged group depends on its location in the complex. Interestingly, close proximity of a charged side-chain to a phosphate of the RNA backbone does not necessarily correspond to a strong contribution as other factors such as flexibility or solvent accessibility play a role; and vice versa, some charged residues that are rather far away from the RNA can still have a strong electrostatic effect on binding (68,69).
Table 4.
Salt dependence of the association rate constant _k_on, dissociation rate constant _k_off and dissociation constant _K_D of the U1A/U1hpII and Fox-1/UGCAUGU interaction
| [NaCl] | _k_on (M−1 s−1) | Relative decrease | _k_off (s−1) | Relative increase | _K_D (nM) | Relative decrease |
|---|---|---|---|---|---|---|
| U1A N-terminal RRM | ||||||
| Wild type | ||||||
| 150 | 1.22 × 107 | 1 | 4.8 × 10−4 | 1 | 0.040 | 1 |
| 220 | 6.2 × 106 | 2.0 | 4.27 × 10−4 | 0.9 | 0.070 | 1.8 |
| 330 | 2.33 × 106 | 5.3 | 5.4 × 10−4 | 1.1 | 0.23 | 5.8 |
| 500 | 4 × 105 | 28 | 1.31 × 10−3 | 2.7 | 3.2 | 81 |
| K20,22,23R | ||||||
| 150 | 5.6 × 106 | 1 | 5.8 × 10−4 | 1 | 0.103 | 1 |
| 220 | 2.5 × 106 | 2.3 | 8.1 × 10−4 | 1.4 | 0.33 | 3.2 |
| 330 | 5.7 × 105 | 9.8 | 1.21 × 10−3 | 2.1 | 2.1 | 21 |
| 500 | 7.3 × 104 | 77 | 2.91 × 10−3 | 5.0 | 40 | 390 |
| K20,22,23Q | ||||||
| 150 | 3.7 × 106 | 1 | 4.2 × 10−3 | 1 | 1.2 | 1 |
| 220 | 1.1 × 106 | 3.2 | 5.7 × 10−3 | 1.4 | 5.3 | 4.5 |
| 330 | 8.1 × 105 | 4.5 | 1.38 × 10−2 | 3.3 | 17.1 | 15 |
| 500 | 2.9 × 105 | 13 | 2.5 × 10−2 | 6.1 | 87 | 74 |
| K20,22,23E | ||||||
| 150 | 2.7 × 105 | 1 | 1.13 × 10−1 | 1 | 430 | 1 |
| 220 | 3.9 × 105 | 0.7 | 2.2 × 10−1 | 1.9 | 550 | 1.3 |
| 330 | 2.4 × 105 | 1.1 | 1.32 × 10−1 | 1.2 | 590 | 1.4 |
| 500 | 9 × 105 | 0.3 | 2.3 | 20 | 4000 | 8.3 |
| Fox-1 RRM | ||||||
| 75 | 1.5 × 108 | 1 | 9.3 × 10−2 | 1 | 0.062 | 1 |
| 150 | 2.7 × 107 | 5.6 | 1.3 × 10−2 | 1.4 | 0.49 | 7.9 |
| 225 | 1.0 × 107 | 15 | 1.9 × 10−2 | 2.1 | 1.8 | 29 |
| 300 | 5.1 × 106 | 29 | 2.4 × 10−2 | 2.7 | 4.6 | 74 |
| 400 | 2.3 × 106 | 65 | 2.6 × 10−2 | 2.9 | 11 | 177 |
| 500 | 1.9 × 106 | 79 | 3.5 × 10−2 | 3.8 | 18 | 290 |
| 600 | 1.2 × 106 | 125 | 4.2 × 10−2 | 4.7 | 34 | 550 |
The favourable free energy for binding of protein to RNA is believed to originate mainly from an entropic effect. When the binding partners are free in solution, the charges on their surfaces attract counterions that are released into bulk solution when the macromolecules bind to one another and find the countercharges on the surface of the binding partner. The polyanion RNA has a very high charge density and therefore buffer cations are thought to condense on its surface (counterion condensation theory). Binding of a protein that carries positive charges will release some of these cations from the high local concentration around the RNA so that they will fall down a concentration gradient into bulk solution. The bulk salt concentration determines the size of this gradient and hence the entropy gain associated with the binding event will be greater at low buffer salt concentrations [reviewed in (47)].
Kinetics
Interestingly, kinetic measurements on ssRNA binding have shown that the salt dependence of the association rate constant _k_on is larger than of the dissociation rate constant _k_off, suggesting that electrostatic interactions in ssRNA recognition are largely long range effects (23,65,66) (see Fox-1 and U1A wild type in Table 4). Opposite charges on protein and RNA lead to a strong attraction, but once the RNA is bound, the complex seems to be stabilized primarily by other factors, as the salt dependence of the _k_off is rather small, albeit present (23,65,66) (Table 4). In this context, it is also interesting to estimate the _k_on at zero ionic strength. For the Fox-1/RNA complex, extrapolation of a curve of log _k_on versus the ionic strength suggests a _k_on of ∼1010 M−1 s−1 in the absence of salt (23). This is as high as the maximum rate constant for collision of molecules in aqueous solutions, the diffusion-limited association rate (70). Bio-molecules usually have association rates that are considerably smaller, because not every collision leads to a productive encounter [(71) and references therein]. For binding of ssRNA, however, long-range electrostatic attraction and steering (the pre-orienting of binding partners that enhances the rate of productive encounters) seem to allow association rates that reach the diffusion limit. This behaviour has also been observed for protein–protein complexes like the Barnase/Barstar complex in which electrostatics play a highly important role in the recognition process (72,73). Furthermore, for the U1A/U1hpII complex, mutations of lysine side-chains to alanine or glutamine show a slightly reduced salt dependence of the association rate constant _k_on, while the salt dependence of the _k_on of lysine to arginine mutants is similar or even higher as compared to the wild-type protein. For a triple-glutamate mutant, the effect is actually reversed and high salt allows a faster association (65) (Table 4). This confirms the importance of these side-chains for electrostatic attraction of the RNA.
Although the _k_on is strongly salt dependent, it is more or less constant for oligonucleotides of different sequences (74,75). This notion, together with kinetic data on U1A aromatic side-chain mutants (66), suggests that nucleic acid recognition is a two-step process, in which any RNA is attracted approximately equally well. However, if stacking and hydrogen-bond interactions that ‘lock’ the interaction cannot be properly established, the complex re-dissociates fast (large _k_off) which results in an overall weak affinity for RNA oligonucleotides of ‘wrong’ sequence (66).
Many ssRNA-binding proteins recognize sequences that are presented in loops. Laird-Offringa and co-workers (74) have evaluated the association and dissociation differences between U1hpII, in which the U1A binding sequence is presented in a loop, and an RNA containing the same binding sequence in an ssRNA of equal length. The effect on _k_on is moderate (∼3-fold), while the effect on _k_off is substantial (590-fold). Hence, the overall loss in affinity is close to 2000-fold. This might reflect the higher entropy loss when an ssRNA as compared to a stem–loop is bound. Additionally, however, there are certain stabilizing interactions with the stem that might be lost when binding the single-stranded target (74).
Intermolecular hydrogen bonds
A hydrogen bond is defined as the interaction between two electronegative atoms that share a proton. Hence, a hydrogen bond always involves a donor group that contributes the proton, and an acceptor group that comprises a lone electron pair capable of accommodating the proton. Owing to this required complementarity between donor and acceptor, intermolecular hydrogen bonds are important players in establishing sequence-specificity in ssRNA recognition.
Conventional hydrogen bonds
In proteins, the side-chains of tryptophane, lysine and arginine can act as hydrogen-bond donors, aspartate and glutamate can act as hydrogen-bond acceptors, and tyrosine, serine, cysteine, threonine, asparagine, glutamine and histidine can act as both donors and acceptors. Furthermore, each amide linkage in the protein backbone includes a hydrogen-bond donor (NH) and a hydrogen-bond acceptor (C=O). Each RNA base comprises both hydrogen-bond donors and acceptors which are characteristic of each base. The purine bases, for example, can be easily differentiated as adenine features a donor, an acceptor and a CH group at ring positions 6, 1 and 2, respectively, while guanine has an acceptor, donor, donor-pattern at the same positions. Similarly, pyrimidines can be discriminated as cytosine comprises an acceptor and a donor at positions 3 and 4, respectively, while uracil has the opposite arrangement.
The contribution of a hydrogen bond to sequence-specificity can be estimated by disrupting individual intermolecular hydrogen-bonds by either mutating the hydrogen-bonding side-chains of the protein or by using modified ligands in which individual donor or acceptor groups have been removed. Early studies of this kind on tyrosyl-tRNA synthetase/substrate complexes yielded stabilizing energies of 2.1–6.3 kJ/mol for neutral hydrogen bonds, and ∼15–19 kJ/mol for hydrogen bonds in which one partner is charged (76). For neutral hydrogen bonds, this corresponds to a factor of ∼2–15 in specificity, i.e. a ligand that engages in a particular hydrogen bond binds ∼2–15 times more tightly than a ligand that cannot form this hydrogen bond. Similar energies have been measured recently for hydrogen bonds at the interfaces of protein–ssRNA complexes. For the N-terminal RRM of U1A, for example, elimination of a single, neutral, intermolecular hydrogen-bond by using different adenine analogues resulted in free energy differences of ∼4.6–10.5 kJ/mol (51) (Table 3). Similarly, disrupting one and two neutral hydrogen bonds in the Fox-1/RNA complex gave ΔΔ_G_ values of 3.9–5.2 kJ/mol and 13 or 14 kJ/mol, respectively, while disruption of four intermolecular hydrogen bonds, including a charged one to an arginine side-chain, resulted in an elevation of the free energy of the complex of 19 kJ/mol (23) (Table 3). The interpretation of affinity constants measured when several hydrogen bonds that recognize one base are disrupted can be tricky, however, since in these cases the base and the protein side-chains in the complex might rearrange. Nevertheless, these data show that individual neutral hydrogen bonds at protein–RNA interfaces are worth 4–10 kJ/mol and hence can sometimes have only small effects on specificity. A whole hydrogen-bond network, however, gives a substantial contribution to binding affinity differences between different RNA sequences and hence to sequence-specificity. It should be kept in mind, however, that the energies measured are not the energies of hydrogen bonds themselves, but rather ‘discrimination energies’ between a complex that features a particular hydrogen bond and a complex that does not (77). Hydrogen-bond interactions in an aqueous surrounding always have to be considered as exchange reactions: hydrogen bonds to water are given up for hydrogen bonds in the complex. This is the reason why they are often associated with rather small energies. Why they are associated with favourable energies at all has been attributed to the fact that upon formation of an intermolecular hydrogen bond, the water molecules that were hydrogen bonded to the donor and acceptor groups of protein and RNA are released into bulk solution, which is entropically favoured (76,77). However, part of the reason might also be that the strength of a hydrogen bond depends on the hydrophobicity of the environment. Hydrogen bonds in the hydrophobic core of a protein seem to be associated with significantly higher energies than those in more accessible parts of the protein (78). Hence, H-bonds that are buried at the protein–RNA interface might be enthalpically more favourable than those to water. Furthermore, a statistical analysis shows that there exist strong geometrical preferences for hydrogen-bonds at protein–RNA interfaces, which in turn suggests that the precise energy of a hydrogen bond depends strongly on the exact relative orientation of donor and acceptor (79). Hence, exact complementarity is required for effective binding, which in turn enhances sequence-specificity.
A method to screen for RNA functional groups that are important for protein binding is the so-called nucleotide analogue interference mapping (NAIM) technique (80). In NAIM, nucleotide analogues are randomly incorporated into an RNA molecule and a screen is performed to identify those RNA molecules that bind to the protein of interest less effectively than the wild-type RNA. Though this method has so far not been extensively employed for ssRNA, it was successfully applied on the U1 snRNP particle and could confirm some of the interactions observed in the U1A/U1hpII co-crystal structure (81). This implies that NAIM might be an effective tool to identify those functional groups within ssRNA oligonucleotides that mediate protein binding and hence to get a detailed insight into protein–ssRNA interactions in the absence of a high-resolution structure.
The CH…O hydrogen bond
The importance of the conventional hydrogen bonds described above for biomolecular recognition has been well established. However, even though the existence of hydrogen bonds involving a CH as a donor group had been evidenced by crystal structures of organic molecules more than 40 years ago (82), the importance of these unconventional hydrogen bonds for biomolecular stability and recognition has been recognized only recently, again due to the analysis of crystal structures [reviewed in (83)]. It is believed that the strongest hydrogen bond in that group is the CH…O bond formed between a CH donor group and an oxygen acceptor. However, the energies of these unconventional hydrogen bonds depend on the acidity of the hydrogen and are particularly strong when the CH group is adjacent to a nitrogen atom. Recently, the importance of CH…O hydrogen bonds in protein–RNA recognition has been pointed out by a computational study: in a structural analysis of 45 protein–RNA complex structures, the authors find that 33% of all potential intermolecular hydrogen bonds are of the CH…O type (84). Interestingly, a large number of these intermolecular CH…O bonds originate from the sugars, in particular from C4′ and C5′ atoms. Within the bases, by far the highest number of CH…O bonds are provided by the C2 of adenine, as it is observed, e.g. at the protein–RNA interface of Pumilio and PABP. In Pumillio, the contact is made between the adenine bound to repeat 3 and the thiol group of a cysteine side-chain (Figure 1A), while in PABP RRM1, the adenine in the N1 position is hydrogen bonded to a carbonyl of the protein main-chain (5,25). Strikingly, however, in ∼70% of the cases observed, the adenine H2 contact is made with the hydroxyl group of a serine side-chain (84). The C8 of adenine and guanine, as well as the C6 of uracil and cytosine are potent CH…O hydrogen-bond donors as well, but are not frequently involved in hydrogen bonds with the protein as they tend to hydrogen bond with the O5′ of their own ribose when they are in the anti conformation (84).
Surface complementarity
Though the experimentally determined binding affinities described above indicate an important role for hydrogen bonds in providing sequence-specificity, it should not be forgotten that surface complementarity in general is an extremely important prerequisite for sequence-specific recognition. In the case that the RNA perfectly fits into binding pockets provided by the protein, favourable dispersion interactions (van der Waals bonding) are maximized. On the other hand, if there are holes, possibly filled with highly constrained and entropically unfavourable water, or steric clashes, which lead to too close contacts that are strongly disfavoured by van der Waals repulsion, the binding affinity will be reduced and the binding partner will be disadvantaged as compared to a ligand that has a perfectly complementary binding surface. Shape recognition plays a particularly important role in the binding of structured RNA molecules and has been reviewed elsewhere (63).
TOWARDS A CODE FOR ssRNA RECOGNITION
Two ways to recognize RNA sequence-specifically
In analysing the molecular basis of how protein domains recognize ssRNA, one can differentiate two basic modes for how sequence-specificity is achieved. For some protein domains, hydrogen bonds to the RNA bases originate from the protein main-chain carbonyl and amide groups and therefore the fold of the protein domain determines the RNA sequence-specificity. This is the case, for example, for the tandem CCCH zinc fingers of Tis11d (7), where each finger recognizes a UAUU sequence. Such an arrangement provides a very rigid and hence highly specific scaffold for RNA binding. However, it also means that small variations in the amino acid sequence could indirectly influence the backbone architecture and change the RNA binding specificity. This makes it virtually impossible to predict which RNA sequence is recognized by these proteins in the absence of a structure.
For other proteins, like Pumilio, sequence-specificity is exclusively provided by hydrogen bonds between the protein side-chains and the RNA bases (5). With such a recognition mode, predicting the RNA sequence that is bound based on the protein primary sequence appears possible. As mentioned earlier, the recognition mode of Pumilio is highly modular. Each Puf repeat recognizes one base and in addition serves as a binding platform for the following base. In each repeat, three amino acid side-chains, all located in helix two, are crucial for RNA recognition (Figure 1A). Different combinations of the amino acids in positions 3, 4 and 7 of this helix specify the binding to the bases, which makes it possible to design a Pumilio-derived specific binder for ssRNAs of distinct sequence. A first attempt of this kind was made by Wang et al. (5) who mutated the asparagine, tyrosine and glutamine at α-helix positions 3, 4 and 7 of repeat 6 (Figure 1A) into serine, asparagine and glutamic acid, respectively, to generate a repeat that specifically recognizes a guanine instead of a uracil. Indeed, the mutant protein binds a U-to-G mutant RNA at least 12 times more strongly than the wild-type RNA.
Role of the protein main-chain of KH and RRM in sequence-specific recognition
The other RNA-binding domains described here (RRM, KH and the MMLV nucleocapsid) achieve sequence-specificity with a combination of both binding modes, i.e. with hydrogen bonds to both the protein main-chain and side-chains. In the KH and nucleocapsid domains, one of the four bound nucleotides is recognized specifically by the protein main-chain. In the MMLV nucleocapsid, this is the guanine at the 3′ end (8,9) (Figure 1C), while in KH domains, the adenine or cytosine in the N3 position is recognized by the backbone of the β2 strand for type I KH domains (15,17,18,21) or the β3 strand for type II KH domains (16) (Figure 2A and B). This indicates that the MMLV nucleocapsid protein and the KH domain have within their fold an inherent preference for specific nucleotide types in one of their binding pockets.
In the case of the RRM, proteins with binding specificity for A-, G- or pyrimidine tracts have been observed. Nevertheless, in examining all known RRM–RNA complex structures, one can see a bias towards particular nucleotide types at certain positions (Table 2). In position N1 of the RRM, a cytosine is found seven times, adenine six times, uracils or thymines four times and guanines only twice. In position N2, on the other hand, guanine and uracil occur five times, adenine four times and cytosine only once. In position N0, there is a strong preference for uracils (11 U or T found). Finally, in position N4, uracils are the most common nucleotide (five times), but the other bases are found at least twice as well. Although not enough complex structures have been solved to make a proper statistical analysis, one can see a certain bias toward a uracil at N0, a cytosine or adenine in N1 and a guanine or a uracil in N2. In fact, a U/G-A/C dinucleotide bound at N1–N2 is never observed, whereas five A/C-U/G sequences are bound in these positions.
A detailed analysis of the interactions in position N1 and N2 partly explains the origin of this sequence bias (Figure 8). Recognition of the RNA base N1 involves one or two hydrogen bonds between the Watson–Crick edge of the base and the main-chain atoms of the last β4 residue and of the residues just C-terminal to it. For almost all cytosines and adenines, the carbonyl oxygen of the last β4 residue [e.g. Y86 in U1A (30)] is hydrogen bonded with one amino proton of the base and the backbone amide two residues after (β4+2, e.g. K88 in U1A) is hydrogen bonded to N3 of cytosine or N1 of adenine (Figure 3B, 5C and 8B). If N1 is a uracil, it is also contacted by atoms of the protein main-chain (Figure 3A), but with more variations in the binding mode (23,26,33). Binding of a guanine in N1 is also quite different in the two RRMs where such an interaction is found, namely CBP20 (28,34) and Sex-lethal RRM2 (26). From this analysis, it appears that the N1 binding pocket of an RRM is readily shaped for binding a C or an A, whereas adaptations seem to be necessary when binding a U or a G.
Figure 8.
Recognition of AG by hnRNPA1 RRM1. (A) Details of the non-sequence-specific contacts to the RNA. (B) Sequence-specific contacts mediated by the protein main-chain. (C) Sequence-specific contacts mediated by the protein side-chains. The colour scheme is as in Figures 2 and 3. PDB accession code is 2UP1. Figures were generated with MOLMOL (88).
Recognition of the RNA base identity in position N2 can also involve hydrogen bonds from the protein main-chain but only when a guanine is bound. In all five complexes with a guanine bound in N2, the base adopts a syn conformation that is stabilized by two hydrogen bonds between the carbonyl oxygen in position β4+2 and both the 2-amino proton and the imino H1 of the guanine (23,35–37). In this syn conformation, the guanine is further stabilized by an intramolecular hydrogen bond between its 2-amino and one of the phosphate oxygens (Figure 3A). As the guanine base is the only base that can engage in these two hydrogen bonds, one could speculate that the default binding sequence for an RRM might be a dinucleotide A/C-G located in N1-N2. When binding A/C-G, no side-chain needs to be involved in the recognition and yet four intermolecular hydrogen bonds with the RNA bases would be formed (Figure 8B). This suggests that the RRM fold might have an inherent binding preference for certain RNA bases, just like the KH domain or the MMLV nucleocapsid zinc knuckle.
Role of the protein side-chains of KH and RRM in sequence-specific recognition
The protein side-chains in the RRM, the KH and the MMLV nucleocapsid zinc knuckle clearly play the major role for discriminating different RNA sequences. For the N1 nucleotide in the RRM, the main side-chains involved in discriminating between different bases appears to be the penultimate residue of β4 (β4-1) and the first residue following β4 (β4+1). Residue β4−1 helps discriminate between A/C and G/U, as E, Q or M side-chains are found in this position hydrogen bonded with an A or a C amino proton, whereas K or R are found in this position hydrogen bonded to uracil O4 or Guanine O6. Residue β4+1 appears to help discriminate between A and C. Indeed, an Ala that interacts with A H2 is found in this position in several complexes (Figure 8C) (36,37) while a Ser correlates with the presence of a C (Figure 3B, contact to O2) (29). However, there are exceptions to this rule as PABP RRM1 has a Ser in the β4+1 position and still accommodates an adenine in N1 (25). Similarly, U1A (30) and U2B″ (31) both contain an alanine in the β4+1 position although a cytosine is bound in N1.
If guanine is bound as N2 on the RRM, specific binding is usually further stabilized by contacts to R or K side-chains from the most N-terminal residue of β1 or from β2 that interact with the O6 and N7 of the guanine (Figures 3B and 8C). It was indeed proven by several crystal structures of hnRNPA1 in complex with various RNAs that an R or K at this position is the determining side-chain for selecting a guanine at N2 (85). For all uracils bound to N2, the most N-terminal residue of β1 is always an asparagine that interacts with O4 of the U. In addition, an arginine of β2 interacts with the O2 of the uracil [in all RRMs except sex-lethal RRM2, where a glutamine of β2 is contacting the U O2 (26)]. Binding of adenine in N2 appears to be more versatile, as the base is not in the same position in the different complexes. In U1A (30) and U2B″ (31), the adenine bound in N2 is recognized by a hydrophobic residue (L or V) of β2 that contacts the A H2 and by a serine located five residues after the end of β4 that interact with both a 6-amino proton and N1 of the adenine. In PABP, however, binding specificity for adenine in N2 is achieved quite differently (25). In RRM1, N58 of β3 is hydrogen-bonded with both the N1 and one of the 4-amino protons of the adenine Watson–Crick edge, whereas in RRM2, N100 from β1 is hydrogen bonded with the N7 and one 4-amino proton of the Hoogsteen edge of the A. In the only case where a cytosine is located in N2, it is recognized by two hydrogen bonds with an arginine side-chain of β2. All in all, it appears that a guanine can be considered the default binding nucleotide in the binding pocket for N2, because it involves the β4+2 backbone carbonyl. Yet, with the presence of an asparagine at the beginning of β1 and of an arginine or lysine in β2 a uracil would be preferred while with a hydrophobic side-chain (L, V or I) in β2 or an asparagine in β3, an adenine would be preferred. There is an exception to this suggestion as an adenine is recognized in PABP RRM2 with an aspargine in β1 and a lysine in β2 but in this case the stacking of the adenine over the aromatic ring of the RNP1 motif is quite reduced (25). This indicates that the binding pocket for N2 is very adaptable.
As discussed earlier, the N0 and N3 binding pockets in the RRM take on several forms, which makes predictions for these binding sites rather difficult. Furthermore, binding specificity in the N0 position can be influenced by neighbouring RNA bases through intramolecular RNA hydrogen bonds. Examples for this are found in PTB, where the uracil in N0 interacts with the cytosine in N1 (29) (Figure 3B), in Fox where the adenine in N0 forms a base pair with the guanine in position N−2 (23) (Figure 3A) or in U1A and U2B″ where a guanine in N0 interacts with a uracil in N−2 (30,31).
In proteins containing KH domains, the side-chains are important to discriminate nucleotide base identity in positions N1, N2 and N4. Although only a few KH domain structures in complex with RNA or DNA are available as compared to RRMs, one can still see where specific side-chains play an important role in sequence recognition. For example, when N2 is a cytosine, such as in Nova1, hnRNPK KH3 and PCBP2 KH1, the base is contacted via two hydrogen bonds by an arginine side-chain from the central β-strand (R54 in Figure 2A). In the other KH domains, this arginine is absent. The identity of N4 that stacks over N3 appears to be discriminated by side-chains from β2 in type I KH domains (β3 in type II, see Figure 2A and B), but no clear rules are apparent from the different structures. The same is true for N1. An interesting additional feature is found in NusA KH3 (16). The Adenine in position N5 folds back and forms a similar H-bond interaction with the β-strand backbone as the adenine in N3. Therefore, an extensive network of polar interactions is created between the three nucleotides N3, N4 and N5 and the β-strand (Figure 2B).
Engineering a specific binder based on RRM or KH scaffolds
Based on the above analysis, it is obvious that rational design of an RRM or KH domain with a novel and defined sequence-specificity based on structural analysis is not as straight-forward as it has proven to be with Pumilio (5). Nevertheless, the set of binding rules proposed above might represent a basis for attempts along this line and a solution to the problem might become even more tractable as more RRM and KH domain structures in complex with RNA will be available.
Alternative approaches to the design of novel RNA binders could be computational design or in vitro selection techniques. Both approaches have in principle been successfully applied to the U1A protein. More than 10 years ago, Laird-Offringa and Belasco could successfully identify amino acid residues important for the specific interaction of U1A with its natural target, the U1hpII RNA, using phage display (86). Interestingly, they were able to generate U1A-derived proteins with an affinity that was even higher than that of wild-type U1A. Hence, repeating this in vitro selection process with a foreign RNA might lead to the generation of novel proteins with high affinity and specificity for any given RNA sequence. Furthermore, this approach might also be applied to derive further binding rules.
More recently, the Rosetta Design algorithm has been used to generate a protein that reproduces the U1A backbone structure to within <1 Å (root mean square deviation) while sharing only ∼30% sequence identity. The design of this U1A-mimic was based on the backbone coordinates of U1A and consequently, the RNA-binding properties of U1A were not retained (87). In the future, it might however become possible to extend such an approach to protein/RNA interfaces and hence to design novel RNA binders in silico.
CONCLUSIONS
The most important chemical interactions that guide ssRNA recognition by proteins are stacking, electrostatics and hydrogen bonding. Generally, stacking and electrostatic interactions play a role in providing affinity (Figure 8A), whereas hydrogen bonds contribute to sequence-specificity as well as affinity (Figure 8B and C). However, although electrostatics are responsible for the initial attraction that brings RNA and protein together, stacking and hydrogen bonds lock the RNA in its proper orientation within the complex. Interestingly, specific hydrogen bonds can be provided either by the backbone or the side-chains. Specificity established by the backbone implies that the overall fold of the protein is readily shaped for the recognition of an RNA of specific sequence. This inherent sequence-specificity of the fold can be seen, for example, for the two zinc-binding domains of Tis11d described in this review (7). On the other hand, the protein Pumilio establishes sequence-specificity solely via side-chains, which allows RNA binding of almost any single-stranded sequence (5). RRMs and KH domains represent an intermediate, where specificity is provided by both the main-chain and side-chains of the domains. Hence, these folds have an inherent preference for certain bases at specific positions but this intrinsic specificity is modulated by additional side-chain interactions which enlarge the spectrum of possible bases recognized. Nature has apparently favoured this latter mode of binding since RRMs and KH domains are the two most common types of RNA-binding domains. The reason for this might be that these RNA binding domains are extremely versatile. In particular, the core RRM domain contains just two consensus binding pockets, which can recognize any given nucleotide, while the rest of the protein is highly adaptable. Furthermore, several of these relatively small domains can be combined within a single polypeptide chain, can be separated by linkers of varying length and structure, and can be employed to recognize short ssRNA stretches within loops. Despite these variations, one can distill some of the rules that determine RNA recognition by RRM and KH domains. This is exciting because it promises that in the future, when we will have access to more structures of protein–RNA complexes, we might be able to predict which RNA sequences are bound by RRM or KH domains and to possibly design novel RNA-binding proteins with defined sequence-specificity.
Acknowledgments
The authors would like to thank Dr Ite Laird-Offringa (University of Southern California) for critical reading of the manuscript and Dr Hong Li (Florida State University) and Dr Winfried Weissenhorn (EMBL, Grenoble) for providing the coordinates of their protein–RNA complex. This investigation was supported by a Predoctoral-Fellowship from the Roche Research Fund for Biology to F.C.O. and grants from the Swiss National Science Foundation, Structural Biology National Center of Competence in Research and from the Roche Research Fund for Biology at the ETH Zurich to F.H.T.A. F.H.T.A. is an EMBO Young Investigator. Funding to pay the Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klug A. Towards therapeutic applications of engineered zinc finger proteins. FEBS Lett. 2005;579:892–894. doi: 10.1016/j.febslet.2004.10.104. [DOI] [PubMed] [Google Scholar]
- 2.Messias A.C., Sattler M. Structural basis of single-stranded RNA recognition. Acc. Chem. Res. 2004;37:279–287. doi: 10.1021/ar030034m. [DOI] [PubMed] [Google Scholar]
- 3.Spassov D.S., Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 4.de Moor C.H., Meijer H., Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., McLachlan J., Zamore P.D., Hall T.M. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Zamore P.D., Hall T.M. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 7.Hudson B.P., Martinez-Yamout M.A., Dyson H.J., Wright P.E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nature Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza V., Summers M.F. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 9.Dey A., York D., Smalls-Mantey A., Summers M.F. Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus. Biochemistry. 2005;44:3735–3744. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 10.De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 11.Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K.J., Wu Z.R., Summers M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem–loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 12.Schuler W., Dong C., Wecker K., Roques B.P. NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes. Biochemistry. 1999;38:12984–12994. doi: 10.1021/bi990378d. [DOI] [PubMed] [Google Scholar]
- 13.Grishin N.V. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siomi H., Matunis M.J., Michael W.M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis H.A., Musunuru K., Jensen K.B., Edo C., Chen H., Darnell R.B., Burley S.K. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 16.Beuth B., Pennell S., Arnvig K.B., Martin S.R., Taylor I.A. Structure of a Mycobacterium tuberculosis NusA–RNA complex. EMBO J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Luyten I., Bottomley M.J., Messias A.C., Houngninou-Molango S., Sprangers R., Zanier K., Kramer A., Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 18.Backe P.H., Messias A.C., Ravelli R.B., Sattler M., Cusack S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Braddock D.T., Baber J.L., Levens D., Clore G.M. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braddock D.T., Louis J.M., Baber J.L., Levens D., Clore G.M. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 21.Du Z., Lee J.K., Tjhen R., Li S., Pan H., Stroud R.M., James T.L. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 A. J. Biol. Chem. 2005;280:38823–38830. doi: 10.1074/jbc.M508183200. [DOI] [PubMed] [Google Scholar]
- 22.Maris C., Dominguez C., Allain F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 23.Auweter S.D., Fasan R., Reymond L., Underwood J.G., Black D.L., Pitsch S., Allain F.H. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allain F.H., Gubser C.C., Howe P.W., Nagai K., Neuhaus D., Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 25.Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 26.Handa N., Nureki O., Kurimoto K., Kim I., Sakamoto H., Shimura Y., Muto Y., Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 27.Johansson C., Finger L.D., Trantirek L., Mueller T.D., Kim S., Laird-Offringa I.A., Feigon J. Solution structure of the complex formed by the two N-terminal RNA-binding domains of Nucleolin and a pre-rRNA target. J. Mol. Biol. 2004;337:799–816. doi: 10.1016/j.jmb.2004.01.056. [DOI] [PubMed] [Google Scholar]
- 28.Mazza C., Segref A., Mattaj I.W., Cusack S. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 30.Oubridge C., Ito N., Evans P.R., Teo C.H., Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 31.Price S.R., Evans P.R., Nagai K. Crystal structure of the spliceosomal U2B′-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 32.Varani L., Gunderson S.I., Mattaj I.W., Kay L.E., Neuhaus D., Varani G. The NMR structure of the 38 kDa U1A protein–PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nature Struct. Biol. 2000;7:329–335. doi: 10.1038/74101. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Tanaka Hall T.M. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nature Struct. Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 34.Calero G., Wilson K.F., Ly T., Rios-Steiner J.L., Clardy J.C., Cerione R.A. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nature Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 35.Allain F.H., Bouvet P., Dieckmann T., Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding J., Hayashi M.K., Zhang Y., Manche L., Krainer A.R., Xu R.-M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enokizono Y., Konishi Y., Nagata K., Ouhashi K., Uesugi S., Ishikawa F., Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 38.Allain F.H., Howe P.W., Neuhaus D., Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5772. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagai K., Oubridge C., Jessen T.H., Li J., Evans P.R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 40.Vitali F., Henning A., Oberstrass F.C., Hargous Y., Auweter S.D., Erat M., Allain F.H. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westover K.D., Bushnell D.A., Kornberg R.D. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 42.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 43.Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 44.Albertini A.A., Wernimont A.K., Muziol T., Ravelli R.B., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W. Crystal structure of the rabies virus nucleoprotein–RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 45.Green T.J., Zhang X., Wertz G.W., Luo M. Structure of the vesicular stomatitis virus nucleoprotein–RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 46.Sponer J., Hobza P. Molecular interactions of nucleic acid bases. A review of quantum-chemical studies. Collection Czechoslovak Chem. Commun. 2003;68:2231–2282. [Google Scholar]
- 47.Bloomfield V.A., Crothers D.M., Tinoco J.I. Nucleic Acids: Structures, properties and Functions. Sausalito, CA: University Science Books; 2000. [Google Scholar]
- 48.Cozzi F., Siegel J.S. Interaction between stacked aryl groups in 1,8-diarylnaphthalenes—dominance of polar/pi over charge-transfer effects. Pure Appl. Chem. 1995;67:683–689. [Google Scholar]
- 49.Allers J., Shamoo Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 50.LeCuyer K.A., Behlen L.S., Uhlenbeck O.C. Mutagenesis of a stacking contact in the MS2 coat protein–RNA complex. EMBO J. 1996;15:6847–6853. [PMC free article] [PubMed] [Google Scholar]
- 51.Nolan S.J., Shiels J.C., Tuite J.B., Cecere K.L., Baranger A.M. Recognition of an essential adenine at a protein–RNA interface: comparison of the contributions of hydrogen bonds and a stacking interaction. J. Am. Chem. Soc. 1999;121:8951–8952. [Google Scholar]
- 52.Kranz J.K., Lu J., Hall K.B. Contribution of the tyrosines to the structure and function of the human U1A N-terminal RNA binding domain. Protein Sci. 1996;5:1567–1583. doi: 10.1002/pro.5560050812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deardorff J.A., Sachs A.B. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A) binding protein. J. Mol. Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 54.Nobeli I., Laskowski R.A., Valdar W.S.J., Thornton J.M. On the molecular discrimination between adenine and guanine by proteins. Nucleic Acids Res. 2001;29:4294–4309. doi: 10.1093/nar/29.21.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallivan J.P., Dougherty D.A. Cation–pi interactions in structural biology. Proc. Natl Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao L., Wang Y., Liu Y., Hu X. Multiple intermolecular interaction modes of positively charged residues with adenine in ATP-binding proteins. J. Am. Chem. Soc. 2003;125:14216–14217. doi: 10.1021/ja036096p. [DOI] [PubMed] [Google Scholar]
- 57.Biot C., Buisine E., Rooman M. Free-energy calculations of protein-ligand cation–pi and amino–pi interactions: from vacuum to proteinlike environments. J. Am. Chem. Soc. 2003;125:13988–13994. doi: 10.1021/ja035223e. [DOI] [PubMed] [Google Scholar]
- 58.Wintjens R., Lievin J., Rooman M., Buisine E. Contribution of cation–pi interactions to the stability of protein–DNA complexes. J. Mol. Biol. 2000;302:395–410. doi: 10.1006/jmbi.2000.4040. [DOI] [PubMed] [Google Scholar]
- 59.Xue S., Calvin K., Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell J.B.O., Nandi C.L., Mcdonald I.K., Thornton J.M., Price S.L. Amino/aromatic interactions in proteins—is the evidence stacked against hydrogen-bonding. J. Mol. Biol. 1994;239:315–331. doi: 10.1006/jmbi.1994.1370. [DOI] [PubMed] [Google Scholar]
- 61.Thornton J.M., Macarthur M.W., Mcdonald I.K., Jones D.T., Mitchell J.B.O., Nandi C.L., Price S.L., Zvelebil M.J.J.M. Protein structures and complexes—what they reveal about the interactions that stabilize them. Philos. Trans. R. Soc. Lond. A—Math. Phys. Eng. Sci. 1993;345:113–129. [Google Scholar]
- 62.Guallar V., Borrelli K.W. A binding mechanism in protein–nucleotide interactions: implication for U1A RNA binding. Proc. Natl Acad. Sci. USA. 2005;102:3954–3959. doi: 10.1073/pnas.0500888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stefl R., Skrisovska L., Allain F.H.T. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 2005;6:33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oberstrass F.C., Lee A., Stefl R., Janis M., Chanfreau G., Allain F.H. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nature Struct. Mol. Biol. 2006;13:160–167. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 65.Law M.J., Linde M.E., Chambers E.J., Oubridge C., Katsamba P.S., Nilsson L., Haworth I.S., Laird-Offringa I.A. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 2006;34:275–285. doi: 10.1093/nar/gkj436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katsamba P.S., Myszka D.G., Laird-Offringa I.A. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J. Biol. Chem. 2001;276:21476–21481. doi: 10.1074/jbc.M101624200. [DOI] [PubMed] [Google Scholar]
- 67.Zuberek J., Jemielity J., Jablonowska A., Stepinski J., Dadlez M., Stolarski R., Darzynkiewicz E. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus. Biochemistry. 2004;43:5370–5379. doi: 10.1021/bi030266t. [DOI] [PubMed] [Google Scholar]
- 68.Garcia-Garcia C., Draper D.E. Electrostatic interactions in a peptide–RNA complex. J. Mol. Biol. 2003;331:75–88. doi: 10.1016/s0022-2836(03)00615-6. [DOI] [PubMed] [Google Scholar]
- 69.GuhaThakurta D., Draper D.E. Contributions of basic residues to ribosomal protein L11 recognition of RNA. J. Mol. Biol. 2000;295:569–580. doi: 10.1006/jmbi.1999.3372. [DOI] [PubMed] [Google Scholar]
- 70.Berg O.G., Vonhippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 71.Northrup S.H., Erickson H.P. Kinetics of protein protein association explained by Brownian dynamics computer-simulation. Proc. Natl Acad. Sci. USA. 1992;89:3338–3342. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schreiber G., Fersht A.R. Interaction of Barnase with its polypeptide inhibitor Barstar studied by protein engineering. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- 73.Schreiber G., Fersht A.R. Rapid, electrostatically assisted association of proteins. Nature Struct. Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 74.Law M.J., Rice A.J., Lin P., Laird-Offringa I.A. The role of RNA structure in the interaction of U1A protein with U1 hairpin II RNA. RNA. 2006;12:1168–1178. doi: 10.1261/rna.75206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hart D.J., Speight R.E., Cooper M.A., Sutherland J.D., Blackburn J.M. The salt dependence of DNA recognition by NF-kappaB p50: a detailed kinetic analysis of the effects on affinityand specificity. Nucleic Acids Res. 1999;27:1063–1069. doi: 10.1093/nar/27.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fersht A.R., Shi J.P., Knill-Jones J., Lowe D.M., Wilkinson A.J., Blow D.M., Brick P., Carter P., Waye M.M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 77.Fersht A.R. The hydrogen-bond in molecular recognition. Trends Biochem. Sci. 1987;12:301–304. [Google Scholar]
- 78.Deechongkit S., Nguyen H., Powers E.T., Dawson P.E., Gruebele M., Kelly J.W. Context-dependent contributions of backbone hydrogen bonding to beta-sheet folding energetics. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y., Kortemme T., Robertson T., Baker D., Varani G. A new hydrogen-bonding potential for the design of protein–RNA interactions predicts specific contacts and discriminates decoys. Nucleic Acids Res. 2004;32:5147–5162. doi: 10.1093/nar/gkh785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ryder S.P., Strobel S.A. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 81.McConnell T.S., Lokken R.P., Steitz J.A. Assembly of the U1 snRNP involves interactions with the backbone of the terminal stem of U1 snRNA. RNA. 2003;9:193–201. doi: 10.1261/rna.2136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sutor D. The CH…O hydrogen bond in crystals. Nature. 1962;195:68–69. [Google Scholar]
- 83.Wahl M.C., Sundaralingam M. C-H…O hydrogen bonding in biology. Trends Biochem. Sci. 1997;22:97–102. doi: 10.1016/s0968-0004(97)01004-9. [DOI] [PubMed] [Google Scholar]
- 84.Treger M., Westhof E. Statistical analysis of atomic contacts at RNA–protein interfaces. J. Mol. Recognit. 2001;14:199–214. doi: 10.1002/jmr.534. [DOI] [PubMed] [Google Scholar]
- 85.Myers J.C., Shamoo Y. Human UP1 as a model for understanding purine recognition in the family of proteins containing the RNA recognition motif (RRM) J. Mol. Biol. 2004;342:743–756. doi: 10.1016/j.jmb.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 86.Laird-Offringa I.A., Belasco J.G. Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target. Proc. Natl Acad. Sci. USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dobson N., Dantas G., Baker D., Varani G. High-resolution structural validation of the computational redesign of human U1A protein. Structure. 2006;14:847–856. doi: 10.1016/j.str.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 88.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 89.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]