An ADAR that edits transcripts encoding ion channel subunits functions as a dimer (original) (raw)
Abstract
In this report, we establish that Drosophila ADAR (adenosine deaminase acting on RNA) forms a dimer on double-stranded (ds) RNA, a process essential for editing activity. The minimum region required for dimerization is the N-terminus and dsRNA-binding domain 1 (dsRBD1). Single point mutations within dsRBD1 abolish RNA-binding activity and dimer formation. These mutations and glycerol gradient analysis indicate that binding to dsRNA is important for dimerization. However, dimerization can be uncoupled from dsRNA-binding activity, as a deletion of the N-terminus (amino acids 1–46) yields a monomeric ADAR that retains the ability to bind dsRNA but is inactive in an editing assay, demonstrating that ADAR is only active as a dimer. Different isoforms of ADAR with different editing activities can form heterodimers and this can have a significant effect on editing in vitro as well as in vivo. We propose a model for ADAR dimerization whereby ADAR monomers first contact dsRNA; however, it is only when the second monomer binds and a dimer is formed that deamination occurs.
Keywords: ADAR/dimer/Drosophila/ion channels/RNA editing
Introduction
With the completion of genome sequences, interest has focused on the diversity in gene products arising from alternative splicing and RNA editing (Caceres and Kornblihtt, 2002; Schaub and Keller, 2002). The most common type of RNA editing found in vertebrates and invertebrates is the conversion of specific adenosines to inosine by the ADAR (adenosine deaminase acting on RNA) family of enzymes (Keegan et al., 2001). ADARs bind to an imperfect double-stranded (ds) RNA duplex within the target transcript, formed between the target exon and sequences in a flanking intron, and deaminate specific adenosines usually with <100% efficiency. Inosine is read as if it were guanosine by the translation machinery, so a consequence of editing is that codons can be changed. Most of the transcripts that are edited encode receptors of the central nervous system (CNS), and aberrant editing occurs in various disorders ranging from epilepsy to malignant brain gliomas (Higuchi et al., 2000; Maas et al., 2001; Sodhi et al., 2001). In addition, ADAR knockouts in mice and flies have unambiguously demonstrated that RNA editing is required to create a full repertoire of receptors in the CNS (Seeburg et al., 1998, 2001; Higuchi et al., 2000; Palladino et al., 2000a). ADARs also deaminate long perfect duplex RNA, which potentially opposes the RNA interference process, and ADAR mutations in Caenorhabditis elegans increase transgene-induced gene silencing (Knight and Bass, 2002).
RNA editing can profoundly alter ion selectivity and gating properties of ion channels (Sommer et al., 1991; Higuchi et al., 2000) as well as subunit maturation and transport from the endoplasmic reticulum to the cell membrane (Greger et al., 2002). For example, the calcium permeability of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) class of glutamate receptor is reduced by editing of the glutamate-gated receptor subunit B (GluR-B) transcript at the Q/R site (Sommer et al., 1991). Inclusion of arginine-containing GluR-B subunits within the heterotetrameric AMPA receptor both reduces calcium permeability and also restricts the flow of receptors to synapses. Editing of transcripts encoding the serotonin 5-HT2C receptor occurs at five positions that can produce at least 12 different protein isoforms in humans (Niswender et al., 1999), with some isoforms displaying reduced coupling of the receptor to G protein (Burns et al., 1997). The edited and unedited isoforms of 5-HT2C receptor display differences in sensitivity to lysergic acid diethylamide (LSD) and antipsychotic drugs, implying an effect of editing on responses to drugs that are often used in psychiatric disorders (Berg et al., 2001; Sodhi et al., 2001).
Transcripts that are edited in Drosophila also encode ion channel subunits, but the number of sites per transcript is more than is found in vertebrates. The cacophony (cac) transcript encoding the pore-forming α1 subunit of a voltage-gated calcium channel is edited at 12 sites (Kawasaki et al., 2002). Transcripts encoding the α1 subunit of a voltage-gated sodium channel are also highly edited (Hanrahan et al., 2000), whereas the glutamate-gated chloride channel (_GluCl_α) transcript is edited at five sites in flies (Semenov and Pak, 1999).
All ADAR proteins share a similar domain structure; they contain either two or three dsRNA-binding domains (dsRBDs) and a catalytic deaminase domain in the C-terminus. ADARs are considered members of the cytidine deaminase (CDA) family due to the conservation of key residues believed to chelate zinc within their catalytic domains (Gerber and Keller, 1999). The amino acids important for catalysis in CDA enzymes are conserved in the ADATs (adenosine deaminases acting on tRNA), APOBEC-1 (apoB RNA-editing CDA subunit 1) and ADAR gene families. Indeed, mutagenesis experiments in which the putative catalytic residues in ADARs were targeted demonstrated the similarity between ADARs and CDA gene families (Lai et al., 1995). Many enzymes in the CDA family, including Escherichia coli CDA, T4 bacteriophage CDA, APOBEC-1 and particularly the adenosine deaminases Tad2/Tad3 and TadA, function as homodimers or heterodimers (Betts et al., 1994; Lau et al., 1994; Navaratnam et al., 1998; Gerber and Keller, 1999; Wolf et al., 2002). Purified ADARs are monomers, and the predicted dsRNA structures at specific editing sites do not show any obvious symmetrical structure that would suggest ADAR dimer formation.
The dsRBDs of ADARs are highly homologous to those found in other dsRNA-binding proteins such as PKR (dsRNA-dependent protein kinase), Staufen and RNase III. These proteins also form dimers and do so via their dsRBDs (Cosentino et al., 1995; Wu and Kaufman, 1997; Patel and Sen, 1998; Romano et al., 1998; Lamontagne et al., 2000).
Due to the homology between Drosophila ADAR and other proteins that form dimers, we wondered whether ADARs also function as dimers. In this study, we show that the dADAR protein does dimerize; however, unlike CDA and Tad2/Tad3 proteins, the deaminase domain is not required. Rather, the dimerization of ADAR resembles that of PKR, as the minimum domain required for dimerization is the N-terminus and first dsRBD. Binding to dsRNA is important to form a stable dimer, but these two properties of binding and dimerization can be dissociated. We demonstrate that a member of the ADAR family is active only as a dimer. Flies containing both the endogenous wild-type protein and an inactive isoform expressed from a transgene show less than wild-type levels of editing at specific sites. Different naturally occurring isoforms of ADAR that display different RNA binding as well as editing activities can form heterodimers with important consequences for the final editing activity in vitro. These findings suggest that ADAR isoforms with slightly different editing activities can combine to increase the repertoire of editing events that occur at a specific time or in a specific cell type.
Results
Dimerization of dADAR
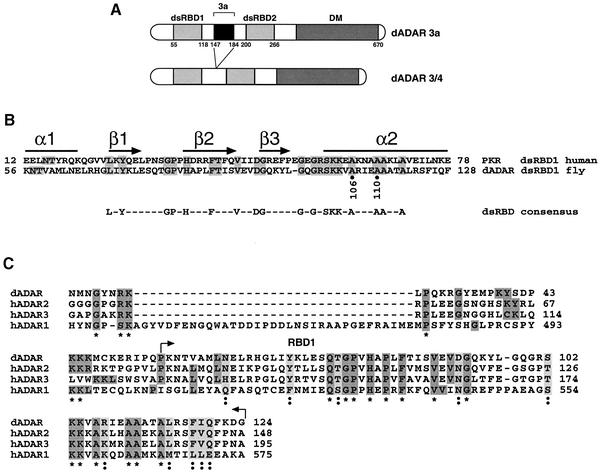
The ADAR 3a isoform (Figure 1A) was used in all experiments in this study unless otherwise indicated. To determine whether ADAR monomers interact, a yeast two-hybrid assay was used. The yeast strain L40 was cotransformed with plasmids encoding the full-length Adar 3a isoform (Figure 1A) fused to either the LexA DNA-binding domain or to the transcription activation domain (AD) of GAL4. The cotransformed yeast L40 cells were plated on selective media with increasing concentrations of 3-aminotriazole (3-AT) to reduce the non-specific background. Positive protein–protein interactions were detected as colonies that turned blue within 20 min in a β-galactoside filter lift assay. Full-length ADAR was found to dimerize in the two-hybrid assay (Figure 2A). This interaction was still observed when the concentration of 3-AT was increased to 5 mM, indicating a strong ADAR dimerization. Only 1.5 mM 3-AT was required to eliminate non-specific background. As controls, each fusion construct was independently tested with the plasmid expressing either the LexA DNA-binding domain or the activation domain of GAL4 alone (Figure 2A).
Fig. 1. Structure of Drosophila ADARs. (A) Schematic representation of the two dADAR isoforms, 3a and 3/4. The enzymes contain two dsRBDs and a deaminase domain (DM). The alternatively spliced exon 3a (111 nucleotides) lies between the two dsRBDs. Amino acid numbers are indicated below the domains. (B) Amino acid sequence comparison between dADAR and hPKR within the first dsRDB. The amino acids that are identical between the two proteins are in grey. A schematic representation of the predicted secondary structure of the dsRBD is shown above. The residues conserved >50% among all dsRBD sequences are shown beneath. (C) Homology within the minimum dimerization domain of the dADAR (AAF63703) to human ADARs (hADAR1-P55265, hADAR2-P78563, hADAR3-Q9NS39). This alignment was compiled with the web site http://ebiac.uk/clustalw. The alignment asterisks below represent identical amino acids and the double dots indicate similar amino acids.

Fig. 2. Dimerization of ADAR protein. (A) Yeast (L40) was cotransformed with plasmids encoding the full-length Adar fused to the LexA binding domain in the yeast expression vector pBTMK or fused to the DNA activation domain of GAL4 in the pACT2 vector. Positive protein–protein interactions were detected as blue colonies after a β-galactosidase assay. Full-length ADAR forms homodimers, and no interaction was detected with just the deaminase domain (DM) or with the controls. The positive control (C+) is the interaction between PAB1-2 and Paip 1, and the negative control (C-) is the absence of interaction between PAB1-2 and IRP (Gray et al., 2000). (B) The ADAR 3a isoform with epitope tags (c-Myc or HA) was transcribed and translated in vitro. The 3/4 isoform was generated with the anti-c-Myc epitope tag. The proteins were mixed and then co-immunoprecipitated with Myc monoclonal antibody. The immunoprecipitated proteins were electrophoresed on 8% SDS–polyacrylamide gel and revealed with an anti-HA polyclonal antibody. Recombinant ADAR 3a is able to form both homodimers and heterodimers.
ADAR dimerization was confirmed by in vitro co-immunoprecipitation of _in vitro_-translated proteins. Plasmid constructs bearing a T7 promoter sequence upstream of full-length Adar 3a sequence fused to either a c-Myc or an HA epitope tag were separately transcribed and translated in vitro with a coupled rabbit reticulocyte system. Equal amounts of the recombinant c-Myc and HA epitope-tagged ADARs were mixed and immunoprecipitated with polyclonal anti-c-Myc antibody coupled to protein G–agarose beads. The immunoprecipitated pro teins were detected by western blot analysis with an anti-HA antibody (Figure 2B). To confirm this result, the reciprocal experiment was also performed in which ADAR was immunoprecipitated with the anti-HA antibody and detected by western blot analysis with an anti-c-Myc antibody (data not shown).
As is the case in mammals, flies generate different ADAR isoforms by alternative splicing (Palladino et al., 2000b). The inclusion of the alternative exon 3a increases the spacing between the two dsRDBs in dADAR (Figure 1A). When the myc-tagged ADAR 3a is replaced with a myc-tagged ADAR 3/4 protein in the experiment described above, a heterodimer (Figure 2B) is formed, indicating that the alternative exon 3a is not important for dimerization. As a control, the ADAR protein was incubated with protein G–agarose beads alone and immunoprecipitated; no cross-reaction was detected. The formation of a heterodimer was also confirmed by the yeast two-hybrid system (data not shown).
Identification of the minimal dimerization domain
To identify and characterize the minimum region required for dimerization, N- or C-terminal portions of ADAR were fused to the DNA-binding domain of LexA and tested in the yeast two-hybrid assay. The deletion endpoints were chosen so as to generate proteins lacking entire conserved domains such as the two dsRBDs, the deaminase domain or the alternatively spliced exon 3a (Figure 3A).
Fig. 3. Minimum dimerization domain in ADAR protein. (A) Schematic representation of Adar deletions expressed in yeast as LexA fusion proteins. Terminal amino acid residue numbers are indicated. All the truncated proteins that retained the N-terminus expressed as LexA fused proteins interacted with full-length ADAR expressed as an AD fusion protein. A weak interaction was detected when the two dsRBDs were expressed as independent domains; blue colour appeared after 7–8 h incubation and is indicated as (–/+), strong interaction with blue colour after 20 min incubation is indicated as (++), and no interaction at any time incubation is indicated as (–). (B) Positive protein–protein interactions between ADAR and truncated ADAR proteins were detected as blue β-galactosidase activity by filter lift assay after 20 min. The amino acid residues encoding the truncated ADAR proteins are indicated. The black frame highlights the ΔN-ADAR mutant.
The truncated fusion proteins that retained the N-terminus of ADAR retained the ability to interact with full-length ADAR that was fused to the GAL4-AD domain (Figure 3). The minimum domain that retained the interaction with full-length ADAR consisted of the N-terminus and the first dsRBD (1–133) (Figure 3), and it displayed a positive interaction in a β-galactosidase filter assay after 20 min, which is similar to that obtained with the full-length proteins. However, the N-terminus alone (amino acids 1–53) and the first dsRBD (amino acids 53–133) did not interact with the full-length ADAR when expressed as independent domains (Figure 3). Neither the second dsRBD (amino acids 196–273) nor the deaminase domain (amino acids 270–670) displayed any interaction when co-expressed with the full-length ADAR (Figure 3). However, if the β-galactosidase incubation time was increased from 20 min to >7 h, the two dsRBDs (amino acids 53–133 and 196–273) displayed a very weak interaction with full-length ADAR (data not shown).
By deleting the N-terminus (amino acids 1–46), ADAR lost the ability to dimerize with the full-length protein (Figure 3). Therefore, it can be concluded that the minimal region of ADAR required to form a stable dimer consists of the N-terminus and the first dsRBD domain (amino acids 1–133).
RNA binding is required for dimerization
Considering that the first dsRBD is required for dimerization, the question of whether RNA was also an essential component arose. dADAR can bind its own RNA (L.Keegan, manuscript in preparation). Therefore, in the yeast two-hybrid assay and in in vitro immunoprecipitation experiments, it can bind to its own transcript. To discover whether RNA was critical for dimer formation, mutant proteins were generated in which the dsRNA-binding activity was disrupted. Advantage was taken of the homology in dsRBD1 to other proteins that bind dsRNA such as PKR (Figure 1B), which also forms dimers that are RNA dependent. PKR is a good model for the dsRBDs of ADAR, as Samuel and colleagues substituted the first two dsRBDs of human ADAR1 with those from PKR and retained partial hADAR1 activity (Liu et al., 2000). Therefore, with the mutational analysis of PKR as a guide, a series of point mutations were carried out in dsRBD1 to eliminate the dsRNA-binding activity of ADAR.
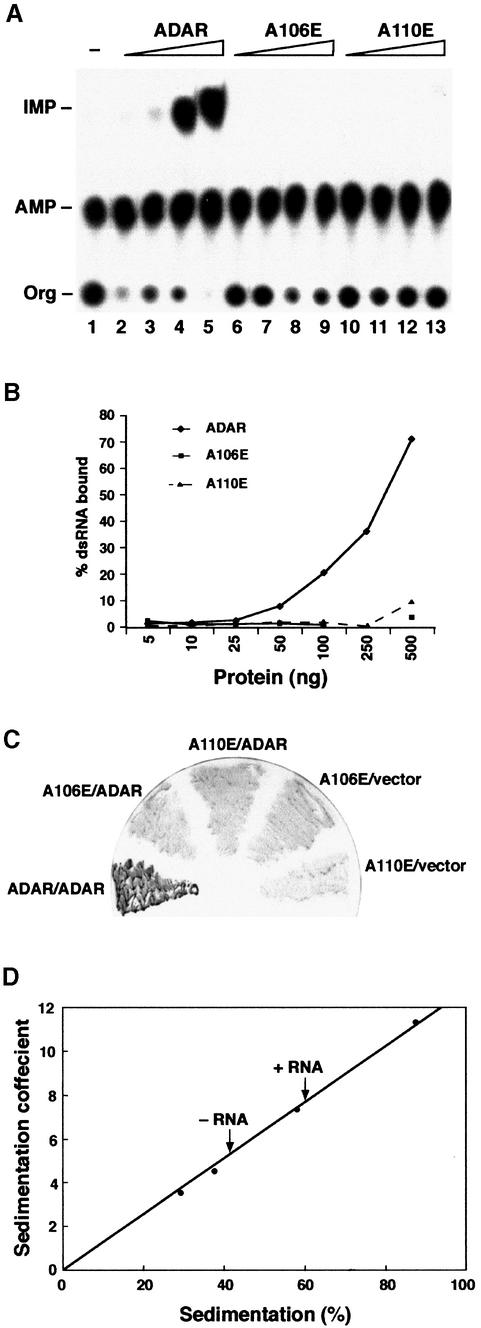
The alanine residues A106 and A110, within the second α helix of the first dsRBD of ADAR, were independently mutated to glutamate (A106E and A110E). To confirm that A106E and A110E are defective in RNA binding, both wild-type and mutant proteins were expressed in Pichia pastoris and purified by Ni2+-NTA affinity chromato graphy. Protein concentrations were normalized, and filter-binding assays were performed with increasing concentrations of protein and a constant amount of 32P-labelled dsRNA. The A106E and A110E mutants are unable to bind to dsRNA (Figure 4B), and no editing was detected in vitro (Figure 4A). As in the case of PKR, the first dsRBD of ADAR is essential for binding to dsRNA regardless of the presence of the intact second dsRBD (Zhang et al., 2001). Neither mutant was able to interact with ADAR in a two-hybrid assay (Figure 4C), demonstrating that the ability to bind dsRNA is required for the formation of dimers in vivo.
Fig. 4. dsRNA is required for ADAR dimerization. Single point mutations in dsRBD1 were generated so that amino acids A106 and A110 were changed to glutamate. (A) An in vitro editing assay was performed with recombinant proteins and chromatographed on a TLC plate. The positions of the origin (Org), adenosine (AMP) and inosine (IMP) are indicated. Increasing amounts of purified proteins were assayed [ADAR 3a (lanes 2–5), the mutant A106E (lanes 6–9) and the mutant A110E (lanes 10–13)], and the protein amounts were 10, 23, 70 and 140 ng, respectively. dsRNA without protein is in lane 1. (B) Filter binding was performed with the same mutant proteins A106E and A110E as well as with ADAR 3a; the protein amounts are indicated on the _x_-axis in nanograms. (C) Two-hybrid interactions between ADAR-LexA and ADAR-AD (positive control) and the two point mutants expressed as LexA fusion proteins and ADAR-AD were detected as blue β-galactosidase activity by filter lift assay. (D) Sedimentation coefficient of Drosophila ADAR 3/4-E/A in the presence or absence of RNA. Protein standards were applied to parallel gradients, and dADAR was detected by western blot analysis. The position of each protein is expressed as a percentage of the total number of fractions recovered from the gradient. The position of dADAR with and without RNA is indicated by arrows.
It is possible that the residue changes in the first dsRBD of ADAR could have affected monomer–monomer interaction as well as interfering with RNA contacts. Therefore, to verify by an independent method that ADAR did not form a dimer in the absence of dsRNA, we determined the sedimentation coefficient of ADAR by centrifugation through glycerol gradients containing 0.1 M KCl in the presence or absence of a specific RNA substrate (Figure 4D). In the absence of RNA, ADAR sediments in the fraction after the BSA standard, as would be expected from its molecular weight; whereas, in the presence of RNA, ADAR sediments in all fractions after aldolase that has a molecular weight of 158 kDa. Therefore, in the absence of RNA, ADAR behaves as a monomer; only in the presence of RNA do we see a shift to a higher molecular weight species.
The sedimentation profiles depicted in Figure 4D were determined using an inactive 3/4 isoform. This was generated by introducing a point mutation in the deaminase domain whereby the glutamate (E330) that has been proposed to be involved in proton shuttling was replaced by alanine (ADAR-E/A) (Lai et al., 1995). Identical sedimentation profiles were obtained with both the inactive and active isoforms (data not shown). This suggests that an active deaminase domain is not required for the formation of a protein–RNA complex.
Dimerization is essential for editing activity
To address the question of whether dimerization is essential for ADAR activity, a deletion mutant lacking the N-terminus (amino acids 1–46) was generated, ΔN-ADAR (amino acids 46–670). This mutant did not interact with full-length ADAR in the two-hybrid assay (Figure 3). Recombinant ΔN-ADAR containing a His6 tag at the C-terminus was overexpressed in P.pastoris, purified and assayed by western blot analysis and by filter-binding assays with dsRNA. Deletion of the N-terminus did not disrupt RNA-binding activity (Figure 5B), demonstrating that the mutant protein was folded correctly. However, ΔN-ADAR failed to edit dsRNA in vitro (Figure 5A). Even with higher concentrations of protein, no editing activity was ever observed (data not shown), suggesting that, even though the protein can bind RNA, this is not sufficient and the active form of ADAR is a dimer.

Fig. 5. ADAR is active as a dimer. (A) The in vitro editing assay was performed with either ADAR or ΔN-ADAR. Increasing amounts (7, 14, 30 and 50 ng) of both proteins were assayed. The products of the assay were chromatographed on a TLC plate and the origin (Org), adenosine (AMP) and inosine (IMP) are indicated. Lane 1 contains 32P-labelled dsRNA incubated without protein. Lanes 2–5 have increasing amounts of ADAR. Lanes 6–9 have increasing amounts of ΔN-ADAR. (B) Filter binding was performed with 32P-labelled dsRNA and increasing amounts of ADAR and ΔN-ADAR. The protein amounts are indicated on the _x_-axis in nanograms. This figure represents the average of two independent experiments.
The formation of heterodimers can influence editing activity
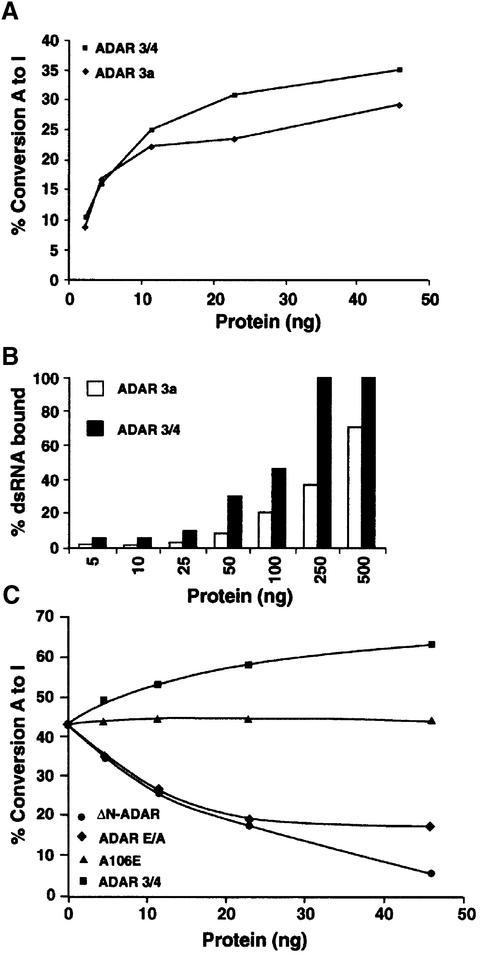
The different ADAR isoforms 3a and 3/4 (Figure 1A) in Drosophila have different binding as well as editing activities on dsRNA (Figure 6A and B), with ADAR 3/4 being the more active isoform. The isoforms can form heterodimers, as the domain involved in the dimerization (NH2–RBD1) is present in both (Figure 2B). To show that heterodimer formation can affect editing activity, inactive ADAR 3/4-E/A was overexpressed in P.pastoris, purified and mixed with ADAR 3a in dsRNA-editing assays. The purified ADAR 3/4-E/A protein has no editing activity but binds dsRNA (data not shown).
Fig. 6. Inactive ADAR that retains the minimum dimerization domain downregulates ADAR activity in vitro. (A) A graph representing the editing activity of purified ADAR 3a and 3/4 on 32P-labelled dsRNA. The 3/4 isoform was more active than the 3a isoform. The protein amounts used are indicated. (B) A graph representing the binding of purified 3a and 3/4 proteins to 32P-labelled dsRNA. The 3/4 binding saturated at 250 ng, whereas the 3a isoform bound less dsRNA under the same experimental conditions. (C) In vitro editing assay was performed with a constant amount of ADAR 3a (7 ng). Increasing amounts of inactive 3/4-E/A (diamonds), or 3/4 wild-type (squares) or A106E (triangles) or ΔN-ADAR (circles) were added to this mixture; the protein amount that was added is indicated. A106E is a negative control as it cannot dimerize or bind to dsRNA, whereas ADAR 3/4 is a positive control.
For protein competition experiments, a concentration of ADAR 3a (7 ng) was chosen so as to give ∼40% conversion of adenosine to inosine in dsRNA (Figure 6C). Increasing concentrations of inactive ADAR 3/4-E/A were added to ADAR 3a. As controls, increasing concentrations of either ADAR 3/4 (positive control) or the ADAR A106E mutant that cannot bind dsRNA or form dimers were also incubated with ADAR 3a. When the 3a and 3/4 isoforms were mixed, an increase in editing activity was observed (Figure 6C, squares), corresponding to the increase in active protein present in the reaction. The editing activity of ADAR 3a was not affected by ADAR A106E (Figure 6C, triangles), showing that there is no interaction between ADARs, even at higher concentrations, without RNA binding.
When the inactive ADAR 3/4-E/A was mixed with 3a, a decrease in editing activity to 50% was observed (Figure 6C, diamonds). Interestingly, we never observed a decrease in editing activity >50% even when inactive 3/4 was added at a 6-fold higher concentration. However, when ΔN-ADAR that binds with high affinity to dsRNA (Figure 5B) was added, a continuing decrease in editing activity was observed with increasing concentrations (Figure 6C, circles). One explanation for this result is that ΔN-ADAR binds dsRNA as a monomer and sequesters the dsRNA.
Decrease in editing of transcripts in transgenic Drosophila that contain both an active and an inactive ADAR isoform
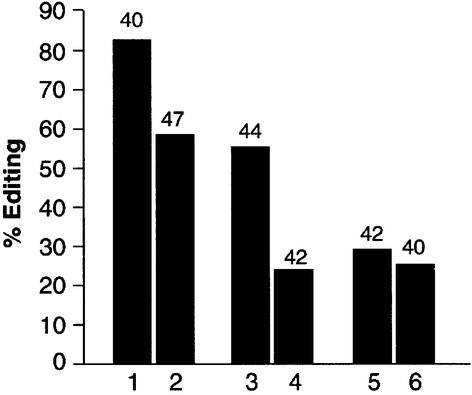
To determine whether the formation of a heterodimer could influence editing in vivo, transgenic Drosophila were generated that contained both the wild-type protein and the inactive isoform ADAR 3/4-E/A. RT–PCR analysis was performed on edited transcripts to measure editing levels. Three sites were analysed in two transcripts: one in the cac transcript; and two in the _GluCl_α transcript. The editing site chosen in cac was S/G (exon 15) and it is edited to 86% in adult flies (Figure 7). The K/R site in the _GluCl_α transcript is also highly edited (55%), and we also examined the N/S site that is edited to a lower level of 29%. For both the cac S/G site and the _GluCl_α K/R site, a decrease in editing of 25–30% was observed in RNA from the transgenic flies that contained the inactive ADAR 3/4 isoform. A slight decrease of ∼4% was observed at the _GluCl_α N/S site. This result is very surprising, as in the same transcript two sites are affected differently in the transgenic flies.
Fig. 7. Formation of a heterodimer in Drosophila influences RNA- editing activity. Two independent RT–PCR reactions were performed on RNA isolated from transgenic flies containing both wild-type and the inactive ADAR 3/4-E/A. RNA editing at three different sites in two transcripts was analysed by sequencing individual clones. Lanes 1, 3 and 5 are sequences from wild-type Canton S, whereas lanes 2, 4 and 6 were obtained from the transgenic flies. Lanes 1 and 2 represent the percentage of editing at the S/G site in exon 15 in the cac transcript, lanes 3 and 4 the editing at the K/R site in the _GluCl_α transcript, and lanes 5 and 6 the _GluCl_α N/S site. The numbers above the bars in the graph are the numbers of clones sequenced.
Discussion
In this report, we establish that dADAR forms a dimer and, most interestingly, that this dimerization is essential for editing activity. The minimum dimerization domain is the N-terminus and dsRBD1 (Figure 3). Single point mutations within dsRBD1 that destroy RNA-binding activity also affect dimer formation, indicating that binding to dsRNA is required for dimerization (Figure 4). However, dimerization can be separated from binding to dsRNA, as a deletion of the N-terminus (amino acids 1–46) yields an ADAR monomer that retains the ability to bind dsRNA but can not dimerize or edit dsRNA (Figure 5). Different isoforms of ADAR can form heterodimers, and this can have a significant effect on editing activity both in vitro (Figure 6C) and in vivo (Figure 7). MacMillan and colleagues reported that human ADAR2 forms a ternary complex only in the presence of RNA; they came to this conclusion after kinetic analysis of the editing of the GluR-B R/G site (Jaikaran et al., 2002). The in vitro experiments in the present study were carried out mainly using dsRNA as a non-specific editing substrate, but the correlation with the results of MacMillan and colleagues convinces us that specific editing sites in ion channel transcripts behave similarly.
Sequence comparison of >100 dsRBDs has identified clusters of highly conserved amino acids, and NMR and X-ray crystallography studies have revealed that the domain has an α1–β1–β2–β3–α2 secondary structure (Figure 1B) (Bycroft et al., 1995; Kharrat et al., 1995; Ryter and Schultz, 1998). The most conserved region of the dsRBD is the second α helix (Figure 1B) that makes contacts with the major groove of the dsRNA, and mutations within this helix can completely destroy RNA binding (McMillan et al., 1995; Zhang et al., 2001). In the present study, the amino acids A106 and A110 of ADAR were independently mutated to glutamate in the α2 helix in dsRBD1 (Figure 1B). These ADAR mutants are unable to bind dsRNA as assayed by filter binding (Figure 4B), do not form dimers with full-length ADAR in a yeast two-hybrid assay (Figure 4C) and do not inhibit RNA editing in mixing experiments with active ADAR isoforms (Figure 6C). These experiments suggest that dsRNA is important for dimer formation and that dsRBD1 from two different monomers must bind dsRNA. However, the N-terminus (amino acids 1–53) or dsRBD1 (amino acids 55–133) were unable to interact with the full-length ADAR when expressed as independent domains (Figure 3), indicating that dsRNA and a single dsRBD1 are not sufficient to form a stable ADAR dimer.
The N-terminus of ADAR is also important for dimerization, as a truncated ADAR lacking the first 46 amino acids failed to interact with the full-length ADAR in vivo despite the presence of an intact dsRBD1 (Figure 3). However, this mutant retained the ability to bind dsRNA better than wild-type ADAR (Figure 5B). More importantly, this mutant was inactive in an in vitro editing assay, demonstrating that ADAR is only active as a dimer (Figure 5A). Apparently, dimerization does not increase the affinity of ADAR for RNA, since the ΔN-ADAR not only binds as well as wild-type protein but even competes efficiently with wild-type ADAR in RNA-editing assays (Figure 6C). This mutant also demonstrates that, in contrast to what occurs in ADATs, dimerization and binding to dsRNA can be separated. Our results do not rule out the possibility that there are also contacts between deaminase domains in ADAR dimers.
The finding that dimerization of ADAR is mediated by an N-terminal region and the first dsRBD is reminiscent of what occurs with other members of the dsRBP gene family, in particular with RNase III and PKR (Wu and Kaufman, 1997; Lamontagne et al., 2000). It has been observed that PKR activity is inhibited by a high concentration of dsRNA, as the binding of each monomer to separate RNA molecules precludes the formation of an active dimer (Chu et al., 1998). ADAR monomers probably behave in a similar manner, as it has been shown that both Xenopus ADAR and hADAR2 are inhibited by excess substrate (Hough and Bass, 1994; Jaikaran et al., 2002). The concentration of both dsRNA and ADAR isoforms are probably critical for the equilibrium between the inactive monomer and the active dimer and this could be a way to regulate RNA-editing activity in vivo. The presence of a high concentration of PKR pushes the equilibrium from a monomer to a dimer in the absence of RNA (Nanduri et al., 1998). However, gel filtration analysis of partially purified mammalian ADAR1 and ADAR2 shows that the proteins elute as monomers (O’Connell and Keller, 1994; Maas et al., 1996), and the sedimentation coefficient of dADAR in the absence of RNA suggests it is a monomer (Figure 4D).
To analyse whether the formation of a heterodimer can affect the efficiency of editing, we mixed ADAR 3a (2 nM dimers) with inactive isoforms in the presence of a 600 base pair dsRNA. If every editable adenosine in this substrate is a separate editing site, the concentration of sites is 8 nM. Consistent with our idea that a minimal editing site must be large enough to bind an ADAR dimer, the effective concentration of editing sites is probably lower than 6 nM. This is clear because ΔN-ADAR competes for sites at the lowest concentration added and competes further at high concentrations. When increasing amounts of the inactive ADAR 3/4-E/A isoform were added to the 3a active protein, a progressive reduction in editing activity to 50% was observed (Figure 6C), but activity did not decrease further with additional protein. The inactive isoform that is capable of forming a heterodimer gave this 50% inhibition. Our interpretation of the mixing experiment with ADAR 3/4-E/A is that the ADAR 3a–ADAR 3/4-E/A heterodimer is only half as active as the ADAR 3a homodimer. This loss in activity is not compensated by the heterodimer being twice as abundant as the homodimer, because of the competition for sites. Because the heterodimer is more abundant, the decrease of >50% at higher concentrations that is seen with ΔN-ADAR may be difficult to observe with ADAR 3/4-E/A, although we do not fully understand why this further decrease is not seen. Interestingly, a similar downregulation effect occurs when wild-type and ADAR 3/4-E/A isoforms are co-expressed together in Drosophila. A greater decrease in activity was observed in vitro than in vivo. An explanation may be that there is more wild-type protein than the inactive 3/4-E/A protein present or that different editing sites may require specific homodimers or heterodimers.
The formation of heterodimers between these isoforms can have an important impact on RNA editing in Drosophila, considering that the ratio of ADAR 3/4 to ADAR 3a isoforms increases during development (Palladino et al., 2000b). Recently, a study of Drosophila Adar identified a new isoform, highly expressed in embryo, that is truncated within the deaminase domain (Ma et al., 2002). Interestingly, this inactive isoform is expressed predominantly during early stages of development, and its abundance correlates with the low level of editing observed during this developmental stage (Palladino et al., 2000b; Ma et al., 2002). We believe that any active ADAR present would be sequestered by this truncated protein. High expression of inactive hADAR2 isoforms with truncated C-termini has been also observed (Gerber et al., 1997; Lai et al., 1997). It is possible that inactive ADAR isoforms may be required to downregulate ADAR activity by forming heterodimers.
A remarkable feature of the dsRBDs is their ability to mediate both RNA–protein and protein–protein interactions with other members of the dsRBP family (Fierro-Monti and Mathews, 2000); An example is the formation of heterodimers between the viral E3L protein and PKR; this interaction inhibits PKR activity in vivo (Romano et al., 1998). Interestingly, the N-terminus of the dADAR can interact with other proteins (A.Gallo, unpublished results). As yet, it is uncertain whether these proteins can sequester ADAR from the active dimer and/or form a larger protein complex that modulates editing activity. In Drosophila, an intriguing link between editing and an RNA helicase maleless (mle), another dsRBP, has already been found (Reenan et al., 2000). This RNA helicase contains two dsRBDs, and it will be interesting to determine whether ADAR can interact directly with MLE and bring it to an editing site where it is required.
How can we interpret the evidence for ADAR dimerization in relation to what is known about editing targets? Most ADAR substrates may be edited on only one side of the putative ADAR dimer, and the complex is probably pseudo-symmetric, as proposed for substrate recognition by APOBEC (Navaratnam et al., 1998; Hersberger et al., 1999). A dimeric model for ADAR predicts that in some cases symmetrical editing may be seen if there are target adenosine residues on opposite strands at a fixed distance apart. In fact, a possible symmetric editing event has been reported (Herbert and Rich, 2001) in which five adenosine residues are edited in a substrate containing the top strand sequence AA-(N15)-TTT. We propose that the ADAR dimer has active sites positioned 16 base pairs apart and that each active site flips out and edits a number of adenosines in succession. Hybrid proteins made between vertebrate ADAR1 and ADAR2 show that ADAR deaminase domains contribute to the different target site specificities of those proteins (Wong et al., 2001). The dimer model predicts that deaminase domain recognition of RNA sequence or structure 16 bases away from the edited adenosine may contribute to specifying the editing site. MacMillan and colleagues showed that a GluR-B R/G site which has only 14 bases of predicted duplex 3′ to the edited base is not edited and forms only a monomeric complex with ADAR (Jaikaran et al., 2002).
We have shown that the dimerization of ADAR enzymes is essential for deamination and is a key step in regulating RNA editing. These findings are important in understanding the enzymatic reaction of ADARs. The RNA structures formed at editing sites must now be re-examined for evidence of symmetry. dADAR is a good model for mammalian ADARs, as it is very homologous in domain structure and sequence to hADARs, particularly to hADAR2 (Palladino et al., 2000b). hADAR2 is able to rescue locomotion defects in Adar mutant Drosophila (L.Keegan, unpublished results), indicating that mechanisms of site recognition in editing of ion channel transcripts are conserved.
Materials and methods
Plasmids and yeast strains
The analysis of ADAR interactions in yeast was performed by subcloning the full-length Adar cDNA and deletions of it in frame with BD-lexA in the pBTMK vector. The selectable markers on this plasmid are Trp1 for yeast and _Kan_R for E.coli. The full-length Adar was also subcloned in frame with AD-Gal4 in the pACT2 vector (Clontech) that contains the leu2 and _Amp_R markers. All clones were sequenced to ensure that the correct fusion protein was generated. The lithium acetate method was used to cotransform the plasmids in the Saccharomyces cerevisiae strain L40 [MATa; HIS200; trp1-901; leu2-3,112; ade2; LYS2::(lexAop)4-HIS3; URA3::(lexAop)8-lacZ; GAL4] (Sambrook et al., 1989; Gietz et al., 1992). The cells containing both plasmids were selected on SD medium lacking leucine, tryptophan and histidine in the presence of increasing concentrations of 3-AT. The activation of the reporter genes (HIS3 and lacZ) was analysed by growth on selective media and by β-galactosidase filter lift assay. To confirm that the recombinant dADAR proteins were expressed in yeast, whole cell protein extracts were prepared from yeast (Printen and Sprague, 1994). Western blot analysis was performed with anti-LexA monoclonal antibody (Clontech) and anti-Gal4-AD monoclonal antibody (Clontech) and visualized with the enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia Biotech).
Protein purification
See Supplementary data available at The EMBO Journal Online.
Site-directed mutagenesis
See Supplementary data.
Generation of deletion mutants
Serial deletions from the C- and N-termini of ADAR were carried out to define the minimum region of the protein capable of forming a dimer with the full-length protein in yeast. Restriction sites were included in the oligonucleotides so as to facilitate the subcloning of Adar in frame with the DNA-binding domain of lexA in the pBTMK vector; _Eco_RI restriction sites were introduced for the forward oligonucleotides and _Bam_HI sites for the reverse oligonucleotides. The number in the name of the oligonucleotide corresponds to nucleotide position in the Adar sequence (DDBJ/EMBL/GenBank accession No. AAF63702): Fw1–21_Eco_RI (5′-CATGAATTCATGTTAAACAGCGCTAATAAC-3′); Fw139–168_Eco_RI (5′-CATGAATTCATGTGCAAGGAGCGCATTC CC-3′); Fw163–178_Eco_RI (5′-CATGAATTCCCGAAGAACACGG TGG-3′); Fw597–615_Eco_RI (5′-CATGAATTCGATAAGGGTCCTG TCATG-3′); Fw810–828_Eco_RI (5′-CATGAATTCATGGTGGTGCC ACAGAAG-3′); Rev2010–1995_Bam_HI (5′-CGCGGATCCTCATTC GGCAAGACCG-3′); Rev821–800_Bam_HI (5′-CGCGGATCCGGCAC CACCATTGGACTGTAGG-3′); Rev596–579_Bam_HI (5′-CGCGGATC CGGAACCTTCTTCTGACCG-3′); Rev400–379_Bam_HI (5′-CGCGGA TCCCGCCGGCTTCAGAGGCGACAG-3′); Rev160–140_Bam_HI (5′- CGCGGATCCGGGAATGCGCTCCTTGCAC-3′).
The nucleotide sequence of all constructs was confirmed by sequence analysis, and western blot analysis was performed with the anti-lexA monoclonal antibody to ensure the protein was of the expected size in yeast.
In vitro transcription and translation
Different Adar fusion constructs with either c-Myc or hemagglutinin (HA) epitope sequences were transcribed and translated in vitro from PCR products with a rabbit reticulocyte coupled transcription–translation kit (Promega). Adar 3a and 3/4 with the c-Myc epitope sequence at the N-terminus was generated by PCR, and the DNA template was Adar-c-Myc subcloned in the pGBKT7 two-hybrid vector (Clontech). An oligonucleotide encoding both the T7 promoter and the c-Myc epitope was used in combination with the reverse oligonucleotide Rev2010–1995_Bam_HI (see above). Adar 3a with the HA epitope sequence was generated by PCR, and the DNA template was Adar subcloned in the pACT2 vector (Clontech). The forward oligonucleotide encoded both the T7 promoter and the HA epitope tag 5′ of the Adar sequence and the reverse oligonucleotide was AD-Rev primer. The sequences of the oligonucleotides are: c-Myc-Fw primer, 5′-AAAATTGTAATAC GACTCACTATAGGGCGAGCCGCCACCATGGAGGAGCAGAAG CTGATC-3′; HA-Fw primer, 5′-AAAATTGTAATACGACTCACTA TAGGGCGAGCCGCCACCATGTACCCATACGACGTTCCAGATT ACGC-3′; and AD-Rev primer, 5′-ACTTGCGGGGTTTTTCAGTA TCTACGAT-3′.
PCR reactions were performed in a thermal cycler with Expand Taq (Roche) and analysed by electrophoresis on an agarose gel. The crude PCR products (usually 4 µl) were added to 40 µl of the TNT T7 PCR Quick Master Mix (Promega) in the presence of 2 µl [35S]methionine (1000 Ci/mmol at 10 mCi/ml) in a final volume of 50 µl and incubated at 30°C for 90 min. The translated proteins were analysed by 8% SDS–PAGE and autoradiography on X-ray film. Western blot analysis was also performed with either anti-HA polyclonal or anti-Myc monoclonal antibodies and revealed by ECL.
Glycerol gradient sedimentation
See Supplementary data.
In vitro protein–protein interaction assay
Equal amounts of the _in vitro_-translated proteins (either 3a-myc and 3a-HA or 3/4-myc and 3a-HA) were mixed and incubated for 1 h at 30°C. Then 30 µl of protein G–agarose beads (Roche), 15 µl of anti-c-Myc monoclonal antibody (or anti-HA polyclonal antibody) and 470 µl of immunoprecipation buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1% Triton X-100, 20% glycerol, 0.5 mM PMSF, 0.7 µg/ml pepstatin and 0.4 µg/mg leupeptin) were added to the proteins and mixed by rotating at 4°C for 90 min. The beads were pelleted by centrifugation at 14 000 r.p.m. for 2 min in a microcentrifuge at 4°C. The beads were washed three times by pipetting with 500 µl of immunoprecipitation buffer. The washed beads were boiled in 15 µl of Laemmli buffer for 2 min, and 7 µl of this was analysed by western blot with either anti-HA-polyclonal (or anti-c-Myc monoclonal antibodies; data not shown).
Analysis of adenosine-to-inosine RNA-editing activity in vitro
See Supplementary data.
Filter-binding assay
See Supplementary data.
Mixing experiment
Purified proteins were normalized by Bradford assay and mixed with dsRNA containing 200 fmol of 32P-labelled adenosine. A constant amount of ADAR 3a (7 ng) and increasing concentrations of different isoforms [either inactive ADAR 3/4-E/A, ADAR 3/4 (wild-type), A106E or ΔN-ADAR] were mixed on ice, and the editing assay was performed at 37°C for 1 h.
Fly strains and RNA editing in vivo
The inactive ADAR 3/4-E/A has an essential glutamate in the active site of the deaminase domain changed to alanine. The protein bears a FLAG tag at the N-terminus and a His6 tag at the C-terminus. The UAS-Adar3/4 EA line was generated by coinjection of a pUAST-Adar3/4 EA construct and helper plasmid into w1118 eggs followed directly by balancing of stable transformant lines using standard procedures. To look for a dominant inhibitory effect of overexpressing ADAR 3/4-E/A in a fly containing normal levels of ADAR protein, male flies of the w; UAS-Adar 3/4 EA 4–1/TM3 Sb line were crossed to females of the genotype y, Adar 1F4, w / w, FM6; (mini w+) actin-5C-Gal4/SM5 Cy. Male progeny from this cross of the genotype w, FM6; (mini w+) actin-5C-Gal4; (mini w+)UAS-Adar3/4 EA were selected for RNA extraction to compare RNA-editing levels with those seen in wild-type Canton-S flies. Individual clones were sequenced from two independent RT–PCR reactions from rescue and control flies.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank J.Brindle for technical assistance; N.Gray for reagents; A.MacMillan for sharing unpublished work; and H.Cooke and J.Rosenthal for comments on the manuscript. This work was supported by the MRC and a grant from the British Heart Foundation (PG/98086).
References
- Berg K.A., Cropper,J.D., Niswender,C.M., Sanders-Bush,E., Emeson,R.B. and Clarke,W.P. (2001) RNA-editing of the 5-HT2C receptor alters agonist-receptor-effector coupling specificity. Br. J. Pharmacol., 134, 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts L., Xiang,S., Short,S.A., Wolfenden,R. and Carter,C.W.Jr (1994) Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol., 235, 635–656. [DOI] [PubMed] [Google Scholar]
- Burns C.M., Chu,H., Rueter,S.M., Hutchinson,L.K., Canton,H., Sanders-Bush,E. and Emeson,R.B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature, 387, 303–308. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Grunert,S., Murzin,A.G., Proctor,M. and St Johnston,D. (1995) NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J., 14, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F. and Kornblihtt,A.R. (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet., 18, 186–193. [DOI] [PubMed] [Google Scholar]
- Chu W.M., Ballard,R., Carpick,B.W., Williams,B.R. and Schmid,C.W. (1998) Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol., 18, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino G.P., Venkatesan,S., Serluca,F.C., Green,S.R., Mathews,M.B. and Sonenberg,N. (1995) Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl Acad. Sci. USA, 92, 9445–9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-Monti I. and Mathews,M.B. (2000) Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci., 25, 241–246. [DOI] [PubMed] [Google Scholar]
- Gerber A.P. and Keller,W. (1999) An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science, 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- Gerber A., O’Connell,M.A. and Keller,W. (1997) Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA, 3, 453–463. [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St. Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency tansformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K., Coller,J.M., Dickson,K.S. and Wickens,M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J., 19, 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I.H., Khatri,L. and Ziff,E.B. (2002) RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron, 34, 759–772. [DOI] [PubMed] [Google Scholar]
- Hanrahan C.J., Palladino,M.J., Ganetzky,B. and Reenan,R.A. (2000) RNA editing of the Drosophila para Na+ channel transcript. Evolutionary conservation and developmental regulation. Genetics, 155, 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. and Rich,A. (2001) The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl Acad. Sci. USA, 98, 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersberger M., Patarroyo-White,S., Arnold,K.S. and Innerarity,T.L. (1999) Phylogenetic analysis of the apolipoprotein B mRNA-editing region. Evidence for a secondary structure between the mooring sequence and the 3′ efficiency element. J. Biol. Chem., 274, 34590–34597. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Maas,S., Single,F.N., Hartner,J., Rozov,A., Burnashev,N., Feldmeyer,D., Sprengel,R. and Seeburg,P.H. (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature, 406, 78–81. [DOI] [PubMed] [Google Scholar]
- Hough R.F. and Bass,B.L. (1994) Purification of the Xenopus laevis dsRNA adenosine deaminase. J. Biol. Chem., 269, 9933–9939. [PubMed] [Google Scholar]
- Jaikaran D.C., Collins,C.H. and MacMillan,A.M. (2002) Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J. Biol. Chem., 277, 37624–37629. [DOI] [PubMed] [Google Scholar]
- Kawasaki F., Collins,S.C. and Ordway,R.W. (2002) Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J. Neurosci., 22, 5856–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L.P., Gallo,A. and O’Connell,M.A. (2001) The many roles of an RNA editor. Nat. Rev. Genet., 2, 869–878. [DOI] [PubMed] [Google Scholar]
- Kharrat A., Macias,M.J., Gibson,T.J., Nilges,M. and Pastore,A. (1995) Structure of the dsRNA binding domain of E.coli RNase III. EMBO J., 14, 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass,B.L. (2002) The role of RNA editing by ADARs in RNAi. Mol. Cell, 10, 809–817. [DOI] [PubMed] [Google Scholar]
- Lai F., Drakas,R. and Nishikura,K. (1995) Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem., 270, 17098–17105. [DOI] [PubMed] [Google Scholar]
- Lai F., Chen,C.-X., Carter,K.C. and Nishikura,K. (1997) Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol., 17, 2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne B., Tremblay,A. and Abou Elela,S. (2000) The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol., 20, 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P.P., Zhu,H.J., Baldini,A., Charnsangavej,C. and Chan,L. (1994) Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc. Natl Acad. Sci. USA, 91, 8522–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lei,M. and Samuel,C.E. (2000) Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR. Proc. Natl Acad. Sci. USA, 97, 12541–12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E., Tucker,M.C., Chen,Q. and Haddad,G.G. (2002) Developmental expression and enzymatic activity of pre-mRNA deaminase in Drosophila melanogaster. Brain Res. Mol. Brain Res., 102, 100–104. [DOI] [PubMed] [Google Scholar]
- Maas S., Melcher,T., Herb,A., Seeburg,P.H., Keller,W., Krause,S., Higuchi,M. and O’Connell,M.A. (1996) Structural requirements for RNA editing in glutamate receptor pre-mRNA by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem., 271, 12221–12226. [DOI] [PubMed] [Google Scholar]
- Maas S., Patt,S., Schrey,M. and Rich,A. (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl Acad. Sci. USA, 98, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan N.A., Carpick,B.W., Hollis,B., Toone,W.M., Zamanian-Daryoush,M. and Williams,B.R. (1995) Mutational analysis of the double-stranded RNA (dsRNA) binding domain of the dsRNA-activated protein kinase, PKR. J. Biol. Chem., 270, 2601–2606. [DOI] [PubMed] [Google Scholar]
- Nanduri S., Carpick,B.W., Yang,Y., Williams,B.R. and Qin,J. (1998) Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J., 17, 5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam N. et al. (1998) Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol., 275, 695–714. [DOI] [PubMed] [Google Scholar]
- Niswender C.M., Copeland,S.C., Herrick-Davis,K., Emeson,R.B. and Sanders-Bush,E. (1999) RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem., 274, 9472–9478. [DOI] [PubMed] [Google Scholar]
- O’Connell M.A. and Keller,W. (1994) Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc. Natl Acad. Sci. USA, 91, 10596–10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino M.J., Keegan,L.P., O’Connell,M.A. and Reenan,R.A. (2000a) A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell, 102, 437–449. [DOI] [PubMed] [Google Scholar]
- Palladino M.J., Keegan,L.P., O’Connell,M.A. and Reenan,R.A. (2000b) dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA, 6, 1004–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.C. and Sen,G.C. (1998) Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol. Cell. Biol., 18, 7009–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printen J.A. and Sprague,G.F.Jr (1994) Protein–protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics, 138, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R.A., Hanrahan,C.J. and Barry,G. (2000) The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron, 25, 139–149. [DOI] [PubMed] [Google Scholar]
- Romano P.R., Zhang,F., Tan,S.L., Garcia-Barrio,M.T., Katze,M.G., Dever,T.E. and Hinnebusch,A.G. (1998) Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol., 18, 7304–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter J.M. and Schultz,S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook S., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schaub M. and Keller,W. (2002) RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie, 84, 791–803. [DOI] [PubMed] [Google Scholar]
- Seeburg P.H., Higuchi,M. and Sprengel,R. (1998) RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res. Brain Res. Rev., 26, 217–229. [DOI] [PubMed] [Google Scholar]
- Seeburg P.H., Single,F., Kuner,T., Higuchi,M. and Sprengel,R. (2001) Genetic manipulation of key determinants of ion flow in glutamate receptor channels in the mouse. Brain Res., 907, 233–243. [DOI] [PubMed] [Google Scholar]
- Semenov E.P. and Pak,W.L. (1999) Diversification of Drosophila chloride channel gene by multiple posttranscriptional mRNA modifications. J. Neurochem., 72, 66–72. [DOI] [PubMed] [Google Scholar]
- Sodhi M.S., Burnet,P.W., Makoff,A.J., Kerwin,R.W. and Harrison,P.J. (2001) RNA editing of the 5-HT2C receptor is reduced in schizophrenia. Mol. Psychiatry, 6, 373–379. [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler,M., Sprengel,R. and Seeburg,P.H. (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell, 67, 11–19. [DOI] [PubMed] [Google Scholar]
- Wolf J., Gerber,A.P. and Keller,W. (2002) tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J., 21, 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Sato,S. and Lazinski,D.W. (2001) Substrate recognition by ADAR1 and ADAR2. RNA, 7, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. and Kaufman,R.J. (1997) A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem., 272, 1291–1296. [DOI] [PubMed] [Google Scholar]
- Zhang F., Romano,P.R., Nagamura-Inoue,T., Tian,B., Dever,T.E., Mathews,M.B., Ozato,K. and Hinnebusch,A.G. (2001) Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J. Biol. Chem., 276, 24946–24958. [DOI] [PubMed] [Google Scholar]