Architecture of initiation-competent 12-subunit RNA polymerase II (original) (raw)
Abstract
RNA polymerase (Pol) II consists of a 10-polypeptide catalytic core and the two-subunit Rpb4/7 complex that is required for transcription initiation. Previous structures of the Pol II core revealed a “clamp,” which binds the DNA template strand via three “switch regions,” and a flexible “linker” to the C-terminal repeat domain (CTD). Here we derived a model of the complete Pol II by fitting structures of the core and Rpb4/7 to a 4.2-Å crystallographic electron density map. Rpb4/7 protrudes from the polymerase “upstream face,” on which initiation factors assemble for promoter DNA loading. Rpb7 forms a wedge between the clamp and the linker, restricting the clamp to a closed position. The wedge allosterically prevents entry of the promoter DNA duplex into the active center cleft and induces in two switch regions a conformation poised for template-strand binding. Interaction of Rpb4/7 with the linker explains Rpb4-mediated recruitment of the CTD phosphatase to the CTD during Pol II recycling. The core–Rpb7 interaction and some functions of Rpb4/7 are apparently conserved in all eukaryotic and archaeal RNA polymerases but not in the bacterial enzyme.
RNA polymerase (Pol) II synthesizes all eukaryotic mRNA in the course of gene transcription. The regulation of gene transcription by Pol II underlies cell proliferation and differentiation. Regulation occurs mainly at the level of transcription initiation, when Pol II assembles with the general transcription factors TFIIB, -D, -E, -F, and -H into the initiation complex on promoter DNA (1–3). In addition to the 10-subunit Pol II core, initiation requires the heterodimeric Rpb4/7 complex that can dissociate from core (4, 5).
Crystallographic structures are available for the Saccharomyces cerevisiae Pol II core (6, 7), which is sufficient for RNA elongation (4, 5). In the core structures, the two large subunits form opposite sides of a central “cleft,” and the eight small subunits are arrayed around the periphery. The cleft contains the active center and is constricted at one end by a protein “wall.” One side of the cleft is formed by a mobile “clamp,” which adopts open states in two crystal forms of the Pol II core (6, 7) but is closed in a further structure of a core elongation complex with bound template DNA and product RNA (8). Mobility of the clamp relies on five protein “switch” regions, which connect the clamp to the remainder of the enzyme (7, 8). A flexible “linker” emerges from the core surface below the clamp and connects to the C-terminal repeat domain (CTD) of the largest Pol II subunit, which is disordered in all core structures.
Counterparts of the Rpb4/7 complex exist in the two other eukaryotic nuclear RNA polymerases (9–13) and the archaeal RNA polymerase (14). The structure of an archaeal Rpb4/7 counterpart revealed an extended complex, spanned by the Rpb7 homolog (15). To understand Rpb4/7 function and elucidate the mechanism of transcription initiation, we studied the 12-subunit Pol II by x-ray crystallography.
Materials and Methods
The yeast Pol II core was purified from a yeast rpb4 deletion strain essentially as described (16). Yeast Rpb4 and Rpb7 were coexpressed in Escherichia coli as described (17), purified to homogeneity, and concentrated to 5–10 mg/ml. For reconstitution of stoichiometric 12-subunit Pol II, 0.5 mg of core was incubated for 1 h at 20°C with a 5-fold excess of Rpb4/7, and unbound Rpb4/7 was removed by gel filtration (Superose 6, Amersham Biosciences). Fractions containing the 12-subunit Pol II were concentrated to 4 mg/ml. Crystals were grown at 20°C with the hanging-drop method by using a reservoir solution of 8% PEG 20000/360 mM NaH2PO4/(NH4)2HPO4, pH 6.0/50 mM dioxane/5 mM DTT. Crystals grew to a maximum size of 0.4 × 0.2 × 0.1 mm. Crystals were transferred stepwise to reservoir solutions with 5%, 10%, 18%, and 22% ethylene glycol or glycerol, incubated at 4°C overnight, mounted in cryoloops, and dispersed and stored in liquid nitrogen. Diffraction was observed beyond 4-Å resolution, but radiation damage and anisotropy limited complete data to 4.2-Å resolution. The highest peak in molecular replacement with AMORE (18) was obtained by using as search model the core elongation complex structure (correlation coefficient = 45.1/18.4 for first/second peak, 10–6.5 Å) (8). This structure was used without nucleic acids as a model for the core. Eight additional residues of the Rpb1 linker were included. The archaeal Rpb4/7 counterpart structure was manually fitted to 2_F_obs – _F_calc and _F_obs – _F_calc electron density maps phased with the core model. Manual rigid-body adjustments accounted for slight changes in the relative position of three domains (Rpb4, yeast Rpb7 residues 1–80, and yeast Rpb7 residues 81–169). Residues 117 and 174–187 of the archaeal Rpb7 counterpart were deleted, and yeast Rpb7 residues 52, 53, 155, and 156 were inserted. An insertion between α-helices H1 and H2 in yeast Rpb4 could not be modeled. Although an additional helical density was observed, the connectivity was uncertain. No continuous density was observed for an N-terminal extension of yeast Rpb4. Despite weak sequence similarity, the overall fold of the C-terminal half of yeast Rpb4 corresponds to that of its archaeal counterpart and is confirmed by the location of selenomethionine 145 in yeast Rpb4, which corresponds to residue 38 in helix 2 of the archaeal Rpb4 counterpart structure. At the resolution of the data, no refinement was carried out, and the fitted structures were reduced to Cα backbones. Ordering of the switches was confirmed with a simulated annealing omit map, calculated with a model lacking switches 1–3, with the program CNS (19).
Results and Discussion
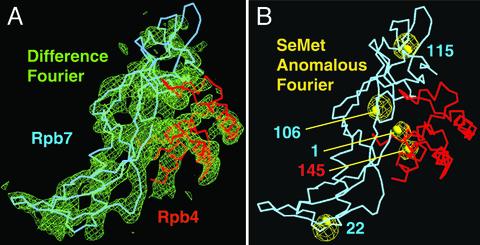
Reconstitution and Structural Analysis of Pol II. Endogenous yeast Pol II contains substoichiometric amounts of Rpb4/7, which impede crystallization. To overcome this difficulty, yeast Pol II was reconstituted from endogenous core and recombinant Rpb4/7 (compare Materials and Methods). The reconstituted 12-subunit polymerase could be purified by size-exclusion chromatography and formed large single crystals within several days. With the use of cryocooling and synchrotron radiation, complete diffraction data to 4.2-Å resolution were obtained (Table 1), which allowed for structure solution by molecular replacement. Initial electron density maps were phased with a truncated core model that lacked the clamp. These maps showed positive difference density for the clamp and for an additional mass on the core surface, which could be fitted with the structure of the archaeal Rpb4/7 counterpart (Fig. 1_A_; ref. 15). The orientation of Rpb4/7 was additionally confirmed by labeling Rpb4/7 with selenomethionine (Fig. 1_B_). The resulting backbone model reveals the location of amino acid residues in all 12 Pol II subunits.
Table 1. X-ray diffraction data.
| Data set* | Native | Rpb4/7 SeMet |
|---|---|---|
| Unit cell axis,† Å | 220.4, 391.4, 282.2 | 223.0, 394.5, 284.4 |
| Wavelength, Å | 0.9185 | 0.9795 (Se peak) |
| Resolution, Å (highest shell) | 50-4.2 (4.35-4.2) | 50-6.5 (6.7-6.5) |
| Unique reflections (highest shell) | 83,431 (7,893) | 25,009 (2,452) |
| Completeness, % (highest shell) | 93.8 (89.5) | 99.9 (100.0) |
| Redundancy | 2.9 | 13.4 |
| I/σI | 6.5 | 9.4 |
| _R_sym, % (highest shell) | 9.3 (36.3) | 7.5 (25.8) |
| No. of peaks in anomalous Fourier | - | 13 (5 Se, 8 Zn) |
Fig. 1.
Structural analysis of the 12-subunit Pol II. (A) Fit of the Rpb4/7 counterpart structure (15) to the initial 4.2-Å difference Fourier map (green). The map was phased with the Pol II core structure and is contoured at 2σ. Cα backbones for Rpb4 and Rpb7 are shown in pink and blue, respectively. Minor adjustments were made to the structure to account for yeast-specific sequence features. The view corresponds to the “front” view of Pol II (6). (B) Selenomethionine (SeMet) anomalous difference Fourier map (yellow). The Fourier map was calculated with anomalous data from a crystal with selenomethionine-labeled Rpb4/7 (Table 1) and phases from the core model and is contoured at 6σ. Selenomethionine was incorporated as described (52). Five selenium peaks with heights between 20.5σ and 10.6σ coincide with the location of methionine side chains. Indicated as yellow spheres are sulfur atoms in methionine side chains after the corresponding archaeal residues were replaced. No peaks were observed for Rpb4 methionines 1 and 114, which are in flexible regions. The figure was prepared with o (53).
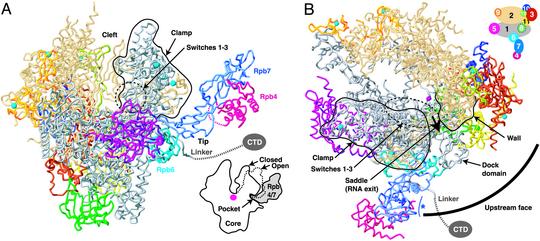
Overall Structure. The model shows a single Rpb4/7 complex on the outside of the core protruding from the base of the clamp (Fig. 2_A_). The location of Rpb4/7 agrees with that in cryoelectron microscopic reconstructions of Pol II (20). However, the orientation of Rpb4/7 differs. Whereas electron microscopy suggested that Rpb4 binds to the Pol II core, our data unambiguously show that instead Rpb7 binds to the Pol II core. The Rpb4/7 location also agrees with that of the Rpb4/7 counterpart in Pol I, also observed by electron microscopy (11, 21). Most of the Rpb4/7 surface is exposed and accessible for interactions with proteins or nucleic acids. Exiting RNA may interact with a potential nucleic acid-binding surface of Rpb7 (15, 22) that faces the Pol II “saddle,” from which RNA emerges (Fig. 2_B_). Such interactions may slightly change the Rpb4/7 orientation that is fixed here by crystal packing.
Fig. 2.
Architecture of the 12-subunit Pol II, coupling of Rpb4/7 binding and clamp closure, and upstream interaction face. (A) Ribbon model of Pol II. The view is as described for Fig. 1. Cyan spheres and a pink sphere depict eight zinc ions and an active-site magnesium ion, respectively. A black line circles the clamp. The linker to the CTD is indicated as a dashed line. In the lower-right corner, a schematic cut-away view is shown. A dashed line indicates the open clamp position observed in form 2 of the Pol II core structure (7). (B) Pol II upstream interaction face. Shown in a view of the model from the “top” (6). The circle segment is centered at the active site and has a radius that corresponds to the minimal distance between the TATA box and the transcription start site (85 Å, ≈25 bp). The saddle between the wall and the clamp and the assumed direction of RNA exit are indicated. A blue asterisk indicates a potential RNA-binding face of Rpb7 (15, 22). A key to subunit color is shown in the upper right corner. The figure was prepared with RIBBONS (54).
Rpb4/7 binds to the Pol II core with the N-terminal ribonucleoprotein-like domain of Rpb7 (15), termed here the “tip” (Fig. 2 A). The remainder of Rpb4/7 extends far from the core surface, explaining why mutations in this region do not impair core binding (22). Consistent with the core–Rpb7 interaction, Rpb7 alone can bind to core (23), and Rpb7 is essential for yeast growth (24), whereas Rpb4 is not (25). Deletion of the rpb4 gene in yeast facilitates dissociation of Rpb7 from core (5). Our model suggests that loss of the Rpb4–Rpb7 interface after Rpb4 deletion destabilizes Rpb7 and facilitates Rpb7 dissociation. Indeed, the Rpb4–Rpb7 interface is conserved (15) such that stable chimeric heterodimers with Rpb4 and Rpb7 from various species can be formed (14, 17, 26). Consistent with our model, Rpb7 overexpression can suppress Rpb4 deletion defects (23, 27). In contrast to our findings, Rpb7 dissociation after Rpb4 deletion was explained earlier by core interaction primarily through Rpb4 (5, 20, 28).
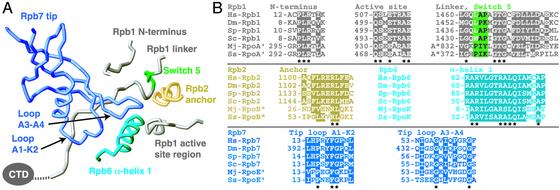
Rpb7 Forms a Conserved Wedge That Restrains the Clamp. The Rpb7 tip forms a wedge between the clamp, the linker, and the core subunit Rpb6 (Fig. 2_A_). The tip partially fills a surface “pocket,” which corresponds to the end of the previously identified potential RNA exit groove 1 (6). The pocket is lined by five protein regions: three in Rpb1, and one each in Rpb2 and Rpb6 (Fig. 3). Rpb4/7 binding to the pocket thus holds together three subunits and may stabilize the Pol II subunit assembly. The pocket–Rpb7 interface is partially hydrophobic and conserved among eukaryotes (Fig. 3), explaining why human Rpb4/7 can functionally replace its yeast counterpart (29).
Fig. 3.
Pocket–tip interaction. (A) Ribbon model of the Rpb7 tip binding with its two outermost loops (15) to the five protein regions (7) that line the pocket. The view is from the “back,” roughly the reverse of that shown in Figs. 1 and 2_A_. Colors are as described for Fig. 2 except that switch region 5 is green. (B) Sequence alignments of protein regions are as described for A. Hs, Homo sapiens; Dm, Drosophila melanogaster; Sp, Schizosaccharomyces pombe; Sc, S. cerevisiae; Mj, Methanococcus jannaschii; Ss, Sulfolobus solfataricus. Conserved residues are highlighted. The stars below the alignments indicate invariant residues. The figure was prepared with RIBBONS (54).
A corresponding pocket–tip interaction apparently exists in eukaryotic Pol I and Pol III and in the archaeal RNA polymerase. The tip domain is conserved in the Rpb7 homologs of Pol I (10, 11), Pol III (9, 12), and archaeal polymerase (15). Similar to Rpb7, the Rpb7 homolog of Pol I binds the common subunit Rpb6 (11) and can dissociate from the core in a Pol I mutant (30). The Rpb7 homolog of Pol III is also dissociable, and disruption of its tip domain is lethal (9). Because core subunits are either shared or homologous in all three eukaryotic enzymes (31), the Rpb4/7 counterparts most likely bind similarly to the cores. Thus the yeast 12-subunit Pol II is a good model for Pol I and Pol III and for the enzymes in higher eukaryotes, because Pol II subunits are highly conserved in sequence and function (32).
In the 12-subunit Pol II, the clamp adopts the closed conformation observed in the core elongation complex structure (8), consistent with electron microscopy in solution (20). Except for the clamp closure, there are no gross changes in core structure after Rpb4/7 binding. Modeling the clamp in the open states observed in the free Pol II core structures (7) results in a clash with the Rpb7 tip (Fig. 2 A). Thus Rpb4/7 can only bind when the clamp is closed, and the clamp can only open after Rpb4/7 has dissociated from the Pol II core. Residual space between the closed clamp and Rpb4/7 only allows for slight changes in the clamp position.
Implications for Transcription Initiation. Coupling between Rpb4/7 binding and clamp closure is relevant for transcription initiation when promoter DNA is loaded onto polymerase. Given that the DNA duplex would be loaded deeply into the cleft, between the clamp and the wall as suggested (7), Rpb4/7 must dissociate from the Pol II core for sufficient clamp opening. However, Rpb4/7 could rebind after DNA melting and clamp closure. Although Rpb4/7 can dissociate from the Pol II core in vitro (5), it is unclear whether it dissociates in vivo. Our model reveals a small core–Rpb7 interface, consistent with a transient interaction, but the core–Rpb4/7 complex is more stable in other species (17, 26, 28).
In an alternative scenario for initiation, Rpb4/7 persistently bound to core would prevent promoter entry to the cleft, and DNA could only bind far above the active center. After DNA melting, the template strand, however, could pass the clamp, slip into the cleft, and bind to the site formed by switch regions 1–3 as observed in the core elongation complex (8). Except for a central part in switch 3, around Rpb2 residues 1120–1127, switches 1–3 adopt a similar conformation in the 12-subunit Pol II and are no longer flexible as in the free core structures (7). Thus, interaction of Rpb4/7 with the Pol II core induces partial formation of the binding site for the DNA template strand. The flexible part of switch 3 may only get ordered after template-strand binding or formation of an early DNA–RNA hybrid. Induced folding of the central part of switch 3 by the growing hybrid could underlie a transition that stabilizes the early elongation complex.
Promoter loading minimally requires assembly of TFIIB, TFIIF, and the TATA box-binding protein (TBP) to DNA regions upstream of the transcription start site (33, 34). Topological considerations predict that these factors interact with the “upstream face” of Pol II around the “dock” domain (Fig. 2_B_; ref. 7). Rpb4/7 dramatically extends the upstream interaction face (Fig. 2_B_), consistent with a role of Rpb4/7 in initiation complex assembly. Indeed, Rpb4/7 stabilizes a minimal initiation complex (35). Both Rpb4/7 and TFIIB bind to the Pol II linker (Figs. 2 and 3; ref. 36), which may be a scaffold for initiation complex assembly. Adjacent binding of Rpb4/7 and TFIIB is consistent with an interaction between the archaeal homologs of TFIIB and Rpb6 (37). In Pol III, the Rpb4 homolog binds to a region corresponding to the linker (12), and it also interacts with a TFIIB-related initiation factor (38). The Rpb7 homolog of Pol I also binds an initiation factor (39, 40). Thus Rpb4/7 and its counterparts bridge the polymerase core with initiation factors, and differences between them could contribute to promoter specificity. The roles of Rpb4/7 in initiation and a possible structural role (see above) can account for transcription shut off in an rpb4 deletion strain of yeast at high temperature (41–43).
It is likely that promoter loading is topologically similar during bacterial transcription initiation, which requires only the σ factor and the core polymerase. The structural conservation of bacterial and eukaryotic core RNA polymerases (44, 45) enables a comparison of our model with recent structures of bacterial RNA polymerase bound to σ (46, 47) and bound to σ and upstream promoter DNA (48). In these structures, the σ factor interacts with regions corresponding to the Pol II upstream face where TFIIB, TFIIF, and TATA box-binding protein are proposed to assemble. Although the σ factor shows sequence similarity to Rpb4 (25), it is not a functional counterpart of Rpb4/7, because Rpb4/7 and σ bind to different sites on the polymerase surfaces. The structure of bacterial polymerase with bound promoter shows a closed clamp and ordered switches, as observed in our model of the initiation-competent Pol II. Additionally, the bacterial complex shows promoter DNA outside the cleft, consistent with our alternative scenario for initiation, in which duplex DNA is loaded and melted above the cleft.
The specific occurrence of an Rpb4/7 complex in Archaea and eukaryotes but not in bacteria is reflected in its functions during transitions within the transcription cycle. During initiation, the Pol II CTD gets phosphorylated and remains phosphorylated during RNA elongation. Dephosphorylation of the CTD, however, is required for Pol II recycling after termination, because only unphosphorylated Pol II can rejoin an initiation complex. CTD dephosphorylation is carried out by the phosphatase Fcp1 (49). Fcp1 is apparently recruited to the phosphorylated CTD by Rpb4/7, because Rpb4 binds Fcp1 (50), and Rpb7 binds the linker to the CTD, which is disordered in our crystals (Figs. 2 and 3). Fcp1 inhibits initiation complex assembly (51), maybe because Fcp1 and a general factor bind to the upstream face in a mutually exclusive manner, ensuring complete CTD dephosphorylation before transcription reinitiation.
Conclusion
The 12-subunit Pol II model explains and suggests functional roles of the Rpb4/7 complex and begins to extend our understanding of eukaryotic transcription toward the mechanism of initiation. Our work further shows that the architecture of a dissociable multiprotein complex can be determined by x-ray analysis even at moderate resolution. The methods used and developed here can be applied to complexes of Pol II with proteins and nucleic acids to address mechanistic questions raised by the complete Pol II model.
Acknowledgments
We thank C. Schulze-Briese and T. Tomizaki for help at the protein crystallography beamline X06SA of the Swiss Light Source; M. Kimura and A. Ishihama for the Rpb4/7 expression plasmid; C. Carles, M. Siaut, and P. Thuriaux for sending manuscripts before publication; K.-P. Hopfner for help with mass spectrometry; A. Meinhart, C. Buchen, and other laboratory members for help; and Q. Eastman, D. Eick, R. Grosschedl, K.-P. Hopfner, M. Meisterernst, R. Sachdev, and members of the laboratory for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Research Grant CR117-2/1, the European Molecular Biology Organization Young Investigator Program, and the Fonds der Chemischen Industrie.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NT9).
Abbreviations: Pol, RNA polymerase; CTD, C-terminal repeat domain.
See commentary on page 6893.
References
- 1.Roeder, R. G. (1998) Cold Spring Harbor Symp. Quant. Biol. 63**,** 201–218. [DOI] [PubMed] [Google Scholar]
- 2.Lee, T. I. & Young, R. A. (2000) Annu. Rev. Genet. 34**,** 77–137. [DOI] [PubMed] [Google Scholar]
- 3.Lemon, B. & Tjian, R. (2000) Genes Dev. 14**,** 2551–2569. [DOI] [PubMed] [Google Scholar]
- 4.Ruet, A., Sentenac, A., Fromageot, P., Winsor, B. & Lacroute, F. (1980) J. Biol. Chem. 255**,** 6450–6455. [PubMed] [Google Scholar]
- 5.Edwards, A. M., Kane, C. M., Young, R. A. & Kornberg, R. D. (1991) J. Biol. Chem. 266**,** 71–75. [PubMed] [Google Scholar]
- 6.Cramer, P., Bushnell, D. A., Fu, J., Gnatt, A. L., Maier-Davis, B., Thompson, N. E., Burgess, R. R., Edwards, A. M., David, P. R. & Kornberg, R. D. (2000) Science 288**,** 640–649. [DOI] [PubMed] [Google Scholar]
- 7.Cramer, P., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292**,** 1863–1876. [DOI] [PubMed] [Google Scholar]
- 8.Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. (2001) Science 292**,** 1876–1882. [DOI] [PubMed] [Google Scholar]
- 9.Sadhale, P. P. & Woychik, N. A. (1994) Mol. Cell. Biol. 14**,** 6164–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shpakovski, G. V. & Shematorova, E. K. (1999) Russ. J. Bioorgan. Chem. 25**,** 700–704. [Google Scholar]
- 11.Peyroche, G., Levillain, E., Siaut, M., Callebaut, I., Schultz, P., Sentenac, A., Riva, M. & Carles, C. (2002) Proc. Natl. Acad. Sci. USA 99**,** 14670–14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siaut, M., Zaros, C., Levivier, E., Ferri, M. L., Court, M., Werner, M., Callebaut, I., Thuriaux, P., Sentenac, A. & Conesa, C. (2003) Mol. Cell. Biol. 23**,** 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, P., Wu, S., Sun, Y., Yuan, C. C., Kobayashi, R., Myers, M. P. & Hernandez, N. (2002) Mol. Cell. Biol. 22**,** 8044–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner, F., Eloranta, J. J. & Weinzierl, R. O. (2000) Nucleic Acids Res. 28**,** 4299–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todone, F., Brick, P., Werner, F., Weinzierl, R. O. & Onesti, S. (2001) Mol. Cell 8**,** 1137–1143. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, A. M., Darst, S. A., Feaver, W. J., Thompson, N. E., Burgess, R. R. & Kornberg, R. D. (1990) Proc. Natl. Acad. Sci. USA 87**,** 2122–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakurai, H., Mitsuzawa, H., Kimura, M. & Ishihama, A. (1999) Mol. Cell. Biol. 19**,** 7511–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navaza, J. (1994) Acta Crystallogr. A 50**,** 157–163. [Google Scholar]
- 19.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54**,** 905–921. [DOI] [PubMed] [Google Scholar]
- 20.Craighead, J. L., Chang, W. H. & Asturias, F. J. (2002) Structure (Cambridge, Mass.) 10**,** 1117–1125. [DOI] [PubMed] [Google Scholar]
- 21.Bischler, N., Brino, L., Carles, C., Riva, M., Tschochner, H., Mallouh, V. & Schultz, P. (2002) EMBO J. 21**,** 4136–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlicky, S. M., Tran, P. T., Sayre, M. H. & Edwards, A. M. (2001) J. Biol. Chem. 276**,** 10097–10102. [DOI] [PubMed] [Google Scholar]
- 23.Sheffer, A., Varon, M. & Choder, M. (1999) Mol. Cell. Biol. 19**,** 2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKune, K., Richards, K. L., Edwards, A. M., Young, R. A. & Woychik, N. A. (1993) Yeast 9**,** 295–299. [DOI] [PubMed] [Google Scholar]
- 25.Woychik, N. A. & Young, R. A. (1989) Mol. Cell. Biol. 9**,** 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin, R. M. & Guilfoyle, T. J. (1998) J. Biol. Chem. 273**,** 5631–5637. [DOI] [PubMed] [Google Scholar]
- 27.Tan, Q., Li, X., Sadhale, P. P., Miyao, T. & Woychik, N. A. (2000) Mol. Cell. Biol. 20**,** 8124–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khazak, V., Estojak, J., Cho, H., Majors, J., Sonoda, G., Testa, J. R. & Golemis, E. A. (1998) Mol. Cell. Biol. 18**,** 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khazak, V., Sadhale, P. P., Woychik, N. A., Brent, R. & Golemis, E. A. (1995) Mol. Biol. Cell 6**,** 759–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smid, A., Riva, M., Bouet, F., Sentenac, A. & Carles, C. (1995) J. Biol. Chem. 270**,** 13534–13540. [DOI] [PubMed] [Google Scholar]
- 31.Sentenac, A., Riva, M., Thuriaux, P., Buhler, J.-M., Treich, I., Carles, C., Werner, M., Ruet, A., Huet, J., Mann, C., et al. (1992) Yeast RNA Polymerase Subunits and Genes (Cold Spring Harbor Lab. Press, Plainview, NY).
- 32.Woychik, N. A. (1998) Cold Spring Harbor Symp. Quant. Biol. 63**,** 311–317, [DOI] [PubMed] [Google Scholar]
- 33.Killeen, M., Coulombe, B. & Greenblatt, J. (1992) J. Biol. Chem. 267**,** 9463–9466. [PubMed] [Google Scholar]
- 34.Ebright, R. H. (1998) Cold Spring Harbor Symp. Quant. Biol. 63**,** 11–20. [DOI] [PubMed] [Google Scholar]
- 35.Jensen, G. J., Meredith, G., Bushnell, D. A. & Kornberg, R. D. (1998) EMBO J. 17**,** 2353–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, H., Friesen, J. D. & Lis, J. T. (1994) Mol. Cell. Biol. 14**,** 7507–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magill, C. P., Jackson, S. P. & Bell, S. D. (2001) J. Biol. Chem. 276**,** 46693–46696. [DOI] [PubMed] [Google Scholar]
- 38.Ferri, M. L., Peyroche, G., Siaut, M., Lefebvre, O., Carles, C., Conesa, C. & Sentenac, A. (2000) Mol. Cell. Biol. 20**,** 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peyroche, G., Milkereit, P., Bischler, N., Tschochner, H., Schultz, P., Sentenac, A., Carles, C. & Riva, M. (2000) EMBO J. 19**,** 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, X., Zhao, J., Zentgraf, H., Hoffmann-Rohrer, U. & Grummt, I. (2002) EMBO Rep. 3**,** 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maillet, I., Buhler, J. M., Sentenac, A. & Labarre, J. (1999) J. Biol. Chem. 274**,** 22586–22590. [DOI] [PubMed] [Google Scholar]
- 42.Miyao, T., Barnett, J. D. & Woychik, N. A. (2001) J. Biol. Chem. 276**,** 46408–46413. [DOI] [PubMed] [Google Scholar]
- 43.Pillai, B., Sampath, V., Sharma, N. & Sadhale, P. (2001) J. Biol. Chem. 276**,** 30641–30647. [DOI] [PubMed] [Google Scholar]
- 44.Darst, S. A. (2201) Curr. Opin. Struct. Biol. 11**,** 155–162. [DOI] [PubMed] [Google Scholar]
- 45.Cramer, P. (2002) Curr. Opin. Struct. Biol. 12**,** 89–97. [DOI] [PubMed] [Google Scholar]
- 46.Murakami, K. S., Masuda, S. & Darst, S. A. (2002) Science 296**,** 1280–1284. [DOI] [PubMed] [Google Scholar]
- 47.Vassylyev, D. G., Sekine, S., Laptenko, O., Lee, J., Vassylyeva, M. N., Borukhov, S. & Yokoyama, S. (2002) Nature 417**,** 712–719. [DOI] [PubMed] [Google Scholar]
- 48.Murakami, K. S., Masuda, S., Campbell, E. A., Muzzin, O. & Darst, S. A. (2002) Science 296**,** 1285–1290. [DOI] [PubMed] [Google Scholar]
- 49.Lin, P. S., Marshall, N. F. & Dahmus, M. E. (2002) Prog. Nucleic Acid Res. Mol. Biol. 72**,** 333–365. [DOI] [PubMed] [Google Scholar]
- 50.Kimura, M., Suzuki, H. & Ishihama, A. (2002) Mol. Cell. Biol. 22**,** 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chambers, R. S., Wang, B. Q., Burton, Z. F. & Dahmus, M. E. (1995) J. Biol. Chem. 270**,** 1–8. [DOI] [PubMed] [Google Scholar]
- 52.Budisa, N., Steipe, B., Demange, P., Eckerskorn, C., Kellermann, J. & Huber, R. (1995) Eur. J. Biochem. 230**,** 788–796. [DOI] [PubMed] [Google Scholar]
- 53.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47**,** 110–119. [DOI] [PubMed] [Google Scholar]
- 54.Carson, M. (1997) Methods Enzymol. 277**,** 493–505. [PubMed] [Google Scholar]