Establishment and Maintenance of Genomic Methylation Patterns in Mouse Embryonic Stem Cells by Dnmt3a and Dnmt3b (original) (raw)
Abstract
We have previously shown that the DNA methyltransferases Dnmt3a and Dnmt3b carry out de novo methylation of the mouse genome during early postimplantation development and of maternally imprinted genes in the oocyte. In the present study, we demonstrate that Dnmt3a and Dnmt3b are also essential for the stable inheritance, or “maintenance,” of DNA methylation patterns. Inactivation of both Dnmt3a and Dnmt3b in embryonic stem (ES) cells results in progressive loss of methylation in various repeats and single-copy genes. Interestingly, introduction of the Dnmt3a, Dnmt3a2, and Dnmt3b1 isoforms back into highly demethylated mutant ES cells restores genomic methylation patterns; these isoforms appear to have both common and distinct DNA targets, but they all fail to restore the maternal methylation imprints. In contrast, overexpression of Dnmt1 and Dnmt3b3 failed to restore DNA methylation patterns due to their inability to catalyze de novo methylation in vivo. We also show that hypermethylation of genomic DNA by Dnmt3a and Dnmt3b is necessary for ES cells to form teratomas in nude mice. These results indicate that genomic methylation patterns are determined partly through differential expression of different Dnmt3a and Dnmt3b isoforms.
DNA methylation is essential for mammalian development and plays crucial roles in a variety of biological processes, such as genomic imprinting and X chromosome inactivation (26). DNA methylation patterns are established during embryonic development through a highly orchestrated process that involves demethylation and de novo methylation and can be inherited in a clonal fashion through the action of maintenance methyltransferase activity (8, 26, 32). During preimplantation development, both paternal and maternal genomes undergo a wave of demethylation, which erases most of the methylation patterns inherited from the gametes. Shortly after implantation, the embryo undergoes a wave of de novo methylation, which establishes a genome-wide hypermethylation pattern (19, 22, 29, 40). De novo methylation also occurs during gametogenesis in both male and female germ cells and is believed to play a critical role in the establishment of genomic imprinting, an epigenetic process that results in differential modification of paternal and maternal alleles during gametogenesis and monoallelic expression of a small set of genes, known as imprinted genes, in the offspring (20, 26, 33). De novo methylation activity is present mainly in embryonic stem (ES) cells and embryonal carcinoma cells, early postimplantation embryos, and developing germ cells, whereas it is largely suppressed in differentiated somatic cells (22, 24, 41, 43). Therefore, ES cells can be a good model system for studying the mechanisms of de novo methylation.
Three active DNA cytosine methyltransferases—Dnmt1, Dnmt3a, and Dnmt3b—have been identified in human and mouse (4, 31, 48). Dnmt1 is ubiquitously expressed in proliferating cells and localizes to DNA replication foci (25). Purified Dnmt1 protein methylates hemimethylated DNA substrates more efficiently than unmethylated DNA in vitro (5). Despite its activity in vitro, Dnmt1 has not been convincingly shown to be able to initiate de novo methylation in vivo. Moreover, inactivation of Dnmt1 in ES cells and mice leads to extensive demethylation of all sequences examined (24, 27). All of these findings suggest that Dnmt1 functions primarily as a maintenance methyltransferase that is responsible for copying the parental-strand methylation pattern onto the daughter strand after each round of DNA replication. In contrast, Dnmt3a and Dnmt3b are strongly expressed in ES cells, early embryos, and developing germ cells but are expressed at low levels in differentiated somatic cells (10, 31). Indeed, genetic studies have demonstrated that Dnmt3a and Dnmt3b are essential for de novo methylation in ES cells and postimplantation embryos, as well as for de novo methylation of imprinted genes in the germ cells (18, 30). Although Dnmt3a and Dnmt3b function primarily as de novo methyltransferases to establish methylation patterns, they may also play a role in maintaining methylation patterns. We have previously shown that some genomic sequences, such as the differentially methylated region 2 (DMR2) of Igf2 and the 5′ region of Xist, are almost completely demethylated and an L1-like repeat is partially demethylated in mutant ES cells that lack Dnmt3a and Dnmt3b (28, 30). It is not clear, however, whether Dnmt3a and Dnmt3b are also required for maintaining global methylation patterns.
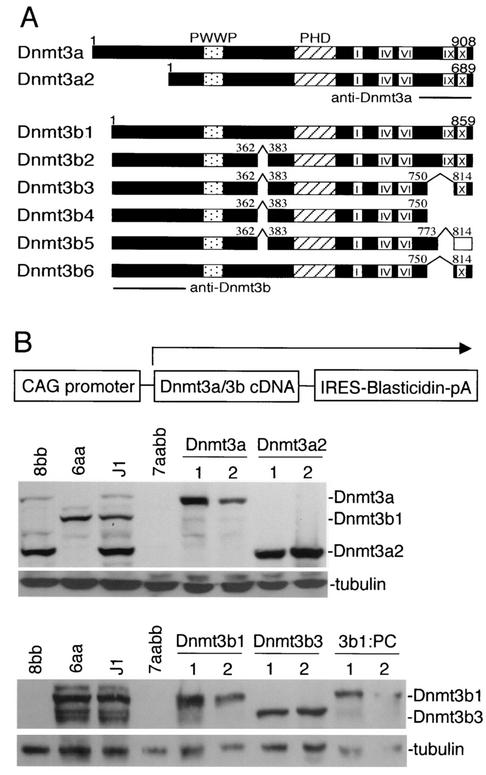
At least two Dnmt3a and six Dnmt3b isoforms have been identified (see Fig. 2A) (10, 17, 31, 37, 48). Dnmt3a and Dnmt3a2 are encoded by transcripts initiated from two different promoters. Dnmt3a2 lacks the N-terminal region of the full-length Dnmt3a and, as a result, they exhibit different subcellular localization patterns. Whereas Dnmt3a is concentrated in heterochromatic foci, Dnmt3a2 localizes diffusely in the nucleus (10). Unlike the Dnmt3a isoforms, all of the known Dnmt3b isoforms are derived from alternative splicing. Dnmt3b1 and Dnmt3b2 are enzymatically active, as shown by in vitro methyltransferase assays, whereas Dnmt3b3, which lacks part of motif IX, appears to be inactive (1, 31). Dnmt3b4, Dnmt3b5, and Dnmt3b6 are also presumably inactive because they lack either part of motif IX (Dnmt3b6) or both motifs IX and X (Dnmt3b4 and Dnmt3b5) (10, 17, 37). Like Dnmt3a, Dnmt3b1 has been shown to localize to heterochromatin (2). These Dnmt3a and Dnmt3b isoforms show different expression patterns during development. Dnmt3a2 and Dnmt3b1 are strongly expressed in ES cells and germ cells but are almost undetectable in most somatic tissues, whereas Dnmt3a and Dnmt3b3 are expressed at low levels in almost all somatic tissues and cell lines examined (3, 10). The biochemical properties and biological functions of different Dnmt3a/3b isoforms remain largely unknown.
FIG. 2.
Stable expression of Dnmt3a and Dnmt3b isoforms in late-passage 7aabb cells. (A) Schematic diagram of Dnmt3a and Dnmt3b isoforms. The conserved PWWP and PHD domains, the methyltransferase motifs (I, IV, VI, IX, and X), and the sites of alternative splicing are indicated (the C-terminal 45 amino acids of Dnmt3b5 are out of frame and shown as an open bar). The locations of the epitopes for the Dnmt3a and Dnmt3b antibodies are also shown. (B) cDNAs encoding Dnmt3a/3b isoforms were subcloned in an expression vector (schematically shown at the top), and these constructs were individually electroporated into late-passage (P70) 7aabb cells, which were subsequently selected in blasticidin-containing medium for 7 days. Blasticidin-resistant clones were analyzed with immunoblotting with anti-Dnmt3a (middle panel) or anti-Dnmt3b (bottom panel) antibodies. As a loading control, the same membranes were immunoblotted with anti-tubulin antibody.
In the present study, we introduced various Dnmt3a/3b isoforms individually back into Dnmt3a−/− Dnmt3b−/− mutant ES cells and showed that these isoforms have both shared and specific genomic targets. In addition, we demonstrated that Dnmt3a and Dnmt3b are required for stable inheritance of global DNA methylation patterns in ES cells and that maintenance of genomic methylation above a threshold level, but not the presence of Dnmt3a and Dnmt3b proteins, is essential for ES cell differentiation and teratoma formation.
MATERIALS AND METHODS
ES cell culture.
Wild-type J1 and mutant ES cells were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 15% fetal bovine serum (HyClone), 0.1 mM nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol, 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 500 U of leukemia inhibitory factor (Invitrogen)/ml. The cells were normally grown on gelatin-coated petri dishes without feeder cells. For long-term culture, the cells were treated with trypsin and passaged every other day, and the passage numbers were recorded.
DNA constructions.
The plasmid vectors expressing Dnmt1, Dnmt3a, Dnmt3a2, Dnmt3b1, Dnmt3b3, and Dnmt3b1:PC (a mutant Dnmt3b1 with the proline-cysteine dipeptide at the active site substituted with glycine-threonine) were generated by subcloning the corresponding cDNAs into pCAG-IRESblast, an expression vector that contains a CAG promoter (a synthetic promoter that includes the chicken β-actin promoter and the human cytomegalovirus immediate early enhancer). pCAG-IRESblast was constructed by replacing the _Eco_RI-_Xho_I fragment of pCAGN2-R(H1)-S3H-I-ZF3 (gift from R. Jaenisch) with an internal ribosome entry site-blasticidin cassette.
The Dnmt3b1 targeting vector, in which a 2-kb region containing exons 21 and 22 was replaced by the PGK-puromycin cassette, was generated by sequentially subcloning Dnmt3b genomic fragments (the 8-kb 5′ arm and 3.3-kb 3′arm were both obtained from a bacterial artificial chromosome clone), the PGK-puromycin cassette, and the PGK-DTA cassette into pBluescript II SK. The identities of all constructs were verified by DNA sequencing.
Stable expression of DNA methyltransferases in ES cells.
Expression vectors encoding Dnmt3a and Dnmt3b isoforms or Dnmt1 were electroporated into Dnmt3a−/− Dnmt3b−/− or Dnmt1−/− ES cells (24, 30), which were subsequently selected in blasticidin-containing medium for 7 days. Blasticidin-resistant colonies were examined for protein expression by immunoblotting analysis with the following antibodies: monoclonal anti-Dnmt3a (clone 64B1446; Imgenex) (10), polyclonal anti-Dnmt3b (10), or polyclonal anti-Dnmt1 (a gift from S. Tajima). As loading controls, the levels of α-tubulin in these samples were determined by immunoblotting with monoclonal anti-tubulin antibody (Ab-1; Oncogene Research Products). Expression of the intended Dnmt proteins was observed in ∼90% of the colonies, most of which maintained the expression level after 4 weeks of culture in blasticidin-containing medium.
Targeted disruption of Dnmt3b1 in ES cells.
The Dnmt3b1 targeting vector was transfected into Dnmt3b+/− or Dnmt3a−/− Dnmt3b+/− ES cells (30) via electroporation, and transfected cells were selected with puromycin. Genomic DNA isolated from puromycin-resistant colonies was digested with _Eco_RV and analyzed by Southern hybridization with a probe 3′ external to the targeting construct. The targeting frequencies for the wild-type alleles in Dnmt3b+/− and Dnmt3a−/− Dnmt3b+/− cells were 4/150 and 6/200, respectively.
DNA methylation analysis.
Genomic DNA isolated from various ES cell lines was digested with methylation-sensitive restriction enzymes and analyzed by Southern hybridization as previously described (24). Probes used for methylation analysis include the following: pMO for endogenous C-type retroviruses (GenBank accession NC_001501) (27), pMR150 for minor satellite repeats (accession no. X14469 and accession no. X07949) (9), intracisternal A particle (IAP; accession no. AF303453) (47), the 3′ region of β_-globin_ cDNA (accession numbers J00413, K01748, and K03545) (12), the 5′ region of Pgk-1 cDNA (accession no. M18735) (12), the coding region of Pgk-2 cDNA (accession no. NM_031190) (12), the 5′ region of Xist cDNA (accession no. AJ421479, gift from T. Sado), the H19 upstream region (accession no. U19619) (45), DMR2 or “probe 6” for Igf2 (accession no. NM_010514) (15), the Igf2r region 2 probe (accession no. NM_010515) (44), Peg1 (accession no. NM_008590) (23), Snrpn DMR1 (accession no. NM_013670) (42), and an oligonucleotide probe (5′-TAT GGC GAG GAA AAC TGA AAA AGG TGG AAA ATT TAG AAA TGT CCA CTG TAG GAC GTG GAA TAT GGC AAG-3′) specific to major satellite repeats. Bisulfite sequencing analysis was performed as described previously (11). We analyzed six of the eight CpG sites within the major satellite repeating unit. The sequences of the PCR primers were: 5′-AAA TCT AGA AAT GTT TAT TGT AGG A-3′ and 5′-TTC GGA TCC TAA AAT ATA TAT TTC TCA T-3′ (the _Xba_I and _Bam_HI sites used for cloning are underlined).
RESULTS
Inactivation of Dnmt3a and Dnmt3b results in progressive loss of DNA methylation in ES cells.
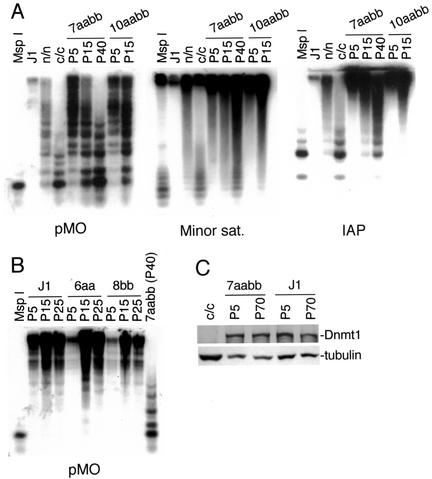
Genetic studies have demonstrated that Dnmt3a and Dnmt3b carry out de novo methylation of the mouse genome during early embryonic development (30). To investigate whether these enzymes are also involved in maintaining global DNA methylation patterns, we cultured Dnmt3a−/− Dnmt3b−/− ES cells (30) continuously for various periods of time and examined the methylation status of various genomic sequences by using methylation-sensitive restriction enzymes. The endogenous C-type retroviruses and IAP repeats, which are interspersed in the mouse genome with about 100 and 1,000 copies per haploid genome, respectively, are normally highly methylated in ES cells (27, 30). These sequences became progressively demethylated in two independent Dnmt3a−/− Dnmt3b−/− cell lines (7aabb and 10aabb), as indicated by increasing sensitivity to _Hpa_II digestion (Fig. 1A). Similar results were obtained when DNA methylation of the major and minor satellite repeats was analyzed (Fig. 1A and data not shown). The major and minor satellite repeats are located in the pericentromeric and centromeric regions at copy numbers of 700,000 and of 50,000 to 100,000, respectively. After prolonged culture of Dnmt3a−/− Dnmt3b−/− ES cells for about 5 months, DNA methylation in both repeats and unique genes examined was almost completely depleted (see below). No significant change in global methylation was observed when wild-type (J1) and Dnmt3a−/− (6aa) or Dnmt3b−/− (8bb) single-mutant ES cells were grown in culture for the same periods of time (Fig. 1B; also, see below). Loss of methylation in Dnmt3a−/− Dnmt3b−/− ES cells was not due to reduced expression of Dnmt1 since immunoblotting analysis indicated that early-passage and late-passage cells had similar levels of Dnmt1 protein (Fig. 1C). These results suggested that the Dnmt3 family of methyltransferases is required for stable inheritance of global DNA methylation patterns in ES cells and that Dnmt3a and Dnmt3b have largely redundant functions in this respect.
FIG. 1.
Inactivation of Dnmt3a and Dnmt3b results in progressive loss of DNA methylation in ES cells. (A) Genomic DNA from Dnmt3a−/− Dnmt3b−/− ES cells (7aabb and 10aabb) that had been grown in culture for 5 to 40 passages, as well as wild-type (J1) and Dnmt1 mutant (n/n and c/c) ES cells, was digested with _Hpa_II and hybridized to probes for endogenous C-type retrovirus repeats (pMO), minor satellite repeats, and IAP repeats. As a control for complete digestion, DNA from J1 cells was digested with _Msp_I. The Dnmt1n allele (the “n” stands for N-terminal disruption) is a partial loss-of-function mutation (27) and the Dnmt1c allele (the “c” stands for disruption of the catalytic or C-terminal domain) is a null mutation (24). (B) Genomic DNA from J1, Dnmt3a−/− (6aa), or Dnmt3b−/− (8bb) ES cells that had been grown in culture for 5 to 25 passages, as well as 7aabb (P40), was digested with _Hpa_II and hybridized to the pMO probe. (C) Lysates from the indicated ES cell lines were immunoblotted with anti-Dnmt1 and anti-tubulin antibodies.
Stable expression of Dnmt3a and Dnmt3b in Dnmt3a−/− Dnmt3b−/− ES cells restores DNA methylation.
Dnmt3a and Dnmt3b isoforms show distinct expression profiles and cellular localization patterns (2, 10), raising the possibility that they may methylate different sets of sequences in the genome. To investigate whether the demethylated state of the Dnmt3a−/− Dnmt3b−/− ES cell genome is reversible and whether different Dnmt3a and Dnmt3b isoforms have distinct specificities in reestablishing methylation patterns, we introduced cDNAs encoding Dnmt3a, Dnmt3a2, Dnmt3b1, Dnmt3b3, and Dnmt3b1:PC (Dnmt3b1 with its PC motif mutated) into late-passage 7aabb ES cells (30) by random integration. Each cDNA was subcloned in a plasmid vector in which a CAG promoter drives the expression of a bicistronic transcript that encodes both the intended Dnmt protein and the selection marker, blasticidin S deaminase (Fig. 2B, top panel). After selection with blasticidin, we were able to obtain individual clones that express various levels of Dnmt3a or Dnmt3b proteins, as determined by immunoblotting analysis (Fig. 2B). The monoclonal Dnmt3a antibody, which recognizes the C-terminal region of Dnmt3a (Fig. 2A), strongly reacts with Dnmt3a and Dnmt3a2 and weakly reacts with Dnmt3b1 and Dnmt3b2 but not the other Dnmt3b isoforms (10). The polyclonal Dnmt3b antibody, which was raised against the N-terminal region of Dnmt3b (Fig. 2A), is Dnmt3b specific and recognizes all known Dnmt3b isoforms (10). For each construct, we chose two independent clones for methylation analysis. The relative levels of Dnmt3a/3b proteins expressed in these clones, compared to the levels of the corresponding endogenous Dnmt3a/3b isoforms in wild-type ES cells (J1, 100%), were roughly estimated based on the intensity of the bands: Dnmt3a (clone 1, 500%; clone 2, 200%), Dnmt3a2 (clone 1, 150%; clone 2, 200%), Dnmt3b1 (clone 1, 150%; clone 2, 80%), Dnmt3b3 (clone 1, 400%; clone 2, 500% [compared to endogenous Dnmt3b6]), and Dnmt3b1:PC (clone 1, 80%; clone 2, 50% [compared to endogenous Dnmt3b1]) (Fig. 2B). We also confirmed by immunoblotting analysis that there was no cross-contamination between the control ES cell lines (J1, 6aa, 8bb, and 7aabb) during the course of long-term passage (Fig. 2B, middle and bottom panels, lanes 1 to 4).
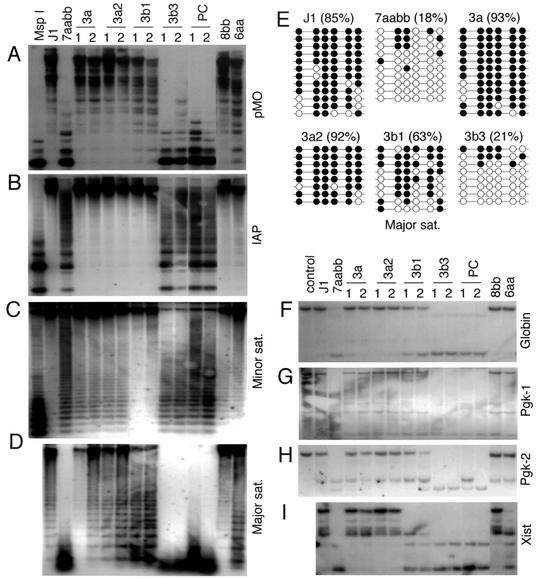
We first examined whether repetitive elements could be remethylated by the expressed Dnmt3a/3b proteins in 7aabb cells. As shown in Fig. 3A to D, expression of Dnmt3a, Dnmt3a2, or Dnmt3b1 substantially restored the methylation levels of the endogenous C-type retroviral DNA, the IAP repeats, and the major and minor satellite repeats, whereas expression of Dnmt3b3 or Dnmt3b1:PC had no effect. Although the two Dnmt3a isoforms showed similar efficiency in methylating these repetitive sequences, Dnmt3a/3a2 and Dnmt3b1 exhibited distinct sequence preferences. Compared to Dnmt3a/3a2, Dnmt3b1 was substantially more efficient in methylating the minor satellite repeats and less efficient in methylating the major satellite repeats and the endogenous C-type retroviral DNA. These enzymes were equally efficient in methylating the IAP repeats and restored the methylation level to normal. To confirm these results, we analyzed genomic DNA from late-passage 6aa and 8bb ES cells and showed that the methylation patterns in these sequences were consistent with those observed in the corresponding Dnmt3a/3b stable clones. The results were further verified with bisulfite sequencing analysis (Fig. 3E). The unit sequence of the major satellite repeats consists of 234 bp and contains eight CpG sites. We examined the methylation status of six of these sites (the other two sites were not in the amplified region). In wild-type J1 cells, 85% of the analyzed CpG sites were methylated. Only 18% of these sites remained methylated in 7aabb cells. Expression of Dnmt3a, Dnmt3a2, and Dnmt3b1 in 7aabb cells restored the methylation levels to 93, 92, and 63%, respectively, whereas expression of Dnmt3b3 had no effect (21%).
FIG. 3.
Expression of Dnmt3a/3b proteins in 7aabb cells restores DNA methylation. (A to D) Methylation of repetitive sequences. Genomic DNA from the indicated ES cell lines was digested with _Hpa_II (A to C) or _Mae_II (D) and hybridized to the indicated probes. DNA from J1 cells digested with _Msp_I was used as a control for complete digestion. (E) Analysis of the methylation status of the major satellite repeating unit by bisulfite sequencing. Genomic DNA from J1 and 7aabb cells, as well as from stable cell lines expressing Dnmt3a, Dnmt3a2, Dnmt3b1, and Dnmt3b3, was analyzed. The methylation status of six CpG sites from 8 to 12 individual clones is shown schematically (black circles represent methylated sites), and the percentages of methylated CpG sites are indicated in parenthesis. (F to I) Methylation of unique genes. The genomic DNA samples described in panels A to D were digested with _Bam_HI and _Hha_I (F and H), _Eco_RI and _Hpa_II (G), or _Eco_RV and Hha_I (I) and hybridized to probes corresponding to the 3′ region of β_-globin (F), the 5′ region of Pgk-1 (G), an exon of Pgk-2 (H), or the 5′ region of Xist (I). DNA from J1 cells digested with _Bam_HI alone (F and H) or _Eco_RI alone (G) was used as controls.
To determine whether expression of Dnmt3a/3b proteins in 7aabb cells also affects methylation of unique genes, a number of specific genomic loci were examined. β-globin and the phosphoglycerate kinase 2 (Pgk-2) gene are highly methylated autosomal genes that show tissue-specific expression patterns. Pgk-1 and Xist, two other highly methylated genes, are located on the X chromosome. The methylation-sensitive sites examined were located in the 5′ region (Pgk-1 and Xist), the coding region (Pgk-2), or the 3′ region (β_-globin_) of the genes. All four loci were highly methylated in the wild-type ES cells (J1) and became substantially demethylated in late-passage 7aabb cells (Fig. 3F to I). With expression of Dnmt3a, Dnmt3a2, or Dnmt3b1, but not of Dnmt3b3 or Dnmt3b1:PC, in 7aabb cells, the examined regions in β_-globin_, Pgk-1, and Pgk-2 genes were completely or partially remethylated. These results were in agreement with the fact that methylation of these loci was maintained in 8bb and 6aa cells (Fig. 3F to H). Interestingly, Dnmt3a or Dnmt3a2 was able to restore methylation of the Xist promoter region to normal, but Dnmt3b1 was not (Fig. 3I). Consistently, inactivation of Dnmt3a alone in ES cells (6aa) resulted in demethylation of the Xist promoter region, whereas inactivation of Dnmt3b alone (8bb) had no effect (Fig. 3I), suggesting that Dnmt3a, but not Dnmt3b, can establish and is required for maintaining methylation of this particular region. Taken together, these data demonstrate that methylation of the highly demethylated genome of Dnmt3a−/− Dnmt3b−/− ES cells can be largely reestablished by Dnmt3a and Dnmt3b and that these enzymes have both shared and specific DNA targets.
Methylation of imprinted genes.
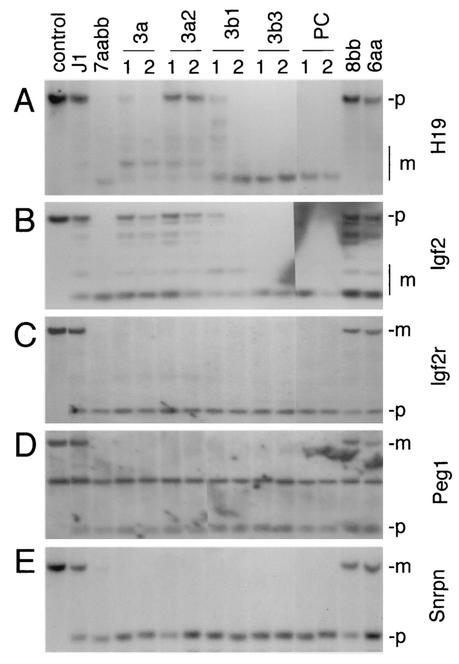
We have previously shown that methylation of some imprinted genes, such as H19 and the Igf2 receptor gene (Igf2r), is maintained in early-passage Dnmt3a−/− Dnmt3b−/− ES cells (30). To determine whether methylation imprints can be stably maintained, the methylation status of a number of imprinted genes was examined at their differentially methylated regions (DMRs) by using genomic DNA from late-passage 7aabb cells. As shown in Fig. 4, all examined loci, including the 5′ upstream region of H19, region 2 of Igf2r, the DMR of Peg1, and DMR1 of Snrpn, became completely demethylated in late-passage 7aabb cells but not in wild-type (J1), 6aa, or 8bb cells. These observations suggested that Dnmt3a and Dnmt3b not only are involved in de novo methylation of imprinted genes in male and female germ cells but may also play a role in maintaining the methylation imprints in the zygote.
FIG. 4.
Expression of Dnmt3a and Dnmt3b proteins in 7aabb cells fails to restore maternal methylation imprints. The same DNA samples described in Fig. 3 were digested with _Sac_I and _Hha_I (A), _Bam_HI and _Hpa_II (B), _Pvu_II and _Hpa_II (C and D), or _Xba_I and _Hha_I (E) and hybridized to probes corresponding to the 5′ upstream region of H19 (A), the DMR2 of Igf2 (B), region 2 of Igf2r (C), the DMR of Peg1 (D), or the DMR1 of Snrpn (E). As controls, DNA from J1 cells was digested with the corresponding enzymes without _Hha_I or _Hpa_II. The fragments derived from the paternal (p) and maternal (m) alleles are indicated.
We then examined whether expression of Dnmt3a/3b proteins in 7aabb cells could restore methylation imprints. The 5′ upstream region of H19, which includes the DMR that regulates expression of Igf2 and H19, is methylated when it is inherited from the father but unmethylated when it is inherited from the mother. Digestion with the methylation-sensitive enzyme _Hha_I resulted in a fully methylated paternal band and several weaker undermethylated smaller bands from the maternal allele in wild-type (J1) ES cells. Demethylation of this region in 7aabb cells resulted in several lower-molecular-weight bands. We found that Dnmt3a2 almost fully remethylated this region, whereas Dnmt3a and Dnmt3b1 caused only minimal remethylation, and Dnmt3b3 and Dnmt3b1:PC showed no activity at all (Fig. 4A). Using similar strategies, we examined several other imprinted genes. DMR2 of Igf2, another paternally methylated region, was fully or partially remethylated by Dnmt3a, Dnmt3a2, or Dnmt3b1 but not remethylated by Dnmt3b3 or Dnmt3b1:PC (Fig. 4B). The intensity of the methylated and unmethylated bands suggested that one allele (presumably the paternal allele) was remethylated and the other allele remained unmethylated, although we could not rule out the possibility that the methylated band resulted from partial methylation of both alleles. In contrast to H19 and Igf2, none of the maternally methylated genes (Igf2r, Peg1, and Snrpn) could be remethylated at their DMRs by overexpression of Dnmt3a/3b proteins (Fig. 4C to E). These observations indicate that the maternal methylation imprints, once lost, cannot be restored in ES cells by de novo methylation.
Dnmt3b6 has no enzymatic activity in vivo.
Consistent with previous results from in vitro DNA methyltransferase assays (1, 31), our rescue experiments showed that Dnmt3b3 had no enzymatic activity. We predict that Dnmt3b4, Dnmt3b5, and Dnmt3b6 are also enzymatically inactive because, like Dnmt3b3, they all lack part of the conserved motif IX due to alternative splicing of exons 21 and 22 (Fig. 2A). To determine whether these isoforms have any activity in vivo, we deleted exons 21 and 22 from the wild-type allele in Dnmt3b+/− and Dnmt3a−/− Dnmt3b+/− ES cells (30) by gene targeting. A PGK-puromycin cassette was inserted in the opposite orientation of Dnmt3b transcription to avoid truncation of the Dnmt3b transcripts (Fig. 5A). Since the major Dnmt3b isoforms expressed in ES cells are Dnmt3b1 and Dnmt3b6 (10), we expected that removal of exons 21 and 22 would eliminate Dnmt3b1 but not Dnmt3b6. A number of clones with a deletion of the wild-type allele were obtained from both Dnmt3b+/− and Dnmt3a−/− Dnmt3b+/− cells, and these clones were referred to as Dnmt3b1KO/− and Dnmt3a−/− Dnmt3b1KO/−, respectively (Fig. 5B). Immunoblotting analysis confirmed that Dnmt3b1 protein was abolished and, concomitantly, the level of Dnmt3b6 protein increased in these cells (Fig. 5C). We examined the methylation status of various repetitive sequences and unique genes in these cells. Unlike the parental Dnmt3b+/− cell line, Dnmt3b1KO/− cells showed significant demethylation of the minor satellite repeats and the methylation pattern was identical to that in Dnmt3b−/− cells (Fig. 5E). Similarly, all sequences examined showed substantial loss of methylation in Dnmt3a−/− Dnmt3b1KO/− cells and exhibited methylation patterns indistinguishable from those observed in Dnmt3a−/− Dnmt3b−/− cells (Fig. 5D and E and data not shown). In addition, Dnmt3a−/− Dnmt3b1KO/− cells failed to methylate newly integrated proviral DNA after infection with a recombinant retrovirus, MoMuLVsup-1, whereas the parental Dnmt3a−/− Dnmt3b+/− cell line showed efficient de novo methylation activity (data not shown). These data provide genetic evidence that exons 21 and 22 are essential for Dnmt3b activity. We conclude that all Dnmt3b isoforms that lack motif IX have no methyltransferase activity in vivo.
FIG. 5.
Dnmt3b6 has no enzymatic activity in vivo. (A) Strategy of targeted deletion of Dnmt3b exons 21 and 22. The top line shows the Dnmt3b genomic structure with exons represented by vertical bars. The targeting vector (second line) was constructed by replacing exons 21 and 22 with a PGK-puromycin cassette. A PGK-DTA cassette was introduced for negative selection to increase the targeting frequency. (B) Southern analysis of the genotype of ES cell lines. Genomic DNA was digested with _Eco_RV and hybridized to a 3′ external probe, as shown in (A). The 16-kb wild-type allele, the 5-kb Dnmt3b1 targeted allele, and the 14-kb Dnmt3b null allele (30) are indicated. (C) Lysates from the indicated cell lines were immunoblotted with anti-Dnmt3b (top), anti-Dnmt3a (middle), and anti-tubulin (bottom) antibodies. (D and E) Genomic DNA from the indicated ES cell lines was digested with _Hpa_II and hybridized to probes for endogenous C-type retrovirus repeats (D) and minor satellite repeats (E).
Dnmt3a/3b-induced remethylation rescues the capacity of Dnmt3a−/− Dnmt3b−/− ES cells to form teratomas in nude mice.
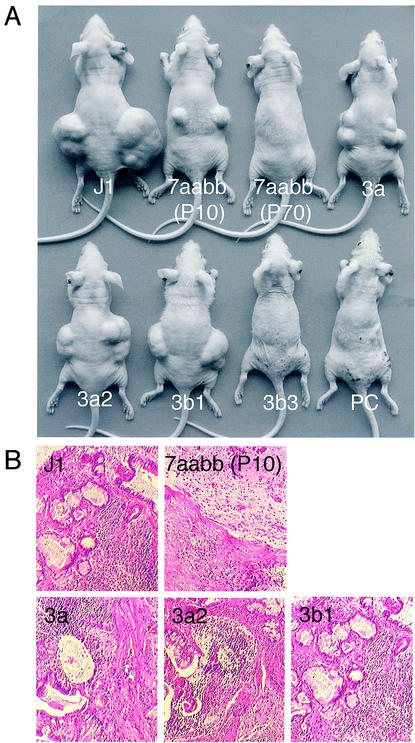
It has been reported that _Dnmt1_-null ES cells die upon induction of differentiation and cannot form teratomas (24, 46). It is not known, however, whether the differentiation defects are caused by loss of methylation or lack of Dnmt1 protein. Unlike _Dnmt1_-null cells, which lose methylation very quickly, Dnmt3a−/− Dnmt3b−/− ES cells show gradual demethylation during the course of continuous passage, which makes it possible to address the relationship between genomic methylation and cellular differentiation. We injected early-passage (passage 10 [P10]) and late-passage (P70) 7aabb cells into nude mice and tested their ability to induce teratomas. While late-passage cells failed to form palpable teratomas (0 of 3) within 4 weeks, early-passage cells retained the ability to induce teratomas (2 of 3) despite their much smaller size compared to those induced by wild-type J1 cells (3 of 3) (Fig. 6A and B). These results indicated that the ability of ES cells to form teratomas is dependent on the level of genomic methylation, but not the presence of Dnmt3a and Dnmt3b proteins.
FIG. 6.
Active Dnmt3a/3b isoforms rescue the capacity of late-passage 7aabb cells to form terotomas in nude mice. (A) The indicated ES cell lines were injected into nude mice subcutaneously on both sides (three to four mice for each cell line, 5 × 105 cells per site), and the mice were examined for teratomas after 4 weeks. A typical representation of the size of the teratomas derived from each cell line is shown. (B) Histological sections of teratomas derived from J1, early-passage (P10) 7aabb, and Dnmt3a, Dnmt3a2, and Dnmt3b1 stable clones showing the presence of multiple types of differentiated cells.
We then asked whether expression of Dnmt3a/3b proteins in late-passage 7aabb cells could rescue the capacity of these cells to induce teratomas. Consistent with their methylation level, stable lines expressing Dnmt3a (3 of 4), Dnmt3a2 (4 of 4), or Dnmt3b1 (4 of 4) were able to induce teratomas in nude mice, whereas those expressing Dnmt3b3 (0 of 4) or Dnmt3b1:PC (0 of 4) were not (Fig. 6A). Although the teratomas induced by these stable lines did not reach the size of those induced by J1 cells (presumably because expression of any one isoform could not fully restore the methylation level), histological analysis revealed that all of these teratomas contained multiple differentiated cell types (epithelial tissue, cartilage, muscle, etc.) with no obvious differences (Fig. 6B).
Overexpression of Dnmt1 fails to restore global DNA methylation in the absence of Dnmt3a and Dnmt3b.
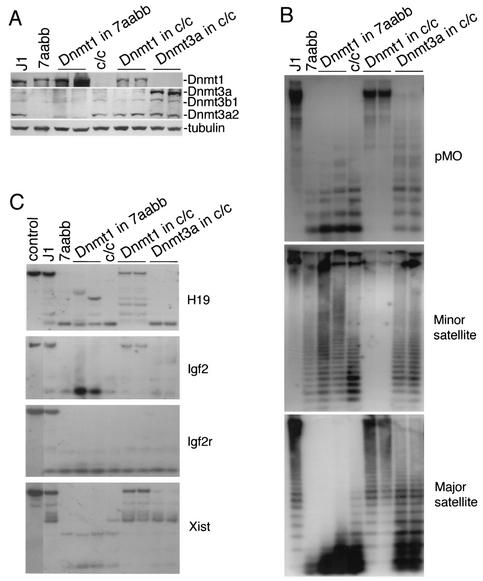
It has been recently reported that overexpression of Dnmt1 in ES cells results in genomic hypermethylation (6). To determine whether Dnmt1 could induce de novo methylation in the absence of Dnmt3a and Dnmt3b, we overexpressed Dnmt1 in late-passage 7aabb cells and, as a control, in _Dnmt1_-null (c/c) ES cells (Fig. 7A). As shown in Fig. 7B and C, introduction of Dnmt1 back into _Dnmt1_-null cells significantly restored methylation of all repetitive sequences and single-copy genes examined except for the maternally imprinted gene Igf2r, a finding consistent with previous observations (6, 46). However, overexpression of Dnmt1 in 7aabb cells had little effect on global methylation compared to the parental cell line, although methylation of a few CpG sites may have occurred in the 5′ region of H19. In contrast, overexpression of Dnmt3a in _Dnmt1_-null cells resulted in slight (but significant) increases in methylation of all sequences examined except for imprinted genes, suggesting that Dnmt3a is able to induce de novo methylation in the absence of Dnmt1, but it cannot maintain methylation at high levels. Taken together, these data provide strong evidence that Dnmt1 alone is not capable of methylating genomic DNA de novo, and both Dnmt1 and Dnmt3 families of methyltransferases are required for the establishment and stable maintenance of hypermethylation of the genome.
FIG. 7.
Dnmt1 and Dnmt3 proteins function cooperatively in maintaining methylation patterns. (A) Dnmt1 or Dnmt3a was overexpressed in 7aabb (P70) or Dnmt1−/− (c/c) ES cells as indicated and stable clones were examined for protein expression by immunoblotting with anti-Dnmt1 (top), anti-Dnmt3a (middle), and anti-tubulin (bottom) antibodies. (B and C) Genomic DNA from the indicated ES cell lines was analyzed for methylation of repetitive sequences (B) and unique genes (C) with the indicated probes.
DISCUSSION
Maintenance methylation is a key process that ensures stable inheritance of tissue-specific DNA methylation patterns from cell to cell during mitosis. It was previously thought that Dnmt1 is solely responsible for the maintenance of DNA methylation patterns since Dnmt1 has strong preference for hemimethylated CpG sites and is localized to DNA replication foci, and inactivation of Dnmt1 by gene targeting in mice results in genome-wide loss of methylation (24, 27). However, there is no evidence that Dnmt1 alone is sufficient to maintain all methylation in the genome. In contrast, our initial studies of ES cells lacking the Dnmt3 family methyltransferases suggest that maintenance of methylation of some sequences such as the DMR2 region of Igf2 and the 5′ region of Xist requires Dnmt3a and Dnmt3b (30). In the present study, we extended our findings and showed that Dnmt3a and Dnmt3b are involved in maintaining global DNA methylation patterns. We demonstrated that inactivation of Dnmt3a and Dnmt3b in ES cells resulted in progressive demethylation of all sequences examined, including repetitive elements, imprinted genes, and nonimprinted genes. These results indicate that Dnmt1 alone is not sufficient for stable inheritance of DNA methylation patterns in ES cells.
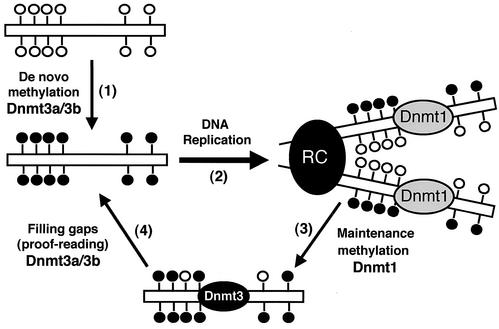
We propose that Dnmt1 is the major maintenance methyltransferase which, in association with the DNA replication machinery, methylates hemimethylated CpG sites with high efficiency but not absolute accuracy, while Dnmt3a and Dnmt3b, via their de novo methylation activity, function as “proofreaders” to fill the gaps of the hemimethylated CpG sites missed by Dnmt1 (Fig. 8). Consistent with this model is the observation that Dnmt1−/− and Dnmt3a−/− Dnmt3b−/− ES cells exhibit very different kinetics of demethylation. Complete inactivation of Dnmt1 resulted in an almost 90% reduction of total methyl CpG in the genome immediately after Dnmt1−/− cell lines were established (at 106 cells or the first passage) (24). In contrast, inactivation of Dnmt3a and Dnmt3b resulted in gradual loss of methylation in most genomic sequences, and it took more than 70 passages to reach a 90% reduction of global methylation.
FIG. 8.
Model for the distinct roles of Dnmt1 and Dnmt3a/3b in de novo and maintenance methylation. (Step 1) Dnmt3a and Dnmt3b establish new DNA methylation patterns by de novo methylation of symmetric CpG dinucleotides (lollipops). (Step 2) Upon DNA replication, the new synthesized DNA becomes hemimethylated at CpG sites. (Step 3) Dnmt1, which is localized to the replication complex (RC), restores full methylation by methylating hemimethylated DNA. However, some CpG sites are left untouched by Dnmt1 and remain hemimethylated. (Step 4) Dnmt3a and Dnmt3b, which may also localize to the RC, recognize unmethylated CpG sites and restore methylation via de novo methylation. In summary, Dnmt1 is the major maintenance methyltransferase, and it has little or no de novo methylation activity in vivo. Dnmt3a and Dnmt3b possess active de novo methyltransferase activity in vivo and are essential for the establishment and maintenance of DNA methylation patterns.
Recent studies have created controversies with regard to the roles of DNMT1 and DNMT3B genes in maintaining methylation patterns in human cancer cell lines. Rhee et al. showed that targeted disruption of DNMT1 or DNMT3B in HCT116 colorectal carcinoma cells caused only minor reduction in CpG methylation, whereas disruption of both DNMT1 and DNMT3B resulted in severe loss of methylation (34, 35), suggesting that the two enzymes have redundant functions in maintaining CpG methylation. In contrast, Robert et al. showed that selective depletion of DNMT1 alone by using antisense or siRNA in HCT116 and other human cancer cell lines resulted in global and gene-specific demethylation (36), a finding consistent with the current view that DNMT1 is the major maintenance enzyme. In the present study, we demonstrated that both Dnmt1 and Dnmt3 families of methyltransferases are required for stable maintenance of global methylation patterns in mouse ES cells. Our observation that neither overexpression of Dnmt1 in Dnmt3a−/− Dnmt3b−/− cells nor overexpression of Dnmt3a in Dnmt1−/− cells could restore methylation to normal levels suggests that these two types of enzymes have distinct and nonredundant functions and that they act cooperatively to maintain hypermethylation of the genome. It also confirms that Dnmt1 has little or no de novo methylation activity in vivo.
How DNA methylation patterns are generated during development remains a mystery. Since the Dnmt1 and Dnmt3 families of methyltransferases do not appear to have any sequence specificity beyond CpG dinucleotides (14, 31, 49), several chromatin-based mechanisms have been proposed to explain how DNA methyltransferases may find their targets in the genome (7). One explanation is that chromosomal regions are not equally accessible to DNA methyltransferases. Consistent with this notion, recent studies of two SNF2 family helicases, ATRX and Lsh, have shown that proteins with chromatin remodeling and DNA helicase activities can modulate DNA methylation in mammalian cells (12, 16). Similarly, the SNF2-like protein DDM1 has been shown to be essential for methylation of both CpG and CpNpG sites in the plant Arabidopsis thaliana (21). Another explanation is that accessory factors (proteins, RNA, etc.) recruit DNA methyltransferases to specific genomic sequences or chromatin structures. A number of proteins, including PCNA, DMAP1, HDAC1, HDAC2, and pRB, have been shown to interact with Dnmt1 and may recruit Dnmt1 to highly methylated heterochromatin during the late S phase (38). The PML-RAR fusion protein and Dnmt3L have been shown to interact with Dnmt3a and may recruit this enzyme to RAR response elements and imprinted genes, respectively (13, 18). In the present study, we provide the first evidence that DNA methylation patterns could also be regulated by expressing different isoforms of Dnmt3a and Dnmt3b. We showed that various Dnmt3a and Dnmt3b isoforms appear to have both common and preferred DNA targets during the process of reestablishing DNA methylation patterns in highly demethylated Dnmt3a−/− Dnmt3b−/− mutant ES cells. Dnmt3a, Dnmt3a2, and Dnmt3b1 exhibited substantial activity toward all of the repetitive sequences examined, but they clearly had sequence preferences, with Dnmt3a/3a2 and Dnmt3b1 preferentially methylating the major and minor satellite repeats, respectively. These enzymes also showed notable differences in methylating certain unique genes. Dnmt3a and Dnmt3a2 were able to methylate the 5′ region of Xist, but Dnmt3b1 was not. Similarly, Dnmt3a2 almost fully restored the methylation status of the 5′ region of H19, whereas Dnmt3a and Dnmt3b1 showed little effect. Given that Dnmt3a and Dnmt3b isoforms show distinct cellular localization patterns (2, 10), their preferences for different genomic sequences may reflect their differences in chromatin accessibility. It is also conceivable that other factors may interact with various Dnmt3a and Dnmt3b isoforms and target them to different genomic regions. It should be noted that the target specificity of different isoforms was determined by overexpression of each isoform in ES cells, although the results are largely consistent with those obtained from Dnmt3a−/− or Dnmt3b−/− single-mutant cells. Genetic studies by inactivating specific isoforms in mice will be necessary to confirm their specificity in development.
During development, both the overall level of Dnmt3a/3b proteins and the ratio between different isoforms show dynamic changes. In early embryos, Dnmt3a and Dnmt3b are highly expressed, and the major isoforms are Dnmt3a2 and Dnmt3b1, respectively. In most somatic tissues, Dnmt3a and Dnmt3b are expressed at low levels, and the only detectable isoforms are usually Dnmt3a and Dnmt3b3 (10). We speculate that Dnmt3a2 and Dnmt3b1 carry out de novo methylation in early postimplantation embryos to establish the initial methylation pattern, and Dnmt3a, in cooperation with Dnmt1, is involved in maintaining tissue-specific methylation patterns. Although Dnmt3b3 has no enzymatic activity, it may function as a regulator of DNA methylation in normal and tumor cells. A recent study has shown that overexpression of Dnmt3b4, another “inactive” isoform, may lead to hypomethylation of pericentromeric satellite regions in human hepatocellular carcinoma (39).
Genetic studies have shown that Dnmt3a and Dnmt3b are involved in the establishment of methylation imprints during gametogenesis (18). Our finding that late-passage 7aabb cells show complete loss of methylation of DMRs of imprinted genes suggests that these enzymes may also play a role in the maintenance of imprinted methylation patterns during embryogenesis. Compared to repetitive sequences, imprinted genes were more resistant to demethylation caused by inactivation of Dnmt3a and Dnmt3b (data not shown). It is possible that maintenance methylation by Dnmt1 is more accurate for single-copy genes than for repetitive elements. While the paternally imprinted H19 and Igf2 genes are susceptible to remethylation by ectopically expressed Dnmt3 proteins in mutant ES cells, maternally imprinted genes are completely resistant to remethylation. We speculate that some essential factors required for the establishment of maternal imprints are present in female germ cells but not in ES cells.
An interesting observation is that early-passage Dnmt3a−/− Dnmt3b−/− ES cells, which still contain significant levels of DNA methylation, are capable of forming teratomas in nude mice, whereas late-passage cells, which are more extensively demethylated, completely lose this capacity. This clearly indicates that the presence of Dnmt3a and Dnmt3b methyltransferases (and, thus, de novo methylation activity) is not required for ES cell differentiation and subsequent cellular proliferation. Rather, these processes are dependent on the level of DNA methylation. In keeping with this notion, expression of enzymatically active Dnmt3 proteins (Dnmt3a, Dnmt3a2, and Dnmt3b1), but not the inactive forms (Dnmt3b3 and Dnmt3b1:PC), rescued the capacity of late-passage mutant cells to form teratomas. Our results are consistent with previous studies showing that Dnmt1 mutant ES cells undergo apoptosis upon differentiation (24, 46). Failure to differentiate and proliferate may account, at least in part, for the early embryonic lethality observed in _Dnmt1_- and _Dnmt3a/3b_-null mutant embryos. A threshold level of DNA methylation may be required for some essential developmental processes. Further studies are necessary to determine how DNA methylation regulates cell proliferation and differentiation.
Acknowledgments
This work was supported by grants CA82389 and GM52106 from the National Institutes of Health (to E.L.). T.C. is a recipient of a long-term fellowship from the Human Frontier Science Program. Y.U. is supported by the Japan Society for the Promotion of Science. J.D. is supported by an institutional NRSA postdoctoral fellowship award to the Cardiovascular Research Center at Massachusetts General Hospital.
We thank H. Lei for excellent technical assistance, R. Jaenisch for the vector pCAGN2-R(H1)-S3H-I-ZF3, S. Tajima for anti-Dnmt1 antibody, and K. Muegge for information on primers for the β_-globin_, Pgk-1, and Pgk-2 probes.
REFERENCES
- 1.Aoki, A., I. Suetake, J. Miyagawa, T. Fujio, T. Chijiwa, H. Sasaki, and S. Tajima. 2001. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 29**:**3506-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276**:**32282-32287. [DOI] [PubMed] [Google Scholar]
- 3.Beaulieu, N., S. Morin, I. C. Chute, M.-F. Robert, H. Nguyen, and A. R. MacLeod. 2002. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J. Biol. Chem. 277**:**28176-28181. [DOI] [PubMed] [Google Scholar]
- 4.Bestor, T., A. Laudano, R. Mattaliano, and V. Ingram. 1988. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203**:**971-983. [DOI] [PubMed] [Google Scholar]
- 5.Bestor, T. H. 1992. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 11**:**2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biniszkiewicz, D., J. Gribnau, B. Ramsahoye, F. Gaudet, K. Eggan, D. Humpherys, M.-A. Mastrangelo, Z. Jun, J. Walter, and R. Jaenisch. 2002. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 22**:**2124-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16**:**6-21. [DOI] [PubMed] [Google Scholar]
- 8.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression: belts, braces, and chromatin. Cell 99**:**451-454. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, V., L. Forrester, J. Sanford, N. Hastie, and J. Rossant. 1984. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature 307**:**284-286. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T., Y. Ueda, S. Xie, and E. Li. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277**:**38746-38754. [DOI] [PubMed] [Google Scholar]
- 11.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22**:**2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, K., T. Fan, T. Geiman, Q. Yan, and K. Muegge. 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15**:**2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Croce, L., V. A. Raker, M. Corsaro, F. Fazi, M. Fanelli, M. Faretta, F. Fuks, F. Lo Coco, T. Kouzarides, C. Nervi, S. Minucci, and P. G. Pelicci. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295**:**1079-1082. [DOI] [PubMed] [Google Scholar]
- 14.Dodge, J., B. H. Ramsahoye, Z. G. Wo, M. Okano, and E. Li. 2002. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene 289**:**41-48. [DOI] [PubMed] [Google Scholar]
- 15.Feil, R., J. Walter, N. D. Allen, and W. Reik. 1994. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 120**:**2933-2943. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons, R. J., T. L. McDowell, S. Raman, D. M. O'Rourke, D. Garrick, H. Ayyub, and D. R. Higgs. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24**:**368-371. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, R. S., C. Wijmenga, P. Luo, A. M. Stanek, T. K. Canfield, C. M. Weemaes, and S. M. Gartler. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96**:**14412-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129**:**1983-1993. [DOI] [PubMed] [Google Scholar]
- 19.Howlett, S. K., and W. Reik. 1991. Methylation levels of maternal and paternal genomes during preimplantation development. Development 113**:**119-127. [DOI] [PubMed] [Google Scholar]
- 20.Jaenisch, R. 1997. DNA methylation and imprinting: why bother? Trends Genet. 13**:**323-329. [DOI] [PubMed] [Google Scholar]
- 21.Jeddeloh, J. A., T. L. Stokes, and E. J. Richards. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22**:**94-97. [DOI] [PubMed] [Google Scholar]
- 22.Kafri, T., M. Ariel, M. Brandeis, R. Shemer, L. Urven, J. McCarrey, H. Cedar, and A. Razin. 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 6**:**705-714. [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre, L., S. Viville, S. C. Barton, F. Ishino, and M. A. Surani. 1997. Genomic structure and parent-of-origin-specific methylation of Peg1. Hum. Mol. Genet. 6**:**1907-1915. [DOI] [PubMed] [Google Scholar]
- 24.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed]
- 25.Leonhardt, H., A. W. Page, H.-U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71**:**865-873. [DOI] [PubMed] [Google Scholar]
- 26.Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3**:**662-673. [DOI] [PubMed] [Google Scholar]
- 27.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69**:**915-926. [DOI] [PubMed] [Google Scholar]
- 28.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22**:**480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monk, M., M. Boubelik, and S. Lehnert. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99**:**371-382. [DOI] [PubMed] [Google Scholar]
- 30.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99**:**247-257. [DOI] [PubMed] [Google Scholar]
- 31.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19**:**219-220. [DOI] [PubMed] [Google Scholar]
- 32.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science 293**:**1089-1093. [DOI] [PubMed] [Google Scholar]
- 33.Reik, W., and J. Walter. 2001. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2**:**21-32. [DOI] [PubMed] [Google Scholar]
- 34.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416**:**552-556. [DOI] [PubMed] [Google Scholar]
- 35.Rhee, I., K. W. Jair, R. W. Yen, C. Lengauer, J. G. Herman, K. W. Kinzler, B. Vogelstein, S. B. Baylin, and K. E. Schuebel. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404**:**1003-1007. [DOI] [PubMed] [Google Scholar]
- 36.Robert, M. F., S. Morin, N. Beaulieu, F. Gauthier, I. C. Chute, A. Barsalou, and A. R. MacLeod. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33**:**61-65. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27**:**2291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson, K. D., and A. P. Wolffe. 2000. DNA methylation in health and disease. Nat. Rev. Genet. 1**:**11-19. [DOI] [PubMed] [Google Scholar]
- 39.Saito, Y., Y. Kanai, M. Sakamoto, H. Saito, H. Ishii, and S. Hirohashi. 2002. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 99**:**10060-10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanford, J. P., H. J. Clark, V. M. Chapman, and J. Rossant. 1987. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1**:**1039-1046. [DOI] [PubMed] [Google Scholar]
- 41.Santos, F., B. Hendrich, W. Reik, and W. Dean. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241**:**172-182. [DOI] [PubMed] [Google Scholar]
- 42.Shemer, R., Y. Birger, A. D. Riggs, and A. Razin. 1997. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc. Natl. Acad. Sci. USA 94**:**10267-10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, C. L., H. Stuhlmann, D. Jahner, and R. Jaenisch. 1982. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 79:4098-4102. [DOI] [PMC free article] [PubMed]
- 44.Stoger, R., P. Kubicka, C. G. Liu, T. Kafri, A. Razin, H. Cedar, and D. P. Barlow. 1993. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73**:**61-71. [DOI] [PubMed] [Google Scholar]
- 45.Tremblay, K. D., J. R. Saam, R. S. Ingram, S. M. Tilghman, and M. S. Bartolomei. 1995. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9**:**407-413. [DOI] [PubMed] [Google Scholar]
- 46.Tucker, K. L., D. Talbot, M. A. Lee, H. Leonhardt, and R. Jaenisch. 1996. Complementation of methylation deficiency in embryonic stem cells by DNA methyltransferase minigene. Proc. Natl. Acad. Sci. USA 93**:**12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20**:**116-117. [DOI] [PubMed] [Google Scholar]
- 48.Xie, S., Z. Wang, M. Okano, M. Nogami, Y. Li, W. W. He, K. Okumura, and E. Li. 1999. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 236**:**87-95. [DOI] [PubMed] [Google Scholar]
- 49.Yoder, J. A., N. S. Soman, G. L. Verdine, and T. H. Bestor. 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues: studies with a mechanism-based probe. J. Mol. Biol. 270**:**385-395. [DOI] [PubMed] [Google Scholar]