Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants (original) (raw)
Abstract
The success of targeting kinases in cancer with small molecule inhibitors has been tempered by the emergence of drug-resistant kinase domain mutations. In patients with chronic myeloid leukemia treated with ABL inhibitors, BCR-ABL kinase domain mutations are the principal mechanism of relapse. Certain mutations are occasionally detected before treatment, suggesting increased fitness relative to wild-type p210 BCR-ABL. We evaluated the oncogenicity of eight kinase inhibitor-resistant BCR-ABL mutants and found a spectrum of potencies greater or less than p210. Although most fitness alterations correlate with changes in kinase activity, this is not the case with the T315I BCR-ABL mutation that confers clinical resistance to all currently approved ABL kinase inhibitors. Through global phosphoproteome analysis, we identified a unique phosphosubstrate signature associated with each drug-resistant allele, including a shift in phosphorylation of two tyrosines (Tyr253 and Tyr257) in the ATP binding loop (P-loop) of BCR-ABL when Thr315 is Ile or Ala. Mutational analysis of these tyrosines in the context of Thr315 mutations demonstrates that the identity of the gatekeeper residue impacts oncogenicity by altered P-loop phosphorylation. Therefore, mutations that confer clinical resistance to kinase inhibitors can substantially alter kinase function and confer novel biological properties that may impact disease progression.
Keywords: chronic myelogenous leukemia, kinase inhibitor resistance, phosphoproteomics, imatinib, dasatinib
Point mutations in the kinase domain of BCR-ABL are primarily responsible for resistance to the ABL inhibitor imatinib (Gleevec) in chronic myelogenous leukemia (CML) patients. The majority of imatinib-resistant BCR-ABL point mutations (of >50 distinct examples now reported clinically) impair drug binding by restricting flexibility of the enzyme, precluding adoption of the inactive conformation required for imatinib binding (1–4). The second generation Abl inhibitor dasatinib is effective against imatinib-resistant CML because it binds the BCR-ABL kinase domain regardless of activation loop conformation (5–7). As a result, the number of BCR-ABL mutations capable of conferring resistance to dasatinib is small and is limited almost exclusively to direct contact sites (8, 9). One mutation, T315I, confers resistance to imatinib, dasatinib, and the imatinib-related compound nilotinib (AMN-107) (10, 11).
Although discovered in the context of drug resistance, there is growing evidence that these mutations may confer other fitness advantages to BCR-ABL. First, the T315I and E255K mutants were each detected by our group in imatinib-naïve CML blast crisis patients by direct sequencing of total BCR-ABL cDNA and are therefore estimated to account for at least 20% of the CML tumor burden in these patients (1). In addition, these and other kinase domain mutations have been identified before treatment in CML patients using mutation-specific quantitative PCR (12–18). Furthermore, the analogous mutation to T315I in the epidermal growth factor receptor (EGFR), T790M, causes resistance to EGFR inhibitors in lung cancer and has been detected in lung cancer patients before treatment and in the germ line of a family pedigree with several cases of lung cancer (19–21). The fact that clones bearing these mutants can expand relative to the wild-type allele in the absence of drug treatment suggest that they confer an oncogenic fitness advantage over the wild-type allele.
To test this hypothesis, we measured the oncogenic potency of a panel of clinically relevant, drug resistant BCR-ABL mutations. This analysis revealed a spectrum that included potencies greater or less than p210 and matched fitness predictions from the epidemiologic observations cited above. In most cases oncogenicity correlated with kinase activity. An important exception is BCR-ABL T315I, which displayed increased oncogenic potency but reduced kinase activity toward classical BCR-ABL substrates, raising the possibility of altered substrate recognition. Using a global mass spectrometry-based phosphotyrosine profiling technique, we found that each drug-resistant mutant has a unique phosphoprotein signature, indicating that single amino acid substitutions in the kinase domain exert effects on substrate recognition. In the case of T315I, a mutation-specific phosphorylation pattern in the phosphate binding loop of BCR-ABL explains the difference in oncogenicity relative to wild-type BCR-ABL and the closely related T315A mutation. These studies demonstrate that kinase domain mutations associated with drug resistance confer unanticipated enzymological and biological properties that may affect disease progression through altered downstream signaling. In addition, phosphoproteome profiling is a powerful tool to rapidly identify candidates for functional validation.
Results
Transformation Potency of Most Kinase Inhibitor-Resistant BCR-ABL Mutants Is Correlated with Kinase Activity.
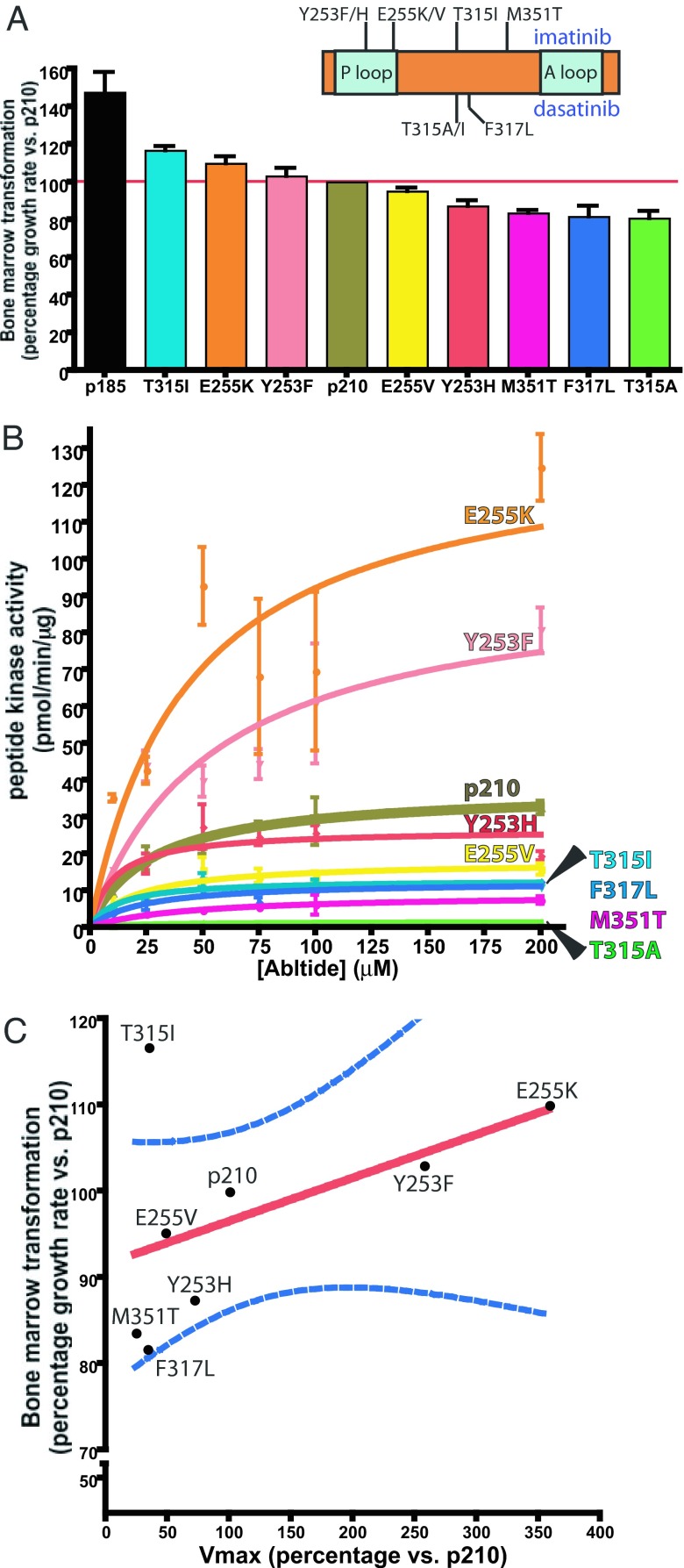
To compare the relative oncogenic potency of eight different kinase inhibitor resistant BCR-ABL p210 mutants (Fig. 1A), we used a modified Whitlock–Witte pre-B cell transformation assay (22–25) that was previously shown to correlate with in vivo CML mouse models and can distinguish subtle and quantifiable differences in potency between different BCR-ABL isoforms (26). These include two detected pretreatment in patients (T315I and E255K) and one documented to disappear after discontinuation of kinase inhibitor therapy (Y253H) (27). We also included examples associated with resistance to imatinib only (E255K, Y253F, E255V, Y253H, and M351T), dasatinib only (T315A), or both drugs (T315I and F317L). After infection with mutant or wild-type p210 BCR-ABL retrovirus, primary mouse bone marrow cells were plated at serial dilutions then monitored for oncogenic transformation as described (26). Potency was quantified by measuring doubling times for each mutant to calculate growth rate relative to wild-type p210 BCR-ABL (hereafter called p210) [Fig. 1A and supporting information (SI) Fig. 4 A_–_C]. The highly oncogenic p185 isoform served as a positive control for the assay (23, 28).
Fig. 1.
Transformation potency of T315I is not correlated with kinase activity. (A Inset) Cartoon of kinase inhibitor-resistant BCR-ABL kinase domain mutations used in this study. After a 21-day growth assay with BCR-ABL mutant-infected primary bone marrow cells, doubling time for each mutant was determined (SI Fig. 4 A_–_C). Data from the transformation assay was modeled to illustrate the growth rate and corresponding transformation potential of each mutant normalized to p210. (B) Kinase activity of each mutant toward various concentrations of biotinylated Abltide was performed in triplicate for 15 min. Velocity is plotted in a Michaelis–Menten graph. (C) The percentage mutant _V_max vs. p210 _V_max (x axis) is plotted against percentage mutant bone marrow growth rate vs. p210 growth rate (y axis). The overall trend of correlation between kinase activity and oncogenic potential is illustrated by the best fit line (red) and the 95% confidence interval curves (blue) for the best fit line, although this relationship need not be linear.
Three groups emerged from this analysis. T315I and E255K consistently showed a 10–20% increase in potency relative to p210, demonstrating that they are indeed gain-of-fitness mutants as suggested by clinical evidence of detection pretreatment. The second group included Y253F and E255V, which displayed potencies similar to p210. The final group contained four mutants (Y253H, M351T, F317L, and T315A) with substantially weaker transforming activity. Of note, the most weakly oncogenic mutation in this assay (T315A) is leukemogenic in mice (29) and has been linked to leukemia relapse in a dasatinib-resistant CML patient in blast crisis.¶¶
Because the increased kinase activity of p185 BCR-ABL has been implicated as the reason for increased potency (28, 30), we hypothesized that the altered oncogenic fitness of the kinase inhibitor resistant BCR-ABL mutants would similarly be associated with a parallel change in kinase activity. First, we examined kinase activity of immunoprecipitated full-length BCR-ABL protein against a number of exogenous substrates using gel-based in vitro kinase assays, using p185 as a positive control (30). With the exception of M351T and T315A, most kinase inhibitor-resistant mutants showed comparable kinase activity to p210 (SI Fig. 4_D_). T315A was particularly striking because it failed to phosphorylate any substrates in this assay despite its ability to autophosphorylate.
To enhance our ability to detect more subtle changes, we repeated these analyses using biotinylated Abltide as substrate to permit quantitative assessment of Michaelis–Menten kinetics, _V_max, and _K_m for each mutant (31–34). Two mutants (E255K and Y253F) had greater kinase activity than p210 (as measured by _V_max), whereas five (Y253F, T315I, E255V, F317L, and M351T) had reduced activity (Fig. 1B and SI Fig. 4_E_). As expected, T315A did not recognize the Abltide substrate and could not be scored. To determine the relationship between kinase activity and oncogenicity, we plotted _V_max versus the potency score determined in the bone marrow assays. These two variables were positively correlated for most of the kinase inhibitor-resistant mutants with one notable exception (Fig. 1C). The T315I mutant was 20% more potent than p210 in bone marrow transformation despite a nearly 50% reduction in _V_max.
System-Wide Tyrosine Phosphorylation Profiling Reveals Unique Mutant-Specific Phosphorylation Events.
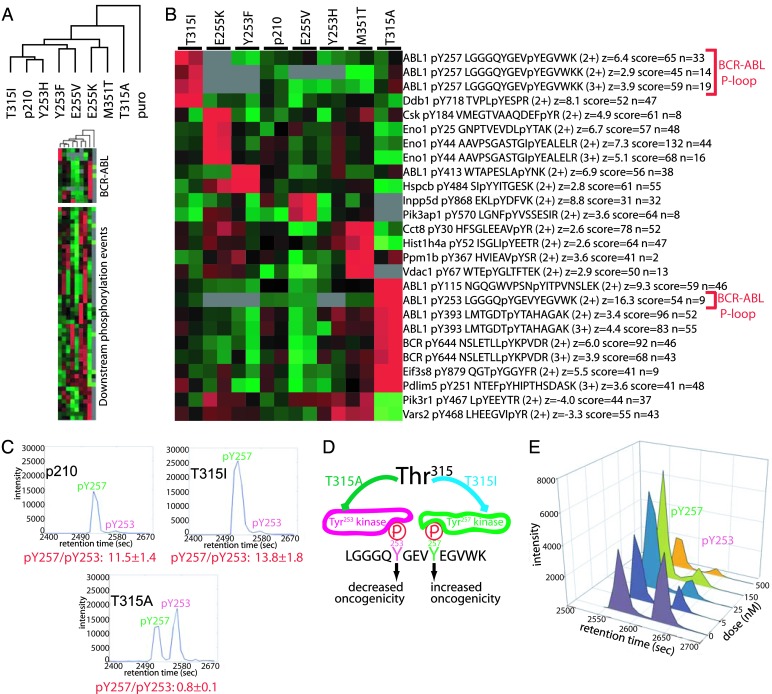
The discordance between oncogenic fitness and _V_max observed with T315I BCR-ABL and the inability of T315A BCR-ABL to recognize conventional ABL substrates suggested that mutation at this highly conserved threonine residue can impact substrate recognition. Therefore, we reasoned that the enhanced transformation potency of T315I might be mediated through phosphorylation of novel substrates. To explore this hypothesis, we used a quantitative, mass spectrometry-based global tyrosine phosphorylation profiling approach to search for unique phosphotyrosine-containing proteins in a panel of Ba/F3 cells stably expressing p210 and the panel of kinase inhibitor resistant BCR-ABL mutants. Furthermore, this assay measures endogenous phosphorylation in an unbiased fashion, and thus both compliments the in vitro kinase assay and moves beyond the restrictions of using a limited set of exogenous substrates. Initial examination of the data set by unsupervised clustering showed that cells expressing the T315A mutant were clearly distinct, but there were no obvious principal components associated with oncogenicity or kinase activity of the other mutants (Fig. 2A and SI Fig. 5). However, supervised analysis revealed several tyrosine phosphorylated peptides were uniquely and reproducibly elevated in cells expressing each of the kinase inhibitor resistant mutants (Fig. 2B), indicative of mutation-specific phosphotyrosine signatures. Phosphoproteome analysis also identified several peptides from BCR-ABL itself that were highly phosphorylated in T315A-expressing cells, consistent with the essentially normal levels of autophosphorylation observed in gel-based kinase assays.
Fig. 2.
System-wide phosphoproteomics reveals unique phosphopeptide profiles in cells expressing drug-resistant BCR-ABL mutants. Lysates from Ba/F3 cells stably expressing imatinib-resistant BCR-ABL mutants or control Ba/F3 cells (harboring empty expression plasmid) were enriched for phosphotyrosine containing tryptic peptides using the antiphosphotyrosine antibody 4G10 and identified and quantitated by mass spectrometry. (A) Global tyrosine phosphoprofiling identifies the T315A mutant as distinct from the other BCR-ABL variants. Relative levels of phosphotyrosine containing peptides (rows) are represented in the heat map with red boxes reflecting relatively high levels and green boxes low levels in the corresponding BCR-ABL variant (columns). Gray boxes indicate peptides that were undetectable in the control sample (MSCV puro, labeled puro). The similarity between mutants is indicated by the unsupervised hierarchical clustering dendrogram. Note that control cells have much fewer detectable phosphorylation events, indicating that the majority of the phosphopeptides in our profiles are direct and indirect targets of BCR-ABL. Identities of the phosphopeptides are presented in SI Fig. 5. (B) Drug resistance conferring point mutations result in unique phosphosignatures. The most uniquely elevated phosphotyrosine events were identified by calculating the z score for the phosphopeptide abundance level in each individual mutant relative to the mean and SD of the peptide level in the other BCR-ABL mutants and p210. Note that the phosphopeptides in B were elevated 2- to 10-fold in the indicated sample, but were not absent in the other mutants. For each phosphorylation event, the Entrez Gene abbreviation, residue of phosphorylation, tryptic peptide, charge state of the mass spectrometry ion, abundance level z score (z), best Mascot sequence assignment score (score), and number of times the peptide was sequenced (n) are indicated. Relative abundances and error estimates for a subset of the peptides in A and B are shown in SI Fig. 6. (C) Phosphorylation patterns of Tyr253 and Tyr257 of the BCR-ABL kinase P-loop correlate with transformation potential. Chromatograms plotting mass spectrometry signal intensity versus liquid chromatography elution time illustrate that Tyr257 is highly phosphorylated in cells expressing T315I (high transformer) and Tyr253 is highly phosphorylated in T315A (low transformer). Quantitative ratios of phospho-Tyr257 and phospho-Tyr253 from Ba/F3 cells expressing p210, T315A, and T315I are below the plot. (D) The identity of the gatekeeper residue influences BCR-ABL P-loop phosphorylation. A cartoon illustrating results from C is shown. (E) Tyr253 is a putative dasatinib-sensitive kinase phosphorylation site. Ba/F3 cells stably expressing T315A were treated with various concentrations of dasatinib for 2 h before phosphopeptide enrichment and mass spectrometric analysis. Quantitative levels of Tyr253 and Tyr257 phosphorylation at each dasatinib concentration are illustrated.
To address the lack of correlation between kinase activity and oncogenicity with T315I, we noted that one of the phosphopeptides specifically associated with this mutation was derived from the ATP binding loop (P-loop) of ABL (Fig. 2B). This observation caught our attention because mutation of Tyr253 in the ABL P-loop to phenylalanine allows the normally nononcogenic c-ABL to transform cells (35). T315I-expressing cells differed from those expressing other mutations due to increased phosphorylation at Tyr257 in the P-loop (not Tyr253). In contrast, Tyr253 (but not Tyr257) was highly phosphorylated relative to other mutants in T315A-expressing cells, as determined by collision induced dissociation (CID) mass spectrometry sequencing (SI Fig. 7), as well as by a consistent relative spacing in liquid chromatography retention time (Fig. 2C). We were unable to determine the stoichiometry of P-loop peptide phosphorylation because unphosphorylated peptides are lost after antiphosphotyrosine immunoprecipitation of the trypsin-digested lysate. Although the tryptic peptide that encompasses the P-loop includes both Tyr253 and Tyr257, we never detected both tyrosines phosphorylated at the same time despite control experiments demonstrating that our assay can enrich and detect a synthetic pTyr253/pTyr257 peptide, albeit with reduced efficiency relative to pTyr253 or pTyr257 alone (SI Table 1). This result suggests that these phosphorylation events are mutually exclusive, but we cannot rule out that a small fraction of Abl P-loop peptide below our detection limit is dually phosphorylated. In summary, T315I-expressing cells favor Tyr257 phosphorylation, whereas T315A-expressing cells favor Tyr253 phosphorylation (Fig. 2D).
Scansite analysis of the P-loop indicated that Tyr253 and Tyr257 both occur in the context of SRC family kinase recognition sites. Because the T315A substitution renders BCR-ABL resistant to dasatinib (a dual ABL/SRC kinase inhibitor) (9), we were able to address the possibility that SRC kinases are upstream of P-loop phosphorylation in T315A-expressing cells by treating cells with concentrations that will inhibit SRC, but not T315A (5 nM). Phosphorylation at Tyr253 was reduced in T315A cells exposed to 5 nM dasatinib whereas Tyr257 phosphorylation was unaffected (Fig. 2E); 500 nM dasatinib was required to block Tyr257 phosphorylation, implicating two distinct mechanisms of BCR-ABL P-loop phosphorylation.
Phosphorylation of the BCR-ABL P-Loop Regulates Oncogenicity.
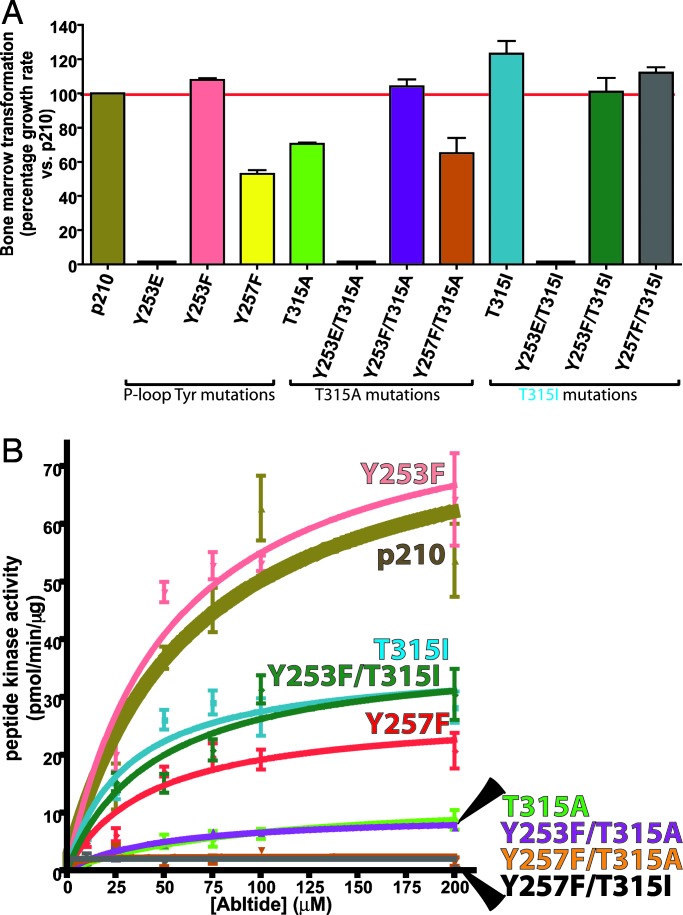
Based on prior work implicating Tyr253 as a negative regulatory site in the context of c-Abl (35), we asked whether the increase in Tyr253 phosphorylation in T315A-expressing cells is responsible for its reduced activity in the bone marrow transformation assay. Remarkably, mutation of this site to phenylalanine (Y253F) rescued the transformation activity of T315A BCR-ABL to wild-type levels (Fig. 3A and SI Fig. 8_A_), providing clues that phosphorylation at this site negatively impacts transformation. The same mutation in the context of T315I or wild-type had a less pronounced effect, consistent with the low levels of Tyr253 phosphorylation detected in these cells. In addition, reciprocal experiments using phosphomimetic mutations (Y253E or Y253D) provided additional evidence for the importance of Tyr253 phosphorylation in transformation potency. Both Y253E and Y253D substitutions suppressed the transforming activity of wild-type and T315I BCR-ABL in the bone marrow assay (Fig. 3A and SI Fig. 8_A_) and in IL-3-dependent Ba/F3 cells (data not shown). These data strongly suggest that phosphorylation of the P-loop tyrosines plays a significant role in BCR-ABL mutant transformation. Note that, although we did not determine the stoichiometry of phosphorylated/unphosphorylated P-loop tyrosines in the entire BCR-ABL pool, the mutatgenesis data suggest that whatever the fraction of phosphorylated P-loop, this fraction is adequate to influence transformation.
Fig. 3.
Phosphorylation of the P-loop tyrosines directs the subsequent oncogenicity of BCR-ABL mutants. (A) The growth rate and corresponding transformation potential of each mutant is normalized to p210. Note that mutation of Tyr253 in the context of T315A restores transformation potency to the level of p210 or the Y253F single mutant. Y253D single and compound mutants were also tested in the bone marrow assay with identical results to Y253E. (B) Kinase activity of BCR-ABL single and compound P-loop tyrosine/gatekeeper mutants.
It is well established that the kinase activity of the serine/threonine kinase Cdk2 is negatively regulated by phosphorylation of an analogous tyrosine residue in its P-loop (36, 37). However, the Y253F mutation did not appreciably alter the kinase activity of wild-type, T315I, or T315A BCR-ABL toward Abltide (Fig. 3B and SI Fig. 8_B_), indicating that the mechanism of negative regulation by Tyr253 phosphorylation likely differs from that of Cdk2.
We also examined the consequences of Tyr257 mutation. Y257F substitution substantially impaired the oncogenicity of wild-type BCR-ABL and dampened kinase activity by 3-fold, suggesting that Tyr257 may play a positive regulatory role, opposite to Tyr253. However, the same mutation had minimal effect on T315I oncogenicity yet caused a 15-fold drop in kinase activity (Fig. 3B). This result is surprising for two reasons. First, the large decrease in T315I kinase activity without a commensurate change in oncogenicity suggests that Abltide is not an optimal substrate for Tyr257 mutants, particularly in the context of T315I. Second, it challenges the simple notion that Tyr257 plays a positive regulatory role, in opposition to Tyr253. One possibility is that Tyr257 regulates access of relevant kinases to Tyr253, which then functions as the primary P-loop determinant of oncogenicity (Fig. 2D). In any case, our findings demonstrate that the principal variables controlling Tyr253 phosphorylation in drug resistant BCR-ABL mutants are: (i) the identity of the substituted residue at Thr315 (Ala favors pTyr253, Ile favors pTyr257), (ii) the status of Tyr257 phosphorylation (either pTyr257 or pTyr253).
Discussion
Clinical resistance to small molecule kinase inhibitors used in cancer therapy occurs primarily through outgrowth of subclones bearing mutations in the target kinase that interfere with drug binding. Initially reported in CML, this mechanism is broadly applicable to other cancers such as gastrointestinal stromal tumors and lung cancer (38, 39). These insights have led to the rapid development of second-generation inhibitors with activity against mutants resistant to first generation drugs and will likely form the basis for combination therapy cocktails to prevent relapse. Although this cancer drug development strategy resembles that used in infectious diseases, there are important differences between drug-resistant mutants isolated from cancers versus infectious agents. Drug-resistant HIV strains are typically crippled in their virulence and only develop in the context of selective pressure caused by drug treatment (40). In contrast, many drug-resistant kinase mutants isolated from cancer patients are detected before drug exposure and, over time, can become the dominant malignant clone in the absence of further selective pressure (1, 19). These more virulent, drug-resistant kinase oncogenes must be detected early to ensure long-term cancer control using targeted therapy.
Here we have catalogued the oncogenicity and enzymology of drug-resistant BCR-ABL mutants that define a spectrum of biological activity, with examples of increased or decreased fitness relative to the wild-type enzyme. Similar conclusions were recently reported from a study of a smaller panel of five imatinib-resistant mutants that was independent of our work (41). In most cases, we found that fitness change correlates with kinase activity. The most notable exception was the T315I mutation, resistant to all of the clinically approved ABL kinase inhibitors, that displays enhanced oncogenic fitness despite reduced kinase activity for classical ABL substrates. We investigated this apparent paradox by a quantitative global phosphoproteome profiling method and found mutation-specific phosphosignatures, indicating that each drug-resistant allele functions as a distinct enzyme with unique biological properties and substrate recognition profiles. In the case of the two drug-resistant gatekeeper threonine mutants (T315I and T315A), the altered phosphorylation profile detected by mass spectrometry allowed identification of changes in P-loop phosphorylation that are functionally linked to oncogenic fitness. It will be of interest to conduct a similar analysis of the drug-resistant EGF receptor T790M mutation that, like T315I, has been detected in patients pretreatment and is implicated as a potential gain-of-fitness allele (19, 42).
In addition to its essential importance in drug resistance, our findings reveal a previously unanticipated role of the gatekeeper residue Thr315 in substrate recognition. One striking observation was the failure of the T315A allele to phosphorylate any classical ABL substrates in vitro. This same threonine to alanine substitution has been engineered into BCR-ABL (and other kinases) to render the enzyme uniquely sensitive to artificial ATP analogues, thereby allowing highly specific identification of downstream substrates and assessment of kinase dependence (29, 31, 43, 44). Our results suggest that the signaling characteristics of these mutant enzymes should be carefully evaluated as they may not always closely mimic the wild-type allele. The identity of the mutant gatekeeper Thr315 residue (Ala or Ile) also impacts the pattern of Tyr253 and Tyr257 phosphorylation in the BCR-ABL P-loop, which impacts oncogenicity (Fig. 2D). The mutagenesis data provide evidence that Tyr253 phosphorylation suppresses oncogenicity, whereas Tyr257 plays a more complex role in promoting oncogenicity. Tyr257 is unlikely to function directly as a positive regulator (to counterbalance negative regulation by Tyr253) because Y257F substitution impairs the oncogencity of wild-type BCR-ABL but not T315I. One model, based on the assumption that both sites are not phosphorylated in tandem, is that pTyr257 precludes pTyr253 (perhaps by limiting access to Tyr253 kinases). The Y257F mutant in combination with T315I presumably escapes the negative impact of this substitution because the efficiency of phosphorylation at the Tyr253 site is reduced by the Ile315 substitution. Much additional work is required to evaluate these and other potential models, including structural studies to understand how Thr315 substitution impacts substrate recognition and to identify the kinases and phosphatases that regulate Tyr253/Tyr257 phosphorylation. The correlation between P-loop phosphorylation site occupancy ratios and oncogenicity, together with the mutagenesis results, strongly implicate BCR-ABL P-loop phosphorylation status in the modulation of BCR-ABL transformation potential and implicate the gatekeeper residue as impacting the P-loop phosphorylation pattern.
CML patients are now screened for drug-resistant BCR-ABL mutations when they fail imatinib therapy using standard sequencing. Mutations that enhance oncogenicity could be prioritized for development of ultrasensitive detection assays so that these alleles are found before clinical evidence of disease progression. It should also be possible to prospectively evaluate the fitness of drug resistant mutations for new kinase inhibitor targets to define the most worrisome alleles in advance of clinical development. Our work also demonstrates the utility of phosphoproteome profiling as a tool for biomarker discovery. Each drug-resistant mutation has a unique phosphopeptide signature that could be used for mutation-specific pharmacodynamic monitoring of target inhibition and, possibly, for identifying novel downstream drug targets. Finally, it may be possible to exploit the important role of Tyr253 phosphorylation in regulating BCR-ABL oncogenicity for therapeutic benefit, particularly in the context of the multiply drug-resistant T315I allele. One goal might be to enhance Tyr253 phosphorylation by inhibiting the relevant phosphatase and/or by complete blockade of Tyr257 phosphorylation with an appropriate kinase inhibitor.
Materials and Methods
Constructs and Cells.
All MSCV-BCR-ABL single mutant constructs and Ba/F3 stable cell lines used in these studies have been reported (1, 9). The Y253D/E and compound Tyr253/Thr315 mutations were created with the QuikChange XL kit (Stratagene, La Jolla, CA). Sequences for the Abltides have been published (31). Oligonucleotides corresponding to these sequences were synthesized for cloning into pGEX-3X (Amersham Pharmacia, Piscataway, NJ), and proteins were expressed as in ref. 45.
Primary Bone Marrow Transformation.
Infection and subsequent growth of murine bone marrow containing BCR-ABL mutants was performed as described (9, 26) with modifications listed in SI Text.
BCR-ABL Kinetics Studies.
Gel-based and Abltide in vitro kinase assays were performed as described in refs. 34 and 45 with modifications detailed in SI Text.
Phospho Enrichment, Mass Spectrometry, and Data Analysis.
Immunoaffinity-based enrichment of digested cellular lysates for tyrosine phosphorylated peptides was performed in a similar manner to ref. 46, and mass spectrometry analysis was performed as in refs. 47 and 48 with modifications as detailed in SI Text.
Supplementary Material
Supporting Information
Acknowledgments
We are grateful to John Wongvipat, Randy Chen, and Yanan Yang for technical assistance, Igor Vivanco for experimental insights, Maureen McMahon for statistical help, and Benno Schwikowski for sharing computational algorithms before their publication. This work was supported by grants from the National Human Genome Research Institute (to T.G.G.) and the Leukemia and Lymphoma Society (to C.L.S.). T.G.G. is an Alfred P. Sloan Research Fellow. C.L.S. is a Doris Duke Distinguished Clinical Scientist and received support as an Investigator of the Howard Hughes Medical Institute.
Abbreviation
CML
chronic myelogenous leukemia.
Footnotes
The authors declare no conflict of interest.
¶¶
Shah, N. P., Nicoll, J., Branford, S., Hughes, T. P., Paquette, R., Talpaz, M., Nicaise, C., Huang, F., Sawyers, C. L. (2005) Blood 106:1093 (abstr.).
References
- 1.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 2.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 3.Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J. Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 4.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 5.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 6.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, et al. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 7.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, et al. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 8.Azam M, Latek RR, Daley GQ. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 9.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Proc Natl Acad Sci USA. 2005;102:3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, et al. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.von Bubnoff N, Manley PW, Mestan J, Sanger J, Peschel C, Duyster J. Blood. 2006 doi: 10.1182/blood-2005-12-010132. in press. [DOI] [PubMed] [Google Scholar]
- 12.Roche-Lestienne C, Lai JL, Darre S, Facon T, Preudhomme C. N Engl J Med. 2003;348:2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- 13.Roche-Lestienne C, Preudhomme C. Semin Hematol. 2003;40:80–82. doi: 10.1053/shem.2003.50046. [DOI] [PubMed] [Google Scholar]
- 14.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, Lai JL, Philippe N, Facon T, Fenaux P, Preudhomme C. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 15.Hato T, Yamanouchi J, Tamura T, Hojo N, Niiya Y, Kohno M, Bando S, Yakushijin Y, Takada K, Sakai I, et al. Int J Hematol. 2004;80:62–66. doi: 10.1532/ijh97.04033. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann WK, Komor M, Wassmann B, Jones LC, Gschaidmeier H, Hoelzer D, Koeffler HP, Ottmann OG. Blood. 2003;102:659–661. doi: 10.1182/blood-2002-06-1756. [DOI] [PubMed] [Google Scholar]
- 17.Barthe C, Gharbi MJ, Lagarde V, Chollet C, Cony-Makhoul P, Reiffers J, Goldman JM, Melo JV, Mahon FX. Br J Haematol. 2002;119:109–111. doi: 10.1046/j.1365-2141.2002.03708.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu WH, Makrigiorgos GM. Leuk Res. 2003;27:979–982. doi: 10.1016/s0145-2126(03)00066-3. [DOI] [PubMed] [Google Scholar]
- 19.Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J, et al. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 20.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 22.McLaughlin J, Chianese E, Witte ON. Proc Natl Acad Sci USA. 1987;84:6558–6562. doi: 10.1073/pnas.84.18.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin J, Chianese E, Witte ON. Mol Cell Biol. 1989;9:1866–1874. doi: 10.1128/mcb.9.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daley GQ, Van Etten RA, Baltimore D. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 25.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, et al. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 26.Smith KM, Yacobi R, Van Etten RA. Mol Cell. 2003;12:27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller MC, Lahaye T, Hochhaus A. Dtsch Med Wochenschr. 2002;127:2205–2207. doi: 10.1055/s-2002-34939. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong S, McLaughlin J, Cheng D, Zhang C, Shokat KM, Witte ON. Proc Natl Acad Sci USA. 2004;101:17456–17461. doi: 10.1073/pnas.0407061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Witucki LA, Shah K, Bishop AC, Shokat KM. Biochemistry. 2000;39:14400–14408. doi: 10.1021/bi000437j. [DOI] [PubMed] [Google Scholar]
- 32.Till JH, Annan RS, Carr SA, Miller WT. J Biol Chem. 1994;269:7423–7428. [PubMed] [Google Scholar]
- 33.Songyang Z, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 34.Brasher BB, Van Etten RA. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 35.Allen PB, Wiedemann LM. J Biol Chem. 1996;271:19585–19591. doi: 10.1074/jbc.271.32.19585. [DOI] [PubMed] [Google Scholar]
- 36.Gould KL, Moreno S, Tonks NK, Nurse P. Science. 1990;250:1573–1576. doi: 10.1126/science.1703321. [DOI] [PubMed] [Google Scholar]
- 37.Gould KL, Nurse P. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 38.Haber DA, Bell DW, Sordella R, Kwak EL, Godin-Heymann N, Sharma SV, Lynch TJ, Settleman J. Cold Spring Harbor Symp Quant Biol. 2005;70:419–426. doi: 10.1101/sqb.2005.70.043. [DOI] [PubMed] [Google Scholar]
- 39.Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, Leversha MA, Jeffrey PD, Desantis D, Singer S, et al. Clin Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 40.Bates M, Wrin T, Huang W, Petropoulos C, Hellmann N. Curr Opin Infect Dis. 2003;16:11–18. doi: 10.1097/00001432-200302000-00003. [DOI] [PubMed] [Google Scholar]
- 41.Griswold IJ, Macpartlin M, Bumm T, Goss VL, O'Hare T, Lee KA, Corbin AS, Stoffregen EP, Smith C, Johnson K, et al. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- 45.Kabak S, Skaggs BJ, Gold MR, Affolter M, West KL, Foster MS, Siemasko K, Chan AC, Aebersold R, Clark MR. Mol Cell Biol. 2002;22:2524–2535. doi: 10.1128/MCB.22.8.2524-2535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 47.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, Wong DT. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 48.Prakash A, Mallick P, Whiteaker J, Zhang H, Paulovich A, Flory M, Lee H, Aebersold R, Schwikowski B. Mol Cell Proteomics. 2006;5:423–432. doi: 10.1074/mcp.M500133-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information