SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting (original) (raw)
Abstract
The signal recognition particle (SRP) cotranslationally targets proteins to cell membranes by coordinated binding and release of ribosome-associated nascent polypeptides and a membrane-associated SRP receptor. GTP uptake and hydrolysis by the SRP-receptor complex govern this targeting cycle. Because no GTPase-activating proteins (GAPs) are known for the SRP and SRP receptor GTPases, however, it has been unclear whether and how GTP hydrolysis is stimulated during protein trafficking in vivo. Using both biochemical and genetic experiments, we show here that SRP RNA enhances GTPase activity of the SRP–receptor complex above a critical threshold required for cell viability. Furthermore, this stimulation is a property of the SRP RNA tetraloop. SRP RNA tetraloop mutants that confer defective growth phenotypes can assemble into SRP–receptor complexes, but fail to stimulate GTP hydrolysis in these complexes in vitro. Tethered hydroxyl radical probing data reveal that specific positioning of the RNA tetraloop within the SRP–receptor complex is required to stimulate GTPase activity to a level sufficient to support cell growth. These results explain why no external GAP is needed and why the phylogenetically conserved SRP RNA tetraloop is required in vivo.
Keywords: signal recognition particle, SRP receptor, GTPase activating protein, 4.5S RNA, GNRA tetraloop
INTRODUCTION
The signal recognition particle (SRP) is a GTP-regulated ribonucleoprotein that targets secretory or membrane proteins during translation. In Escherichia coli, the SRP comprises the Ffh protein and 4.5S RNA, which together recognize hydrophobic signal peptides as they emerge from translating ribosomes. The SRP targets these ribosome-nascent chain complexes (RNCs) to the inner membrane by interacting with the SRP receptor (SR), FtsY (Doudna and Batey 2004; Shan and Walter 2005a). Both Ffh and FtsY are GTPases that associate into a heterodimer upon GTP binding and deliver the RNC to a transmembrane pore—a translocon—at the inner membrane. Upon GTP hydrolysis, SRP and FtsY dissociate, and the SRP is primed for a new round of nascent-chain recognition and trafficking.
Although GTP binding and hydrolysis govern the protein targeting cycle, the mechanism of GTPase regulation in vivo has not been elucidated. Unlike other GTPases, no external GTPase activating proteins (GAPs) or guanine exchange factors (GEFs) have been found for the SRP. Instead, the N-terminal (N) and GTPase (G) domains of Ffh and FtsY, when associated with each other, catalyze GTP hydrolysis at least 1000-fold faster than the basal rate observed for either protein alone (Powers and Walter 1995; Peluso et al. 2001). Crystal structures of the NG–NG heterodimer revealed two composite GTP binding sites at the protein–protein interface near several catalytically critical amino acids, providing a compelling model for GTPase stimulation in the complex (Egea et al. 2004; Focia et al. 2004). Extensive site-directed mutagenesis and kinetic analyses verified that multiple catalytic groups from each protein facilitate hydrolysis of the GTP at each active site (Shan and Walter 2003, 2005b; Egea et al. 2004; Shan et al. 2004).
Nonetheless, these results do not address the role of the 4.5S RNA, which also stimulates GTPase activity in the SRP–FtsY complex in vitro and is essential for SRP function in vivo (Brown and Fournier 1984; Peluso et al. 2001; Sagar et al. 2004; Gu et al. 2005). A truncated form of the 114-nucleotide (nt) RNA, including just the distal 44 nt at the hairpin end (domain IV) of the molecule, is sufficient to support cell growth (Fig. 1; Batey et al. 2000). Within this region, two evolutionarily conserved segments are known to be critical for SRP activity in vivo. One of these, comprising the two internal loops near the hairpin end of the RNA, forms the binding site for the signal peptide-binding M domain of Ffh (Batey et al. 2000). The other, the 4-nt GGAA tetraloop adjacent to the Ffh recognition site, has been proposed to be required for SRP binding to FtsY based on the behavior of 4.5S RNA tetraloop mutants in vivo and in vitro (Jagath et al. 2001). In contrast, however, recent data showed that a 4.5S RNA tetraloop mutant with a lethal phenotype in vivo can associate into an SRP–FtsY complex at physiologically relevant GTP concentrations in vitro (Spanggord et al. 2005). Importantly, this mutant RNA failed to stimulate GTP hydrolysis in the SRP–FtsY complex above the rate observed in the absence of RNA (Spanggord et al. 2005). A structural model of the SRP–FtsY complex placing the 4.5S RNA at the interface of the Ffh–FtsY heterodimer and the tetraloop near the G domains of Ffh and FtsY is consistent with tetraloop-mediated GTPase stimulation (Spanggord et al. 2005).
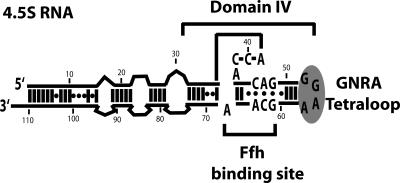
FIGURE 1.
Wild-type 4.5S RNA and sequence of the tetraloop mutants. Secondary structure of the wild-type 4.5S RNA with tetraloop sequence GGAA. Non-Watson–Crick base pairs are indicated by (•). The Ffh binding site indicates the two conserved internal loops. Domain IV is the universally conserved region of SRP RNA. Gray oval highlights the conserved GNRA tetraloop of 4.5S RNA.
To determine whether 4.5S RNA-stimulated GTPase activity in the SRP–FtsY complex is physiologically essential, we analyzed the ability of five different 4.5S RNA mutants to stimulate GTPase activity in vitro and to complement wild-type 4.5S RNA depletion in vivo. The results reveal that mutants unable to stimulate GTPase activity in SRP–FtsY complexes above that observed in the absence of 4.5S RNA produce severe growth phenotypes in in vivo complementation experiments. Furthermore, site-directed hydroxyl radical probing shows that specific positioning of the 4.5S RNA tetraloop at the Ffh–FtsY interface is essential to enhance the rate of GTP hydrolysis to a level sufficient for cell viability. The SRP RNA tetraloop thus functions to enhance the GTPase activity of the SRP–FtsY complex, explaining why no external GAP is needed and why the phylogenetically conserved tetraloop is required in vivo.
RESULTS
Binding affinity of 4.5S RNA tetraloop mutants to Ffh
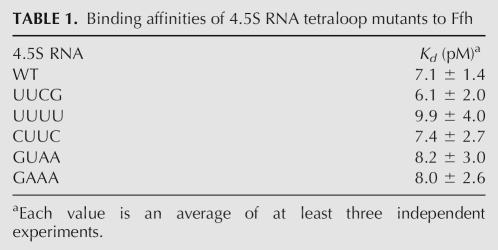
Previous studies suggested that the tetraloop of 4.5S RNA does not contribute to the binding affinity of SRP RNA for Ffh. Footprinting of 4.5S RNA in the presence of Ffh showed little or no protection at the tetraloop (Lentzen et al. 1996). Crystal structures and structural models obtained by FRET showed that the tetraloop of 4.5S RNA does not make any direct contacts with Ffh (Batey et al. 2000; Buskiewicz et al. 2005). Furthermore, the tetraloop mutant UUCG and wild-type 4.5S RNA had the same binding affinity for Ffh as assayed by nitrocellulose filter binding (Jagath et al. 2001), and 4.5S RNA variants lacking the tetraloop also bind to Ffh with affinities similar to that of the wild-type 4.5S RNA (Batey et al. 2001; Batey and Doudna 2002). Nevertheless, we performed nitrocellulose filter-binding assays with the 4.5S tetraloop mutants used in this study to ensure that mutations did not affect the interaction between Ffh and the RNA. As shown in Table 1, all of the 4.5S RNA tetraloop mutants have binding affinities for Ffh in the range of 6–10 pM, similar to the 7 pM value measured for wild-type 4.5S RNA. This indicates that, as expected, the tetraloop does not affect the interaction between 4.5S RNA and Ffh.
TABLE 1.
Binding affinities of 4.5S RNA tetraloop mutants to Ffh
GTPase activity of SRP–FtsY complexes containing 4.5S RNA mutants
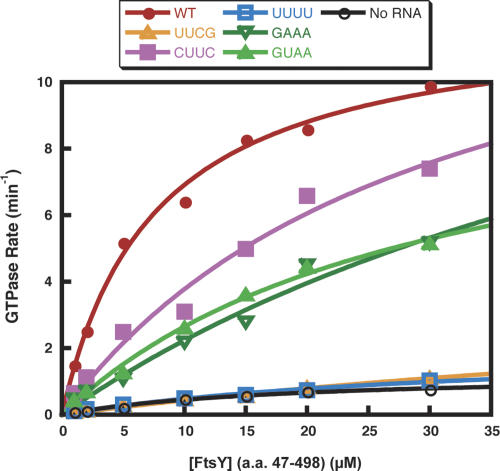
Two previous studies have led to different conclusions about the function of the phylogenetically conserved GNRA tetraloop of 4.5S RNA. One set of results suggested that the tetraloop affects SRP binding to its receptor protein FtsY (Jagath et al. 2001), whereas the other implied that the tetraloop regulates GTPase activity of the SRP–FtsY complex (Spanggord et al. 2005). To investigate the biochemical and physiological role of this region of 4.5S RNA, we first tested the GTPase activities of SRP–FtsY complexes containing five different tetraloop mutants: UUCG, UUUU, CUUC, GUAA, and GAAA (Fig. 2). Similar to previous results, an SRP–FtsY complex containing wild-type RNA catalyzed GTP hydrolysis sixfold faster than an Ffh–FtsY complex lacking the RNA, or an SRP–FtsY complex containing the UUCG mutant RNA (Fig. 2; Peluso et al. 2001; Spanggord et al. 2005). Interestingly, an SRP–FtsY complex containing 4.5S RNA with a UUUU tetraloop sequence also catalyzed GTP hydrolysis at a rate similar to that observed in the absence of 4.5S RNA (Fig. 2). SRP–FtsY complexes containing each of the other three 4.5S RNA tetraloop mutants, CUUC, GAAA, and GUAA, showed twofold reductions in GTPase rates relative to wild-type complexes (Fig. 2).
FIGURE 2.
GTPase activites of SRP–FtsY complex. Fixed concentrations of SRP or Ffh with varying amounts of FtsY (amino acids 47–498) in the presence of 100 μM GTP and trace amount of γ-32P-GTP were used in this assay. The concentrations of wild type (•), CUUC (▪), GAAA (∇), and GUAA (▴) SRP were 0.2 μM whereas the concentrations of UUCG (Δ) and UUUU (□) SRP were 2 μM. In the absence of RNA (○), only 1 μM of Ffh was used. The extrapolated maximal rate constants for wild type, CUUC, GAAA, GUAA, UUCG, UUUU, and no RNA are 13.4 ± 4.1, 15.2 ± 5.1, 10.9 ± 2.8, 8.4 ± 2.5, 3.2 ± 1.4, 1.3 ± 0.4, and 2.2 ± 1.5 min−1, respectively. The maximal rate constants and values of the data points on the curves are averages of at least three independent experiments.
SRP–FtsY complex stability in the presence of 4.5S RNA mutants
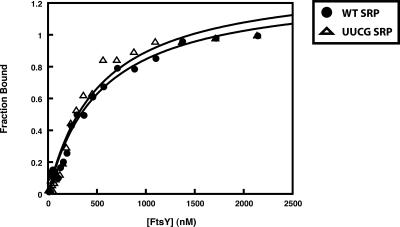
A possible explanation for the observed partial or complete loss of GTPase stimulation in SRP–FtsY complexes containing 4.5S RNA tetraloop mutants is that these RNAs prevent stable FtsY binding, as previously proposed (Jagath et al. 2001). To test this possibility, SRP–FtsY ternary complex formation for wild-type and UUCG 4.5S RNA was monitored by enzyme-linked immunosorbent assay (ELISA) using single-chain antibodies (scFv) IA02 and IB11 (Wu et al. 2006). The antibody scFv IA02 specifically recognizes the SRP–FtsY complex, and not SRP, Ffh, or FtsY alone (Wu et al. 2006). This recognition is independent of the RNA since IA02 can detect Ffh–FtsY complexes (data not shown). However, during the 20-min incubation of this assay, Ffh–FtsY complexes are not stably formed in the absence of the 4.5S RNA (Shepotinovskaya et al. 2003). scFv IB11 is specific to Ffh alone or in the SRP complex, but does not recognize FtsY (Wu et al. 2006). Both antibodies’ specificities are unaffected by the GNRA tetraloop since they show similar levels of detection for complexes containing either the wild-type or UUCG 4.5S RNA (data not shown). Given the specificities of IB11 for SRP alone and IA02 for the ternary complex, the extent of SRP–FtsY complex formation with either wild-type or UUCG 4.5S RNA was monitored by ELISA. The amount of SRP–FtsY complex formed was determined by normalizing the ELISA signal of IA02 against the signal from IB11. As shown in Figure 3, wild-type and UUCG 4.5S RNA have similar levels of SRP–FtsY complex formation. In comparison, the GTPase assay mentioned above uses significantly higher amounts of SRP and FtsY (see Materials and Methods), supporting the conclusion that the loss of GTPase stimulation by the UUCG mutant is not due to complex instability.
FIGURE 3.
ELISA of SRP–FtsY complex. SRP–FtsY complex formation with wild-type (•) and UUCG (Δ) 4.5S RNA was detected by ELISA using 50 nM of wild-type or UUCG SRP, 0.5 mM GMPPNP, and titrating concentrations of FtsY (amino acids 47–497). scFv IA02, which recognizes only the complex, was used to determine the amount of complex formed, and scFv IB11 was used to detect the total amount of SRP in each binding reaction. These data are representatives of three independent experiments.
Structural probing of SRP–FtsY complexes containing wild-type versus mutant 4.5S RNA
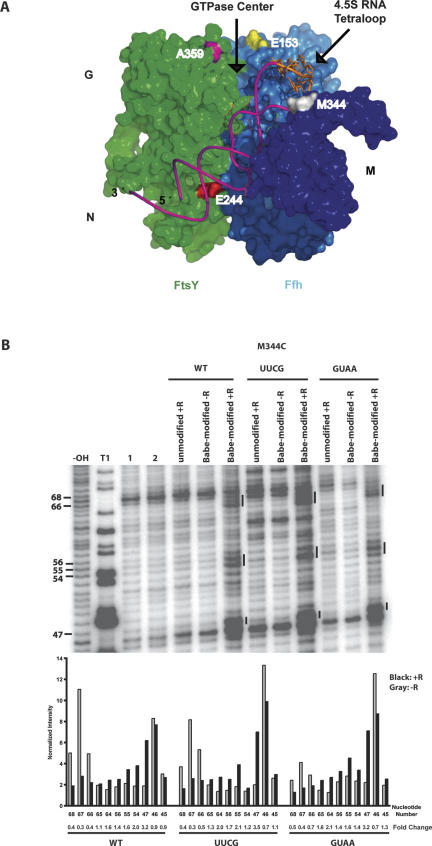
A recent model of the wild-type SRP–FtsY complex suggests that the 4.5S RNA tetraloop localizes near the GTPase domains of the Ffh–FtsY heterodimer upon SRP–FtsY association (Fig. 4A; Spanggord et al. 2005). To test the possibility that tetraloop mutations perturb positioning of the 4.5S RNA at this protein–protein interface, tethered hydroxyl radical probing experiments were used to assess the localization of wild-type versus mutated forms of 4.5S RNA in SRP–FtsY complexes. We probed SRP–FtsY complexes containing the UUCG or GUAA 4.5S RNA tetraloop mutation, which have a complete or partial loss of GTPase stimulation relative to wild-type 4.5S RNA-containing complexes, respectively. Chemically modified proteins were prepared by mutating single residues in either Ffh or FtsY to cysteine, enabling site-specific derivatization with bromoacetamidobenzyl-EDTA-Fe (BABE-Fe) (Fig. 4A; Spanggord et al. 2005). In the presence of hydrogen peroxide and sodium ascorbate, the BABE-Fe moiety generates hydroxyl radicals that cleave the backbone of the RNA (Joseph and Noller 2000).
FIGURE 4.
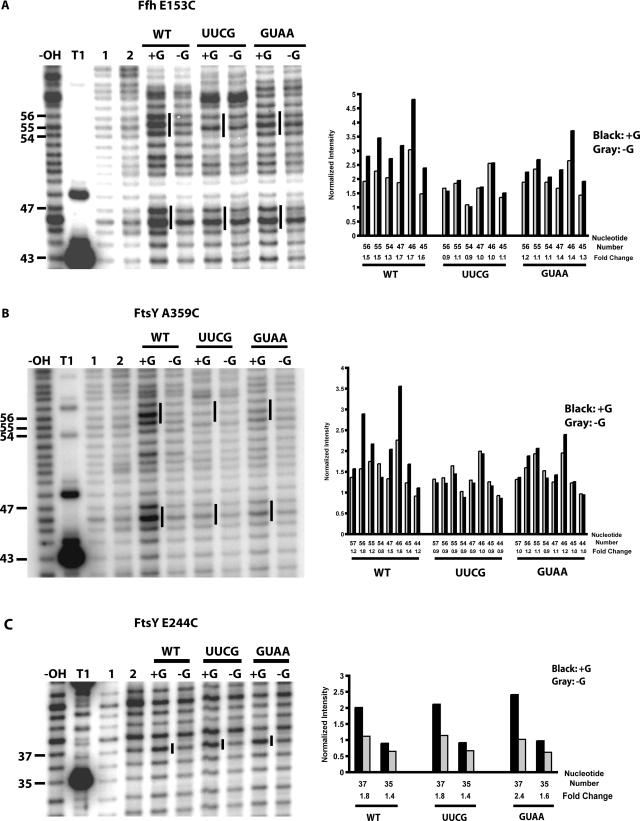
Model of the SRP–FtsY complex and hydroxyl radical probing of full-length wild type, UUCG, and GUAA 4.5S RNA in a saturable SRP–FtsY (amino acids 197–498) complex derivatized by BABE-Fe at positions 344 of Ffh. (A) Model of the SRP–FtsY complex based on previous hydroxyl radical probing data (Spanggord et al. 2005). G, N, and M indicate the GTPase, N-terminal, and M domains for Ffh and FtsY. FtsY is shown in green. NG and M domains of Ffh are shown in blue and purple, respectively. 4.5S RNA is shown in pink and the nucleotides of tetraloop is depicted in orange. Residues A359 and E244 of FtsY and E153 and M344 of Ffh are positions of BABE-Fe. (B) Lanes labeled −OH, T1, 1, and 2 indicate 4.5S RNA alkaline hydrolysis, partial T1 RNase guanosine cleavage, 4.5S RNA alone in buffer, or in the presence of cleaving reagents, respectively. All remaining lanes are hydroxyl radical probing reactions performed in the presence of GMPPCP and FtsY. +R and −R indicate the presence and absence of cleaving reagents. Numbers on the left indicate nucleotide positions of 4.5S RNA and bars indicate regions of cleavage sites. Quantitations of specific nucleotides are shown with normalization for loading differences between the +G and −G lanes for the three RNA constructs. Fold change indicates the intensity of the nucleotide in the +G reaction relative to the −G reaction. These data are representatives of three independent experiments.
BABE-Fe-modified position 344 of the Ffh M-domain was used to test for possible tetraloop-dependent structural changes in Ffh–4.5S RNA binding. The similarity of cleavage patterns and fold changes in the wild-type, UUCG, and GUAA 4.5S RNAs is consistent with the observation that tetraloop mutations do not affect Ffh–RNA binding (Table 1; Fig. 4B). BABE-Fe-modified positions 153 of Ffh and 359 of FtsY (Fig. 4A) were used to detect any structural perturbations of the tetraloop mutant RNAs in the SRP–FtsY complex because in previous work they each cleaved the tetraloop region of the wild-type 4.5S RNA only when the SRP–FtsY complex forms (Spanggord et al. 2005). When wild-type 4.5S RNA was used to form SRP–FtsY complexes containing a BABE-Fe moiety at residues 153 of Ffh or 359 of FtsY, pronounced cleavages as previously observed were detected within the RNA tetraloop and asymmetric loop at nucleotides 47–45 (cf. lanes +G and −G for wild type in Fig. 5A,B, respectively; Spanggord et al. 2005). In contrast, similar complexes formed with 4.5S RNA mutants bearing a UUCG tetraloop sequence produced no significant cleavage of the RNA over background (cf. nucleotides 57–44 fold changes for wild type and UUCG in Fig. 5A,B, respectively). Subtle cleavages above background were detected for the GUAA mutant (cf. nucleotides 57–44 fold changes for for wild type and GUAA in Fig. 5A,B, respectively). The changes in cleavage of the UUCG and GUAA mutant RNAs were not due to inhibition of SRP–FtsY complex formation with the modified proteins, as confirmed by native gel mobility shift assays (data not shown).
FIGURE 5.
Tethered hydroxyl radical probing of SRP–FtsY complexes. Strand scission of full-length wild-type (WT), UUCG, and GUAA 4.5S RNA in a saturable SRP–FtsY complex derivatized by BABE-Fe at positions Ffh E153 (A), FtsY A359 (B), and FtsY E244 (C). Lanes labeled −OH, T1, 1, and 2 indicate 4.5S RNA alkaline hydrolysis, partial T1 RNase guanosine cleavage, 4.5S RNA alone in buffer, or in the presence of cleaving reagents, respectively. Lanes labeled −G and +G indicate the absence and presence of GMPPCP, respectively. Nucleotide positions of 4.5S RNA are indicated on the left. Regions of cleavage sites for wild type and equivalent sites for UUCG and GUAA are indicated by bars. Quantitations of specific nucleotides are shown with normalization for loading differences between the +G and −G lanes for the three RNA constructs. Fold change indicates the intensity of the nucleotide in the +G reaction relative to the −G reaction. These data are representatives of three independent experiments.
The lack of cleavage above background in the tetraloop region of the two mutant RNAs could result from a failure of the entire 4.5S RNA helix to localize correctly within the SRP–FtsY complex, or it could be a consequence of more subtle perturbations in tetraloop positioning in relation to the BABE-Fe-modified residues. To distinguish between these possibilities, the position of the distal stem region of the wild-type and mutant 4.5S RNAs was probed using SRP–FtsY complexes containing a BABE-Fe modification at residue 244 of FtsY (Fig. 4A; Spanggord et al. 2005). BABE-Fe at residue 244 of FtsY induced similar cleavages in the wild-type 4.5S RNA and the two tetraloop mutants, consistent with RNA helix localization near the Ffh–FtsY heterodimer interface in each case (Fig. 5C). These results indicate that the tetraloop sequence does not affect global 4.5S RNA positioning in the SRP–FtsY complex.
Together, the data are consistent with 4.5S RNA tetraloop mutations causing local disruption of the loop position relative to the BABE-Fe-modified residues in the SRP–FtsY complex.
In vivo complementation with 4.5S RNA tetraloop mutants
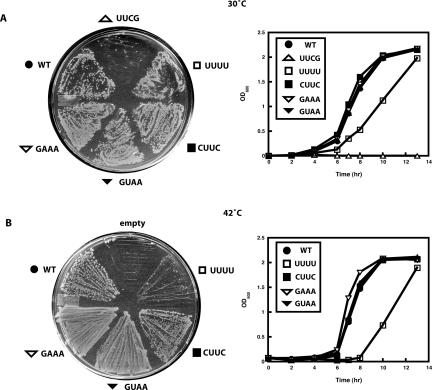
The above results show that in vitro, the tetraloop of 4.5S RNA regulates the GTP hydrolysis activity of the SRP–FtsY complex. Might this activity be important physiologically? To answer this question, 4.5S RNA tetraloop mutants were tested for complementation of bacterial cells lacking the wild-type 4.5S RNA (Fig. 6). Each mutant was analyzed in E. coli strain S1610, a lambda lysogen in which the endogenous wild-type 4.5S RNA allele is maintained and expressed at 30°C, but deleted at 42°C (Brown and Fournier 1984). The 4.5S RNA mutants were expressed under the control of the native 4.5S RNA promoter (Brown et al. 1989; Batey et al. 2000). When transformants expressing each tetraloop mutant were tested at 30°C, all except UUCG and UUUU grew similarly to wild type both on solid media and in LB liquid culture (Fig. 6A). In contrast to wild-type cells, transformants expressing the UUCG 4.5S RNA mutants produced relatively fewer and smaller colonies when grown on agar plates (Fig. 6A). No growth was observed for cells expressing the UUCG mutant in LB liquid media, whereas cells expressing the UUUU mutant grew slower than wild type (Fig. 6A). In contrast to a previous study (Jagath et al. 2001), the growth curve at 30°C observed here reveals that the UUCG allele of 4.5S RNA leads to cell death despite the presence of the wild-type 4.5S RNA (Brown and Fournier 1984). Because the UUCG tetraloop variant has a dominant lethal phenotype, it was not tested for complementation of cells after removal of the wild-type version of the 4.5S RNA gene.
FIGURE 6.
In vivo complementation with 4.5S RNA mutants. E. coli strain S1610 transformed with plasmids encoding wild-type (•),UUCG (Δ), CUUC (▪), UUUU (□), GUAA (▾), and GAAA (∇) 4.5S RNA were grown at (A) 30°C and (B) 42°C. E. coli transformed with the specified 4.5S RNA construct was streaked onto an agar plate with ampicillin at 30°C. Liquid cultures of the specified 4.5S RNA constructs grown at 30°C were streaked onto an agar plate with ampicillin at 42°C. Growth of the E. coli harboring wild-type, UUCG, UUUU, CUUC, GUAA, and GAAA 4.5S RNA in LB media with ampicillin was monitored over time at the designated temperatures. No data were available for the UUCG mutant at 42°C since there was no growth observed at 30°C. These data are representatives of three independent experiments.
When tested at 42°C, a condition in which the chromosomal (wild-type) 4.5S RNA gene is lost, cells expressing each of the four remaining 4.5S RNA mutants, respectively, showed similar growth relative to wild type in all but one case (Fig. 6B). Cells expressing the UUUU tetraloop mutant produced very few colonies at 42°C on solid media and grew slowly in liquid (LB) media (Fig. 6B). Hence, all of the tetraloop mutants tested can complement 4.5S RNA deletion in vivo except for UUCG, and the UUUU mutant exhibits a slow-growth phenotype. In comparison, a previous study (Jagath et al. 2001) showed that both UUUU and CUUC mutants grew slower than wild type even though the UUUU mutant was the only one that showed a loss of binding to FtsY. Taken together with the biochemical and structural data presented above, these complementation data indicate that an essential in vivo function of 4.5S RNA is to stimulate GTP hydrolysis by the SRP–FtsY complex above a critical threshold.
DISCUSSION
A physiologically essential role of SRP RNA is to stimulate GTP hydrolysis by the SRP–receptor complex
The signal recognition particle undergoes regulated cycles of GTP-dependent receptor binding and hydrolysis that are integral to its function in cotranslational targeting of hydrophobic proteins to the translocon. Analysis of the activities and structural properties of SRP RNA tetraloop mutants in vitro and in vivo reveals that a physiologically essential function of the RNA is to stimulate the rate of GTP turnover in SRP–receptor complexes. 4.5S RNA variants containing mutations in the functionally critical tetraloop sequence have varying abilities to stimulate GTP hydrolysis in SRP–FtsY complexes. Two mutants, UUCG and UUUU, abolish RNA enhancement of GTP hydrolysis completely, whereas other mutants produce intermediate effects. Importantly, the ELISA experiment shows that SRP containing the UUCG mutant forms a stable complex with FtsY at concentrations well below those used to measure defective GTPase activity. This supports the conclusion that the differences in GTPase activities of the different tetraloop mutant-containing complexes are not caused by defects in complex formation. Instead, changes in tethered hydroxyl radical cleavage patterns provide evidence that the structures of SRP–FtsY complexes containing 4.5S RNA tetraloop mutants are perturbed. In vivo SRP RNA complementation results correlate with the ability of each 4.5S RNA variant to stimulate GTP hydrolysis by SRP–FtsY complexes in vitro. These in vivo results differ from previous in vivo data, perhaps due to the nonendogenous promoter used, and hence potentially higher level of 4.5S RNA expression induced, in the prior work (Jagath et al. 2001). Together, the results presented here suggest that a threshold rate of GTP hydrolysis by the SRP–receptor complex, conferred by the 4.5S RNA tetraloop, is required for cell viability. Furthermore, the threshold may be rather narrow; a twofold defect in GTPase rate is tolerated without obvious effect on cell growth, while a sixfold effect, equivalent to the absence of 4.5S RNA, is lethal.
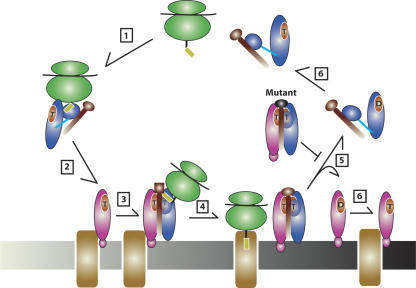
The dominant lethal phenotype of the 4.5S RNA variant containing a UUCG tetraloop may result from a combination of diminished GTPase stimulation in the SRP–receptor complex and the unusual stability of the UUCG tetraloop itself. RNA hairpins containing UUCG versus GAAA tetraloops have _Tm_s of 71.7°C and 65.9°C, respectively (Antao et al. 1991), suggesting that the UUCG mutant may have a significantly increased half-life relative to the wild type or other loop variants of 4.5S RNA. The natural three- to fivefold excess of 4.5S RNA over Ffh (Jensen and Pedersen 1994) and the high affinity of the 4.5S RNA–Ffh interaction (Batey and Doudna 2002) would effectively trap signal recognition particles containing highly stable UUCG mutant RNA in dysfunctional FtsY-bound complexes (Fig. 7).
FIGURE 7.
Model of the mechanism of 4.5S RNA in E. coli SRP-dependent protein translocation. Step 1, cytoplasmic SRP bound with GTP is loaded with a RNC containing a signal peptide. Step 2, the RNC is translocated to the inner membrane by SRP binding to its receptor (FtsY) bound with GTP. Step 3, interaction of SRP with FtsY induces a conformational rearrangement in the SRP as previously observed (Spanggord et al. 2005). Presence of the signal peptide in the M domain perturbs interaction of the tetraloop of 4.5S RNA and the Ffh–FtsY heterodimer (indicated by the box) and inhibits GTP hydrolysis. Step 4, signal peptide release from the SRP and docking of the RNC with translocon. Step 5, tetraloop of 4.5S RNA interacts with the G domains of Ffh and FtsY and stimulates GTP hydrolysis of the SRP–FtsY complex and dissociation of SRP and FtsY. Tetraloop mutants (black oval) inhibit GTP hydrolysis and prevent recycling of SRP and FtsY. Step 6, exchange of GDP to GTP for SRP and FtsY and primed for a new round of protein translocation.
Differential effects of 4.5S RNA mutants suggest roles for both RNA structure and sequence in SRP function
Our data show that the tetraloop of SRP RNA functions to stimulate the catalytic activity of the SRP–receptor complex to a physiologically useful level. What features of the RNA tetraloop may achieve this rate enhancement? The conserved tetraloop in bacteria is a GNRA tetraloop (Rosenblad et al. 2003). Comparison of GNRA and UUCG tetraloop structures show that the loop conformations are different, possibly contributing to the differences in GTPase enhancement and hydroxyl radical probing that were observed (Cheong et al. 1990; Allain and Varani 1995; Batey et al. 2000; Ennifar et al. 2000). This suggests that a GNRA tetraloop conformation is necessary for 4.5S RNA to enhance GTP hydrolysis activity of the SRP–FtsY complex in E. coli. In addition, the tetraloop sequence is needed for maximum hydrolysis activity. Both GAAA and GUAA tetraloops have the GNRA conformation (Jucker et al. 1996; Butcher et al. 1997; Batey et al. 2000), but stimulated GTPase activities of the SRP–FtsY complexes by threefold, compared to the sixfold stimulation conferred by the wild-type tetraloop (GGAA). Both of these variant 4.5S RNAs can complement wild-type 4.5S RNA deletion in vivo. Other 4.5S RNA tetraloop mutants including UUAA, GUUA, GGUU, UGAA, GGUA, and GGAU also have no obvious in vivo growth defects, despite likely differences in some of the tetraloop conformations relative to wild-type 4.5S RNA (data not shown; Heus and Pardi 1991; Jucker et al. 1996).
There is also evidence suggesting the stem closing the tetraloop may be important in stimulating GTP hydrolysis of the SRP–FtsY complex. A mutant recovered in a genetic screen for E. coli variants defective in inner membrane protein insertion contained a change of GG to AA in the 4.5S RNA gene (Tian and Beckwith 2002). These two guanosines participate in the two C-G base pairs that close the tetraloop. Based on the structure of 4.5S RNA bound to the M domain (Batey et al. 2000; Batey et al. 2001), it is unlikely that this mutation affects the binding of 4.5S RNA to Ffh. Instead, these mutations in the stem may destabilize the tetraloop (Williams and Hall 2000), possibly leading to changes in the loop structure and loss of stimulation in GTP hydrolysis. The hydroxyl radical probing of the tetraloop mutants also hints at the possibility that other regions near the tetraloop also affect GTP hydrolysis. Probing of mutant RNAs with proteins that were BABE-Fe modified at residues E153 of Ffh and A359 of FtsY showed a loss of cleavage at residues 47–44 relative to wild type, suggesting that positions of these residues relative to the BABE-Fe moiety have changed relative to the wild-type tetraloop. Intriguingly, a recent crystal structure of the Ffh–FtsY heterodimer revealed a GMP bound at the heterodimer interface where the 4.5S RNA is predicted to bind, suggesting a possible site of interaction for nucleotides in or near the 4.5S RNA tetraloop (Focia et al. 2006).
The role of 4.5S RNA in SRP-dependent translocation
The data presented here strongly suggest that a key physiological role of 4.5S RNA in E. coli is to stimulate GTP hydrolysis of the SRP–FtsY complex. Such a rate enhancement presumably maintains the necessary rate of SRP cycling required for normal cell growth, but how is the enhancement triggered? During cotranslational protein targeting, SRP binds to a translating ribosome (RNC) bearing an N-terminal signal sequence in the nascent peptide (Fig. 7). Structures of Ffh bound to 4.5S RNA show the signal sequence binding site of Ffh adjacent to the RNA tetraloop (Batey et al. 2000; Rosendal et al. 2003), and in the SRP–FtsY complex model they both abut the GTPase center of the heterodimer (Spanggord et al. 2005). Hence, it is possible that the presence of a signal peptide inhibits GTP hydrolysis of the SRP–FtsY complex by perturbing the interaction of the tetraloop with the GTPase domains of the Ffh–FtsY heterodimer. This would prevent premature GTP hydrolysis by the SRP–FtsY complex, ensuring that only SRP–FtsY complexes loaded with RNCs proceed through the cycle and that SRP dissociates from FtsY only after successful delivery of an RNC to the translocon. The observed decreases in GTPase activity of SRP–FtsY complexes containing 4.5S RNA tetraloop mutations presented in this study imply that only subtle perturbation of the tetraloop by the signal peptide would be necessary to inhibit hydrolysis. Thus, the release of signal peptide from the SRP–FtsY complex could trigger the 4.5S RNA tetraloop to enhance GTP hydrolysis by the complex.
In addition to stimulating GTP hydrolysis, 4.5S RNA may also function in vivo to enhance the binding rates of SRP to FtsY as suggested by the 200-fold increase of the on and off rates of Ffh to FtsY in the presence of the RNA (Peluso et al. 2000, 2001). Thus, 4.5S RNA would enhance the rate of ribosomes delivered to the inner membrane and recycling of SRP and FtsY in vivo. Whether the tetraloop itself is responsible for the observed kinetic effects remains to be tested.
The results presented here reveal an essential in vivo role of the SRP RNA tetraloop as an enhancer, stimulating the GTPase activity of the SRP–receptor complex to a physiologically critical level. This explains a central role of RNA in cotranslational protein targeting and expands the functional repertoire of RNA.
MATERIALS AND METHODS
Cloning and purification of 4.5S RNA tetraloop mutants
For the complementation in vivo experiments, the plasmid encoding the full-length wild-type 4.5S RNA was used as a template for QuickChange mutagenesis (Stratagene) to generate the different tetraloop mutants (Brown et al. 1989; Batey et al. 2000). For 4.5S RNA purification, a puc19 plasmid with an insert containing a T7 promoter followed by the hammerhead ribozyme, 4.5S RNA gene, and HDV ribozyme was used. This plasmid was also used to create all tetraloop mutants by QuickChange mutagenesis. Transcription and purification of the wild-type and mutant 4.5S RNA were done as described previously (Batey et al. 2001). The concentration of 4.5S RNA was determined with a spectrophotometer at a wavelength of 260 nm and the extinction coefficient of 1.0×106 L/mol·cm (Integrated DNA Technologies). All 4.5S RNAs were stored in water at −20°C. Prior to use, each 4.5S RNA was folded by resuspending it in a buffer with 1.5 mM [Mg2+], heated to 95°C for 1 min, and then cooled on ice for 1 min.
Filter binding assay
Binding assays were performed as previously described (Batey and Doudna 2002). Less than 2 pM of 5′-γ-32P-end labeled 4.5S RNA were incubated for 1 h at room temperature with varying concentrations of Ffh protein before applying to the membranes. Data were quantitated using ImageQuant (Molecular Dynamics) software.
GTPase assay
The assay was performed similarly to that previously described (Peluso et al. 2001; Spanggord et al. 2005). Briefly, for wild type, CUUC, and GUAA, 0.2 μM of Ffh and 0.4 μM of the respective 4.5S RNA were used to form 0.2 μM of SRP, whereas for GAAA, UUUU, and UUCG, 2 μM of Ffh and 4 μM of 4.5S RNA were used to form 2 μM of SRP. In reactions without 4.5S RNA, only 1 μM of Ffh was used. The concentrations of FtsY (amino acids 47–498) were 1, 2, 5, 10, 15, 20, and 30 μM. The calculated rate constants were normalized to the concentrations of SRP and Ffh, respectively. The GTPase rates reported are averages of at least three separate experiments.
Enzyme-linked immunosorbent assay (ELISA)
Ffh and FtsY (residues 47–498) were purified as previously described (Peluso et al. 2001; Spanggord et al. 2005). GMPPNP was purchased from Sigma and purified as previously mentioned (Karbstein et al. 2005). For the FtsY titration experiment, 100 nM of Ffh and 50 nM of either wild-type or UUCG 4.5S RNA were initially incubated for 5 min at room temperature to ensure SRP formation, then 0.5 mM GMPPNP and varying concentrations of FtsY were added and incubated for another 10 min to allow binding of SRP to FtsY. The buffer condition for the reaction was 50 mM HEPES (pH 7.5), 150 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM DTT, and 0.01% Nikkol. All incubations were done at room temperature. SRP–FtsY complexes were added to a 96-well plate (Nalge Nunc International USA) and incubated for 1 h. The complexes were removed and 3% (w/v) of BSA (Sigma) in PBS was added and incubated for 1 h. After removing the BSA, scFv IA02 (complex specific) or IB11 (Ffh or SRP specific) in PBS was added and incubated for 1 h. scFv IA02 and IB11 were used at dilutions of 1:10 and 1:100, respectively, from a stock of 100 ng/μL. The antibodies were removed and each well was washed three times with 100 μL of PBS-0.1% Tween. Protein-L peroxidase conjugated (Pierce) was added at a dilution of 1:1000 from a 0.5 mg/mL stock in PBS and incubated for 1 h. Each well was washed three times with PBS-0.1% Tween before 50 μL of TMB substrate (Pierce) were added. The substrate was incubated for 20 min and the reaction was stopped with the addition of 1 M sulfuric acid. The absorbance at 450 nm of each well was measured with a Spectra MAX 190 plate reader (Molecular Devices). The fraction of SRP bound with FtsY was determined by the absorbance of IA02 divided by the sum of the absorbances of IA02 and IB11.
Hydroxyl radical probing
All RNAs in this study were 5′-γ-32P-end labeled by incubating them with 40 units of T4 poylnucleotide kinase (NEB) in 10× reaction buffer (500 mM Imidazole-Cl at pH 7.3; 180 mM MgCl2; 50 mM DTT; 1 mM spermine; 1 mM EDTA). In addition, the samples were supplemented with PEG-8000 (6% final concentration). Labeling reactions were allowed to proceed for 1.5 h at 37°C. 5′-end labeled RNAs were phenol/chloroform extracted and ethanol precipitated and purified on a 12% denaturing gel.
Affinity cleaving assays were performed as previously described (Spanggord et al. 2005). Briefly, SRP–FtsY complexes were preformed as above except that the binding reactions were spiked with 5′-γ-32P-end labeled 4.5S RNA. After incubation as above, cleavage of the RNA was initiated by the addition of hydrogen peroxide and sodium ascorbate [final concentration of 0.01% (v/v) and 5 mM, respectively], followed by a 2-min incubation at room temperature. Distilled water was added to each sample, followed by two successive phenol extractions and ethanol precipitation of the RNA. Cleaved 4.5S RNA was resuspended in formamide loading dye and analyzed on a 12% denaturing gel. Data were obtained from the gels using storage phosphor autoradiography and a STORM PhosphorImager (Molecular Dynamics).
Quantitations of specific nucleotide positions were done using the ImageQuant software. To account for loading differences between lanes, the intensity of a cleaved nucleotide position is normalized to the intensity of a noncleaved nucleotide position within the same lane. The fold change for a specific nucleotide position is the normalized intensity in the presence of GMPPCP or cleaving reagents divided by its normalized intensity in the absence of GMPPCP or cleaving reagents.
In vivo complementation
E. coli strain S1610 (Brown et al. 1989) harboring the 4.5S RNA gene on a lambda prophage was transformed with a plasmid encoding either the wild-type or mutant 4.5S RNA. After transformation, the bacteria culture was grown on an ampicillin (50 mg/mL) agar plate at 30°C overnight. A single colony was picked and grown in 5 mL of LB media containing ampicillin for about 10 h at 30°C. In the case of tetraloop mutant UUCG, several small colonies were used since these were the only colonies that grew from the transformation. Bacteria from these cultures were used for the solid media growth assay at 30°C and 42°C. The cell density of the 5 mL culture was determined by measuring the optical density at a wavelength of 600 nm. To ensure that the same amount of bacteria between the different constructs was used for the growth curve assay, the density of each of the 5 mL cultures was diluted to the same optical density. Then 50 μL of the diluted culture were added to 10 mL of fresh LB media containing ampicillin to start the growth curve assay. Cultures were grown in parallel at 30°C and 42°C, and 1 mL aliquots were removed for measurement. The in vivo experiments were repeated at least three times.
ACKNOWLEDGMENTS
We thank Shu-ou Shan for reagents and assistance on the GTPase assay and members of the Doudna laboratory for insightful discussions and help on the manuscript. This work was supported by the N.I.H.
Footnotes
REFERENCES
- Allain, F.H., Varani, G. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- Antao, V.P., Lai, S.Y., Tinoco, I., Jr. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Doudna, J.A. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry. 2002;41:11703–11710. doi: 10.1021/bi026163c. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Lucast, L., Rha, B., Doudna, J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Sagar, M.B., Doudna, J.A. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- Brown, S., Fournier, M.J. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- Brown, S., Thon, G., Tolentino, E. Genetic selection and DNA sequences of 4.5S RNA homologs. J. Bacteriol. 1989;171:6517–6520. doi: 10.1128/jb.171.12.6517-6520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz, I., Peske, F., Wieden, H.J., Gryczynski, I., Rodnina, M.V., Wintermeyer, W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J. Mol. Biol. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Butcher, S.E., Dieckmann, T., Feigon, J. Solution structure of the conserved 16 S-like ribosomal RNA UGAA tetraloop. J. Mol. Biol. 1997;268:348–358. doi: 10.1006/jmbi.1997.0964. [DOI] [PubMed] [Google Scholar]
- Cheong, C., Varani, G., Tinoco, I., Jr. Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A., Batey, R.T. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- Egea, P.F., Shan, S.O., Napetschnig, J., Savage, D.F., Walter, P., Stroud, R.M. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Nikulin, A., Tishchenko, S., Serganov, A., Nevskaya, N., Garber, M., Ehresmann, B., Ehresmann, C., Nikonov, S., Dumas, P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- Focia, P.J., Shepotinovskaya, I.V., Seidler, J.A., Freymann, D.M. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focia, P.J., Gawronski-Salerno, J., Coon, J.S.T., Freymann, D.M. Structure of a GDP:AlF4 complex of the SRP GTPases Ffh and FtsY, and identification of a peripheral nucleotide interaction site. J. Mol. Biol. 2006;360:631–643. doi: 10.1016/j.jmb.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, S.Q., Jockel, J., Beinker, P., Warnecke, J., Semenkov, Y.P., Rodnina, M.V., Wintermeyer, W. Conformation of 4.5S RNA in the signal recognition particle and on the 30S ribosomal subunit. RNA. 2005;11:1374–1384. doi: 10.1261/rna.7219805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus, H.A., Pardi, A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Jagath, J.R., Matassova, N.B., de Leeuw, E., Warnecke, J.M., Lentzen, G., Rodnina, M.V., Luirink, J., Wintermeyer, W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, C.G., Pedersen, S. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: The stability of Ffh protein is dependent on the concentration of 4.5S RNA. J. Bacteriol. 1994;176:7148–7154. doi: 10.1128/jb.176.23.7148-7154.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, S., Noller, H.F. Directed hydroxyl radical probing using iron(II) tethered to RNA. Methods Enzymol. 2000;318:175–190. doi: 10.1016/s0076-6879(00)18052-8. [DOI] [PubMed] [Google Scholar]
- Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H., Pardi, A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- Karbstein, K., Jonas, S., Doudna, J.A. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Lentzen, G., Moine, H., Ehresmann, C., Ehresmann, B., Wintermeyer, W. Structure of 4.5S RNA in the signal recognition particle of Escherichia coli as studied by enzymatic and chemical probing. RNA. 1996;2:244–253. [PMC free article] [PubMed] [Google Scholar]
- Peluso, P., Herschlag, D., Nock, S., Freymann, D.M., Johnson, A.E., Walter, P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- Peluso, P., Shan, S.O., Nock, S., Herschlag, D., Walter, P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- Powers, T., Walter, P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- Rosenblad, M.A., Gorodkin, J., Knudsen, B., Zwieb, C., Samuelsson, T. SRPDB: Signal recognition particle database. Nucleic Acids Res. 2003;31:363–364. doi: 10.1093/nar/gkg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendal, K.R., Wild, K., Montoya, G., Sinning, I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar, M.B., Lucast, L., Doudna, J.A. Conserved but nonessential interaction of SRP RNA with translation factor EF-G. RNA. 2004;10:772–778. doi: 10.1261/rna.5266504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, S.O., Walter, P. Induced nucleotide specificity in a GTPase. Proc. Natl. Acad. Sci. 2003;100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, S.O., Walter, P. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 2005a;579:921–926. doi: 10.1016/j.febslet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Shan, S.O., Walter, P. Molecular crosstalk between the nucleotide specificity determinant of the SRP GTPase and the SRP receptor. Biochemistry. 2005b;44:6214–6222. doi: 10.1021/bi0500980. [DOI] [PubMed] [Google Scholar]
- Shan, S.O., Stroud, R.M., Walter, P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepotinovskaya, I.V., Focia, P.J., Freymann, D.M. Crystallization of the GMPPCP complex of the NG domains of Thermus aquaticus Ffh and FtsY. Acta Crystallogr. 2003;59:1834–1837. doi: 10.1107/s0907444903016573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanggord, R.J., Siu, F., Ke, A., Doudna, J.A. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat. Struct. Mol. Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- Tian, H., Beckwith, J. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 2002;184:111–118. doi: 10.1128/JB.184.1.111-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, D.J., Hall, K.B. Experimental and computational studies of the G[UUCG]C RNA tetraloop. J. Mol. Biol. 2000;297:1045–1061. doi: 10.1006/jmbi.2000.3623. [DOI] [PubMed] [Google Scholar]
- Wu, S., Ke, A., Doudna, J.A. A fast and efficient procedure to produce scFvs specific for large macromolecular complexes. J. Immunol. Methods. 2006 doi: 10.1016/j.jim.2006.10.005. ePub # 17126854. [DOI] [PMC free article] [PubMed] [Google Scholar]