Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma (original) (raw)
Abstract
The cancer stem cell hypothesis suggests that malignant growth depends on a subset of tumor cells with stem cell-like properties of self-renewal. Because hedgehog (Hh) signaling regulates progenitor cell fate in normal development and homeostasis, aberrant pathway activation might be involved in the maintenance of such a population in cancer. Indeed, mutational activation of the Hh pathway is associated with medulloblastoma and basal cell carcinoma; pathway activity is also critical for growth of other tumors lacking such mutations, although the mechanism of pathway activation is poorly understood. Here we study the role and mechanism of Hh pathway activation in multiple myeloma (MM), a malignancy with a well defined stem cell compartment. In this model, rare malignant progenitors capable of clonal expansion resemble B cells, whereas the much larger tumor cell population manifests a differentiated plasma cell phenotype that pathologically defines the disease. We show that the subset of MM cells that manifests Hh pathway activity is markedly concentrated within the tumor stem cell compartment. The Hh ligand promotes expansion of MM stem cells without differentiation, whereas the Hh pathway blockade, while having little or no effect on malignant plasma cell growth, markedly inhibits clonal expansion accompanied by terminal differentiation of purified MM stem cells. These data reveal that Hh pathway activation is heterogeneous across the spectrum of MM tumor stem cells and their more differentiated progeny. The potential existence of similar relationships in other adult cancers may have important biologic and clinical implications for the study of aberrant Hh signaling.
Keywords: cancer stem cells, cancer therapy, cyclopamine
The hedgehog (Hh) signaling pathway is a highly conserved system for the regulation of patterning and progenitor cell fates in animal development (1). In mammals, three Hh ligands (sonic, Indian, and desert) mediate signaling through morphogen gradients as dual lipid-modified signaling peptides (2, 3). Without ligand, the predominant Hh receptor patched1 (Ptch1) maintains tonic inhibition of Hh signaling through catalytic inhibition of smoothened (Smo) (4), a seven-transmembrane domain protein essential for Hh pathway activation (5). Smo is the molecular target of cyclopamine, a plant-derived alkaloid that is a naturally occurring Hh pathway antagonist (6, 7). Activation of Smo results in activation of the Gli transcription factors that trigger the Hh transcriptional program, including up-regulation of Ptch1 and Gli1 expression (2, 5). In the developing cerebellum, granule cell precursors require sonic Hh (Shh) derived from specialized Purkinje cells for their proliferation during the perinatal period (8). Similarly, the embryonic and adult skin requires the pathway to maintain and expand its progenitor compartment (8). Loss of function mutations in PTCH1 or gain of function mutations in SMO are associated with medulloblastoma and basal cell carcinoma (9, 10). Germ-line mutations in PTCH1 (Gorlin syndrome) result in a marked propensity for both of these cancers, which, along with several well characterized mouse models, show that mutational activation of Hh signaling is sufficient to drive oncogenesis in organs that depend on this pathway for their development (10, 11). Moreover, an ongoing requirement for Hh pathway activation in medulloblastoma is revealed by the effectiveness of cyclopamine and other Smo antagonists as potential tumor therapies in mouse models (12, 13).
Accumulating evidence from several groups suggests that aberrant Hh signaling is a feature of some lung (14–16), foregut (17, 18), and prostate (19, 20) cancers without a requirement for mutations in PTCH1 or SMO. The mechanism of abnormal Hh pathway activation in such “non-Gorlin” tumors remains unclear; however, growth of a significant proportion of these cancers is inhibited by cyclopamine, suggesting that events leading to SMO activation contribute to the malignant phenotype of these tumors (21). Genetic mouse models suggest that Hh signaling may contribute to the initiation and expansion of an aberrant progenitor population (22, 23), which is then maintained in cancer as a tumor “stem cell.” This idea is consistent with the hierarchical theory of cancer stem cells, in which a small number of poorly differentiated, clonally competent progenitors maintain growth and self-renewal of the tumor (8, 22).
To explore the hypothesis that Hh signaling regulates tumor stem cell fate in non-Gorlin cancers, we turned to multiple myeloma (MM), a plasma cell malignancy of the bone marrow, as a model. MM is characterized by abnormal proliferation of plasma cells that express the surface marker CD138 and typically synthesize a monoclonal, light chain-restricted antibody (24). We have shown previously that MM contains two distinct populations, a small number of CD138neg CD19+ stem cells that resemble memory B cells and a large majority of malignant CD138+ CD19neg terminally differentiated plasma cells (25). The CD138neg cells are highly efficient at forming colonies from single cells that give rise to CD138+ progeny and can initiate MM following transplantation into nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (25). By contrast, CD138+ cells have limited clonal potential in cell lines and no clonal potential when purified from bone marrow aspirates from MM patients (25). In this work, we explore the importance of Hh signaling in tumor stem cell biology by using MM as a model. In a subset of both MM cell lines and clinical samples, we demonstrate a marked asymmetry in the level of Hh pathway component expression, Hh reporter activity, and cyclopamine sensitivity between the stem cell and differentiated tumor cell compartments of this tumor. Inhibition of SMO function suggests that Hh signaling is required to maintain MM stem cells in an undifferentiated, clonally proliferative state. This paradigm could form the basis for investigating aberrant Hh signaling in other tumors.
Results and Discussion
Hh Pathway Expression and Activity in MM Cell Lines.
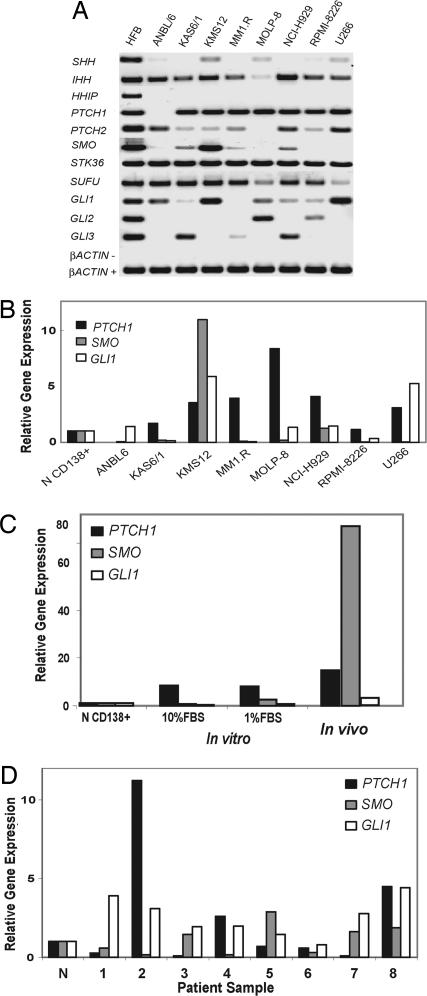
Using a conventional approach to studying Hh signaling in tumors (14, 17), we assayed transcript levels of Hh pathway components in MM cell lines conditioned to grow in low-serum conditions. We included two cell lines in which we have identified stem cell populations, NCI-H929 and RPMI 8226 (25). Qualitative RT-PCR analysis demonstrates Hh pathway component expression was variable across a range of MM cell lines (Fig. 1A). NCI-H929 cells coexpressed INDIAN HEDGEHOG (IHH), PTCH1, SMO, and GLI1. In addition, all of the MM cell lines lacked expression of HHIP, a secreted Hh ligand antagonist that is epigenetically silenced in pancreatic cancer (26). This expression pattern resembles that of other cancers in which Hh signaling may play an important role (14, 17). By contrast, RPMI 8226 cells lack expression of SMO, which is essential for Hh signaling (27), and should therefore render this cell line Hh ligand independent (Fig. 1A). Quantitative expression analysis demonstrated variable up-regulation of PTCH1, SMO, and GLI1 in MM cell lines when compared with normal human CD138+ cells derived from healthy donor bone marrow (Fig. 1B). Both NCI-H929 and KMS-12 lines exhibited up-regulation of SMO (Fig. 1B).
Fig. 1.
Expression profiling of the Hh pathway in MM. (A) Qualitative RT-PCR analysis of Hh pathway gene expression in MM cell lines, and human fetal brain (HFB) used as a positive control. β_ACTIN_− denotes PCR results from mock cDNA synthesis reactions lacking RT. (B) Quantitative real-time RT-PCR analysis of PTCH1, SMO, and GLI1 gene expression in MM cell lines relative to expression in normal bone marrow CD138+ cells (N CD138+). (C) Quantitative RT-PCR analysis of PTCH1, SMO, and GLI1 gene expression in GFP-labeled NCI-H929 cells grown in marrow of NOD/SCID mice. Bony lesions were dissected from transplanted mice by using a fluorescence microscope. Expression is shown relative to that in normal bone marrow normal CD138+ cells (N CD138+) and cells cultured in 1% and 10% FBS. (D) Quantitative real-time RT-PCR analysis of gene expression in CD138+ bone marrow cells from patients with MM relative to expression in normal bone marrow CD138+ cells (N).
Hh Pathway Expression in MM in Vivo.
To address the effect of culture conditions and the tumor microenvironment on the Hh pathway, NCI-H929 cells were labeled with retroviral green fluorescent protein (GFP) expression vector and injected intravenously into sublethally irradiated NOD/SCID mice. Osteolytic engraftment is detected by fluorescent lesions in the spine and skull [supporting information (SI) Fig. 5_A_], characteristic MM histopathology (SI Fig. 5_B_), and cells strongly immunoreactive for GFP (SI Fig. 5_C_). This model is similar to that described by using RPMI 8226 cells in NOD/SCID mice (28). NCI-H929-derived tumors expressed markedly increased levels of SMO when compared with cultured MM cells (Fig. 1C). In addition, we observed significant alterations in Hh pathway gene expression in response to changes in serum concentration in cultured cells (Fig. 1C). These data reveal that the cell microenvironment markedly affects the expression of Hh pathway components. However, we observed only modest up-regulation of SMO, GLI1, and PTCH1 (Fig. 1D) in bone marrow cells from MM patients purified on the basis of CD138 expression. To explain this discrepancy, we considered the possibility that CD138+ bone marrow cells from MM patients lack a pathway active population.
Hh Pathway Expression and Activity in MM Stem Cells.
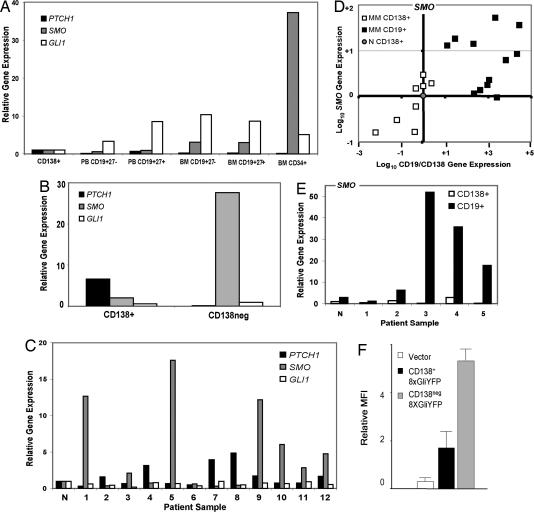
Our earlier studies demonstrated the existence of highly clonogenic CD19+ CD138neg population in MM cell lines and primary bone marrow samples from MM patients (25). We hypothesized that this tumor stem cell population, lacking in clinical CD138+ bone marrow samples, was the locus of Hh pathway activation in MM. Consistent with previous reports (29, 30), quantitative analysis of Hh pathway transcripts in normal human bone marrow populations revealed that CD34+ bone marrow progenitors expressed the highest levels of SMO (Fig. 2A), but showed relatively low expression of PTCH1. In contrast, differentiated cells of the B lymphocyte lineage down-regulated SMO expression in favor of increasing levels of PTCH1 (Fig. 2A). NCI-H929 MM cells sorted by CD138 expression revealed a marked asymmetry in gene expression, with the highest levels of SMO seen in the CD138neg population (Fig. 2B). Although GLI1 expression was evenly distributed, PTCH1 expression was a prominent feature of the CD138+ tumor subset, resembling the pattern seen in normal CD138+ cells.
Fig. 2.
Hh pathway activation in MM. (A) Quantitative expression analysis of PTCH1, SMO, and GLI1 in normal human bone marrow (BM) and peripheral blood (PB) cells normalized to expression levels in normal bone marrow CD138+ cells. (B) Quantitative real-time RT-PCR analysis of Hh pathway gene expression in NCI-H929 cells, flow sorted into CD138+ and CD138neg populations. (C) Quantitative RT-PCR analysis of PTCH1, SMO, and GLI1 expression in purified CD19+ cells from MM patient bone marrow relative to expression in normal bone marrow CD19+ cells (N). (D) Scatter plot of gene expression in purified CD19 or CD138+ bone marrow cells from MM patients, shown as the ratio of CD19 to CD138 gene expression on the x axis and the level of SMO expression on the y axis. (E) Relative expression of SMO in paired CD19+ and CD138+ cell samples obtained from patients with MM. Expression is shown relative to expression in normal CD138 cells. (F) GLI-responsive YFP reporter activity in NCI-H929 cells cotransfected with a CMV-RFP control plasmid and stained for CD138. Mean fluorescence intensity (MFI) for YFP in RFP-positive cells is shown for CD138+ and CD138neg fractions (n = 3).
The B cell compartment of MM patients contains highly clonogenic, light chain-restricted MM stem cells and a variable fraction of residual normal B cells that lack such properties (25). Quantitative transcript analysis of CD19+ CD138neg cells from MM bone marrow aspirates reveals prominent up-regulation of SMO mRNA in many patients when compared with normal bone marrow CD19+ cells (Fig. 2C). Although consistent with the MM cell line data, we cannot exclude the possibility that up-regulation of Hh pathway activity may be occurring in normal B cells, activated as part of a humoral immune response. Similarly, we cannot rule out the possibility that progression of MM leads to increased representation of MM B cell progenitors in the CD19+ compartment, which then contributes to increased levels of Hh pathway transcripts. Despite these variables, our data suggest that up-regulation of SMO expression is a common feature of normal bone marrow stem cells, as well as MM stem cells, in both cell lines and primary tumor samples.
Expression of SMO was then analyzed in CD19+ and CD138+ bone marrow cells from MM patients. To control for the purity of the cell sorts in this experiment, we also measured the expression of CD19 and CD138 mRNA and analyzed the level of SMO expression as a function of the CD19/CD138 ratio in each sample (Fig. 2D). Paired CD19+ and CD138+ purified from the bone marrow of the same patients also revealed that SMO expression was consistently higher in the CD19+ compartment (Fig. 2E). We characterized the CD138neg population as the predominant locus of Hh pathway activation in NCI-H929 cells by using a fluorescence reporter assay (Fig. 2F). In this experiment, the luciferase expression cassette of the 8XGliLuc reporter vector (31) was replaced with cDNA encoding yellow fluorescent protein (YFP). NCI-H929 cells were cotransfected with a vector expressing red fluorescent protein (RFP) under the control of a CMV promoter, and all cells were stained for CD138. FACS analysis of mean fluorescence intensity for YFP was calculated in RFP-expressing cells and segregated according to CD138 expression. Fig. 2F demonstrates that Gli reporter activity was seen predominantly in the CD138neg fraction.
Taken together, these data suggest that SMO expression and activity predominates in the stem cell compartment of MM. However, these data contradict the standard model of Hh signaling in which PTCH1 expression is used as a marker of Hh pathway expression. In contrast, we find that in cells from normal and MM bone marrow, SMO is highly expressed in the progenitor compartment, whereas PTCH expression predominates in more differentiated cells. These data suggest that stem cell populations may express high levels of SMO and inadequate levels of PTCH1, resulting in a significant level of basal pathway activation, which can be further augmented by ligand stimulation. This finding also suggests a mechanism by which organ-specific progenitors might regulate Hh signaling in both homeostasis and cancer.
A Requirement for Hh Signaling in MM Stem Cells.
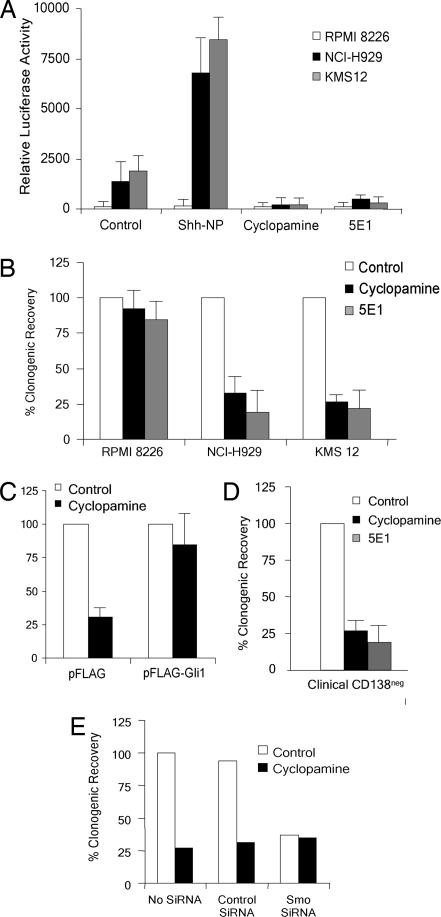
Using an Hh pathway-specific luciferase reporter (31), we next showed that the Hh pathway was active in NCI-H929 and KMS-12 cells (Fig. 3A). This activity could be increased by Hh ligand (ShhNp), as well as inhibited by cyclopamine or the neutralization of Hh ligand with the monoclonal antibody 5E1 (Fig. 3A). By contrast, RPMI 8226 cells, which lack SMO expression, exhibited no significant reporter activity. Activation of Hh signaling by ligand implies that the downstream Hh signaling pathway is intact because pathway activity in PTCH1 mutant cells is ligand-independent (17). Down-regulation of signaling by both cyclopamine and 5E1 implies that constitutive, ligand-driven Hh signaling is present in NCI-H929 and KMS-12 cells (17).
Fig. 3.
Clonogenic capacity of MM cells in vitro. (A) GLI-responsive luciferase reporter activity in three MM cell lines treated with purified, dual lipid-modified Shh ligand (Shh-NP, 4 nM), cyclopamine (5 μM), or 5E1 (10 μg/ml). (B) Clonogenic capacity of MM cell lines treated with cyclopamine (5 μM) or 5E1 (10 μg/ml), expressed as % of control ± SEM (n = 5). (C) Clonogenic capacity of NCI-H929 cells transiently transfected with a control expression vector or a vector overexpressing FLAG-Gli1 followed by cyclopamine treatment, expressed as % of control ± SEM. (D) Clonogenic capacity of CD19+ CD34neg bone marrow cells purified from patients with MM (n = 5) treated with cyclopamine or 5E1, expressed as % of control ± SEM. (E) Clonogenic capacity of NCI-H929 cells treated with combinations of siRNAs and cyclopamine.
In conventional cell growth assays measuring cell number or viability, we observed little or no effect of any of these treatments on the growth of whole MM cell cultures over a 10-day period (data not shown), demonstrating that the majority of these cells were not dependent on Hh signaling for survival or short-term replication. To test the requirement for Hh signaling in tumor self-renewal, we studied three MM cell lines (NCI-H929, KMS-12, and RPMI 8226) with similar clonogenic capacity in vitro (data not shown). Cells were treated with cyclopamine or 5E1 for 3 days, washed free of drug or antibody, and then plated in equal numbers of live cells in cloning assays as described (25). Inhibition of Hh signaling with cyclopamine or 5E1 markedly inhibited the clonal capacity of NCI-H929 and KMS12 cells (Fig. 3B). By contrast, RPMI 8226 cells, which lack SMO expression, were unaffected (Fig. 3B). NCI-H929 cells transiently overexpressing GLI1 were resistant to the inhibitory effects of cyclopamine on clonal growth (Fig. 3C).
The clonogenic capacity of CD19+ CD27+ cells obtained from five different MM patients' bone marrow aspirates was also inhibited by both cyclopamine and 5E1 treatment (Fig. 3D). Because normal CD19+ CD27+ bone marrow cells are not clonogenic in this assay (25), the effect of the Hh pathway blockade is entirely due to effects on MM stem cells, suggesting that Hh signaling is required for the clonal expansion of MM progenitors isolated from the human bone marrow. To further confirm the specificity of our strategies to inhibit Hh signaling, we generated an siRNA directed against SMO, which resulted in a 10-fold reduction in transcript levels in NCI-H929 cells (data not shown). Mock transfection, or transfection with a noncoding siRNA, had no effect on clonogenic growth, whereas the addition of cyclopamine inhibited clonal growth in both cases (Fig. 3E). By contrast, _SMO_-specific siRNA effectively reduced clonogenic capacity, but no additional inhibition was observed with the addition of cyclopamine to these cells (Fig. 3E). These data suggest that, in NCI-929 cells, cyclopamine and SMO siRNA have identical effects on stem cell function. The specificity of cyclopamine is demonstrated by the absence of additional growth effects in cells lacking its molecular target.
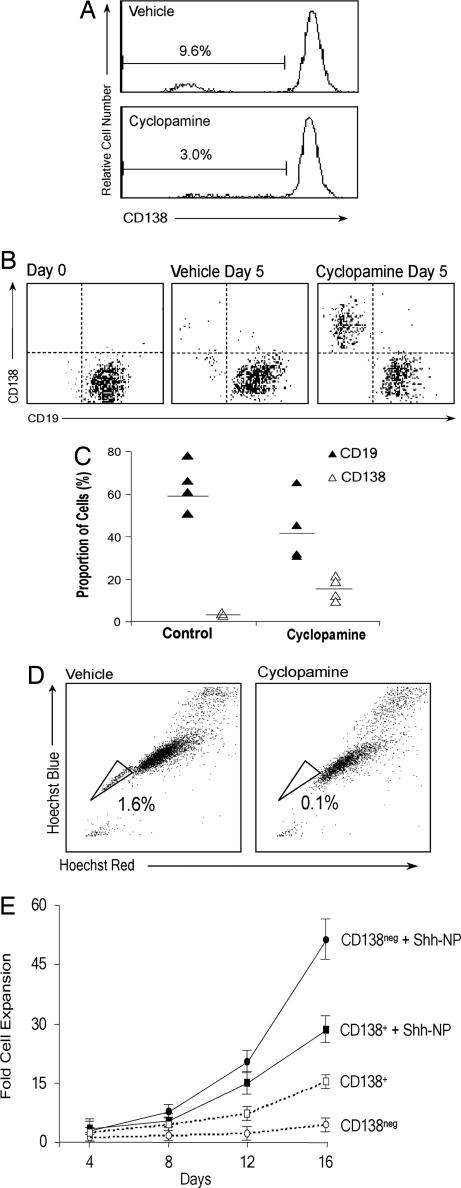
Inhibition of Hh signaling may limit clonal growth of MM by depleting the stem cell population through cell death or differentiation, or it may act to block expansion of a transient amplifying population. To address this issue, we studied the effects of the Hh pathway blockade on the stem cell and plasma cell compartments of MM. Treatment of NCI-H929 cells with cyclopamine resulted in a relative decrease in the CD138neg population (Fig. 4A). Cyclopamine treatment of purified CD19+ CD27+ bone marrow cells from MM patients induced significant plasma cell differentiation, accompanied by a similar depletion of the CD19+ population (Fig. 4 B and C). We recently demonstrated that a Hoechst 33342 negative side population in MM correlates functionally and phenotypically with our previously defined MM stem cell population (25). Depletion of the side population fraction in NCI-H929 (Fig. 4D) and KMS12 (data not shown) cells treated with cyclopamine supports our contention that the MM stem cell compartment is Hh pathway-dependent and mirrors recent similar findings in embryonal brain tumors by using Notch pathway inhibitors (32).
Fig. 4.
Effects of Hh signaling on growth and differentiation of MM stem cells in vitro. (A) FACS analysis of CD138 expression in NCI-H929 cells treated with vehicle or cyclopamine (5 μM). The percentage of CD138+ cells is indicated. (B) Representative FACS analysis of CD19 and CD138 expression in purified MM patient bone marrow cells at initial purification and after 5 days of culture treated with vehicle or cyclopamine (5 μM). (C) Graphical data from five patient samples as described in B. (D) FACS analysis of Hoechst 33342-stained MM cells treated with vehicle or cyclopamine (5 μM). The % side population is indicated in each panel. (E) Clonal expansion assay of CD138neg and CD138+ cells from the NCI-H929 cell line treated with ShhNp (4 nM). Data are shown as mean ± SEM (n = 5).
Activation of Hh signaling in CD138neg progenitors isolated from NCI-H929 cells with ShhNp induced a marked expansion of this progenitor population (Fig. 4E). The maintenance, by Hh stimulation, of CD138neg cells through 12 days of continuous growth (SI Fig. 6) is consistent with prior studies demonstrating the capacity of Hh ligand stimulation to expand undifferentiated bone marrow stem cells in vitro (29, 33). Taken together, our results are most consistent with a model in which Hh pathway inhibition depletes the clonogenic fraction in MM through terminal differentiation rather than cytotoxicity, and that activation of Hh signaling has the capacity to expand MM stem cells without triggering terminal differentiation.
Discussion
We have explored the potential role of Hh signaling in cancer stem cell biology by studying MM, a hematologic malignancy with a phenotypically distinct progenitor population (25). This model allowed us to explore how Hh signaling might function in cancers lacking mutations in PTCH1 or SMO that result in clonal deregulation of Hh signaling. In our model, we find that the stem cell compartment expresses SMO and GLI1, but little PTCH1, whereas the differentiated plasma cell compartment appears to gain PTCH1 and IHH expression at the expense of SMO. These findings are supported by recent studies identifying high-level expression of Hh pathway components in benign and malignant breast epithelial progenitors (34), although these authors found coexpression of SMO, PTCH1, GLI1, and IHH within the tumor-initiating spheres, perhaps reflecting a more heterogeneous progenitor population than in the MM stem cell model. Our data also suggest that, in stem cell populations, down-regulation of PTCH1 in the presence of high-level SMO expression renders such cells sensitive to Hh ligand and results in pathway activation. Consistent with this idea is our demonstration that Hh ligand can markedly expand the MM stem cell population when delivered exogenously. Because normal bone marrow stromal cells are a potential source of Hh ligand (29, 30), it is also possible that MM progenitors activate Hh signaling through interactions with the bone marrow microenvironment. Testing this concept in lung, pancreas, and prostate cancers will require definitive identification of clonogenic progenitors. Nevertheless, if our demonstration of Hh pathway activation in the stem cell compartment of a non-Gorlin tumor can be confirmed in these tumors, our findings would have significant implications for the investigation of Hh pathway signaling in cancer, which include: (i) the possibility that Hh pathway activity may be present in a very small percentage of tumor cells, necessitating selection or isolation of tumor cell populations for optimal examination of Hh pathway activity; (ii) the marked effect of the tumor microenvironment on Hh signaling; (iii) the potential need to measure responses to Hh pathway inhibition by measuring clonogenic capacity rather than overall cell growth; (iv) the possibility that PTCH1 transcription may be modulated by mechanisms other than GLI signaling in tumor stem cells; and (v) the potential need to shift the emphasis of preclinical and clinical studies of Hh antagonist therapy toward measures of long-term self-renewal. If SMO antagonists are used to target cancer stem cell self-renewal, the ability to measure Hh pathway activity in a discrete stem cell compartment in vivo would make MM an ideal disease in which to test this concept in the clinic.
Materials and Methods
Animals.
Experiments were conducted in accordance with protocols approved by the Johns Hopkins Institutional Animal Care and Use Committee. Mice were irradiated with 400 rads before tail vein injection of 5 × 106 GFP-labeled NCI-H929 cells.
Cell Biology.
Cell lines were obtained from American Type Tissue Collection except for KMS-12 and MOLP-8 (German Collection of Microorganisms and Cell Cultures), ANBL6, KAS6/1 (gift from Diane Jelinek, Mayo Clinic, Minneapolis, MN), and MM1.R (gift from Nancy Krett and Steven Rosen, Northwestern University, Evanston, IL). Cells were grown in Advanced RPMI (Invitrogen, Carlsbad, CA) with 1% FBS, 10 mM Hepes, and l-glutamine. Flow-sorting of MM cell lines was performed as described (25). Clinical specimens were obtained with written informed consent, approved by the Johns Hopkins Medical Institutes Institutional Review Board, and sorted as described (25). Clonogenic growth assays were performed as described (25). Details are available in SI Materials and Methods. Side population assays were performed as described (35). Detailed methods are available in SI Materials and Methods. Lentiviral EGFP vectors were a gift from Linzhao Chen (The Johns Hopkins University) and were used as described (36).
Immunohistochemistry.
Sections were stained by using the Vectastain ABC system (Vector Laboratories, Burlingame, CA) with the Invitrogen rabbit anti-GFP antibody (A11122).
RT-PCR.
All RNA samples were quality controlled by using a Bio-Rad (Hercules, CA) Experion Bioanalyzer and then subjected to DNase treatment before reverse transcription. Real-time quantitative PCR studies used the Bio-Rad SYBR green system and thermal cycler. All quantitative calculations were performed by using the Δc-t method. Primer sequences are listed in SI Table 1.
Transfections.
Cells were transfected with Gli-responsive and Renilla luciferase expression vectors (pRL-CMV; Promega, Madison, WI) by using the Amaxa Biosystems (Gaithersburg, MD) Nucleofector system and analyzed by using the dual luciferase reporter assay (Promega). A Gli-responsive YFP reporter (p8XGli-YFP) was constructed by replacing the luciferase gene with the coding sequence for YFP cotransfected into NCI-H929 cells with a constitutive RFP expression vector (pMax-RFP; Clontech, Mountain View, CA). FACS analysis was as described (25). siRNAs were electroporated at a concentration of 50 nM. Sequences are available in SI Materials and Methods.
Supplementary Material
Supporting Information
Acknowledgments
We thank Infinity Pharmaceuticals for the gift of cyclopamine, Keith Young for the ShhNp, and Karen McGovern and Julian Adams for helpful discussions. This work was supported by the Commonwealth Foundation, the National Cancer Institute/Specialized Program of Research Excellence (P50CA058184), the National Cancer Institute (K23CA107040), The American Society of Clinical Oncology, the Flight Attendant Medical Research Institute's Young Clinical Scientist Award (no. 01250), and the Sidney Kimmel Cancer Foundation. C.D.P. is partially supported by a gift from Genentech. P.A.B. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
Hh
hedgehog
MM
multiple myeloma
NOD/SCID
nonobese diabetic/severe combined immunodeficient
Ptch
patched
RFP
red fluorescent protein
Shh
sonic Hh
Smo
smoothened
YFP
yellow fluorescent protein.
Footnotes
Conflict of interest statement: C.D.P. is a consultant for Infinity Pharmaceuticals with no financial relationship. C.D.P.'s salary is partially supported by a gift from Genentech. D.N.W. is a consultant for Infinity Pharmaceuticals with no financial relationship. D.N.W. is a former paid consultant for Genentech but has no current financial relationship. P.A.B. is a former consultant for Curis and currently holds shares in Curis.
References
- 1.Ingham PW, McMahon AP. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Hooper JE, Scott MP. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 3.Mann RK, Beachy PA. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- 4.Taipale J, Cooper MK, Maiti T, Beachy PA. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 5.Lum L, Beachy PA. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 6.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 7.Chen JK, Taipale J, Cooper MK, Beachy PA. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Wechsler-Reya R, Scott MP. Annu Rev Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- 10.Corcoran RB, Scott MP. J Neurooncol. 2001;53:307–318. doi: 10.1023/a:1012260318979. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 12.Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, Beachy PA. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 13.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, Curran T. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 15.Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, Jones D, Castro CY, Logrono R, Haque A, et al. Cancer Lett. 2006;244:53–60. doi: 10.1016/j.canlet.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 16.Vestergaard J, Pedersen MW, Pedersen N, Ensinger C, Tumer Z, Tommerup N, Poulsen HS, Larsen LA. Lung Cancer. 2006;52:281–290. doi: 10.1016/j.lungcan.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 18.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, et al. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karhadkar SS, Bova GS, Abdallad N, Dhara S, Gardner D, Maitra A, Isaacs JT, Berman DM, Beachy PA. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Ruiz IAA. Proc Natl Acad Sci USA. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins DN, Peacock CD. Biochem Pharmacol. 2004;68:1055–1060. doi: 10.1016/j.bcp.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 22.Taipale J, Beachy PA. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz i Altaba A, Sanchez P, Dahmane N. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 24.Kuehl WM, Bergsagel PL. Nat Rev Cancer. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 25.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, Smith BD, Civin CI, Jones RJ. Blood. 2004;103:2332–2336. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin ST, Sato N, Dhara S, Chang R, Hustinx SR, Abe T, Maitra A, Goggins M. Cancer Biol Ther. 2005;4:728–733. doi: 10.4161/cbt.4.7.1802. [DOI] [PubMed] [Google Scholar]
- 27.Zhang XM, Ramalho-Santos M, McMahon AP. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- 28.Mitsiades CS, Mitsiades NS, Bronson RT, Chauhan D, Munshi N, Treon SP, Maxwell CA, Pilarski L, Hideshima T, Hoffman RM, Anderson KC. Cancer Res. 2003;63:6689–6696. [PubMed] [Google Scholar]
- 29.Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, Chadwick K, Ling LE, Karanu FN, Bhatia M. Nat Immunol. 2001;2:172–180. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- 30.Kobune M, Ito Y, Kawano Y, Sasaki K, Uchida H, Nakamura K, Dehari H, Chiba H, Takimoto R, Matsunaga T, et al. Blood. 2004;104:1002–1009. doi: 10.1182/blood-2003-09-3347. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Development (Cambridge, UK) 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 32.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, Eberhart CG. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 33.Zon LI. Nat Immunol. 2001;2:142–143. doi: 10.1038/84239. [DOI] [PubMed] [Google Scholar]
- 34.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui Y, Golob J, Kelleher E, Ye Z, Pardoll D, Cheng L. Blood. 2002;99:399–408. doi: 10.1182/blood.v99.2.399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information