Acetylation-Mediated Transcriptional Activation of the ETS Protein ER81 by p300, P/CAF, and HER2/Neu (original) (raw)
Abstract
The regulated expression of the ETS transcription factor ER81 is a prerequisite for normal development, and its dysregulation contributes to neoplasia. Here, we demonstrate that ER81 is acetylated by two coactivators/acetyltransferases, p300 and p300- and CBP-associated factor (P/CAF) in vitro and in vivo. Whereas p300 acetylates two lysine residues (K33 and K116) within the ER81 N-terminal transactivation domain, P/CAF targets only K116. Acetylation of ER81 not only enhances its ability to transactivate but also increases its DNA binding activity and in vivo half-life. Furthermore, oncogenic HER2/Neu, which induces phosphorylation and thereby activation of ER81, was less able to activate acetylation-deficient ER81 mutants, indicating that both acetyltransferase and protein kinase-specific regulatory mechanisms control ER81 activity. Importantly, HER2/Neu overexpression stimulates the ability of p300 to acetylate ER81, likely by inducing phosphorylation of p300 through the Ras→Raf→mitogen-activated protein kinase pathway. This represents a novel mechanism by which oncogenic HER2/Neu, Ras, or Raf may promote tumor formation by enhancing acetylation not only of ER81 but also of other downstream effector transcription factors as well as histones.
A direct correlation between reversible histone acetylation within chromatin and transcriptional competence of eukaryotic target genes has been long recognized. This posttranslational modification is mediated by histone acetyltransferases (HATs) that catalyze the transfer of an acetyl moiety from acetyl coenzyme A onto the ɛ-amino group of specific lysine residues of histones and also nonhistone proteins. At least four human HAT families have been identified (29), and prominent examples of HATs are GCN5 (11), p300 and its homolog CREB-binding protein (CBP) (6, 38), p300- and CBP-associated factor (P/CAF) (50), and TAFII250 (34). Impairment of HAT-encoding genes has been associated with developmental aberrations (45, 48, 51) and certain cancers (16, 29), thereby underscoring the importance of HATs in orchestrating normal cell growth and differentiation.
p300 and CBP are highly homologous global transcriptional coactivators in the cells of higher eukaryotes (17, 25). Depending on promoter and cellular context, p300 and CBP can enhance transcription in several ways: they act as bridging proteins, connecting sequence-specific transcription factors to the basal transcriptional apparatus; they serve as scaffolds to assemble a complex of diverse transcription factors; they acetylate histones in the target promoter, leading to an open, transcriptionally competent chromatin configuration; or they acetylate and thereby functionally regulate specific transcription factors. Compared to many other HATs, p300 and CBP are very potent and versatile enzymes as they readily acetylate all four histones and several nonhistone proteins including p53 (20, 31), MyoD (43), and E2F1 (33).
One of the interaction partners of p300 and CBP is the ER81 protein, which belongs to the ETS family of transcription factors, which bind to a 10-bp DNA sequence with an invariant core motif, 5′-GGA(A/T)-3′ (18). ER81 is important in neuronal function, as it regulates the development of selective connections between sensory and motor neurons in the spinal cord (3). Also, ER81 is overexpressed in metastatic mammary tumors (4, 44) and causes Ewing's sarcoma upon chromosomal translocation with the EWS gene (27). Furthermore, ER81 is a downstream effector of the proto-oncoproteins HER2/Neu and Ras, which regulate ER81 through specific phosphorylation events via mitogen-activated protein kinase (MAPK) pathways (9, 23). Because of the widespread role of ER81 in various biological and pathological processes, it is important to understand further molecular mechanisms governing the activity of ER81.
In the present study we report that p300 and the associated P/CAF protein directly acetylate ER81 and thereby affect ER81 stability, DNA binding, and transactivation. Further, we show how acetylation and phosphorylation jointly regulate ER81 transcriptional activity. In addition, we demonstrate that oncogenic HER2/Neu stimulates ER81 not only via phosphorylation but also through an in vivo enhancement of p300 HAT activity, which may pleiotropically affect tumor formation.
MATERIALS AND METHODS
Plasmids.
The coding sequences for the HAT domains of p300 (amino acids 1195 to 1681) and P/CAF (amino acids 493 to 658) were cloned into pGEX-2T-6His-PL2. The mammalian expression vectors for p300 and p300Δ1430-1504 (13), p300-hemagglutinin (HA) (15), P/CAF and P/CAFΔ497-526 (41), Flag-P/CAF (50), HER2/Neu-V664E (7), BXB (12), and H-Ras-G12V (8) were as reported previously. Coding sequences for Myc-tagged murine ER81 and truncated versions thereof were cloned into the pCS3+-6Myc vector. Wild-type and mutated Gal4-ER811-210 constructs were generated by cloning appropriate ER81 cDNA into the pAB-Gal4-linker vector (5).
Immunoprecipitations with antiacetyllysine antibodies.
293T cells were transiently transfected with 1.5 μg of p300-HA expression plasmid, 0.5 μg of plasmids expressing wild-type or mutated Myc-tagged ER81, and 2 μg of the Flag-histone deacetylase 3 (HDAC3) plasmid. Two days after transfection, the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.1], 30 mM Na4P2O7, 50 mM NaF, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) supplemented with 10 μg of leupeptin/ml, 2 μg of aprotinin/ml, 1 μg of pepstatin A/ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM Na3VO4, and 10 mM sodium butyrate and centrifuged at 20,800 × g for 10 min. Lysates were precleared with protein A-agarose beads (Repligen), and inputs were removed (1 to 3%). Immunoprecipitations were performed with antiacetyllysine antibodies (PAN-AC1, Abcam, and rabbit antiserum no. 06-933; Upstate Biotechnology) or a control anti-HA antibody (12CA5; Roche), followed by incubation with protein A-agarose beads. The immunocomplexes were heated at 100°C for 5 min in RIPA buffer containing 0.5% SDS, reimmunoprecipitated with an anti-Myc (9E10) antibody, followed by binding to protein A-agarose beads. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and revealed by anti-Myc Western blotting. For endogenous ER81, anti-acetyllysine antibodies (rabbit antiserum 06-933; Upstate Biotechnology) or control anti-GAL4 antibodies (DBD; Santa Cruz Biotechnology) coupled to protein A were used, followed by Western blotting with anti-ER81 antibodies (anti-ETV-1 C20; Santa Cruz Biotechnology).
Coimmunoprecipitations.
293T cells were lysed in a mixture containing 2.5 mM Tris-HCl, 7.5 mM Na4P2O7, 12.5 mM NaCl, 12.5 mM NaF, 0.25% Triton X-100, 10 μg of leupeptin/ml, 2 μg of aprotinin/ml, 1 μg of pepstatin A/ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM Na3VO4, and 10 mM sodium butyrate (pH 7.1). Precipitations were then performed with protein A-agarose beads (Repligen) and either anti-Flag (M2; Sigma), anti-HA (12CA5; Roche), or anti-ER81 (anti-ETV-1 C20; Santa Cruz Biotechnology) antibodies, and coimmunoprecipitated proteins were detected by immunoblotting as described above.
Expression and purification of recombinant proteins.
Glutathione _S_-transferase (GST) fusion proteins were purified from the respective Escherichia coli BL21 extracts with glutathione-Sepharose (Pharmacia) according to standard procedures. For GST-ER812-477, the GST moiety was cleaved off with thrombin and ER812-477 was purified with Ni2+-nitrilotriacetic acid agarose (Qiagen), which makes use of a histidine tag inserted between the GST moiety and the ER81 amino acids.
GST pull-down assay.
GST or GST-ER812-477 was bound to glutathione-agarose (Sigma). Lysates from 293T cells transiently transfected with the Flag-P/CAF expression plasmid were prepared in Frackelton buffer (10 mM Tris-HCl, 30 mM Na4P2O7, 50 mM NaCl, 50 mM NaF, 1% Triton X-100 [pH 7.1]) containing a protease inhibitor cocktail. One hundred microliters of cell lysates was incubated in 0.25× Frackelton buffer with protein-bound GST-agarose beads for 2 h at 4°C. The beads were washed four times, boiled in sample buffer, and subjected to SDS-PAGE. Bound P/CAF was detected by anti-Flag Western blotting.
Isolation of RNA and RT-PCR.
Cytoplasmic RNA was isolated from transiently transfected 293T cells and analyzed with the Access reverse transcription-PCR (RT-PCR) kit (Promega) as described before (9). Primers for MMP-1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were as reported previously (9).
In vitro acetylation.
In vitro acetylation assays were performed with 50 mM HEPES (pH 7.4)-10 mM sodium butyrate-0.1 mM phenylmethylsulfonyl fluoride-1 mM dithiothreitol in the presence of 0.25 μCi of [14C]acetyl coenzyme A (57 mCi/mmol; Amersham). Reactions were performed at 30°C for 1 h and were stopped by adding 2× Laemmli sample buffer. The reaction mixture was then subjected to SDS-PAGE, and dried gels were analyzed by autoradiography. Immunoprecipitated p300 was obtained by transiently transfecting 293T cells with 1 μg of p300-HA with or without 1 μg of HER2/Neu-V664E. Twenty-four hours after transfection, serum starvation (0.1% fetal bovine serum) was done for 12 h, followed by immunoprecipitations with anti-HA antibodies and RIPA buffer and finally by two washes in phosphate-buffered saline. In some cases, immunoprecipitates were then incubated with 50 U of calf intestinal alkaline phosphatase (Roche) for 30 min at 37°C and afterwards washed twice with phosphate-buffered saline before usage in in vitro acetylation assays.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as described previously (23). Briefly, 293T cells were transfected with 0.5 μg of wild-type cytomegalovirus (CMV)-ER812-477 or various mutant versions of it with or without p300-HA (3 μg). Cell lysates were equalized for protein amount by Western blotting using anti-ER81 antibodies (anti-ETV-1 C20; Santa Cruz Biotechnology) and incubated with a 32P-labeled double-stranded E74 oligonucleotide probe.
Pulse-chase experiments.
Pulse-chase experiments were performed as described previously (21). 293T cells were transfected with 1 μg of Myc-ER812-477 with or without p300-HA or Flag-P/CAF (3 μg). Thirty-six hours after transfection, the medium was replaced with methionine-free RPMI 1640 (Sigma) supplemented with 2% charcoal-stripped fetal bovine serum, l-glutamine (0.3 g/liter), and l-cysteine-2HCl (0.652 g/liter). Methionine starvation was done for 1 h and was followed by addition of 100 μCi of [35S]methionine (Amersham) to each dish. Cells were incubated at 37°C for 2 h and washed twice with phosphate-buffered saline and then Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, and 0.015 g of methionine/liter was added. At various time points the cells were harvested and immunoprecipitations were performed with anti-Myc antibodies. Quantitation of incorporation of radioactivity was done with a PhosphorImager.
In vivo phosphorylation.
Mv1Lu cells were transiently transfected with HA-tagged p300 with or without HER2/Neu-V664E. Thirty-six hours after transfection, cells were metabolically labeled with 32Pi (0.1 mCi/ml) for 4 h, and p300 was then immunoprecipitated with anti-HA antibodies (12CA5; Roche). The immunoprecipitates were subjected to SDS-PAGE followed by anti-HA immunoblotting and additionally autoradiography.
Luciferase assays.
Cells were seeded out on 6-cm-diameter dishes and transiently transfected by the calcium phosphate coprecipitation method (23). Transfections utilized 1 μg of TORU-luc, Gal42-tk80-luc, or MMP-1 (−525 to +15) promoter luciferase reporter plasmid in conjunction with mammalian expression plasmids.
RESULTS
ER81-dependent transcription is regulated by acetylation.
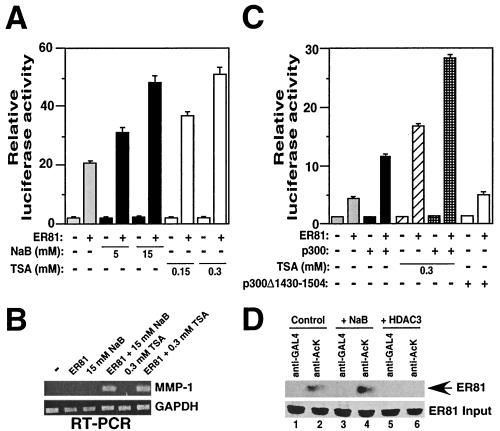
ER81 forms a complex with the ubiquitously expressed p300 and CBP proteins, but it has remained unresolved whether HAT activity of p300 and CBP is required for coactivation of ER81-mediated gene transcription (39). To determine whether the transcriptional activity of ER81 is regulated by acetylation, the ER81-responsive TORU-luc luciferase reporter construct (35) was employed to transfect Mv1Lu cells, which were subsequently treated with the HDAC inhibitor sodium butyrate or trichostatin A (TSA) (36, 53). As shown in Fig. 1A, both HDAC inhibitors enhanced the transcriptional activity of cotransfected ER81 in a dose-dependent manner up to 2.5-fold. In contrast, no effect on TORU-luc activity was observable in the absence of ER81, suggesting that acetylation specifically stimulates ER81-dependent transcription.
FIG. 1.
Augmentation of ER81-mediated transcription by HDAC inhibitors and p300. (A) Mv1Lu cells were cotransfected with the TORU-luc reporter construct and either 0.5 μg of CMV-ER812-477 or empty expression vector pEV3S. Cells were treated with sodium butyrate (NaB) or TSA for 12 h prior to lysis. (B) Detection of MMP-1 and GAPDH gene expression in 293T cells by RT-PCR. Cells were transiently transfected with either 0.5 μg of CMV-ER812-477 or pEV3S and treated with the indicated HDAC inhibitors 12 h prior to lysis. (C) CV-1 cells were cotransfected with TORU-luc, CMV-ER812-477, or pEV3S vector (0.2 μg) and p300 or p300Δ1430-1504 (0.4 μg) as indicated. (D) 293T cell extracts were immunoprecipitated with no antibody or protein A-coupled anti-GAL4 or antiacetyllysine antibodies (06-933; Upstate Biotechnology). Subsequently, immunoblotting was performed with anti-ER81 antibodies (anti-ETV-1 C20; Santa Cruz Biotechnology). Input levels of ER81 were detected by immunoblotting ER81 immunoprecipitated with anti-ETV-1 antibodies. Where indicated, cells were treated with sodium butyrate or cotransfected with 1 μg of the HDAC3 expression plasmid.
We have previously shown that ER81 activates a matrix metalloproteinase gene, the MMP-1 gene, in 293T cells (9). Thus, we analyzed by RT-PCR whether HDAC inhibitors could also promote transcription of the endogenous MMP-1 gene in 293T cells. Transfection of ER81 did not result in detectable MMP-1 mRNA levels, nor did the utilization of the HDAC inhibitor sodium butyrate or TSA (Fig. 1B). However, when ER81 was transfected into 293T cells and simultaneously HDAC inhibitors were employed, significant expression of MMP-1 was observed. In contrast, no change of GAPDH levels were detected, indicating the specificity of MMP-1 gene activation by ER81 and HDAC inhibitors. These results further imply that acetylation of ER81 activates ER81's ability to upregulate target gene transcription.
To further explore this conjecture, ER81 was coexpressed with p300 in CV-1 cells that express very low levels of endogenous p300 or CBP (55). ER81-mediated luciferase activity of the TORU-luc reporter increased approximately 2.5-fold in the presence of p300, and no effect of p300 was detectable in the absence of ER81 (Fig. 1C). In contrast, a HAT-deficient mutant protein, p300Δ1430-1504 (13), had no effect on ER81 activity. Furthermore, TSA enhanced ER81-dependent transcription in the presence and absence of p300. Altogether, these results imply that ER81 target gene expression is regulated by p300-mediated acetylation, which might entail direct acetylation of ER81 by p300.
To demonstrate that endogenous ER81 is indeed acetylated in vivo, we immunoprecipitated a 293T cell extract with antiacetyllysine antibodies or control anti-GAL4 antibodies. Indeed, immunoblotting with anti-ER81 antibodies revealed that endogenous ER81 was specifically immunoprecipitated by antiacetyllysine antibodies (Fig. 1D, lane 2). Furthermore, addition of sodium butyrate enhanced the acetylation of ER81, whereas coexpression of HDAC3 inhibited it (Fig. 1D, lanes 4 and 6). Thus, ER81 exists as an acetylated protein in vivo and its degree of acetylation is modulated by HDAC activity.
ER81 is acetylated in vitro and in vivo by p300.
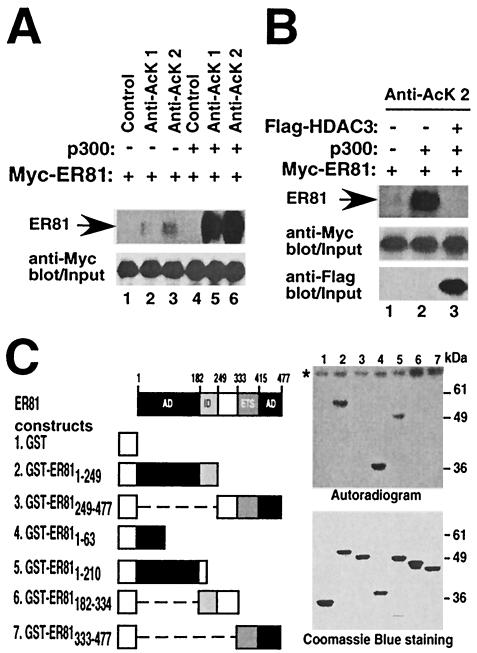
To further prove that ER81 is acetylated in vivo, immunoprecipitations with two different antiacetyllysine antibodies were performed using extracts of 293T cells transfected with Myc-tagged ER81. Ectopically expressed ER81 appears to be poorly acetylated (Fig. 2A, lanes 2 and 3), probably due to the limiting amounts of endogenous p300 in 293T cells. However, coexpression of p300 resulted in robust acetylation of ER81 (Fig. 2A, lanes 5 and 6), indicating that ER81 is acetylated in vivo by p300. Control anti-Myc Western blotting showed that similar amounts of ER81 protein levels were present (Fig. 2A, bottom).
FIG. 2.
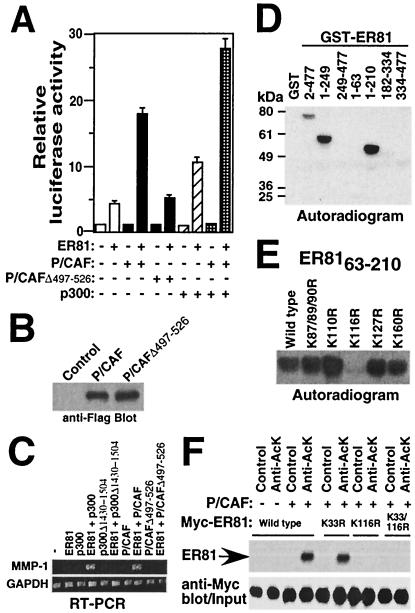
ER81 is acetylated by p300. (A) 293T cells were cotransfected with Myc-tagged ER81 without or with the p300-HA expression plasmid. Immunoprecipitations were performed with either a control antibody or antiacetyllysine antibodies (AcK 1, PAN-AC1 and Abcam; AcK 2, 06-933; Upstate Biotechnolgy), followed by anti-Myc Western blotting (top). ER81 protein levels are shown by anti-Myc Western blotting (bottom). (B) Antiacetyllysine immunoprecipitations similar to those in panel A in the presence of Flag-HDAC3. (C) In vitro acetylation assays. Various GST-ER81 chimeras were incubated with p300-HAT and [14C]acetyl coenzyme A and then separated by SDS-PAGE. Acetylated ER81 was detected by autoradiography. Asterisk, autoacetylated p300 HAT. (Bottom) Coomassie blue-stained protein gel with purified recombinant proteins. AD, activation domain; ID, inhibitory domain; ETS, DNA binding domain of ER81.
We also coexpressed HDAC3 with p300 and immunoprecipitated ER81 with antiacetyllysine antibodies. HDAC3 suppressed acetylation of ER81 in the presence of p300, indicating that the acetylation status of ER81 is determined by antagonistic HAT and HDAC activities (Fig. 2B). In contrast, coexpression of HDAC1 and HDAC2 did not deacetylate ER81 (data not shown), even though HDAC1, -2, and -3 are all members of the same class I HDAC family. However, it has been shown that HDAC1 and -2 are more closely related to each other than to HDAC3 (49). In conclusion, these data strongly suggest that ER81 is acetylated by p300 in vivo and is specifically deacetylated by HDAC3. However, it is unlikely that HDAC3 is exclusively responsible for ER81 deacetylation, as many different HDAC genes are present in mammalian cells.
Next, we wanted to map the region of ER81 undergoing acetylation. To this end, in vitro acetylation assays were performed using the HAT domain of p300 (residues 1195 to 1681; p300-HAT) and various GST-ER81 fusion proteins as substrates. p300-HAT acetylated GST-ER811-249, GST-ER811-63, and GST-ER811-210 (Fig. 2C; lanes 2, 4, and 5, respectively), whereas amino acids 182 to 477 contained no acetylation site (lanes 3, 6, and 7). As such, the acetylation site(s) is located within the N-terminal transactivation domain of ER81 (amino acids 1 to 182).
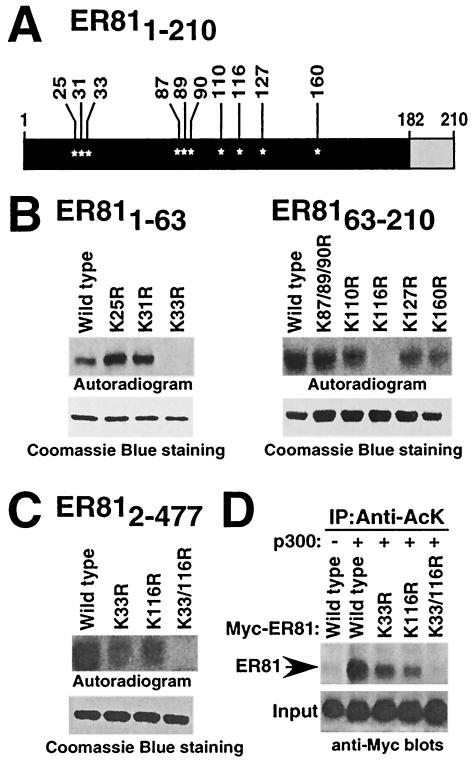
ER81 is acetylated by p300 on K33 and K116.
The N-terminal ER81 activation domain contains a total of 10 lysine residues, any of which might be acetylated by p300 (Fig. 3A). To map the acetylation site(s), we therefore replaced lysine residues with arginines in order to retain the positive charge while preventing acetylation. The respective GST-ER811-63 and GST-ER8163-210 fusion proteins were used as substrates in in vitro acetylation assays. Only mutations K33R and K116R abolished acetylation of GST-ER811-63 and GST-ER8163-210, respectively; other mutations did not have a significant effect on in vitro acetylation (Fig. 3B). Thus, lysines 33 and 116 appear to be the sites of p300 acetylation. To demonstrate that these lysines are also utilized as acetylation sites in full-length ER81, we purified bacterially expressed wild-type and mutated ER812-477 and investigated the ability of p300-HAT to acetylate these ER81 proteins. Whereas both single-mutant proteins (K33R and K116R) were still acetylated by p300-HAT, acetylation of the double-mutant protein K33/116R was completely abolished (Fig. 3C).
FIG. 3.
Lysines 33 and 116 of ER81 are acetylated by p300. (A) All lysine residues in the region between amino acids 1 to 210 of ER81 (asterisks). (B) In vitro acetylation assays of the indicated GST-ER81 mutant proteins. (C) In vitro acetylation of ER812-477. (D) 293T cells were cotransfected with wild-type Myc-tagged ER812-477 or the indicated mutant versions (0.5 μg) and the p300-HA expression plasmid (3 μg). Immunoprecipitations were performed with the antiacetyllysine antibody (06-933; Upstate Biotechnology) as detailed in the legend for Fig. 2A.
To confirm that K33 and K116 are also targeted by p300 in vivo, immunoprecipitations of ER81 coexpressed with p300 were performed with antiacetyllysine antibodies. Compared to wild-type ER81, the mutant proteins K33R and K116R showed lower levels of acetylation, and no acetylation of the K33/116R mutant protein was detected (Fig. 3D). Taken together, our results indicate that ER81 is acetylated on lysines 33 and 116 both in vitro and in vivo by p300.
ER81 and P/CAF are in a complex in vivo.
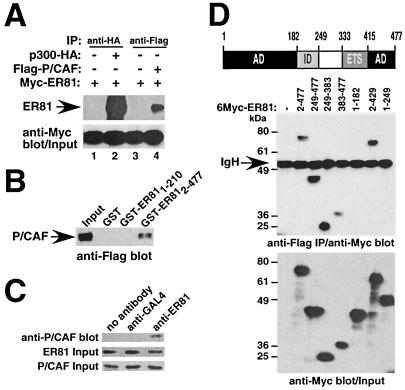
The coactivator P/CAF was originally discovered through its association with p300 and CBP and has intrinsic HAT activity (50). As such, ER81 might, at least indirectly via p300 or CBP, also recruit P/CAF to target gene promoters. To demonstrate complex formation between ER81 and P/CAF in vivo, we cotransfected 293T cells with Myc-tagged ER81 and either p300-HA or Flag-P/CAF. Coimmunoprecipitations were performed using whole-cell lysates with anti-HA or anti-Flag antibodies, followed by anti-Myc Western blotting. As reported previously (39), p300 coimmunoprecipitated with ER81 (Fig. 4A, lane 2). Immunoprecipitations with anti-Flag antibodies also led to the specific detection of ER81 (Fig. 4A, lane 4), suggesting that ER81 and P/CAF bind to each other in vivo. To confirm this interaction between ER81 and P/CAF in vitro, GST pull-down assays were performed. Whereas the GST moiety and GST-ER811-210 did not interact with P/CAF, GST-ER812-477 did (Fig. 4B).
FIG. 4.
ER81 interacts with P/CAF in vivo and in vitro. (A) Coimmunoprecipitations (IP) were done with lysates of 293T cells transfected with Myc-tagged ER812-477 and either p300-HA or Flag-P/CAF. ER81 was detected by anti-Myc Western blotting. (B) GST pull-down assays were carried out by incubating whole-cell extract of 293T cells expressing Flag-P/CAF with glutathione-agarose beads prebound to control GST protein, GST-ER811-210, or GST-ER812-477. (C) Coimmunoprecipitation of endogenous P/CAF and ER81. 293T cell extracts were immunoprecipitated with the indicated antibodies and P/CAF was revealed with anti-P/CAF antibodies (kindly provided by Yoshihiro Nakatani). (D) Mapping the ER81 domain responsible for interaction with P/CAF. Immunoprecipitations were performed with anti-Flag antibodies by using lysates of 293T cells transfected with Flag-P/CAF and various Myc-tagged ER81 truncations (middle). The expression levels of ER81 proteins were assessed by anti-Myc Western blotting (bottom). IgH, immunoglobulin H; AD, ID, and ETS are as defined for Fig. 2.
To further corroborate a physiological interaction between ER81 and P/CAF, we studied the formation of complexes between endogenous ER81 and endogenous P/CAF in 293T cells. To this end, cell extracts were immunoprecipitated with anti-ER81 antibodies or control anti-GAL4 antibodies. Coprecipitated P/CAF was then detected by Western blotting. As shown in Fig. 4C, P/CAF was detectable only in anti-ER81 immunoprecipitates, again indicating that ER81 and P/CAF interact in vivo.
To map the ER81 domain responsible for interacting with P/CAF, 293T cells were transfected with various truncated versions of Myc-tagged ER81 and Flag-P/CAF as indicated (Fig. 4D). Cell lysates were immunoprecipitated with anti-Flag antibodies, and the coimmunoprecipitated ER81 truncations were visualized by Western blotting using anti-Myc antibodies. Besides full-length ER812-477, ER812-429, ER81249-477, ER81249-383, and ER81383-477 were found to interact with P/CAF (Fig. 4D, middle). Thus, ER81 associates with P/CAF through amino acids 249 to 383 and 383 to 477.
Acetylation of ER81 by P/CAF.
Since P/CAF associates with ER81, we investigated whether transcriptional activity of ER81 is modulated by P/CAF. To this end, CV-1 cells were transfected with ER81 along with P/CAF and/or p300. P/CAF potentiated the transcriptional activity of ER81 by fourfold, while the HAT-deficient Δ497-526 mutant protein (41) had no effect (Fig. 5A). Western blotting confirmed comparable levels of expression of P/CAF and the P/CAFΔ497-526 mutant protein (Fig. 5B). No activation of transcription by P/CAF was observed in the absence of ER81. In addition, P/CAF-mediated activation of ER81 was further enhanced in the presence of p300. These results indicate that P/CAF is a coactivator of ER81 and that the HAT activity of P/CAF is required for its ability to coactivate.
FIG. 5.
P/CAF stimulates transcriptional activity of ER81 and acetylates K116. (A) CV-1 cells were cotransfected with TORU-luc, CMV-ER812-477 or pEV3S vector (0.2 μg), p300-HA (0.4 μg), and Flag-P/CAF or Flag-P/CAFΔ497-526 (0.5 μg) as indicated. The resulting luciferase activities were measured. (B) Anti-Flag Western blot of immunoprecipitated Flag-P/CAF and Flag-P/CAFΔ497-526 expressed in CV-1 cells. (C) Expression of MMP-1 and GAPDH assessed by RT-PCR in 293T cells transiently transfected with 0.2 μg of CMV-ER812-477 or empty vector pEV3S and 0.5 μg of the indicated wild-type or mutated cofactors. (D) In vitro acetylation assays were performed with various GST-ER81 fusion proteins and the HAT domain of P/CAF. (E) In vitro acetylation assay of wild-type or mutated GST-ER8163-210 similar to those of panel D. (F) 293T cells were cotransfected with wild-type Myc-tagged ER81 or indicated mutant versions and the P/CAF-Flag expression plasmid. Immunoprecipitations were performed with either the control or antiacetyllysine antibody (06-933; Upstate Biotechnology), followed by anti-Myc Western blotting (top). The input levels of ER81 are shown at the bottom.
We then investigated the impact of p300 and P/CAF on the activation of the endogenous MMP-1 gene by ER81 in 293T cells. Whereas ER81, p300, or P/CAF alone did not elicit detectable MMP-1 gene transcription in our RT-PCR analyses, coexpression of ER81 with either p300 or P/CAF did (Fig. 5C). In contrast, the HAT-deficient mutant proteins p300Δ1430-1504 and P/CAFΔ497-526 were incapable of cooperating with ER81 to induce MMP-1 gene transcription. These data corroborate the notion that the acetyltransferase function of p300 and P/CAF is required for efficient ER81-mediated transcription.
To investigate whether ER81 is acetylated by P/CAF, in vitro acetylation assays were performed. Indeed, the HAT domain of P/CAF acetylated ER812-477, ER811-249, and ER811-210 (Fig. 5D). However, ER811-63, which is a substrate for p300-HAT, did not become acetylated by P/CAF, suggesting that acetylation of ER81 occurs within amino acids 64 to 210. Mutation of lysine 116 to arginine, but not mutation of any of the other lysine residues within ER81 amino acids 63 to 210, completely inhibited the acetylation of GST-ER8163-210 (Fig. 5E). We then confirmed in vivo acetylation of K116 by P/CAF by performing antiacetyllysine immunoprecipitation experiments. P/CAF induced in vivo acetylation of both wild-type and K33R ER81, but none in the K116R and K33/116R mutant proteins (Fig. 5F). Thus, P/CAF solely acetylates K116 in vitro and in vivo whereas p300 additionally acetylates K33.
K116 acetylation augments ER81 DNA binding.
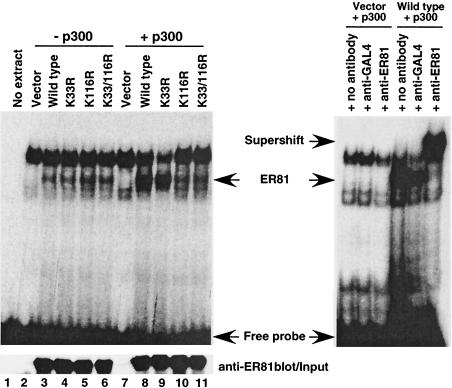
One functional consequence of acetylation of a transcription factor is frequently a change in its DNA binding ability. As such, we examined the ability of ER81 to bind to the E74 probe, a well-characterized DNA binding site for ER81 (23), and proved the identity of the ER81 band by supershifting experiments (Fig. 6, right). As shown in Fig. 6 (left, lanes 3 to 6), acetylation site mutant proteins bound to the E74 probe as avidly as wild-type ER81. Upon p300 coexpression in 293T cells, the DNA binding ability of ER81 was enhanced approximately threefold (compare lanes 3 and 8). Similarly, the K33R mutant protein displayed an enhanced DNA binding activity upon coexpression with p300 (compare lanes 4 and 9), whereas the K116R and K33/116R mutant proteins were basically unaffected by p300 expression. These results indicate that acetylation of ER81 by p300 on K116 enhances the DNA binding ability of ER81. Although K116 is also acetylated by P/CAF, we were unable to observe any effect on DNA binding by coexpressing P/CAF with ER81, the reason being that P/CAF acetylates ER81 at a much lower stoichiometry than p300 in the 293T cells used (data not shown).
FIG. 6.
Acetylation of K116 potentiates DNA binding of ER81. Lysates were prepared from 293T cells transiently transfected with either wild-type or mutant CMV-ER812-477 and with or without p300-HA. The 32P-labeled E74 oligonucleotide was used to perform electrophoretic mobility shift assays. The lysates were normalized for equal expression of ER81 protein by anti-ER81 (anti-ETV-1 C20; Santa Cruz Biotechnology) Western blotting (bottom). (Right) Proof of the identity of ER81 by supershift experiments utilizing anti-ER81 or control anti-GAL4 antibodies.
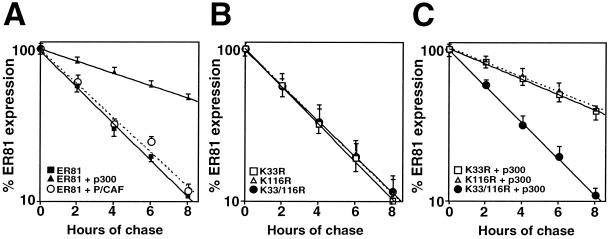
Acetylation stabilizes the ER81 protein.
To investigate if acetylation by p300 or P/CAF had any protein-stabilizing effect, the in vivo half-life (_t_1/2) of the ER81 protein was determined. p300 overexpression enhanced _t_1/2 of ER81 threefold from 2.5 to 7.5 h (Fig. 7A), whereas P/CAF did not significantly affect _t_1/2, again due to a very low stoichiometry of in vivo acetylation. We then assessed the _t_1/2s of various ER81 mutant proteins in the absence and presence of p300. No difference between the _t_1/2s of the wild-type ER81 and the K33R, K116R, or K33/116R mutant protein in the absence of p300 overexpression was observed (Fig. 7B). However, in the presence of p300 (Fig. 7C), the mutant proteins differed slightly (5.9 h for K33R and 6.1 h for K116R versus 7.5 h for wild-type ER81) or strongly (_t_1/2 of K33/116R = 2.5 h) in their stabilities from wild-type ER81. Thus, p300-mediated acetylation on both K33 and K116 contributes to ER81 stability.
FIG. 7.
Acetylation-mediated stabilization of ER81. Pulse-chase experiments were performed with 293T cells transfected with Myc-tagged ER81 plus either p300-HA or Flag-P/CAF (A) or Myc-tagged ER81 mutant proteins K33R, K116R, and K33/116R without (B) or with (C) p300-HA. Cells were pulsed with [35S]methionine and chased for up to 8 h, followed by immunoprecipitations with anti-Myc antibodies. The relative amount of radioactive ER81 (logarithmic scale) is plotted as a function of time with the signal at time zero set to 100%. The results of three independent experiments are shown.
Acetylation of ER81 affects its basal transcriptional activity.
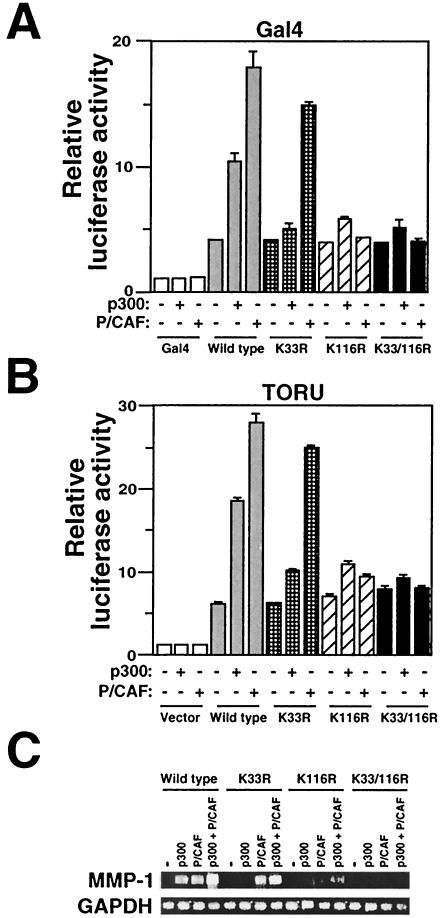
To assess the impact of acetylation on the ER81 transactivation potential, the N-terminal 210 amino acids of ER81, which encompass its strong activation domain as well as the sites of acetylation, were fused to the DNA binding domain of Gal4. The transcriptional activity of the resulting Gal4-ER811-210 fusion proteins was measured with a Gal4 DNA binding site-driven luciferase reporter construct. As shown in Fig. 8A, the K33R, K116R, and K33/116R mutant proteins behaved indistinguishably from wild-type ER81 in the absence of overexpressed p300 or P/CAF. However, whereas p300 enhanced the transactivation potential of the wild-type Gal4-ER811-210 fusion protein, the mutant proteins were barely, if at all, responsive to p300 overexpression. These results are consistent with the notion that acetylation on both K33 and K116 is required for p300 to activate ER81. On the other hand, the K33R mutant protein was stimulated equivalently to wild-type ER81 by P/CAF, whereas the K116R and K33/116R mutant proteins were not at all stimulated by P/CAF, consistent with the fact that K116, but not K33, is a target of P/CAF.
FIG. 8.
Role of K33 and K116 acetylation in p300- and P/CAF-mediated activation of ER81. Wild-type ER81 or the indicated mutant proteins were cotransfected into CV-1 cells either as Gal4-ER811-210 constructs (0.2 μg) with the Gal42-tk80-luc luciferase reporter (A) or as CMV-ER812-477 expression vectors (0.2 μg) with TORU-luc (B). p300 (0.4 μg) and P/CAF (0.5 μg) were coexpressed as indicated, and the resulting luciferase activities were measured. (C) RT-PCR analysis of MMP-1 gene transcription in 293T cells. Where indicated, wild-type or acetylation-deficient ER81 mutant proteins (0.2 μg) were coexpressed in the presence of p300 or P/CAF (0.5 μg).
We then also tested the ability of full-length ER81 to be regulated by acetylation with the TORU-luc reporter. As shown in Fig. 8B, acetylation on both K33 and K116 was required for p300 to activate ER81-dependent transcription but only acetylation on K116 was required in the case of P/CAF. To exclude the possibility that these effects might be due to the up-to-threefold differences in DNA binding or protein stability between wild-type and mutated ER81 (Fig. 6 and Fig. 7), ER81 proteins were expressed at saturating amounts so that these differences would be barely noticeable. In conclusion, a major way in which p300 and P/CAF activate ER81-dependent transcription is by raising ER81's transactivation potential through acetylation of the N-terminal activation domain. However, p300 and P/CAF differ, at least in part, in their mechanisms of activation of ER81-mediated transcription, since the K33R mutant protein was activated by P/CAF but not by p300, although both of these coactivators are still able to acetylate K116 in the K33R mutant protein (Fig. 3 and 5). Thus, K116 acetylation in itself appears to be insufficient to activate ER81-dependent transcription.
We extended our analysis to the stimulation of endogenous MMP-1 gene transcription upon acetylation of ER81. As shown above (Fig. 5C), expression of wild-type ER81 in the presence of p300 or P/CAF resulted in significant levels of MMP-1 mRNA, detectable in our RT-PCR analyses (Fig. 8C). In contrast, the K33R mutant ER81 was stimulated only by P/CAF, whereas the K116R and the K33/116R mutant proteins were stimulated in their activities by neither p300 nor P/CAF, consistent with our results obtained by studying artificial luciferase reporter constructs (Fig. 8A and B). These results indicate the physiological importance of ER81 acetylation on specific lysine residues for the regulation of gene transcription.
HER2/Neu-stimulated transcription activation is impaired in ER81 acetylation mutant proteins.
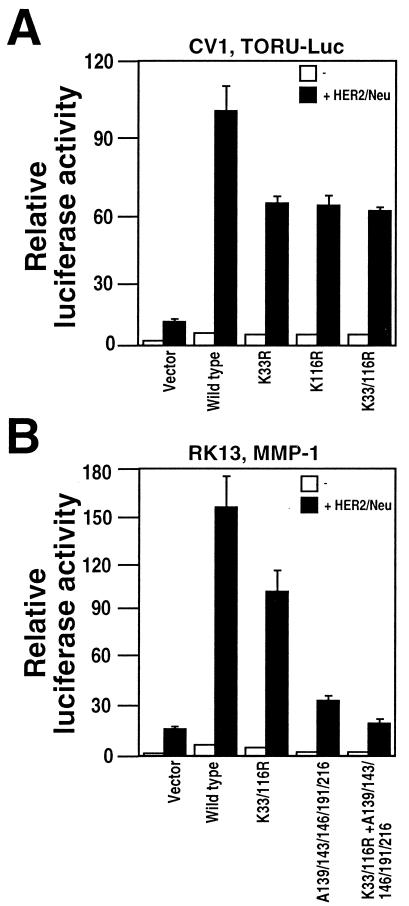
Our previous studies have shown that ER81 is regulated through phosphorylation via MAPK pathways upon overexpression of HER2/Neu (9, 10). Having identified acetylation as a regulator of basal ER81 activity, we wanted to study its impact on HER2/Neu-mediated activation of ER81. Indeed, HER2/Neu-elicited activation of the TORU-luc reporter was significantly reduced (∼40%) with all ER81 acetylation site mutant proteins tested (Fig. 9A). Similar results were obtained with the MMP-1 promoter (Fig. 9B and data not shown). However, mutation of all phosphorylation sites (T139, T143, S146, S191, and S216) that are inducibly phosphorylated upon HER2/Neu overexpression and that regulate ER81 function (9) had a larger impact on ER81's ability to activate the MMP-1 promoter than mutation of the two acetylation sites (Fig. 9B, compare A139/143/146/191/216 to K33/116R). Yet, additional mutation of the two acetylation sites reduced the activity of the A139/143/146/191/216 mutant protein by nearly one-half. These results indicate that both phosphorylation and acetylation of ER81 are required for maximal ER81-dependent transcription. They also point at the possibility that acetylation of ER81 might be induced by HER2/Neu.
FIG. 9.
ER81 mutant proteins K33 and K116 show impaired HER2/Neu-stimulated transactivation. (A) ER81 activity measured in CV-1 cells with the TORU-luc reporter. Cells were transiently transfected with wild-type or mutant CMV-ER812-477 (1 μg) and 0.5 μg of HER2/Neu-V664E. (B) One microgram of wild-type or mutated CMV-ER812-477 expression vectors was cotransfected with the MMP-1 luc reporter and HER2/Neu-V664E (0.5 μg) as indicated, and resulting luciferase activities in RK13 cells were measured.
HER2/Neu expression enhances HAT activity of p300.
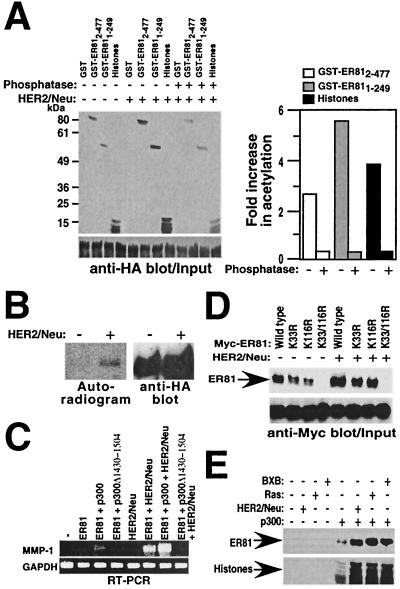
To analyze whether HER2/Neu can stimulate the HAT activity of p300, p300 was coexpressed with or without HER2/Neu, immunoprecipitated, and used to acetylate GST-ER81 fusion proteins or core histones. HER2/Neu enhanced acetylation of GST-ER812-477 and GST-ER811-249 by 2.6- and 5.5-fold, respectively (Fig. 10A). Similarly, acetylation of histones by p300 was increased 3.8-fold upon HER/2Neu coexpression. These results demonstrate that HER2/Neu enhances the HAT activity of p300, most likely by inducing phosphorylation of p300. Consistently, phosphatase treatment of p300 immunoprecipitated from HER2/Neu-overexpressing cells inhibited any stimulation of p300 HAT activity. Rather, p300 HAT activity was reduced to ∼50% of the basal level after the phosphatase treatment, suggesting that p300 is phosphorylated and thereby activated to some degree even in the absence of HER2/Neu overexpression. In contrast, no change in HAT activity of P/CAF was seen upon HER2/Neu expression (data not shown).
FIG. 10.
HER2/Neu potentiates p300 HAT activity. (A) 293T cells were transfected with p300-HA (1 μg) with or without HER2/Neu-V664E (1 μg). Immunoprecipitations were performed with anti-HA antibodies, and products were used either directly or after phosphatase treatment for in vitro acetylation of GST, GST-ER812-477, GST-ER811-249, or histones (upper left). The expression levels of p300 were compared by anti-HA Western blotting (lower left). Changes in HAT activity were determined by measuring the incorporation of radioactivity with a PhosphorImager normalized to densitometric determination of p300 protein amount and are graphically depicted on the right. (B) In vivo phosphorylation of p300. Mv1Lu cells were transfected with 4 μg of p300-HA and 0.5 μg of HER2/Neu-V664E as indicated. After in vivo labeling with 32Pi, p300-HA was immunoprecipitated and incorporation of radioactivity was assessed by autoradiography. (Right) Corresponding anti-HA immunoblot. (C) MMP-1 gene expression evaluated by RT-PCR. 293T cells were transfected with 0.5 μg of CMV-ER812-477 or empty vector pEV3S, 0.4 μg of p300 or p300Δ1430-1504, and 1 μg of HER2/Neu-V664E as indicated. (D) 293T cells were transfected with p300-HA (1 μg), HER2/Neu-V664E (1 μg), and either wild-type Myc-tagged ER812-477 or the indicated mutant versions (0.5 μg). Immunoprecipitations were performed with the antiacetyllysine antibody (06-933; Upstate Biotechnology), followed by Western blotting with anti-Myc antibodies (top). Input levels of Myc-tagged ER81 proteins are shown at the bottom. (E) 293T cells were transfected with or without p300-HA (1 μg) and with or without HER2/Neu-V664E (1 μg), H-Ras-G12V (0.05 μg), or BXB (0.5 μg). Equal amounts of p300 immunoprecipitated with anti-HA antibodies were used for in vitro acetylation of GST-ER812-477 (top) or histones (bottom).
To ascertain that HER2/Neu induces the phosphorylation of p300 in vivo, metabolic 32Pi labeling experiments were performed with Mv1Lu cells transiently transfected with p300-HA and HER2/Neu-V664E. As shown in Fig. 10B (left), cotransfection of HER2/Neu induces the phosphorylation of p300. Western blotting (right) showed comparable levels of p300 protein expression in the presence and absence of HER2/Neu. These results imply that HER2/Neu may regulate p300 HAT activity through the induction of p300 phosphorylation.
Next, we investigated the effect of p300 on HER2/Neu- and ER81-mediated MMP-1 gene transcription. As reported before (9), HER2/Neu and ER81 synergistically activated the endogenous MMP-1 gene (Fig. 10C). Importantly, overexpression of p300 significantly enhanced MMP-1 gene transcription in the presence of ER81 and HER2/Neu, whereas the HAT-deficient mutant protein p300-Δ1430-1504 inhibited MMP-1 gene stimulation. These results indicate that the HAT activity of p300 is essential for HER2/Neu to upregulate MMP-1 gene transcription via ER81, further suggesting that regulation of HAT activity by HER2/Neu may be important in modulating ER81 target gene transcription.
To analyze how HER2/Neu may affect in vivo acetylation of ER81 by p300, Myc-tagged ER81 and its acetylation site mutant proteins were immunoprecipitated with antiacetyllysine antibodies and then detected by anti-Myc Western blotting. Coexpression of HER2/Neu enhanced acetylation of wild-type, K33R, and K116R ER81 by approximately threefold, whereas no acetylation of the double-mutant protein, K33/116R, was observable (Fig. 10D). These results indicate that HER2/Neu can regulate ER81 activity, in part, by acetylation of ER81.
HER2/Neu overexpression leads to the activation of the Ras→Raf→MAPK pathway (52), suggesting that oncogenic Ras or Raf may also stimulate p300 HAT activity. Thus, p300 was coexpressed with an oncogenic mutant Ras, Ras-G12V, or BXB, a constitutively active Raf-1 molecule (12), immunoprecipitated, and used for in vitro acetylation assays. Compared to p300 alone, coexpression of Ras-G12V and BXB led to enhanced acetylation levels for both GST-ER812-477 and core histones, similar to what was observed with HER2/Neu (Fig. 10E). Thus, we report here for the first time that HER2/Neu, Ras, and Raf, most likely via activation of MAPKs, stimulate in vivo the HAT activity of p300.
DISCUSSION
p300 is a versatile coactivator that can interact with multiple transcription factors including ER81 (17, 25). As p300 is a potent HAT modifying histones and thereby loosening chromatin structure, this represents one mechanism by which p300 stimulates ER81-dependent gene transcription. The present study provides biochemical and functional evidence for a hitherto-unrecognized mechanism by which p300 regulates ER81 function at multiple levels (DNA binding, protein stability, and transactivation) by direct acetylation of ER81 itself. Additionally, this study has identified a new cofactor of ER81, the HAT P/CAF. Furthermore, we provide evidence that p300 HAT activity is regulated by HER2/Neu, Ras, and Raf in vivo, pointing to a novel mechanism for how these proto-oncoproteins can affect gene transcription. Finally, we have identified, to our knowledge for the first time, in ER81 a transcription factor whose activity is enhanced by simultaneous phosphorylation and acetylation triggered by a member of the growth factor receptor tyrosine kinase family.
Two in vivo acetylation sites were mapped in the N-terminal transactivation domain of ER81, K33 and K116. While K116 can be acetylated by both p300 and P/CAF, K33 is targeted only by p300. A series of peptide substrates was used to determine that the consensus sequence for p300-mediated acetylation is at residues possessing a positive charge at either position −3 or + 4 or both (46). K33 of ER81 is flanked by arginine residues (underlined) at both of these positions (29-VRKRKFINRD-38) but not K116 (112-SYGEKCLYNV-121). In addition, K116 is not part of the possible recognition motif (GKXXP) for the GCN5-P/CAF family of HATs (40). As such, K116 is one more example where deviations from p300 or P/CAF acetylation consensus motifs occur (46).
Although P/CAF binds to p300 and thus could indirectly form a complex with ER81 in vivo, P/CAF may also bind to ER81 independently of p300. This notion is supported by the fact that ER81 amino acids 249 to 383 strongly interact with P/CAF in vivo but are not sufficient for binding to p300 (39). Furthermore, p300 mutant protein Δ33, which is incapable of binding to P/CAF (41) and whose overexpression can replace that of endogenous p300 from ER81, did not prevent P/CAF from stimulating ER81-dependent transcription (data not shown), again suggesting that P/CAF binds directly to ER81. However, it is highly likely that P/CAF is corecruited by p300 to ER81 and that a multiprotein complex with mutual binding of all members to one another exists in vivo.
Acetylation of transcription factors may exert either stimulatory or inhibitory effects on transcription via a variety of mechanisms. Our study suggests that ER81 function is regulated by acetylation at three different levels, enhancement of DNA binding activity, increase of protein stability, and stimulation of its transactivation potential. Prominently, acetylation has been shown to enhance the DNA binding ability of p53 (20) and E2F1 (33) but disrupts DNA binding by HMG1Y (37). In ER81, acetylation of K116, but not K33, enhances the affinity of ER81 for DNA. Since the DNA binding ETS domain (amino acids 333 to 415) of ER81 does not contain the ER81 acetylation sites, the observed enhancement of DNA binding upon acetylation of K116 seems to be due to a conformational change in ER81 allowing the ETS domain to more avidly bind to DNA.
Acetylation on either K33 or K116 was found to enhance the in vivo _t_1/2 of ER81. A similar observation has been reported for E2F1 (33). One mechanism by which acetylation may stabilize proteins is the prevention of ubiquitination, and thus of ubiquitin-mediated proteasomal degradation, at the same lysine residues (19). However, this is not the case for ER81, since the K33/116R mutant protein had the same _t_1/2 of 2.5 h as wild-type ER81 in the absence of p300, not 7.5 h, as for wild-type ER81 acetylated by p300. Rather, one might speculate that acetylation on lysines 33 and 116 prevents ubiquitination at other lysine residues in ER81 by inducing a conformational change in ER81 or allowing ER81 to interact with proteins that shield it from being recognized by ubiquitin ligases.
A third way in which acetylation furthers ER81-dependent transcription is by increasing the potency of its N-terminal transactivation domain, as shown with the Gal4-ER811-210 fusion proteins. However, the mechanism via which acetylation of ER81 stimulates its transactivation function remains to be studied. One possibility is the recruitment of cofactors or chromatin remodeling complexes that solely interact with acetylated lysine residues via their bromodomains (14).
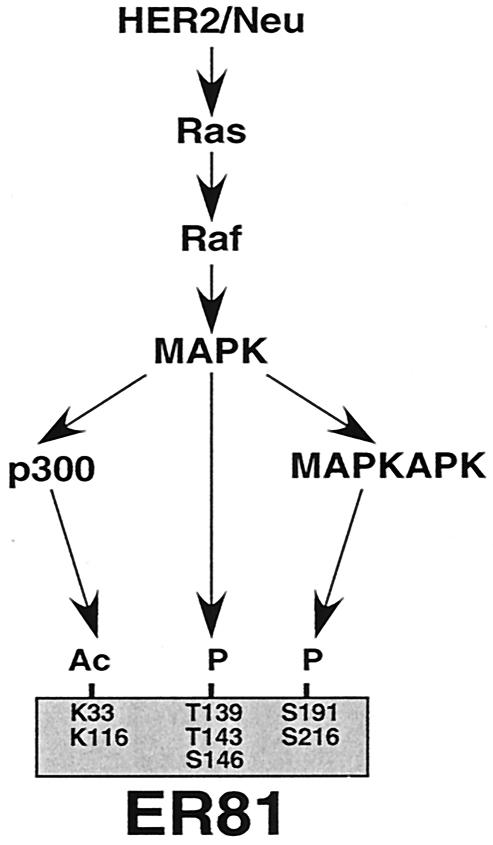
Our previous studies have shown that ER81 is activated through phosphorylation by MAPKs and MAPK-activated protein kinases upon stimulation with HER2/Neu (9, 24, 47). We therefore analyzed the possibility of the existence of a dual regulatory mechanism (acetylation and phosphorylation) for ER81 activity. Indeed, either mutation of the two in vivo acetylation sites or five phosphorylation sites reduced HER2/Neu-mediated activation of ER81, and joint mutation of acetylation and phosphorylation sites nearly eliminated the ability of HER2/Neu to activate ER81-dependent transcription. Thus, both acetylation and phosphorylation of ER81 are required for maximal transactivation (Fig. 11). Similarly, phosphorylation and acetylation appear to jointly activate p53 (42) and B-Myb (28), whereas for human positive coactivator 4 (30) and forkhead transcription factors (32) phosphorylation prevents acetylation and thereby activation of transcription.
FIG. 11.
Model for ER81 as a target of HER2/Neu-induced acetylation and phosphorylation events. MAPKAPK, MAPK-activated protein kinase.
The HAT activity of p300 and CBP can be regulated through phosphorylation. For instance, cyclin E-Cdk2 may phosphorylate CBP and thereby stimulate its HAT activity (2), whereas phosphorylation by protein kinase Cδ appears to inhibit the HAT activity of p300 (54). We showed that HER2/Neu, a receptor tyrosine kinase belonging to the epidermal growth factor receptor family, induces the HAT activity of p300, but not P/CAF, and accordingly also the in vivo acetylation of ER81. Similarly, signaling molecules downstream of HER2/Neu (52) that belong to the Ras→Raf→MAPK pathway, oncogenic Ras-G12V and constitutively active Raf-1, enhanced p300-mediated acetylation of ER81. Thus, the HAT activity of p300 and CBP is likely activated through MAPK phosphorylation upon HER2/Neu expression, consistent with our findings that treating p300 with phosphatase reduces its HAT activity and that HER2/Neu induces in vivo phosphorylation of p300 and also consistent with previous reports showing that ERK1-MAPK phosphorylates CBP and increases CBP HAT activity in vitro (1, 26).
Mutated or overexpressed Ras, Raf, and HER2/Neu are among the most prominent human oncoproteins. Mutations in Ras are found in 30% of all human cancers, whereas overexpression of HER2/Neu has been detected in cancers of the breast, ovary, endometrium, colon, kidney, and lung (22). In particular, HER2/Neu overexpression occurs in ∼30% of all human breast tumors. Interestingly, mice engineered to overexpress HER2/Neu in breast tissue develop mammary tumors displaying enhanced expression of ER81 (44). In addition, ER81 is expressed in human breast tumor specimens and even overexpressed in some human breast tumor cell lines (4, 9). Also, HER2/Neu-activated ER81 may be part of a positive-feedback loop stimulating the expression of HER2/Neu (10). Further, both HER2/Neu-mediated phosphorylation (9) and acetylation of ER81 induce the ability of ER81 to activate gene transcription. Taken together, ER81 appears to be an ideal downstream executioner of the oncogenic potential of HER2/Neu in breast tumors. However, ER81 may also be involved in tumor formation elicited by oncogenic Ras and Raf.
The fact that p300 HAT activity is increased by oncogenic Ras, Raf, HER2/Neu, and probably other growth factor receptors signaling via the MAPK pathway may be relevant not only to ER81 but also to other transcription factors regulated by acetylation. In addition, chromatin can be loosened by histone acetylation upon stimulation of p300 HAT activity. Thus, HER2/Neu, Ras, and Raf may pleiotropically affect gene transcription, unraveling a potential mechanism whereby these signal transduction proteins elicit changes in gene transcription profile necessary for normal ontogenesis and also tumor formation.
Acknowledgments
We thank T. Kouzarides, Y. Nakatani, and P. Overbeek for generously providing reagents.
This work was supported by grant CA085257 from the National Cancer Institute, the Mayo Foundation, and a scholarship (to R.J.) from the Sidney Kimmel Foundation for Cancer Research.
REFERENCES
- 1.Ait-Si-Ali, S., D. Carlisi, S. Ramirez, L. C. Upegui-Gonzalez, A. Duquet, P. Robin, B. Rudkin, A. Harel-Bellan, and D. Trouche. 1999. Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 262**:**157-162. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396**:**184-186. [DOI] [PubMed] [Google Scholar]
- 3.Arber, S., D. R. Ladle, J. H. Lin, E. Frank, and T. M. Jessell. 2000. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell 101**:**485-498. [DOI] [PubMed] [Google Scholar]
- 4.Baert, J. L., D. Monte, E. A. Musgrove, O. Albagli, R. L. Sutherland, and Y. de Launoit. 1997. Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int. J. Cancer 70**:**590-597. [DOI] [PubMed] [Google Scholar]
- 5.Baniahmad, A., A. C. Kohne, and R. Renkawitz. 1992. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 11**:**1015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384**:**641-643. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Levy, R., H. F. Paterson, C. J. Marshall, and Y. Yarden. 1994. A single autophosphorylation site confers oncogenicity to the Neu/ErbB-2 receptor and enables coupling to the MAP kinase pathway. EMBO J. 13**:**3302-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block, C., R. Janknecht, C. Herrmann, N. Nassar, and A. Wittinghofer. 1996. Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat. Struct. Biol. 3**:**244-251. [DOI] [PubMed] [Google Scholar]
- 9.Bosc, D. G., B. S. Goueli, and R. Janknecht. 2001. HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene 20**:**6215-6224. [DOI] [PubMed] [Google Scholar]
- 10.Bosc, D. G., and R. Janknecht. 2002. Regulation of Her2/neu promoter activity by the ETS transcription factor, ER81. J. Cell. Biochem. 86**:**174-183. [DOI] [PubMed] [Google Scholar]
- 11.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84**:**843-851. [DOI] [PubMed] [Google Scholar]
- 12.Bruder, J. T., G. Heidecker, and U. R. Rapp. 1992. Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 6**:**545-556. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Q., D. H. Dowhan, D. Liang, D. D. Moore, and P. A. Overbeek. 2002. CREB-binding protein/p300 co-activation of crystallin gene expression. J. Biol. Chem. 277**:**24081-24089. [DOI] [PubMed] [Google Scholar]
- 14.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399**:**491-496. [DOI] [PubMed] [Google Scholar]
- 15.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8**:**869-884. [DOI] [PubMed] [Google Scholar]
- 16.Gayther, S. A., S. J. Batley, L. Linger, A. Bannister, K. Thorpe, S. F. Chin, Y. Daigo, P. Russell, A. Wilson, H. M. Sowter, J. D. Delhanty, B. A. Ponder, T. Kouzarides, and C. Caldas. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 24**:**300-303. [DOI] [PubMed] [Google Scholar]
- 17.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14**:**1553-1577. [PubMed] [Google Scholar]
- 18.Graves, B. J., and J. M. Petersen. 1998. Specificity within the ets family of transcription factors. Adv. Cancer Res. 75**:**1-55. [DOI] [PubMed] [Google Scholar]
- 19.Gronroos, E., U. Hellman, C. H. Heldin, and J. Ericsson. 2002. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10**:**483-493. [DOI] [PubMed] [Google Scholar]
- 20.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90**:**595-606. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, F., F. Martelli, D. M. Livingston, and Z. Wang. 1996. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 10**:**2949-2959. [DOI] [PubMed] [Google Scholar]
- 22.Hynes, N. E., and D. F. Stern. 1994. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim. Biophys. Acta 1198**:**165-184. [DOI] [PubMed] [Google Scholar]
- 23.Janknecht, R. 1996. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol. Cell. Biol. 16**:**1550-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janknecht, R. 2001. Cell type-specific inhibition of the ETS transcription factor ER81 by mitogen-activated protein kinase-activated protein kinase 2. J. Biol. Chem. 276**:**41856-41861. [DOI] [PubMed] [Google Scholar]
- 25.Janknecht, R. 2002. The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol. Histopathol. 17**:**657-668. [DOI] [PubMed] [Google Scholar]
- 26.Janknecht, R., and A. Nordheim. 1996. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun. 228**:**831-837. [DOI] [PubMed] [Google Scholar]
- 27.Jeon, I. S., J. N. Davis, B. S. Braun, J. E. Sublett, M. F. Roussel, C. T. Denny, and D. N. Shapiro. 1995. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 10**:**1229-1234. [PubMed] [Google Scholar]
- 28.Johnson, L. R., T. K. Johnson, M. Desler, T. A. Luster, T. Nowling, R. E. Lewis, and A. Rizzino. 2002. Effects of B-Myb on gene transcription: phosphorylation-dependent activity and acetylation by p300. J. Biol. Chem. 277**:**4088-4097. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides, T. 1999. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9**:**40-48. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, B. R., V. Swaminathan, S. Banerjee, and T. K. Kundu. 2001. p300-mediated acetylation of human transcriptional coactivator PC4 is inhibited by phosphorylation. J. Biol. Chem. 276**:**16804-16809. [DOI] [PubMed] [Google Scholar]
- 31.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19**:**1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmud, D. L., M. G-Amlak, D. K. Deb, L. C. Platanias, S. Uddin, and A. Wickrema. 2002. Phosphorylation of forkhead transcription factors by erythropoietin and stem cell factor prevents acetylation and their interaction with coactivator p300 in erythroid progenitor cells. Oncogene 21**:**1556-1562. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19**:**662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87**:**1261-1270. [DOI] [PubMed] [Google Scholar]
- 35.Monte, D., L. Coutte, J. L. Baert, I. Angeli, D. Stehelin, and Y. de Launoit. 1995. Molecular characterization of the ets-related human transcription factor ER81. Oncogene 11**:**771-779. [PubMed] [Google Scholar]
- 36.Morrow, C. S., M. Nakagawa, M. E. Goldsmith, M. J. Madden, and K. H. Cowan. 1994. Reversible transcriptional activation of mdr1 by sodium butyrate treatment of human colon cancer cells. J. Biol. Chem. 269**:**10739-10746. [PubMed] [Google Scholar]
- 37.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 2**:**457-467. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87**:**953-959. [DOI] [PubMed] [Google Scholar]
- 39.Papoutsopoulou, S., and R. Janknecht. 2000. Phosphorylation of ETS transcription factor ER81 in a complex with its coactivators CREB-binding protein and p300. Mol. Cell. Biol. 20**:**7300-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polesskaya, A., A. Duquet, I. Naguibneva, C. Weise, A. Vervisch, E. Bengal, F. Hucho, P. Robin, and A. Harel-Bellan. 2000. CREB-binding protein/p300 activates MyoD by acetylation. J. Biol. Chem. 275**:**34359-34364. [DOI] [PubMed] [Google Scholar]
- 41.Reid, J. L., A. J. Bannister, P. Zegerman, M. A. Martinez-Balbas, and T. Kouzarides. 1998. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 17**:**4469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12**:**2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4**:**725-734. [DOI] [PubMed] [Google Scholar]
- 44.Shepherd, T. G., L. Kockeritz, M. R. Szrajber, W. J. Muller, and J. A. Hassell. 2001. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr. Biol. 11**:**1739-1748. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, Y., I. Naruse, T. Maekawa, H. Masuya, T. Shiroishi, and S. Ishii. 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. USA 94**:**10215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, P. R., H. Kurooka, Y. Nakatani, and P. A. Cole. 2001. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J. Biol. Chem. 276**:**33721-33729. [DOI] [PubMed] [Google Scholar]
- 47.Wu, J., and R. Janknecht. 2002. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J. Biol. Chem. 277**:**42669-42679. [DOI] [PubMed] [Google Scholar]
- 48.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26**:**229-232. [DOI] [PubMed] [Google Scholar]
- 49.Yang, W. M., Y. L. Yao, J. M. Sun, J. R. Davie, and E. Seto. 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 272**:**28001-28007. [DOI] [PubMed] [Google Scholar]
- 50.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382**:**319-324. [DOI] [PubMed] [Google Scholar]
- 51.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93**:**361-372. [DOI] [PubMed] [Google Scholar]
- 52.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2**:**127-137. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265**:**17174-17179. [PubMed] [Google Scholar]
- 54.Yuan, L., J. Soh, and I. Weinstein. 2002. Inhibition of histone acetyltransferase function of p300 by PKCdelta. Biochim. Biophys. Acta 1592**:**205. [DOI] [PubMed] [Google Scholar]
- 55.Zanger, K., S. Radovick, and F. E. Wondisford. 2001. CREB binding protein recruitment to the transcription complex requires growth factor-dependent phosphorylation of its GF box. Mol. Cell 7**:**551-558. [DOI] [PubMed] [Google Scholar]