Medical Sequencing at the Extremes of Human Body Mass (original) (raw)
Abstract
Body weight is a quantitative trait with significant heritability in humans. To identify potential genetic contributors to this phenotype, we resequenced the coding exons and splice junctions of 58 genes in 379 obese and 378 lean individuals. Our 96-Mb survey included 21 genes associated with monogenic forms of obesity in humans or mice, as well as 37 genes that function in body weight–related pathways. We found that the monogenic obesity–associated gene group was enriched for rare nonsynonymous variants unique to the obese population compared with the lean population. In addition, computational analysis predicted a greater fraction of deleterious variants within the obese cohort. Together, these data suggest that multiple rare alleles contribute to obesity in the population and provide a medical sequencing-based approach to detect them.
Obesity is reaching epidemic proportions in developed countries and represents a significant risk factor for hypertension, heart disease, diabetes, and dyslipidemia.1 Although the growing prevalence of obesity in the population is thought to be caused by increasing caloric intake and declining energy expenditure,2 individual susceptibility to obesity is strongly influenced by heredity. Twin, adoption, and family studies have indicated that 40%–70% of interindividual variation in BMI is heritable.3,4 In a limited number of cases, single gene defects have been linked to obesity,5 but the majority of cases are thought to be attributable to complex genetic and/or environmental interactions. In this study, we sought to explore the relationship between sequence variation in multiple candidate genes and the extremes of human body mass.
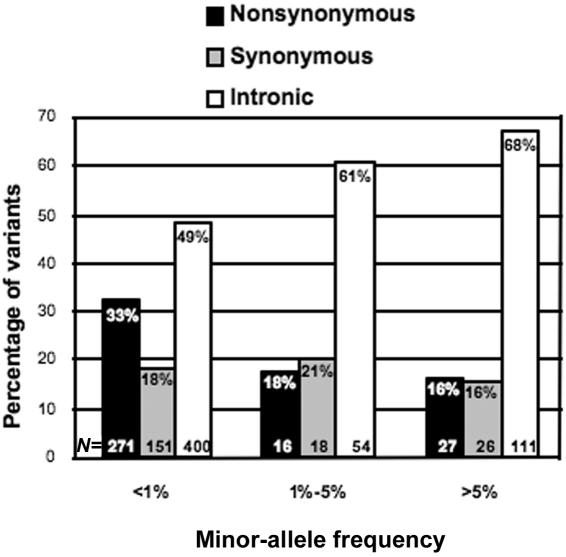
Candidate genes for the study included (a) 21 genes strongly associated with obesity that, when disrupted, lead to monogenic forms of obesity in humans and/or that cause obesity when inactivated in mice and (b) 37 genes involved in regulation of food intake,6 adipogenesis,7 energy expenditure,8 or lipid metabolism (table 1). The coding exons and splice junctions of each gene were sequenced in 379 extremely obese (mean BMI 49.0, >95th percentile adjusted for age and sex; BMI was calculated as weight in kilograms divided by square of height in meters) white men and women ascertained through an obesity clinic at the University of Ottawa and in 378 lean (mean BMI 19.4, <10th percentile adjusted for age and sex) apparently healthy white men and women who participated in a study of leanness at the same institution (table 2). A total of 134 kb (60 kb coding and 74 kb noncoding) was sequenced in each individual, representing 96 Mb of high-quality sequence data, with an average coverage of 734 individuals per exon (table 3). Cumulatively, we identified 1,074 genetic variants (see the tab-delimited ASCII file, which can be imported into a spreadsheet, of data set 1), averaging one variant per 125 bp of the reference human genome sequence. Of the variants, 252 were common polymorphisms (minor-allele frequency >1%), whereas the remaining 822 were rare variants, including 400 noncoding, 150 synonymous, and 272 nonsynonymous variants; the nonsynonymous variants included 3 in-frame indels and 8 severe alleles (6 out-of-frame indels and 2 nonsense changes). In accord with previous large-scale gene-centric sequence analyses,169–171 we observed a paucity of nonsynonymous variants with increasing minor-allele frequency, which is consistent with purifying selection acting on a significant fraction of such DNA sequence changes (fig. 1). Of the 1,074 variants identified in this study, 989 (92%) were not listed in dbSNP (build 124), and, as expected, the majority of these variants (800 [81%] of 989) were rare (i.e., had a minor-allele frequency <1%).
Table 1. .
Summary of Genes and Rare Coding Variants That Are Unique to the Obese or Lean Population
| Obesea | Leana | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Groupand Gene | OMIM | Mouse Knockouts | Mouse Transgenics | Human Mutations | Associations | NS | S | NS | S |
| Monogenic obesity: | |||||||||
| BRS3 | 300107 | Obese9 | None | None | None | 1 | 0 | 3 | 0 |
| CARTPT | 602606 | Obese10 | None | Obese11 | Yes12–14 | 1 | 1 | 0 | 1 |
| FABP4 | 600434 | Obese15 | None | None | Yes16 | 1 | 0 | 2 | 0 |
| HTR2C | 312861 | Obese17 | None | None | Yes18,19 | 1 | 0 | 0 | 0 |
| IL6 | 147620 | Obese20 | None | None | Yes21–27 | 0 | 1 | 0 | 0 |
| LEP | 164160 | Obese28 | Lean29 | Obese30 | Yes31–38 | 0 | 3 | 0 | 1 |
| MC3R | 155540 | Obese39 | None | Obese40 | Yes41,42 | 2 | 0 | 1 | 1 |
| MC4R | 155541 | Obese43 | None | Obese44 | Yes45–49 | 8 | 1 | 2 | 1 |
| NHLH2 | 162361 | Obese50 | None | None | None | 2 | 0 | 1 | 1 |
| NMU | 605103 | Obese51 | None | None | None | 1 | 0 | 1 | 0 |
| NPB | 607996 | Obese52 | None | None | None | 1 | 0 | 2 | 0 |
| NPBWR1 | 600730 | Obese53 | None | None | None | 3 | 0 | 1 | 0 |
| NPY1R | 162641 | Obese54 | None | None | None | 1 | 1 | 2 | 1 |
| NPY2R | 162642 | Obese55 | None | None | Yes56–58 | 2 | 3 | 2 | 0 |
| NPY5R | 602001 | Obese59 | None | None | Yes60 | 1 | 1 | 1 | 0 |
| NR0B2 | 604630 | No apparent phenotype61 | None | Obese62,63 | Yes63–65 | 3 | 0 | 2 | 0 |
| PNPLA2 | 609059 | Obese66 | None | None | None | 3 | 1 | 2 | 2 |
| POMC | 176830 | Obese67 | None | Obese68 | Yes69–77 | 2 | 3 | 1 | 3 |
| PYY | 600781 | Obese78 | None | Obese79 | Yes56–58,80 | 2 | 0 | 1 | 0 |
| SIM1 | 603128 | Obese81 | Lean82 | Obese83,84 | None | 6 | 2 | 0 | 2 |
| UCP3 | 602044 | No apparent phenotype85 | Lean86 | Obese87 | Yes88–99 | 5 | 1 | 2 | 3 |
| Total | 46 | 18 | 26 | 16 | |||||
| Obesity candidate: | |||||||||
| ADIPOQ | 605441 | Insulin resistance100 | None | None | Yes101–107 | 2 | 0 | 2 | 0 |
| AGRP | 602311 | No apparent phenotype108 | Obese109 | None | Yes110,111 | 1 | 1 | 0 | 2 |
| APOA5 | 606368 | Hyperlipidemia112 | Lipid112 | None | Yes113,114 | 1 | 0 | 2 | 1 |
| ARNT2 | 606036 | Lethal115 | None | None | None | 2 | 2 | 3 | 0 |
| ASIP | 600201 | No apparent phenotype116 | Obese117 | None | None | 0 | 0 | 0 | 0 |
| C1QTNF2 | … | None | None | None | None | 1 | 2 | 0 | 2 |
| C3AR1 | 605246 | Hypoallergic118 | None | None | None | 4 | 0 | 4 | 3 |
| CCK | 118440 | No apparent phenotype119 | None | None | None | 0 | 0 | 1 | 0 |
| CPT1B | 601987 | None | None | None | None | 5 | 2 | 7 | 2 |
| CSF2 | 138960 | Pulmonary anomalies120 | None | None | None | 0 | 0 | 0 | 1 |
| DGAT1 | 604900 | Lean121 | None | None | Yes122,123 | 5 | 3 | 2 | 2 |
| DGAT2 | 606983 | Lean124 | None | None | None | 5 | 0 | 3 | 2 |
| GHRL | 605353 | No apparent phenotype125 | None | None | Yes126–129 | 1 | 0 | 0 | 1 |
| GHSR | 601898 | No apparent phenotype130 | None | None | Yes131–133 | 1 | 2 | 2 | 1 |
| HSD11B1 | 600713 | Obesity resistance134 | Obese135 | None | Yes136,137 | 0 | 1 | 1 | 0 |
| HTR7 | 182137 | Hyperthermia138 | None | None | None | 1 | 2 | 1 | 3 |
| INSIG1 | 602055 | None | None | None | None | 0 | 2 | 3 | 0 |
| INSIG2 | 608660 | None | None | None | None | 1 | 2 | 2 | 1 |
| LIPC | 151670 | Hyperlipidemia139 | None | None | Yes140 | 4 | 5 | 2 | 7 |
| NMUR1 | 604153 | None | None | None | None | 4 | 4 | 2 | 1 |
| NMUR2 | 605108 | None | None | None | None | 4 | 0 | 3 | 0 |
| NPBWR2 | 600731 | None | None | None | None | 1 | 2 | 2 | 5 |
| NPY | 162640 | No apparent phenotype141 | None | None | Yes142–145 | 0 | 0 | 0 | 0 |
| NTS | 162650 | No apparent phenotype146 | None | None | None | 0 | 0 | 4 | 0 |
| PPARGC1A | 604517 | Lean147 | None | None | Yes148–153 | 3 | 1 | 4 | 1 |
| PPY | 167780 | None | Lean154 | None | None | 0 | 0 | 1 | 0 |
| PRKAA1 | 602739 | None | None | None | None | 3 | 1 | 4 | 0 |
| PRKAA2 | 600497 | Glucose tolerance155 | None | None | Yes156 | 4 | 2 | 3 | 1 |
| PRKAB1 | 602740 | None | None | None | Yes156 | 0 | 1 | 0 | 0 |
| PRKAB2 | 602741 | None | None | None | Yes156 | 2 | 0 | 1 | 0 |
| PRKAG1 | 602742 | None | None | None | None | 0 | 1 | 0 | 1 |
| PRKAG2 | 602743 | None | None | Heart157 | None | 2 | 0 | 1 | 2 |
| PRKAG3 | 604976 | Glycogen metabolism158 | Glycogen158 | None | None | 10 | 3 | 4 | 1 |
| RETN | 605565 | Gluconeogenesis159 | Obese160 | None | Yes161–166 | 0 | 0 | 1 | 0 |
| SIRT1 | 604479 | Insulin sensitivity167 | None | None | None | 3 | 2 | 1 | 4 |
| TGFBR2 | 190182 | Embryogenesis168 | None | None | None | 1 | 1 | 1 | 1 |
| WDTC1 | … | None | None | None | None | 1 | 0 | 2 | 0 |
| Total | 72 | 42 | 69 | 45 |
Table 2. .
Summary of Individuals Included in This Study
| Variable | Obese Cohort | Lean Cohort |
|---|---|---|
| No. of individuals | 379 | 378 |
| BMIa,b | 49.0 ± 8.8 | 19.4 ± 1.6 |
| BMIa percentile for age and sex | >95th | <10th |
| Ageb (years) | 49.5 ± 10.7 | 45.5 ± 13.0 |
| Female (%) | 63 | 64 |
| Weightb (kg) | 124.8 ± 29.3 | 56.9 ± 9.0 |
| Heightb (cm) | 167.6 ± 10.1 | 170.5 ± 9.2 |
| Waist circumferenceb (cm) | 122.5 ± 20.1 | 75.8 ± 6.5 |
Table 3. .
Sequencing Summary
| Measure | Value |
|---|---|
| No. of genes | 58 |
| No. of exons | 324 |
| Genomic sequence covered (bp): | |
| Total | 134,449 |
| Coding | 60,372 |
| Noncoding | 74,077 |
| Sequence overall (bp): | |
| Total | 96,059,368 |
| Coding | 44,254,489 |
| Noncoding | 51,804,879 |
| Total no. of variants | 1,074 |
| No. of rare variants: | |
| Total | 822 |
| Nonsynonymous | 272 |
| Synonymous | 150 |
| Noncoding | 400 |
| No. of common variants: | |
| Total | 252 |
| Nonsynonymous | 43 |
| Synonymous | 44 |
| Noncoding | 165 |
| No. of novel SNPs covered | 989 |
| No. of known dbSNPs covered | 85 |
| No. of dbSNPs not discovered | 366 |
Figure 1. .
The percentage of nonsynonymous, synonymous, and intronic variants for different minor-allele frequencies. Percentages and the actual number (N) of variants are written inside the bars of the graph.
It has been reported elsewhere that multiple rare variants can have a strong effect on complex traits, especially in the population extremes of a given phenotype.172,173 We therefore examined the frequencies of the nonsynonymous variants in the obese and lean cohorts. Of the 272 rare nonsynonymous changes identified, 213 were unique to one group, with a small excess in the obese population (118 changes) compared with the lean population (95 changes), which did not reach statistical significance. A similar analysis revealed that the prevalence of unique rare synonymous variants, which approximate functionally neutral changes, was essentially identical in the obese and lean cohorts (60 in obese and 61 in lean). We next examined the distributions of nonsynonymous and synonymous variants within each gene individually and found that none of the genes had a statistically significant excess of nonsynonymous variants in the obese or lean group. However, when the genes associated with monogenic forms of obesity were considered together (table 1), unique nonsynonymous variants were significantly more common in the obese group (46 variants in 41 individuals) than in the lean group (26 variants in 27 individuals) (P<.05, by Fisher’s exact test). In contrast, the number of unique synonymous variants in these genes was almost identical among the obese group (18 variants) and lean group (16 variants). It is worth noting that the genes that accounted for the highest difference are MC4R (MIM 155541) (8 variants in obese vs. 2 in lean), SIM1 (MIM 603128) (6 in obese vs. 0 in lean), and UCP3 (MIM 602044) (5 in obese vs. 2 in lean).
The excess of nonsynonymous variants among obese individuals may reflect chance fluctuation in allele frequencies, population stratification, or the accumulation in this group of functional sequence variants that predispose individuals to obesity. Chance fluctuation in allele frequencies seems unlikely, since the excess of nonsynonymous variants in the obese group was not because of an increased number of variants in any single gene, but rather was because of the aggregate contribution of variants at several unlinked loci. Population stratification also seems improbable, since both groups comprised white men and women from the same region (Ottawa, Canada). Furthermore, the number of synonymous variants (table 1) and the allele frequencies of ∼250 common sequence variants (see below) in these genes were similar in the obese and lean groups. Therefore, it seems likely that the excess of rare variants in the obese group represents the accumulation of functional alleles that contribute to the phenotype in these individuals.
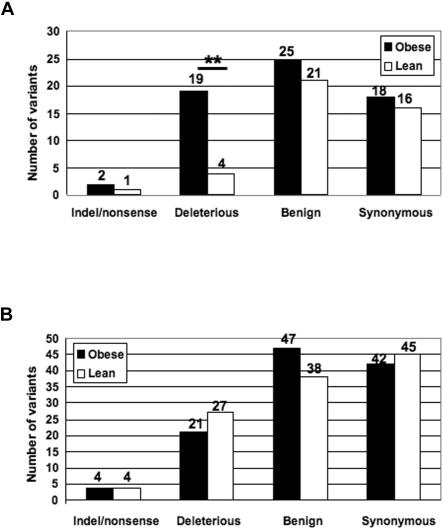
As a first step to assess the functional significance of the nonsynonymous sequence variants identified in the 21 genes associated with monogenic forms of obesity, we used the computer algorithm PolyPhen174 to predict the effects of amino acid substitutions on protein function. We observed that variants identified in the monogenic obesity gene group were more likely to be deleterious in the obese cohort than in the lean cohort (19 in the obese vs. 4 in the lean; P<.002, by exact binomial test) (fig. 2_A_). In comparison, the number of benign variants (25 in the obese and 21 in the lean) and the number of synonymous variants (18 in the obese vs. 16 in lean) in these genes were similar in both cohorts. In contrast, the distribution of synonymous, benign, and deleterious alleles in the 37 candidate genes not associated with monogenic forms of obesity was similar in the obese and lean groups (fig. 2_B_). This finding is consistent with the notion that the excess of nonsynonymous sequence variants among the monogenic obesity genes in the obese cohort reflects the accumulation of functional variants.
Figure 2. .
PolyPhen distribution analysis of variants unique to the obese and lean cohorts. Data are presented for genes with evidence of monogenic involvement in obesity (A) and for genes with a biological plausibility for a role in obesity (B). The number of variants is indicated above each bar of the graph. A double asterisk (**) indicates P<.002.
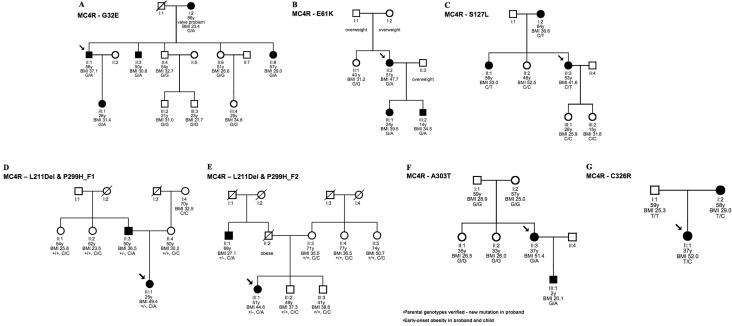
To determine whether nucleotide changes within these genes segregate with BMI, we examined familial segregation for 28 rare variants identified in 14 genes (10 monogenic and 4 candidate genes; see data set 1) in obese kindreds, comprising the proband and all first-degree family members who were available and willing to participate. We used MC4R as a test case, since it is the most common cause of monogenic obesity, estimated to account for 1%–6% of cases of severe obesity.44 In our study, we identified eight nonsynonymous variants that were unique to the obese cohort, compared with two unique variants in the lean cohort (table 1). We found that the mutant MC4R alleles clearly failed to segregate with obesity in three of the six kindreds with six or more family members available (fig. 3), including an allele with a previously characterized frameshift mutation (L211; 4-bp deletion; fig. 3_E_)175 that is almost certainly functional. To corroborate that these MC4R variants were indeed functional, we performed established in vitro functional assays for the novel MC4R variants44 identified in our obese population. Of the six putative mutations analyzed for segregation, five displayed impaired MC4R function (table 4). These findings are consistent with previous studies that also show incomplete correlation between MC4R mutations and obesity,176,177 illustrating the difficulties inherent in determining the correspondence between genotype and phenotype in common complex phenotypes such as obesity. Although several of the kindreds available for study were small, none of the other rare variants examined in 13 additional genes showed significant segregation with BMI in a total of 21 kindreds (data not shown), with the exception of PYY (MIM 600781) Q62P, which we have reported elsewhere.79
Figure 3. .
Familial segregation of MC4R variants and BMI. BMI values are based on the subjects' maximum weight. The arrow indicates the individual sequenced in the cohort. y = years.
Table 4. .
Functional Characterization of MC4R Nonsynonymous Variants in the Obese Cohort
| Results of Functional Studies | |||||||
|---|---|---|---|---|---|---|---|
| Variant | Sequence | n | Known or Novel | alpha-MSH Activation (EC50) | Basal Activity | Summary | Family Segregation Data |
| S30F | tgagt[c/t]ccttg | 1 | Known185 | Not tested alone182 | Not tested alone182 | … | Not tested |
| G32E | ccttg[g/a]aaaag | 1 | Novel | .3 nM | 70% | Minor | Figure 3_A_ |
| E61K | tgttg[g/a]agaat | 1 | Novel | Low | ⩽10% | Severe | Figure 3_B_ |
| S127L | tgact[c/t]ggtga | 1 | Known182 | 29 nM | 80% | Intermediate | Figure 3_C_ |
| L211Dela | ttct[ctct/-]atgt | 2 | Known175 | Truncated receptor | Truncated receptor | Severe | Figure 3_D_ |
| P299Ha | cgatc[c/a]tctga | 2 | Known182 | Negative | ⩽10% | Severe | Figure 3_E_ |
| A303T | tttat[g/a]cactc | 1 | Novel | Low | ⩽10% | Severe | Figure 3_F_ |
| C326R | gcctt[t/c]gtgac | 1 | Novel | .4 nM | 150% | Minor | Figure 3_G_ |
| Wild type | … | … | … | .3 nM | 100% | … | … |
Although the goal of our study was not to perform an exhaustive genetic association study between common variants and BMI, we identified 252 polymorphisms with a frequency >1% and examined the frequency distributions of the variants in the obese and lean cohorts (see data set 1). We found two variants that displayed a significant frequency difference between the two populations: rs6599571 in DGAT1 (MIM 604900) and rs1800832 in NTS (MIM 162650) (both variants are in the 5′ UTR of their gene) (see data set 1). In an attempt to replicate these findings, we compared their frequencies in a second obese cohort (_n_=382; mean BMI 38.6) and a second lean cohort (_n_=381; mean BMI 20.8). For both variants, we observed no significant difference in the allele frequencies between the second cohorts (data not shown), supporting the hypothesis that the initial observation was likely a false-positive discovery or limited to very extreme BMI phenotypes. We should further note that none of the 37 sequenced common variants that were examined elsewhere for their association with BMI displayed a significant frequency difference between our original obese and lean groups (table A1 in appendix A). These results suggest that, in this population, common variants within the coding regions and their proximal exon-intron junctions in this subset of 58 genes are unlikely to contribute appreciably to susceptibility to extreme BMI. However, because we screened primarily the coding sequences and splice junctions of these genes, we cannot exclude the possibility that common sequence variations in noncoding regions that were not sequenced in this study may have significant effects on BMI.
Whereas the heritability of BMI has been firmly established, the identification of genes that contribute to obesity has proved challenging. Genomewide association scans are becoming more feasible, both technologically and economically, and, with them, investigators have begun to systematically explore common variants that influence obesity.178 However, such studies fail to capture rare variants that have also been shown to influence human phenotypes.172,173 Resequencing of candidate genes selected for biological plausibility, in an attempt to capture such rare variants, has, in a few instances, resulted in the identification of obesity-associated variants. For instance, the observation that _Mc4r_-knockout mice are obese43 led to the subsequent finding that mutations in this gene may lead to obesity in humans.175,179 In the present study, we sought to use a similar approach, using a large-scale sequencing strategy with numerous obesity candidate genes in two cohorts with extreme BMI. We did not uncover a large number of novel genes associated with obesity, an endeavor that may have been obstructed by reasons that range from a partial candidate-gene list (58 genes), a large but still limited collection of only white individuals (_n_=∼380 in each group), the sequencing of mainly coding regions, and limited power and availability of subject pedigrees. However, we did identify several genes that warrant further investigation. For instance, we observed a noteworthy rare nonsynonymous variant difference between the obese and lean cohorts for SIM1 (6 variants in obese vs. 0 in lean) and PRKAG3 (10 in obese vs. 4 in lean), suggesting that nonsynonymous variants within these genes may influence susceptibility to obesity. SIM1 is of particular interest because of its strong biological plausibility, including evidence that human chromosomal aberrations within the SIM1 region may lead to obesity,83,84 the observation that Sim1 heterozygous null mice develop obesity,81 and the absence of reported human obesity–associated rare nonsyndromic variants. In addition, we uncovered a significant difference in the total number of nonsynonymous variants in previously characterized monogenic obesity genes between the obese and lean cohorts, indicating that multiple rare variants may have an incomplete effect on this phenotype. Our familial segregation analysis demonstrated that even thoroughly characterized human monogenic obesity genes, such as MC4R, fail to show consistent linkage with BMI, which further suggests that these variants exhibit variable penetrance. Although our analysis encompassed a modest fraction of candidate BMI genes, it strengthens the hypothesis that the majority of genetic etiology that governs obesity is complex and is likely to be influenced by a combination of multiple susceptibility alleles, the majority of which are not independently causative of extreme BMI.
_Subjects.—_Unrelated obese white subjects were recruited from the patient population of the University of Ottawa Weight Management Clinic and the Heart Institute Lipid Clinic by use of criteria reported elsewhere.79 Briefly, inclusion criteria included a BMI >36; a history of obesity for at least 10 years of adult life; no history of treatment with oral glucocorticoids, antipsychotics, or lithium; and no history of medical conditions including major depression, bipolar affective disorder, or psychosis. Unrelated lean subjects of the same ethnic background, with a BMI ⩽10th percentile for age and sex and with no prior history of a BMI >25th percentile for >2 consecutive years were recruited from the Ottawa community (table 2). Subjects were excluded if they had any medical condition that affects weight gain, such as hypo- or hyperthyroidism, eating disorders, major depression, or malabsorption syndromes. The management of phenotypic data was performed using the SAS statistical package (version 9.1 [SAS Institute]). BMI for obese and lean subjects was categorized according to population percentiles for age and sex by use of the Canadian Heart Health Survey data for subjects aged >18 years (data on file; Health Canada) and the National Health and Nutrition Examination Survey data for children.180 This study was approved by the institutional review boards of the University of Ottawa Heart Institute and the Ottawa Hospital, and informed written consent was obtained from all participants. Genomic DNA was extracted from white blood cells by standard methods.181
_Sequencing and data analysis.—_Primers were designed to give a maximum product size of 500 bp and a minimum of 40 bp flanking the splice sites, by use of the Exon Locator and eXtractor for Resequencing program (ELXR Web site). An M13F tag (gttttcccagtcacgacgttgta) and an M13R tag (aggaaacagctatgaccat) were added to forward and reverse primers, respectively. From each sample, 10 ng of DNA was amplified in a 10-μl PCR by use of AmpliTaq Gold (Applied Biosystems) and was cleaned using the PCR product presequencing kit (USB Corporation). Bidirectional sequencing was performed using both of the M13 primers and BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) (JGI Web site), and cleaning was done with tetraethylene glycol before separation on a 3730xl DNA Analyzer (Applied Biosystems). Base calling, quality assessment, and assembly were performed using the Phred, Phrap, Consed (Green Group Web site), and PolyPhred (PolyPhred Web site) software suite. To filter out low-quality sequence, only sequences that had a Phred score ⩾27 were included in the analysis. To minimize false-negative results, we manually analyzed sequence data after PolyPhred analysis at a rank of 5. In addition, every low-quality read was visually examined for indels. All sequence variants identified were verified by manual inspection of the chromatograms, and 156 (99%) of the 157 nonsynonymous variants were verified by a second independent sequencing reaction. All variants were examined by Arlequin (Arlequin Web site), to test for Hardy-Weinberg equilibrium (table A1 in appendix A). Of the 1,074 genetic variants identified, 12 (4 coding and 8 noncoding) had >50% of the data missing in either the lean or the obese panel and thus were removed from further analysis.
_PolyPhen analysis.—_All coding SNPs were subdivided into groups of frameshift/nonsense variants, synonymous variants, and missense variants. Missense variants were further classified with respect to their potential impact on protein structure or function, on the basis of sequence conservation analyzed using a new version of the PolyPhen method.174 PolyPhen relies on the analysis of multiple sequence alignments of homologous proteins, together with functional annotation and structural information if available. The new version of PolyPhen constructs multiple sequence alignments by using a pipeline of several existing programs for alignment of sequences, alignment quality control, and clustering of sequences. Computational prediction methods are statistical in nature; therefore, certain percentages of false-positive (∼10%) and false-negative (20%–30%) predictions are expected. However, application of computational predictions increases power to detect differences in the number of rare functional nonsynonymous variants in candidate genes between populations with different phenotypes.
MC4R _functional analysis.—_Cloning and functional studies of the MC4R mutations were performed as described elsewhere.176,182,183 Briefly, since MC4R is a single-exon gene, mutated alleles were amplified and cloned directly from the genomic DNA of the patient. This also allowed for confirmation of the presence and the nature of the mutations. Human MC4R alleles were cloned into the pcDNA3 expression vector (Invitrogen), to express the native form and the N-terminal FLAG-tagged and/or C-terminal V5His-tagged form of the receptor. All expression vectors were sequenced, to establish the presence of the mutation and the absence of PCR-induced mutations.
For alpha–melanocyte stimulating hormone (alpha-MSH) activation studies, receptors were transiently transfected into an human embryonic kidney (HEK) 293 cell line stably expressing luciferase under the control of a cAMP-responsive promoter.182 Cells were split into 96-well plates 24 h after transfection, and, 36 h after transfection, they were washed and incubated in stimulation medium (Minimum Essential Medium–alpha containing 0.1 mg/ml BSA and 0.25 mM isobutylmethylxanthine) and were stimulated with different concentrations of alpha-MSH (Sigma) for 6 h at 37°C in a 5% CO2 incubator. Luciferase activity, representing cAMP produced in response to alpha-MSH, was assessed using the Steady-Glo Luciferase Assay System (Promega) and a microplate luminescence counter (Packard Instrument). Results were normalized to the maximal stimulation by 8Br-cAMP. Basal activity of the receptors was determined by transient cotransfection with the cAMP-dependent luciferase–expressing plasmid. All experiments were normalized for transfection efficiency by cotransfection of a plasmid encoding the Renilla luciferase–expression plasmid pRL-RSV, to control for transfection efficiency. Data were analyzed using the GraphPad Prism software (GraphPad Software).
_Statistical analysis of common variants.—_Common SNPs were preprocessed to remove triallelic SNPs (one SNP removed) and SNPs for which >50% of the data were missing (three SNPs removed). In addition, we clustered together SNPs that differed in at most three individuals, picking one representative from each such cluster. Standard χ2 tests based on a 3×2 contingency were applied for each of the remaining 252 SNPs, on the basis of a contingency table of genotype-phenotype frequencies. The obtained P values were adjusted for multiple SNP testing by use of the false-discovery-rate procedure.184
Supplementary Material
Data Set 1
Acknowledgments
We thank the Joint Genome Institute’s production sequencing group and Thet Naing, for technical assistance; members of the Rubin lab, for helpful comments on the manuscript; and the many subjects and their families who participated in these studies. Research was conducted at the E. O. Lawrence Berkeley National Laboratory and the Joint Genome Institute, performed under Department of Energy Contract DE-AC0378SF00098, University of California (to L.A.P.). Research performed at the Ottawa Heart Institute was supported by Heart & Stroke Foundation of Ontario grant NA5413 (to R.M.). Subject recruitment was supported in part by a grant from GlaxoSmithKline (to R.M. and R.D.). R.S. was supported by an Alon Fellowship.
Appendix A
Table A1. .
Analysis of Previously Examined Variants for Association with BMI
| No. of Subjects with Genotype | P Value | ||||
|---|---|---|---|---|---|
| Gene and Published Variant (dbSNP) | Sequence | Obese Cohort | Lean Cohort | χ2 Testa | Multiple Testingb |
| ADIPOQ: | |||||
| G15G102–105,107,186 (rs2241766) | cccgg[t/g]catga | TT = 291; GT = 57; GG = 35 | TT = 279; GT = 57; GG = 35 | .969638 | 1 |
| AGRP: | |||||
| A67T110,111 (rs5030980) | aggag[g/a]ctcag | GG = 299; GA = 28; AA = 0 | GG = 323; AG = 26; AA = 0 | .593803 | .935988 |
| APOA5: | |||||
| 5′ UTR114 (rs651821) | agcag[a/g]taatg | AA = 322; AG = 43; GG = 1 | AA = 319; AG = 48; GG = 1 | .594544 | .935988 |
| S16W114 (rs3135506) | gtttt[c/g]ggcca | CC = 331; CG = 38; GG = 1 | CC = 331; CG = 43; GG = 2 | .599993 | .935988 |
| GHRL: | |||||
| R51Q126,127,129 | gcccc[g/a]agctc | GG = 369; GA = 7; AA = 0 | GG = 370; GA = 2; AA = 0 | .0967801 | .633575 |
| L72M126–129 (rs696217) | atgaa[c/a]tggaa | CC = 319; CA = 57; AA = 2 | CC = 323; CA = 48; AA = 1 | .382417 | .935988 |
| GHSR: | |||||
| G57G131,133 (rs495225) | gtggg[t/c]atcgc | TT = 189; TC = 158; CC = 32 | TT = 143; TC = 118; CC = 33 | .478552 | .935988 |
| MC3R: | |||||
| T6K42 (rs3746619) | aaaga[c/a]gtatc | CC = 310; CA = 61; AA = 1 | CC = 311; CA = 59; AA = 2 | .854573 | .980245 |
| V81I41,42 (rs3827103) | ccgag[g/a]ttttc | GG = 314; GA = 62; AA = 1 | GG = 312; GA = 58; AA = 2 | .762315 | .9797 |
| MC4R: | |||||
| I103V46–49 (rs2229616) | ccatt[a/g]tcatc | GG = 369; GA = 10; AA = 0 | GG = 366; GA = 11; AA = 0 | .660644 | .958137 |
| NPY: | |||||
| L7P142,145 (rs16139) | gcgac[t/c]ggggc | TT = 343; TC = 34; CC = 0 | TT = 354; TC = 19; CC = 1 | .0359879 | .633575 |
| S50S144 (rs9785023) | tactc[g/a]gcgct | AA = 95; AG = 209; GG = 73 | AA = 107; AG = 189; GG = 78 | .392291 | .935988 |
| IVS2-116delT144 (rs16134) | (tgaacacctgacaataa/-) | −/− = 95; −/+ = 175; +/+ = 65 | −/− = 95; −/+ = 155; +/+ = 88 | .0974731 | .633575 |
| S68S144 (rs5574) | cgatc[c/t]agccc | TT = 102; TC = 184; CC = 78 | TT = 99; TC = 177; CC = 94 | .44487 | .935988 |
| NPY2R: | |||||
| I195I56–58 (rs1047214) | atcat[t/c]ccgga | TT = 111; TC = 179; CC = 84 | TT = 108; TC = 192; CC = 72 | .493036 | .935988 |
| I312I56,58 (rs2880415) | cacat[t/c]atcgc | TT = 108; TC = 175; CC = 90 | TT = 109; TC = 192; CC = 73 | .277517 | .935988 |
| NPY5R: | |||||
| 3′ UTR60 | ttttg[t/c]taaca | TT = 335; TC = 26; CC = 0 | TT = 317; TC = 41; CC = 1 | .0499633 | .633575 |
| NR0B2: | |||||
| G171A63–65 (rs6659176) | gaaag[g/c]gacca | GG = 320; GC = 37; CC = 2 | GG = 293; GC = 67; CC = 3 | .00666681 | .260006 |
| POMC: | |||||
| C6C76 (rs8192605) | tgctg[c/t]agccg | CC = 364; CT = 7; TT = 0 | CC = 357; CT = 5; TT = 0 | .589687 | .935988 |
| SSG99∧100Ins69,71,73 (rs10654394) | ggc[-/agcagcggc]gca | −/− = 343; −/+ = 31; +/+ = 2 | −/− = 329; −/+ = 46; +/+ = 1 | .0730815 | .633575 |
| A195A76 (rs2071345) | cctgc[c/t]gatga | CC = 374; CT = 0; TT = 0 | CC = 367; CT = 1; TT = 0 | 1 | 1 |
| E214G72 | ggccg[a/g]gaga | AA = 368; AG = 7; GG = 0 | AA = 368; AG = 3; GG = 0 | .208963 | .905505 |
| Y221C77,187 | cccct[a/g]cagga | AA = 374; AG = 1; GG = 0 | AA = 370; AG = 2; GG = 0 | 1 | 1 |
| R236G69,70 | acaag[c/g]gctac | CC = 369; CG = 6; GG = 0 | CC = 366; CG = 6; GG = 0 | .98881 | 1 |
| 3′ UTR74–76 (rs1042571) | gctct[c/t]ccctg | CC = 227; CT = 129; TT = 18 | CC = 236; CT = 117; TT = 18 | .687893 | .958137 |
| PPARGC1A: | |||||
| T394T153 (rs2970847) | aaaac[g/a]gaaat | GG = 226; GA = 107; AA = 14 | GG = 240; GA = 111; AA = 20 | .686798 | .958137 |
| G482S148–153 (rs8192678) | agacc[g/a]gtgaa | GG = 155; GA = 176; AA = 42 | GG = 153; GA = 167; AA = 52 | .519019 | .935988 |
| PYY: | |||||
| R72T56,57,80 (rs1058046) | ggaca[g/c]gcttc | GG = 174; GC = 153; CC = 47 | GG = 166; GC = 173; CC = 34 | .173746 | .847014 |
| IVS3-6156,80 (IVS3+68) (rs162430) | catca[c/t]ttaac | CC = 293; CT = 51; TT = 6 | CC = 299; CT = 67; TT = 5 | .425068 | .935988 |
| RETN: | |||||
| IVS2+39164 (rs3219177) | ggtct[c/t]agaga | CC = 234; CT = 133; TT = 12 | CC = 227; CT = 100; TT = 17 | .138268 | .77035 |
| 3′ UTR162,164 (rs3745368) | tgcgg[g/a]ggagc | GG = 350; GA = 24; AA = 1 | GG = 352; GA = 22; AA = 1 | .760829 | .9797 |
| SIM1: | |||||
| P352T188 (rs3734354) | ccaaa[c/a]cagcc | CC = 277; CA = 96; AA = 8 | CC = 279; CA = 92; AA = 6 | .821095 | .9797 |
| A371V188 (rs3734355) | ggggg[c/t]caaat | CC = 277; CT = 96; TT = 8 | CC = 278; CT = 92; TT = 6 | .828977 | .9797 |
| T653T188 | cccac[c/t]gcact | CC = 369; CT = 14; TT = 0 | CC = 357; CT = 20; TT = 0 | .278331 | .935988 |
| UCP3: | |||||
| Y99Y95,99 (rs1800006) | ctcta[t/c]gactc | TT = 217; TC = 137; CC = 25 | TT = 210; TC = 143; CC = 22 | .81318 | .9797 |
| V102I92,95,96 (rs2734830) | actcc[g/a]tcaag | GG = 378; GA = 1; AA = 0 | GG = 375; GA = 0; AA = 0 | 1 | 1 |
| Y210Y92,95,96,99 (rs2075577) | gacta[t/c]cacct | TT = 191; TC = 107; CC = 85 | TT = 202; TC = 105; CC = 69 | .381994 | .935988 |
Web Resources
The URLs for data presented herein are as follows:
- Arlequin, http://lgb.unige.ch/arlequin/ (for tests of Hardy-Weinberg equilibrium)
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/ (for known SNP analysis)
- ELXR, http://mutation.swmed.edu/ex-lax/ (for primer design)
- Green Group, http://www.phrap.org/ (for Phred, Phrap, and Consed for sequence analysis)
- JGI, http://www.jgi.doe.gov/sequencing/protocols/archive/BigDye3.1auto1.0.doc (for sequencing protocol)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MC4R, SIM1, UCP3, PYY, DGAT1, NTS, and genes in table 1)
- PolyPhen, http://genetics.bwh.harvard.edu/pph/ (for analysis of missense changes)
- PolyPhred, http://droog.mbt.washington.edu/PolyPhred.html (for sequence analysis)
References
- 1.Bell CG, Walley AJ, Froguel P (2005) The genetics of human obesity. Nat Rev Genet 6:221–234 10.1038/nrg1556 [DOI] [PubMed] [Google Scholar]
- 2.French SA, Story M, Jeffery RW (2001) Environmental influences on eating and physical activity. Annu Rev Public Health 22:309–335 10.1146/annurev.publhealth.22.1.309 [DOI] [PubMed] [Google Scholar]
- 3.Comuzzie AG, Allison DB (1998) The search for human obesity genes. Science 280:1374–1377 10.1126/science.280.5368.1374 [DOI] [PubMed] [Google Scholar]
- 4.Friedman JM (2003) A war on obesity, not the obese. Science 299:856–858 10.1126/science.1079856 [DOI] [PubMed] [Google Scholar]
- 5.Perusse L, Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Snyder EE, Bouchard C (2005) The human obesity gene map: the 2004 update. Obes Res 13:381–490 [DOI] [PubMed] [Google Scholar]
- 6.Flier JS (2004) Obesity wars: molecular progress confronts an expanding epidemic. Cell 116:337–350 10.1016/S0092-8674(03)01081-X [DOI] [PubMed] [Google Scholar]
- 7.Harp JB (2004) New insights into inhibitors of adipogenesis. Curr Opin Lipidol 15:303–307 10.1097/00041433-200406000-00010 [DOI] [PubMed] [Google Scholar]
- 8.Lowell BB, Spiegelman BM (2000) Towards a molecular understanding of adaptive thermogenesis. Nature 404:652–660 [DOI] [PubMed] [Google Scholar]
- 9.Ohki-Hamazaki H, Watase K, Yamamoto K, Ogura H, Yamano M, Yamada K, Maeno H, Imaki J, Kikuyama S, Wada E, et al (1997) Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature 390:165–169 10.1038/36568 [DOI] [PubMed] [Google Scholar]
- 10.Asnicar MA, Smith DP, Yang DD, Heiman ML, Fox N, Chen YF, Hsiung HM, Koster A (2001) Absence of cocaine- and amphetamine-regulated transcript results in obesity in mice fed a high caloric diet. Endocrinology 142:4394–4400 10.1210/en.142.10.4394 [DOI] [PubMed] [Google Scholar]
- 11.del Giudice EM, Santoro N, Cirillo G, D’Urso L, Di Toro R, Perrone L (2001) Mutational screening of the CART gene in obese children: identifying a mutation (Leu34Phe) associated with reduced resting energy expenditure and cosegregating with obesity phenotype in a large family. Diabetes 50:2157–2160 10.2337/diabetes.50.9.2157 [DOI] [PubMed] [Google Scholar]
- 12.Challis BG, Yeo GS, Farooqi IS, Luan J, Aminian S, Halsall DJ, Keogh JM, Wareham NJ, O’Rahilly S (2000) The CART gene and human obesity: mutational analysis and population genetics. Diabetes 49:872–875 10.2337/diabetes.49.5.872 [DOI] [PubMed] [Google Scholar]
- 13.Yamada K, Yuan X, Otabe S, Koyanagi A, Koyama W, Makita Z (2002) Sequencing of the putative promoter region of the cocaine- and amphetamine-regulated-transcript gene and identification of polymorphic sites associated with obesity. Int J Obes Relat Metab Disord 26:132–136 10.1038/sj.ijo.0801848 [DOI] [PubMed] [Google Scholar]
- 14.Guerardel A, Barat-Houari M, Vasseur F, Dina C, Vatin V, Clement K, Eberle D, Vasseur-Delannoy V, Bell CG, Galan P, et al (2005) Analysis of sequence variability in the CART gene in relation to obesity in a Caucasian population. BMC Genet 6:19 10.1186/1471-2156-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM (1996) Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 274:1377–1379 10.1126/science.274.5291.1377 [DOI] [PubMed] [Google Scholar]
- 16.Damcott CM, Moffett SP, Feingold E, Barmada MM, Marshall JA, Hamman RF, Ferrell RE (2004) Genetic variation in fatty acid-binding protein-4 and peroxisome proliferator-activated receptor γ interactively influence insulin sensitivity and body composition in males. Metabolism 53:303–309 10.1016/j.metabol.2003.10.010 [DOI] [PubMed] [Google Scholar]
- 17.Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, Dallman MF, Julius D (1995) Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374:542–546 10.1038/374542a0 [DOI] [PubMed] [Google Scholar]
- 18.Pooley EC, Fairburn CG, Cooper Z, Sodhi MS, Cowen PJ, Harrison PJ (2004) A 5-HT2C receptor promoter polymorphism (HTR2C −759C/T) is associated with obesity in women, and with resistance to weight loss in heterozygotes. Am J Med Genet B Neuropsychiatr Genet 126:124–127 10.1002/ajmg.b.20143 [DOI] [PubMed] [Google Scholar]
- 19.McCarthy S, Mottagui-Tabar S, Mizuno Y, Sennblad B, Hoffstedt J, Arner P, Wahlestedt C, Andersson B (2005) Complex HTR2C linkage disequilibrium and promoter associations with body mass index and serum leptin. Hum Genet 117:545–557 10.1007/s00439-005-1328-6 [DOI] [PubMed] [Google Scholar]
- 20.Wallenius V, Wallenius K, Ahren B, Rudling M, Carlsten H, Dickson SL, Ohlsson C, Jansson JO (2002) Interleukin-6-deficient mice develop mature-onset obesity. Nat Med 8:75–79 10.1038/nm0102-75 [DOI] [PubMed] [Google Scholar]
- 21.Berthier MT, Paradis AM, Tchernof A, Bergeron J, Prud’homme D, Despres JP, Vohl MC (2003) The interleukin 6-174G/C polymorphism is associated with indices of obesity in men. J Hum Genet 48:14–19 10.1007/s100380300002 [DOI] [PubMed] [Google Scholar]
- 22.Huang QY, Shen H, Deng HY, Conway T, Davies KM, Li JL, Recker RR, Deng HW (2003) Linkage and association of the CA repeat polymorphism of the IL6 gene, obesity-related phenotypes, and bone mineral density (BMD) in two independent Caucasian populations. J Hum Genet 48:430–437 10.1007/s10038-003-0053-z [DOI] [PubMed] [Google Scholar]
- 23.Kubaszek A, Pihlajamaki J, Punnonen K, Karhapaa P, Vauhkonen I, Laakso M (2003) The C-174G promoter polymorphism of the IL-6 gene affects energy expenditure and insulin sensitivity. Diabetes 52:558–561 10.2337/diabetes.52.2.558 [DOI] [PubMed] [Google Scholar]
- 24.Roth SM, Schrager MA, Lee MR, Metter EJ, Hurley BF, Ferrell RE (2003) Interleukin-6 (IL6) genotype is associated with fat-free mass in men but not women. J Gerontol A Biol Sci Med Sci 58:B1085–B1088 [DOI] [PubMed] [Google Scholar]
- 25.Stephens JW, Hurel SJ, Cooper JA, Acharya J, Miller GJ, Humphries SE (2004) A common functional variant in the interleukin-6 gene is associated with increased body mass index in subjects with type 2 diabetes mellitus. Mol Genet Metab 82:180–186 10.1016/j.ymgme.2004.04.001 [DOI] [PubMed] [Google Scholar]
- 26.Wernstedt I, Eriksson AL, Berndtsson A, Hoffstedt J, Skrtic S, Hedner T, Hulten LM, Wiklund O, Ohlsson C, Jansson JO (2004) A common polymorphism in the interleukin-6 gene promoter is associated with overweight. Int J Obes Relat Metab Disord 28:1272–1279 10.1038/sj.ijo.0802763 [DOI] [PubMed] [Google Scholar]
- 27.Hamid YH, Rose CS, Urhammer SA, Glumer C, Nolsoe R, Kristiansen OP, Mandrup-Poulsen T, Borch-Johnsen K, Jorgensen T, Hansen T, et al (2005) Variations of the interleukin-6 promoter are associated with features of the metabolic syndrome in Caucasian Danes. Diabetologia 48:251–260 10.1007/s00125-004-1623-0 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM (1994) Positional cloning of the mouse obese gene and its human homologue. Nature 372:425–432 10.1038/372425a0 [DOI] [PubMed] [Google Scholar]
- 29.Ogawa Y, Masuzaki H, Hosoda K, Aizawa-Abe M, Suga J, Suda M, Ebihara K, Iwai H, Matsuoka N, Satoh N, et al (1999) Increased glucose metabolism and insulin sensitivity in transgenic skinny mice overexpressing leptin. Diabetes 48:1822–1829 10.2337/diabetes.48.9.1822 [DOI] [PubMed] [Google Scholar]
- 30.Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, Sewter CP, Digby JE, Mohammed SN, Hurst JA, et al (1997) Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature 387:903–908 10.1038/43185 [DOI] [PubMed] [Google Scholar]
- 31.Oksanen L, Ohman M, Heiman M, Kainulainen K, Kaprio J, Mustajoki P, Koivisto V, Koskenvuo M, Janne OA, Peltonen L, et al (1997) Markers for the gene ob and serum leptin levels in human morbid obesity. Hum Genet 99:559–564 10.1007/s004390050406 [DOI] [PubMed] [Google Scholar]
- 32.Butler MG, Hedges L, Hovis CL, Feurer ID (1998) Genetic variants of the human obesity (OB) gene in subjects with and without Prader-Willi syndrome: comparison with body mass index and weight. Clin Genet 54:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li WD, Reed DR, Lee JH, Xu W, Kilker RL, Sodam BR, Price RA (1999) Sequence variants in the 5′ flanking region of the leptin gene are associated with obesity in women. Ann Hum Genet 63:227–234 10.1046/j.1469-1809.1999.6330227.x [DOI] [PubMed] [Google Scholar]
- 34.Le Stunff C, Le Bihan C, Schork NJ, Bougneres P (2000) A common promoter variant of the leptin gene is associated with changes in the relationship between serum leptin and fat mass in obese girls. Diabetes 49:2196–2200 10.2337/diabetes.49.12.2196 [DOI] [PubMed] [Google Scholar]
- 35.Mammes O, Betoulle D, Aubert R, Herbeth B, Siest G, Fumeron F (2000) Association of the G-2548A polymorphism in the 5′ region of the LEP gene with overweight. Ann Hum Genet 64:391–394 10.1017/S0003480000008277 [DOI] [PubMed] [Google Scholar]
- 36.Hoffstedt J, Eriksson P, Mottagui-Tabar S, Arner P (2002) A polymorphism in the leptin promoter region (−2548 G/A) influences gene expression and adipose tissue secretion of leptin. Horm Metab Res 34:355–359 10.1055/s-2002-33466 [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Wilk JB, Borecki I, Williamson S, DeStefano AL, Xu G, Liu J, Ellison RC, Province M, Myers RH (2004) Common variants in the 5′ region of the leptin gene are associated with body mass index in men from the National Heart, Lung, and Blood Institute Family Heart Study. Am J Hum Genet 75:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Lende T, Te Pas MF, Veerkamp RF, Liefers SC (2005) Leptin gene polymorphisms and their phenotypic associations. Vitam Horm 71:373–404 10.1016/S0083-6729(05)71013-X [DOI] [PubMed] [Google Scholar]
- 39.Chen AS, Marsh DJ, Trumbauer ME, Frazier EG, Guan XM, Yu H, Rosenblum CI, Vongs A, Feng Y, Cao L, et al (2000) Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet 26:97–102 10.1038/79254 [DOI] [PubMed] [Google Scholar]
- 40.Lee YS, Poh LK, Loke KY (2002) A novel melanocortin 3 receptor gene (MC3R) mutation associated with severe obesity. J Clin Endocrinol Metab 87:1423–1426 10.1210/jc.87.3.1423 [DOI] [PubMed] [Google Scholar]
- 41.Boucher N, Lanouette CM, Larose M, Perusse L, Bouchard C, Chagnon YC (2002) A +2138InsCAGACC polymorphism of the melanocortin receptor 3 gene is associated in human with fat level and partitioning in interaction with body corpulence. Mol Med 8:158–165 [PMC free article] [PubMed] [Google Scholar]
- 42.Schalin-Jantti C, Valli-Jaakola K, Oksanen L, Martelin E, Laitinen K, Krusius T, Mustajoki P, Heikinheimo M, Kontula K (2003) Melanocortin-3-receptor gene variants in morbid obesity. Int J Obes Relat Metab Disord 27:70–74 10.1038/sj.ijo.0802184 [DOI] [PubMed] [Google Scholar]
- 43.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, et al (1997) Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88:131–141 10.1016/S0092-8674(00)81865-6 [DOI] [PubMed] [Google Scholar]
- 44.Lubrano-Berthelier C, Cavazos M, Dubern B, Shapiro A, Stunff CL, Zhang S, Picart F, Govaerts C, Froguel P, Bougneres P, et al (2003) Molecular genetics of human obesity-associated MC4R mutations. Ann N Y Acad Sci 994:49–57 [DOI] [PubMed] [Google Scholar]
- 45.Chagnon YC, Chen WJ, Perusse L, Chagnon M, Nadeau A, Wilkison WO, Bouchard C (1997) Linkage and association studies between the melanocortin receptors 4 and 5 genes and obesity-related phenotypes in the Quebec Family Study. Mol Med 3:663–673 [PMC free article] [PubMed] [Google Scholar]
- 46.Rosmond R, Chagnon M, Bouchard C, Bjorntorp P (2001) A missense mutation in the human melanocortin-4 receptor gene in relation to abdominal obesity and salivary cortisol. Diabetologia 44:1335–1338 10.1007/s001250100649 [DOI] [PubMed] [Google Scholar]
- 47.Geller F, Reichwald K, Dempfle A, Illig T, Vollmert C, Herpertz S, Siffert W, Platzer M, Hess C, Gudermann T, et al (2004) Melanocortin-4 receptor gene variant I103 is negatively associated with obesity. Am J Hum Genet 74:572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutanen J, Pihlajamaki J, Karhapaa P, Vauhkonen I, Kuusisto J, Moilanen Mykkanen L, Laakso M (2004) The Val103Ile polymorphism of melanocortin-4 receptor regulates energy expenditure and weight gain. Obes Res 12:1060–1066 [DOI] [PubMed] [Google Scholar]
- 49.Heid IM, Vollmert C, Hinney A, Doring A, Geller F, Lowel H, Wichmann HE, Illig T, Hebebrand J, Kronenberg F (2005) Association of the 103I MC4R allele with decreased body mass in 7937 participants of two population based surveys. J Med Genet 42:e21 10.1136/jmg.2004.027011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Good DJ, Porter FD, Mahon KA, Parlow AF, Westphal H, Kirsch IR (1997) Hypogonadism and obesity in mice with a targeted deletion of the Nhlh2 gene. Nat Genet 15:397–401 10.1038/ng0497-397 [DOI] [PubMed] [Google Scholar]
- 51.Hanada R, Teranishi H, Pearson JT, Kurokawa M, Hosoda H, Fukushima N, Fukue Y, Serino R, Fujihara H, Ueta Y, et al (2004) Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat Med 10:1067–1073 10.1038/nm1106 [DOI] [PubMed] [Google Scholar]
- 52.Kelly MA, Beuckmann CT, Williams SC, Sinton CM, Motoike T, Richardson JA, Hammer RE, Garry MG, Yanagisawa M (2005) Neuropeptide B-deficient mice demonstrate hyperalgesia in response to inflammatory pain. Proc Natl Acad Sci USA 102:9942–9947 10.1073/pnas.0503795102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishii M, Fei H, Friedman JM (2003) Targeted disruption of GPR7, the endogenous receptor for neuropeptides B and W, leads to metabolic defects and adult-onset obesity. Proc Natl Acad Sci USA 100:10540–10545 10.1073/pnas.1334189100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kushi A, Sasai H, Koizumi H, Takeda N, Yokoyama M, Nakamura M (1998) Obesity and mild hyperinsulinemia found in neuropeptide Y-Y1 receptor-deficient mice. Proc Natl Acad Sci USA 95:15659–15664 10.1073/pnas.95.26.15659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naveilhan P, Hassani H, Canals JM, Ekstrand AJ, Larefalk A, Chhajlani V, Arenas E, Gedda K, Svensson L, Thoren P, et al (1999) Normal feeding behavior, body weight and leptin response require the neuropeptide Y Y2 receptor. Nat Med 5:1188–1193 10.1038/13514 [DOI] [PubMed] [Google Scholar]
- 56.Hung CC, Pirie F, Luan J, Lank E, Motala A, Yeo GS, Keogh JM, Wareham NJ, O’Rahilly S, Farooqi IS (2004) Studies of the peptide YY and neuropeptide Y2 receptor genes in relation to human obesity and obesity-related traits. Diabetes 53:2461–2466 10.2337/diabetes.53.9.2461 [DOI] [PubMed] [Google Scholar]
- 57.Lavebratt C, Alpman A, Persson B, Arner P, Hoffstedt J (2005) Common neuropeptide Y2 receptor gene variant is protective against obesity among Swedish men. Int J Obes (Lond) 30:453–459 [DOI] [PubMed] [Google Scholar]
- 58.Ma L, Tataranni PA, Hanson RL, Infante AM, Kobes S, Bogardus C, Baier LJ (2005) Variations in peptide YY and Y2 receptor genes are associated with severe obesity in Pima Indian men. Diabetes 54:1598–1602 10.2337/diabetes.54.5.1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marsh DJ, Hollopeter G, Kafer KE, Palmiter RD (1998) Role of the Y5 neuropeptide Y receptor in feeding and obesity. Nat Med 4:718–721 10.1038/nm0698-718 [DOI] [PubMed] [Google Scholar]
- 60.Jenkinson CP, Cray K, Walder K, Herzog H, Hanson R, Ravussin E (2000) Novel polymorphisms in the neuropeptide-Y Y5 receptor associated with obesity in Pima Indians. Int J Obes Relat Metab Disord 24:580–584 10.1038/sj.ijo.0801200 [DOI] [PubMed] [Google Scholar]
- 61.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M (2002) Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell 2:713–720 10.1016/S1534-5807(02)00154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, Kikuchi N, Onigata K, Kojima I, et al (2001) Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci USA 98:575–580 10.1073/pnas.021544398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Echwald SM, Andersen KL, Sorensen TI, Larsen LH, Andersen T, Tonooka N, Tomura H, Takeda J, Pedersen O (2004) Mutation analysis of NR0B2 among 1545 Danish men identifies a novel c.278G>A (p.G93D) variant with reduced functional activity. Hum Mutat 24:381–387 10.1002/humu.20090 [DOI] [PubMed] [Google Scholar]
- 64.Hung CC, Farooqi IS, Ong K, Luan J, Keogh JM, Pembrey M, Yeo GS, Dunger D, Wareham NJ, O'Rahilly S (2003) Contribution of variants in the small heterodimer partner gene to birthweight, adiposity, and insulin levels: mutational analysis and association studies in multiple populations. Diabetes 52:1288–1291 10.2337/diabetes.52.5.1288 [DOI] [PubMed] [Google Scholar]
- 65.Mitchell SM, Weedon MN, Owen KR, Shields B, Wilkins-Wall B, Walker M, McCarthy MI, Frayling TM, Hattersley AT (2003) Genetic variation in the small heterodimer partner gene and young-onset type 2 diabetes, obesity, and birth weight in U.K. subjects. Diabetes 52:1276–1279 10.2337/diabetes.52.5.1276 [DOI] [PubMed] [Google Scholar]
- 66.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al (2006) Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312:734–737 10.1126/science.1123965 [DOI] [PubMed] [Google Scholar]
- 67.Yaswen L, Diehl N, Brennan MB, Hochgeschwender U (1999) Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat Med 5:1066–1070 10.1038/12506 [DOI] [PubMed] [Google Scholar]
- 68.Krude H, Biebermann H, Luck W, Horn R, Brabant G, Gruters A (1998) Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet 19:155–157 10.1038/509 [DOI] [PubMed] [Google Scholar]
- 69.del Giudice EM, Cirillo G, Santoro N, D’Urso L, Carbone MT, Di Toro R, Perrone L (2001) Molecular screening of the proopiomelanocortin (POMC) gene in Italian obese children: report of three new mutations. Int J Obes Relat Metab Disord 25:61–67 10.1038/sj.ijo.0801485 [DOI] [PubMed] [Google Scholar]
- 70.Challis BG, Pritchard LE, Creemers JW, Delplanque J, Keogh JM, Luan J, Wareham NJ, Yeo GS, Bhattacharyya S, Froguel P, et al (2002) A missense mutation disrupting a dibasic prohormone processing site in pro-opiomelanocortin (POMC) increases susceptibility to early-onset obesity through a novel molecular mechanism. Hum Mol Genet 11:1997–2004 10.1093/hmg/11.17.1997 [DOI] [PubMed] [Google Scholar]
- 71.Rosmond R, Ukkola O, Bouchard C, Bjorntorp P (2002) Polymorphisms in exon 3 of the proopiomelanocortin gene in relation to serum leptin, salivary cortisol, and obesity in Swedish men. Metabolism 51:642–644 10.1053/meta.2002.31333 [DOI] [PubMed] [Google Scholar]
- 72.Suviolahti E, Ridderstrale M, Almgren P, Klannemark M, Melander O, Carlsson E, Carlsson M, Hedenbro J, Orho-Melander M (2003) Pro-opiomelanocortin gene is associated with serum leptin levels in lean but not in obese individuals. Int J Obes Relat Metab Disord 27:1204–1211 10.1038/sj.ijo.0802392 [DOI] [PubMed] [Google Scholar]
- 73.Santoro N, del Giudice EM, Cirillo G, Raimondo P, Corsi I, Amato A, Grandone A, Perrone L (2004) An insertional polymorphism of the proopiomelanocortin gene is associated with fasting insulin levels in childhood obesity. J Clin Endocrinol Metab 89:4846–4849 10.1210/jc.2004-0333 [DOI] [PubMed] [Google Scholar]
- 74.Baker M, Gaukrodger N, Mayosi BM, Imrie H, Farrall M, Watkins H, Connell JM, Avery PJ, Keavney B (2005) Association between common polymorphisms of the proopiomelanocortin gene and body fat distribution: a family study. Diabetes 54:2492–2496 10.2337/diabetes.54.8.2492 [DOI] [PubMed] [Google Scholar]
- 75.Chen Y, Snieder H, Wang X, Kaviya B, McCaffrey C, Spector TD, Carter ND, O’Dell SD (2005) Proopiomelanocortin gene variants are associated with serum leptin and body fat in a normal female population. Eur J Hum Genet 13:772–780 10.1038/sj.ejhg.5201407 [DOI] [PubMed] [Google Scholar]
- 76.Sutton BS, Langefeld CD, Williams AH, Norris JM, Saad MF, Haffner SM, Bowden DW (2005) Association of proopiomelanocortin gene polymorphisms with obesity in the IRAS family study. Obes Res 13:1491–1498 [DOI] [PubMed] [Google Scholar]
- 77.Lee YS, Challis BG, Thompson DA, Yeo GS, Keogh JM, Madonna ME, Wraight V, Sims M, Vatin V, Meyre D, et al (2006) A POMC variant implicates β-melanocyte-stimulating hormone in the control of human energy balance. Cell Metab 3:135–140 10.1016/j.cmet.2006.01.006 [DOI] [PubMed] [Google Scholar]
- 78.Boey D, Lin S, Karl T, Baldock P, Lee N, Enriquez R, Couzens M, Slack K, Dallmann R, Sainsbury A, et al (2006) Peptide YY ablation in mice leads to the development of hyperinsulinaemia and obesity. Diabetologia 49:1360–1370 10.1007/s00125-006-0237-0 [DOI] [PubMed] [Google Scholar]
- 79.Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Collier JM, Hebert S, Doelle H, Dent R, Pennacchio LA, McPherson R (2006) A PYY Q62P variant linked to human obesity. Hum Mol Genet 15:387–391 10.1093/hmg/ddi455 [DOI] [PubMed] [Google Scholar]
- 80.Torekov SS, Larsen LH, Glumer C, Borch-Johnsen K, Jorgensen T, Holst JJ, Madsen OD, Hansen T, Pedersen O (2005) Evidence of an association between the Arg72 allele of the peptide YY and increased risk of type 2 diabetes. Diabetes 54:2261–2265 10.2337/diabetes.54.7.2261 [DOI] [PubMed] [Google Scholar]
- 81.Michaud JL, Boucher F, Melnyk A, Gauthier F, Goshu E, Levy E, Mitchell GA, Himms-Hagen J, Fan CM (2001) Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum Mol Genet 10:1465–1473 10.1093/hmg/10.14.1465 [DOI] [PubMed] [Google Scholar]
- 82.Kublaoui BM, Holder JL Jr, Tolson KP, Gemelli T, Zinn AR (2006) SIM1 overexpression partially rescues agouti yellow and diet-induced obesity by normalizing food intake. Endocrinology 147:4542–4549 10.1210/en.2006-0453 [DOI] [PubMed] [Google Scholar]
- 83.Holder JL Jr, Butte NF, Zinn AR (2000) Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum Mol Genet 9:101–108 10.1093/hmg/9.1.101 [DOI] [PubMed] [Google Scholar]
- 84.Faivre L, Cormier-Daire V, Lapierre JM, Colleaux L, Jacquemont S, Genevieve D, Saunier P, Munnich A, Turleau C, Romana S, et al (2002) Deletion of the SIM1 gene (6q16.2) in a patient with a Prader-Willi-like phenotype. J Med Genet 39:594–596 10.1136/jmg.39.8.594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gong DW, Monemdjou S, Gavrilova O, Leon LR, Marcus-Samuels B, Chou CJ, Everett C, Kozak LP, Li C, Deng C, et al (2000) Lack of obesity and normal response to fasting and thyroid hormone in mice lacking uncoupling protein-3. J Biol Chem 275:16251–16257 10.1074/jbc.M910177199 [DOI] [PubMed] [Google Scholar]
- 86.Clapham JC, Arch JR, Chapman H, Haynes A, Lister C, Moore GB, Piercy V, Carter SA, Lehner I, Smith SA, et al (2000) Mice overexpressing human uncoupling protein-3 in skeletal muscle are hyperphagic and lean. Nature 406:415–418 10.1038/35019082 [DOI] [PubMed] [Google Scholar]
- 87.Argyropoulos G, Brown AM, Willi SM, Zhu J, He Y, Reitman M, Gevao SM, Spruill I, Garvey WT (1998) Effects of mutations in the human uncoupling protein 3 gene on the respiratory quotient and fat oxidation in severe obesity and type 2 diabetes. J Clin Invest 102:1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Otabe S, Clement K, Dubois S, Lepretre F, Pelloux V, Leibel R, Chung W, Boutin P, Guy-Grand B, Froguel P, et al (1999) Mutation screening and association studies of the human uncoupling protein 3 gene in normoglycemic and diabetic morbidly obese patients. Diabetes 48:206–208 [PubMed] [Google Scholar]
- 89.Cassell PG, Saker PJ, Huxtable SJ, Kousta E, Jackson AE, Hattersley AT, Frayling TM, Walker M, Kopelman PG, Ramachandran A, et al (2000) Evidence that single nucleotide polymorphism in the uncoupling protein 3 (UCP3) gene influences fat distribution in women of European and Asian origin. Diabetologia 43:1558–1564 10.1007/s001250051569 [DOI] [PubMed] [Google Scholar]
- 90.Otabe S, Clement K, Dina C, Pelloux V, Guy-Grand B, Froguel P, Vasseur F (2000) A genetic variation in the 5′ flanking region of the UCP3 gene is associated with body mass index in humans in interaction with physical activity. Diabetologia 43:245–249 10.1007/s001250050037 [DOI] [PubMed] [Google Scholar]
- 91.Halsall DJ, Luan J, Saker P, Huxtable S, Farooqi IS, Keogh J, Wareham NJ, O’Rahilly S (2001) Uncoupling protein 3 genetic variants in human obesity: the c-55t promoter polymorphism is negatively correlated with body mass index in a UK Caucasian population. Int J Obes Relat Metab Disord 25:472–477 10.1038/sj.ijo.0801584 [DOI] [PubMed] [Google Scholar]
- 92.Lanouette CM, Giacobino JP, Perusse L, Lacaille M, Yvon C, Chagnon M, Kuhne F, Bouchard C, Muzzin P, Chagnon YC (2001) Association between uncoupling protein 3 gene and obesity-related phenotypes in the Quebec Family Study. Mol Med 7:433–441 [PMC free article] [PubMed] [Google Scholar]
- 93.Ukkola O, Tremblay A, Sun G, Chagnon YC, Bouchard C (2001) Genetic variation at the uncoupling protein 1, 2 and 3 loci and the response to long-term overfeeding. Eur J Clin Nutr 55:1008–1015 10.1038/sj.ejcn.1601261 [DOI] [PubMed] [Google Scholar]
- 94.Yanagisawa Y, Hasegawa K, Dever GJ, Otto CT, Sakuma M, Shibata S, Miyagi S, Kaneko Y, Kagawa Y (2001) Uncoupling protein 3 and peroxisome proliferator-activated receptor γ2 contribute to obesity and diabetes in Palauans. Biochem Biophys Res Commun 281:772–778 10.1006/bbrc.2001.4417 [DOI] [PubMed] [Google Scholar]
- 95.Kimm SY, Glynn NW, Aston CE, Damcott CM, Poehlman ET, Daniels SR, Ferrell RE (2002) Racial differences in the relation between uncoupling protein genes and resting energy expenditure. Am J Clin Nutr 75:714–719 [DOI] [PubMed] [Google Scholar]
- 96.Lanouette CM, Chagnon YC, Rice T, Perusse L, Muzzin P, Giacobino JP, Gagnon J, Wilmore JH, Leon AS, Skinner JS, et al (2002) Uncoupling protein 3 gene is associated with body composition changes with training in HERITAGE study. J Appl Physiol 92:1111–1118 [DOI] [PubMed] [Google Scholar]
- 97.Damcott CM, Feingold E, Moffett SP, Barmada MM, Marshall JA, Hamman RF, Ferrell RE (2004) Genetic variation in uncoupling protein 3 is associated with dietary intake and body composition in females. Metabolism 53:458–464 10.1016/j.metabol.2003.11.019 [DOI] [PubMed] [Google Scholar]
- 98.Lindholm E, Klannemark M, Agardh E, Groop L, Agardh CD (2004) Putative role of polymorphisms in UCP1-3 genes for diabetic nephropathy. J Diabetes Complicat 18:103–107 10.1016/S1056-8727(03)00019-9 [DOI] [PubMed] [Google Scholar]
- 99.Liu YJ, Liu PY, Long J, Lu Y, Elze L, Recker RR, Deng HW (2005) Linkage and association analyses of the UCP3 gene with obesity phenotypes in Caucasian families. Physiol Genomics 22:197–203 10.1152/physiolgenomics.00031.2005 [DOI] [PubMed] [Google Scholar]
- 100.Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, et al (2002) Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med 8:731–737 10.1038/nm724 [DOI] [PubMed] [Google Scholar]
- 101.Menzaghi C, Ercolino T, Di Paola R, Berg AH, Warram JH, Scherer PE, Trischitta V, Doria A (2002) A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes 51:2306–2312 10.2337/diabetes.51.7.2306 [DOI] [PubMed] [Google Scholar]
- 102.Stumvoll M, Tschritter O, Fritsche A, Staiger H, Renn W, Weisser M, Machicao F, Haring H (2002) Association of the T-G polymorphism in adiponectin (exon 2) with obesity and insulin sensitivity: interaction with family history of type 2 diabetes. Diabetes 51:37–41 10.2337/diabetes.51.1.37 [DOI] [PubMed] [Google Scholar]
- 103.Yang WS, Tsou PL, Lee WJ, Tseng DL, Chen CL, Peng CC, Lee KC, Chen MJ, Huang CJ, Tai TY, et al (2003) Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J Mol Med 81:428–434 10.1007/s00109-002-0409-4 [DOI] [PubMed] [Google Scholar]
- 104.Filippi E, Sentinelli F, Trischitta V, Romeo S, Arca M, Leonetti F, Di Mario U, Baroni MG (2004) Association of the human adiponectin gene and insulin resistance. Eur J Hum Genet 12:199–205 10.1038/sj.ejhg.5201120 [DOI] [PubMed] [Google Scholar]
- 105.Fumeron F, Aubert R, Siddiq A, Betoulle D, Pean F, Hadjadj S, Tichet J, Wilpart E, Chesnier MC, Balkau B, et al (2004) Adiponectin gene polymorphisms and adiponectin levels are independently associated with the development of hyperglycemia during a 3-year period: the epidemiologic data on the insulin resistance syndrome prospective study. Diabetes 53:1150–1157 10.2337/diabetes.53.4.1150 [DOI] [PubMed] [Google Scholar]
- 106.Gu HF, Abulaiti A, Ostenson CG, Humphreys K, Wahlestedt C, Brookes AJ, Efendic S (2004) Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish Caucasians. Diabetes 53:S31–S35 10.2337/diabetes.53.2007.S31 [DOI] [PubMed] [Google Scholar]
- 107.Hu FB, Doria A, Li T, Meigs JB, Liu S, Memisoglu A, Hunter D, Manson JE (2004) Genetic variation at the adiponectin locus and risk of type 2 diabetes in women. Diabetes 53:209–213 10.2337/diabetes.53.1.209 [DOI] [PubMed] [Google Scholar]
- 108.Qian S, Chen H, Weingarth D, Trumbauer ME, Novi DE, Guan X, Yu H, Shen Z, Feng Y, Frazier E, et al (2002) Neither agouti-related protein nor neuropeptide Y is critically required for the regulation of energy homeostasis in mice. Mol Cell Biol 22:5027–5035 10.1128/MCB.22.14.5027-5035.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS (1997) Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 278:135–138 10.1126/science.278.5335.135 [DOI] [PubMed] [Google Scholar]
- 110.Argyropoulos G, Rankinen T, Neufeld DR, Rice T, Province MA, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C (2002) A polymorphism in the human agouti-related protein is associated with late-onset obesity. J Clin Endocrinol Metab 87:4198–4202 10.1210/jc.2002-011834 [DOI] [PubMed] [Google Scholar]
- 111.Marks DL, Boucher N, Lanouette CM, Perusse L, Brookhart G, Comuzzie AG, Chagnon YC, Cone RD (2004) Ala67Thr polymorphism in the Agouti-related peptide gene is associated with inherited leanness in humans. Am J Med Genet A 126:267–271 10.1002/ajmg.a.20600 [DOI] [PubMed] [Google Scholar]
- 112.Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM (2001) An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294:169–173 10.1126/science.1064852 [DOI] [PubMed] [Google Scholar]
- 113.Aberle J, Evans D, Beil FU, Seedorf U (2005) A polymorphism in the apolipoprotein A5 gene is associated with weight loss after short-term diet. Clin Genet 68:152–154 10.1111/j.1399-0004.2005.00463.x [DOI] [PubMed] [Google Scholar]
- 114.Elosua R, Ordovas JM, Cupples LA, Lai CQ, Demissie S, Fox CS, Polak JF, Wolf PA, D’Agostino RA, O’Donnell CJ (2006) Variants at the APOA5 locus, association with carotid atherosclerosis, and modification by obesity: the Framingham Study. J Lipid Res 47:990–996 10.1194/jlr.M500446-JLR200 [DOI] [PubMed] [Google Scholar]
- 115.Keith B, Adelman DM, Simon MC (2001) Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc Natl Acad Sci USA 98:6692–6697 10.1073/pnas.121494298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Voisey J, van Daal A (2002) Agouti: from mouse to man, from skin to fat. Pigment Cell Res 15:10–18 10.1034/j.1600-0749.2002.00039.x [DOI] [PubMed] [Google Scholar]
- 117.Klebig ML, Wilkinson JE, Geisler JG, Woychik RP (1995) Ectopic expression of the agouti gene in transgenic mice causes obesity, features of type II diabetes, and yellow fur. Proc Natl Acad Sci USA 92:4728–4732 10.1073/pnas.92.11.4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Humbles AA, Lu B, Nilsson CA, Lilly C, Israel E, Fujiwara Y, Gerard NP, Gerard C (2000) A role for the C3a anaphylatoxin receptor in the effector phase of asthma. Nature 406:998–1001 10.1038/35023175 [DOI] [PubMed] [Google Scholar]
- 119.Lacourse KA, Swanberg LJ, Gillespie PJ, Rehfeld JF, Saunders TL, Samuelson LC (1999) Pancreatic function in CCK-deficient mice: adaptation to dietary protein does not require CCK. Am J Physiol 276:G1302–G1309 [DOI] [PubMed] [Google Scholar]
- 120.Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, Dickersin GR, Bachurski CJ, Mark EL, Whitsett JA, et al (1994) Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science 264:713–716 10.1126/science.8171324 [DOI] [PubMed] [Google Scholar]
- 121.Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RV Jr (2000) Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat Genet 25:87–90 10.1038/75651 [DOI] [PubMed] [Google Scholar]
- 122.Ludwig EH, Mahley RW, Palaoglu E, Ozbayrakci S, Balestra ME, Borecki IB, Innerarity TL, Farese RV Jr (2002) DGAT1 promoter polymorphism associated with alterations in body mass index, high density lipoprotein levels and blood pressure in Turkish women. Clin Genet 62:68–73 10.1034/j.1399-0004.2002.620109.x [DOI] [PubMed] [Google Scholar]
- 123.Coudreau SK, Tounian P, Bonhomme G, Froguel P, Girardet JP, Guy-Grand B, Basdevant A, Clement K (2003) Role of the DGAT gene C79T single-nucleotide polymorphism in French obese subjects. Obes Res 11:1163–1167 [DOI] [PubMed] [Google Scholar]
- 124.Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, Farese RV Jr (2004) Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J Biol Chem 279:11767–11776 10.1074/jbc.M311000200 [DOI] [PubMed] [Google Scholar]
- 125.Sun Y, Ahmed S, Smith RG (2003) Deletion of ghrelin impairs neither growth nor appetite. Mol Cell Biol 23:7973–7981 10.1128/MCB.23.22.7973-7981.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ukkola O, Ravussin E, Jacobson P, Snyder EE, Chagnon M, Sjostrom L, Bouchard C (2001) Mutations in the preproghrelin/ghrelin gene associated with obesity in humans. J Clin Endocrinol Metab 86:3996–3999 10.1210/jc.86.8.3996 [DOI] [PubMed] [Google Scholar]
- 127.Hinney A, Hoch A, Geller F, Schafer H, Siegfried W, Goldschmidt H, Remschmidt H, Hebebrand J (2002) Ghrelin gene: identification of missense variants and a frameshift mutation in extremely obese children and adolescents and healthy normal weight students. J Clin Endocrinol Metab 87:2716 10.1210/jc.87.6.2716 [DOI] [PubMed] [Google Scholar]
- 128.Korbonits M, Gueorguiev M, O’Grady E, Lecoeur C, Swan DC, Mein CA, Weill J, Grossman AB, Froguel P (2002) A variation in the ghrelin gene increases weight and decreases insulin secretion in tall, obese children. J Clin Endocrinol Metab 87:4005–4008 10.1210/jc.87.8.4005 [DOI] [PubMed] [Google Scholar]
- 129.Ukkola O, Ravussin E, Jacobson P, Perusse L, Rankinen T, Tschop M, Heiman ML, Leon AS, Rao DC, Skinner JS, et al (2002) Role of ghrelin polymorphisms in obesity based on three different studies. Obes Res 10:782–791 [DOI] [PubMed] [Google Scholar]
- 130.Sun Y, Wang P, Zheng H, Smith RG (2004) Ghrelin stimulation of growth hormone release and appetite is mediated through the growth hormone secretagogue receptor. Proc Natl Acad Sci USA 101:4679–4684 10.1073/pnas.0305930101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang HJ, Geller F, Dempfle A, Schauble N, Friedel S, Lichtner P, Fontenla-Horro F, Wudy S, Hagemann S, Gortner L, et al (2004) Ghrelin receptor gene: identification of several sequence variants in extremely obese children and adolescents, healthy normal-weight and underweight students, and children with short normal stature. J Clin Endocrinol Metab 89:157–162 10.1210/jc.2003-031395 [DOI] [PubMed] [Google Scholar]
- 132.Baessler A, Hasinoff MJ, Fischer M, Reinhard W, Sonnenberg GE, Olivier M, Erdmann J, Schunkert H, Doering A, Jacob HJ, et al (2005) Genetic linkage and association of the growth hormone secretagogue receptor (ghrelin receptor) gene in human obesity. Diabetes 54:259–267 10.2337/diabetes.54.1.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miyasaka K, Hosoya H, Sekime A, Ohta M, Amono H, Matsushita S, Suzuki K, Higuchi S, Funakoshi A (2005) Association of ghrelin receptor gene polymorphism with bulimia nervosa in a Japanese population. J Neural Transm 113:1279–1285 10.1007/s00702-005-0393-2 [DOI] [PubMed] [Google Scholar]
- 134.Kotelevtsev Y, Holmes MC, Burchell A, Houston PM, Schmoll D, Jamieson P, Best R, Brown R, Edwards CR, Seckl JR, et al (1997) 11β-hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid-inducible responses and resist hyperglycemia on obesity or stress. Proc Natl Acad Sci USA 94:14924–14929 10.1073/pnas.94.26.14924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, Flier JS (2001) A transgenic model of visceral obesity and the metabolic syndrome. Science 294:2166–2170 10.1126/science.1066285 [DOI] [PubMed] [Google Scholar]
- 136.Draper N, Echwald SM, Lavery GG, Walker EA, Fraser R, Davies E, Sorensen TI, Astrup A, Adamski J, Hewison M, et al (2002) Association studies between microsatellite markers within the gene encoding human 11β-hydroxysteroid dehydrogenase type 1 and body mass index, waist to hip ratio, and glucocorticoid metabolism. J Clin Endocrinol Metab 87:4984–4990 10.1210/jc.2001-011375 [DOI] [PubMed] [Google Scholar]
- 137.Gelernter-Yaniv L, Feng N, Sebring NG, Hochberg Z, Yanovski JA (2003) Associations between a polymorphism in the 11 beta hydroxysteroid dehydrogenase type I gene and body composition. Int J Obes Relat Metab Disord 27:983–986 10.1038/sj.ijo.0802327 [DOI] [PubMed] [Google Scholar]
- 138.Hedlund PB, Danielson PE, Thomas EA, Slanina K, Carson MJ, Sutcliffe JG (2003) No hypothermic response to serotonin in 5-HT7 receptor knockout mice. Proc Natl Acad Sci USA 100:1375–1380 10.1073/pnas.0337340100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Homanics GE, de Silva HV, Osada J, Zhang SH, Wong H, Borensztajn J, Maeda N (1995) Mild dyslipidemia in mice following targeted inactivation of the hepatic lipase gene. J Biol Chem 270:2974–2980 10.1074/jbc.270.7.2974 [DOI] [PubMed] [Google Scholar]
- 140.St-Pierre J, Miller-Felix I, Paradis ME, Bergeron J, Lamarche B, Despres JP, Gaudet D, Vohl MC (2003) Visceral obesity attenuates the effect of the hepatic lipase −514C>T polymorphism on plasma HDL-cholesterol levels in French-Canadian men. Mol Genet Metab 78:31–36 10.1016/S1096-7192(02)00223-8 [DOI] [PubMed] [Google Scholar]
- 141.Erickson JC, Clegg KE, Palmiter RD (1996) Sensitivity to leptin and susceptibility to seizures of mice lacking neuropeptide Y. Nature 381:415–421 10.1038/381415a0 [DOI] [PubMed] [Google Scholar]
- 142.Karvonen MK, Pesonen U, Koulu M, Niskanen L, Laakso M, Rissanen A, Dekker JM, Hart LM, Valve R, Uusitupa MI (1998) Association of a leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y with high serum cholesterol and LDL cholesterol levels. Nat Med 4:1434–1437 10.1038/4027 [DOI] [PubMed] [Google Scholar]
- 143.Uusitupa MI, Karvonen MK, Pesonen U, Koulu M (1998) Neuropeptide Y: a novel link between the neuroendocrine system and cholesterol metabolism. Ann Med 30:508–510 [DOI] [PubMed] [Google Scholar]
- 144.Bray MS, Boerwinkle E, Hanis CL (2000) Sequence variation within the neuropeptide Y gene and obesity in Mexican Americans. Obes Res 8:219–226 [DOI] [PubMed] [Google Scholar]
- 145.Karvonen MK, Koulu M, Pesonen U, Uusitupa MI, Tammi A, Viikari J, Simell O, Ronnemaa T (2000) Leucine 7 to proline 7 polymorphism in the preproneuropeptide Y is associated with birth weight and serum triglyceride concentration in preschool aged children. J Clin Endocrinol Metab 85:1455–1460 10.1210/jc.85.4.1455 [DOI] [PubMed] [Google Scholar]
- 146.Dobner PR, Fadel J, Deitemeyer N, Carraway RE, Deutch AY (2001) Neurotensin-deficient mice show altered responses to antipsychotic drugs. Proc Natl Acad Sci USA 98:8048–8053 10.1073/pnas.141042198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al (2004) Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121–135 10.1016/j.cell.2004.09.013 [DOI] [PubMed] [Google Scholar]
- 148.Esterbauer H, Oberkofler H, Linnemayr V, Iglseder B, Hedegger M, Wolfsgruber P, Paulweber B, Fastner G, Krempler F, Patsch W (2002) Peroxisome proliferator-activated receptor-γ coactivator-1 gene locus: associations with obesity indices in middle-aged women. Diabetes 51:1281–1286 10.2337/diabetes.51.4.1281 [DOI] [PubMed] [Google Scholar]
- 149.Muller YL, Bogardus C, Pedersen O, Baier L (2003) A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes 52:895–898 10.2337/diabetes.52.3.895 [DOI] [PubMed] [Google Scholar]
- 150.Fanelli M, Filippi E, Sentinelli F, Romeo S, Fallarino M, Buzzetti R, Leonetti F, Baroni MG (2005) The Gly482Ser missense mutation of the peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) gene associates with reduced insulin sensitivity in normal and glucose-intolerant obese subjects. Dis Markers 21:175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sookoian S, Garcia SI, Porto PI, Dieuzeide G, Gonzalez CD, Pirola CJ (2005) Peroxisome proliferator-activated receptor gamma and its coactivator-1 alpha may be associated with features of the metabolic syndrome in adolescents. J Mol Endocrinol 35:373–380 10.1677/jme.1.01837 [DOI] [PubMed] [Google Scholar]
- 152.Ridderstrale M, Johansson LE, Rastam L, Lindblad U (2006) Increased risk of obesity associated with the variant allele of the PPARGC1A Gly482Ser polymorphism in physically inactive elderly men. Diabetologia 49:496–500 10.1007/s00125-005-0129-8 [DOI] [PubMed] [Google Scholar]
- 153.Vimaleswaran KS, Radha V, Anjana M, Deepa R, Ghosh S, Majumder PP, Rao MR, Mohan V (2006) Effect of polymorphisms in the PPARGC1A gene on body fat in Asian Indians. Int J Obes (Lond) 30:884–891 [DOI] [PubMed] [Google Scholar]
- 154.Ueno N, Inui A, Iwamoto M, Kaga T, Asakawa A, Okita M, Fujimiya M, Nakajima Y, Ohmoto Y, Ohnaka M, et al (1999) Decreased food intake and body weight in pancreatic polypeptide-overexpressing mice. Gastroenterology 117:1427–1432 10.1016/S0016-5085(99)70293-3 [DOI] [PubMed] [Google Scholar]
- 155.Viollet B, Andreelli F, Jorgensen SB, Perrin C, Geloen A, Flamez D, Mu J, Lenzner C, Baud O, Bennoun M, et al (2003) The AMP-activated protein kinase α2 catalytic subunit controls whole-body insulin sensitivity. J Clin Invest 111:91–98 10.1172/JCI200316567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sun MW, Lee JY, de Bakker PI, Burtt NP, Almgren P, Rastam L, Tuomi T, Gaudet D, Daly MJ, Hirschhorn JN, et al (2006) Haplotype structures and large-scale association testing of the 5′ AMP-activated protein kinase genes PRKAA2, PRKAB1, and PRKAB1 with type 2 diabetes. Diabetes 55:849–855 10.2337/diabetes.55.03.06.db05-1418 [DOI] [PubMed] [Google Scholar]
- 157.Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, Ahmad F, Lozado R, Shah G, Fananapazir L, et al (2001) Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med 344:1823–1831 10.1056/NEJM200106143442403 [DOI] [PubMed] [Google Scholar]
- 158.Barnes BR, Marklund S, Steiler TL, Walter M, Hjalm G, Amarger V, Mahlapuu M, Leng Y, Johansson C, Galuska D, et al (2004) The 5′-AMP-activated protein kinase γ3 isoform has a key role in carbohydrate and lipid metabolism in glycolytic skeletal muscle. J Biol Chem 279:38441–38447 10.1074/jbc.M405533200 [DOI] [PubMed] [Google Scholar]
- 159.Banerjee RR, Rangwala SM, Shapiro JS, Rich AS, Rhoades B, Qi Y, Wang J, Rajala MW, Pocai A, Scherer PE, et al (2004) Regulation of fasted blood glucose by resistin. Science 303:1195–1198 10.1126/science.1092341 [DOI] [PubMed] [Google Scholar]
- 160.Kim KH, Zhao L, Moon Y, Kang C, Sul HS (2004) Dominant inhibitory adipocyte-specific secretory factor (ADSF)/resistin enhances adipogenesis and improves insulin sensitivity. Proc Natl Acad Sci USA 101:6780–6785 10.1073/pnas.0305905101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Engert JC, Vohl MC, Williams SM, Lepage P, Loredo-Osti JC, Faith J, Dore C, Renaud Y, Burtt NP, Villeneuve A, et al (2002) 5′ flanking variants of resistin are associated with obesity. Diabetes 51:1629–1634 10.2337/diabetes.51.5.1629 [DOI] [PubMed] [Google Scholar]
- 162.Sentinelli F, Romeo S, Arca M, Filippi E, Leonetti F, Banchieri M, Di Mario U, Baroni MG (2002) Human resistin gene, obesity, and type 2 diabetes: mutation analysis and population study. Diabetes 51:860–862 10.2337/diabetes.51.3.860 [DOI] [PubMed] [Google Scholar]
- 163.Azuma K, Oguchi S, Matsubara Y, Mamizuka T, Murata M, Kikuchi H, Watanabe K, Katsukawa F, Yamazaki H, Shimada A, et al (2004) Novel resistin promoter polymorphisms: association with serum resistin level in Japanese obese individuals. Horm Metab Res 36:564–570 10.1055/s-2004-825762 [DOI] [PubMed] [Google Scholar]
- 164.Conneely KN, Silander K, Scott LJ, Mohlke KL, Lazaridis KN, Valle TT, Tuomilehto J, Bergman RN, Watanabe RM, Buchanan TA, et al (2004) Variation in the resistin gene is associated with obesity and insulin-related phenotypes in Finnish subjects. Diabetologia 47:1782–1788 10.1007/s00125-004-1537-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mattevi VS, Zembrzuski VM, Hutz MH (2004) A resistin gene polymorphism is associated with body mass index in women. Hum Genet 115:208–212 10.1007/s00439-004-1128-4 [DOI] [PubMed] [Google Scholar]
- 166.Ukkola O, Kesaniemi YA, Tremblay A, Bouchard C (2004) Two variants in the resistin gene and the response to long-term overfeeding. Eur J Clin Nutr 58:654–659 10.1038/sj.ejcn.1601861 [DOI] [PubMed] [Google Scholar]
- 167.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, et al (2006) Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol 4:e31 10.1371/journal.pbio.0040031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oshima M, Oshima H, Taketo MM (1996) TGF-β receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev Biol 179:297–302 10.1006/dbio.1996.0259 [DOI] [PubMed] [Google Scholar]
- 169.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–238 10.1038/10290 [DOI] [PubMed] [Google Scholar]
- 170.Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22:239–247 10.1038/10297 [DOI] [PubMed] [Google Scholar]
- 171.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, et al (2003) Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA 100:5896–5901 10.1073/pnas.0730857100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH (2004) Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305:869–872 10.1126/science.1099870 [DOI] [PubMed] [Google Scholar]
- 173.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH (2006) Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci USA 103:1810–1815 10.1073/pnas.0508483103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ramensky V, Bork P, Sunyaev S (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30:3894–3900 10.1093/nar/gkf493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yeo GS, Farooqi IS, Aminian S, Halsall DJ, Stanhope RG, O’Rahilly S (1998) A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet 20:111–112 10.1038/2404 [DOI] [PubMed] [Google Scholar]
- 176.Vaisse C, Clement K, Durand E, Hercberg S, Guy-Grand B, Froguel P (2000) Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest 106:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hinney A, Bettecken T, Tarnow P, Brumm H, Reichwald K, Lichtner P, Scherag A, Nguyen TT, Schlumberger P, Rief W, et al (2006) Prevalence, spectrum, and functional characterization of melanocortin-4 receptor gene mutations in a representative population-based sample and obese adults from Germany. J Clin Endocrinol Metab 91:1761–1769 10.1210/jc.2005-2056 [DOI] [PubMed] [Google Scholar]
- 178.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, et al (2006) A common genetic variant is associated with adult and childhood obesity. Science 312:279–283 10.1126/science.1124779 [DOI] [PubMed] [Google Scholar]
- 179.Vaisse C, Clement K, Guy-Grand B, Froguel P (1998) A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet 20:113–114 10.1038/2407 [DOI] [PubMed] [Google Scholar]
- 180.Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z, Wei R, Curtin LR, Roche AF, Johnson CL (2002) 2000 CDC growth charts for the United States: methods and development. Vital Health Stat 11:1–190 [PubMed] [Google Scholar]
- 181.Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lubrano-Berthelier C, Durand E, Dubern B, Shapiro A, Dazin P, Weill J, Ferron C, Froguel P, Vaisse C (2003) Intracellular retention is a common characteristic of childhood obesity-associated MC4R mutations. Hum Mol Genet 12:145–153 10.1093/hmg/ddg016 [DOI] [PubMed] [Google Scholar]
- 183.Lubrano-Berthelier C, Dubern B, Lacorte JM, Picard F, Shapiro A, Zhang S, Bertrais S, Hercberg S, Basdevant A, Clement K, et al (2006) Melanocortin 4 receptor mutations in a large cohort of severely obese adults: prevalence, functional classification, genotype-phenotype relationship, and lack of association with binge eating. J Clin Endocrinol Metab 91:1811–1818 10.1210/jc.2005-1411 [DOI] [PubMed] [Google Scholar]
- 184.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B Met 57:289–300 [Google Scholar]
- 185.Hinney A, Schmidt A, Nottebom K, Heibult O, Becker I, Ziegler A, Gerber G, Sina M, Gorg T, Mayer H, et al (1999) Several mutations in the melanocortin-4 receptor gene including a nonsense and a frameshift mutation associated with dominantly inherited obesity in humans. J Clin Endocrinol Metab 84:1483–1486 10.1210/jc.84.4.1483 [DOI] [PubMed] [Google Scholar]
- 186.Ukkola O, Ravussin E, Jacobson P, Sjostrom L, Bouchard C (2003) Mutations in the adiponectin gene in lean and obese subjects from the Swedish obese subjects cohort. Metabolism 52:881–884 10.1016/S0026-0495(03)00074-X [DOI] [PubMed] [Google Scholar]
- 187.Biebermann H, Castaneda TR, van Landeghem F, von Deimling A, Escher F, Brabant G, Hebebrand J, Hinney A, Tschop MH, Gruters A, et al (2006) A role for β-melanocyte-stimulating hormone in human body-weight regulation. Cell Metab 3:141–146 10.1016/j.cmet.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 188.Hung CC, Luan J, Sims M, Keogh JM, Hall C, Wareham NJ, O’Rahilly S, Farooqi IS (2007) Studies of the SIM1 gene in relation to human obesity and obesity-related traits. Int J Obes (Lond) 31:429–434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set 1