G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis (original) (raw)
Abstract
Covalent modification of histone tails is crucial for transcriptional regulation, mitotic chromosomal condensation, and heterochromatin formation. Histone H3 lysine 9 (H3-K9) methylation catalyzed by the Suv39h family proteins is essential for establishing the architecture of pericentric heterochromatin. We recently identified a mammalian histone methyltransferase (HMTase), G9a, which has strong HMTase activity towards H3-K9 in vitro. To investigate the in vivo functions of G9a, we generated _G9a_-deficient mice and embryonic stem (ES) cells. We found that H3-K9 methylation was drastically decreased in _G9a_-deficient embryos, which displayed severe growth retardation and early lethality. _G9a_-deficient ES cells also exhibited reduced H3-K9 methylation compared to wild-type cells, indicating that G9a is a dominant H3-K9 HMTase in vivo. Importantly, the loss of G9a abolished methylated H3-K9 mostly in euchromatic regions. Finally, G9a exerted a transcriptionally suppressive function that depended on its HMTase activity. Our results indicate that euchromatic H3-K9 methylation regulated by G9a is essential for early embryogenesis and is involved in the transcriptional repression of developmental genes.
Keywords: Euchromatin, heterochromatin, histone H3-K9 methylation, G9a HMTase, mammalian development, transcriptional regulation
In all eukaryotes, the covalent modification of histone N-terminal tails plays an important role in the regulation of transcription, mitosis, and heterochromatin formation. The “histone code” hypothesis (Strahl and Allis 2000; Jenuwein and Allis 2001) predicts that different modifications of specific amino acids in histones or their combinations are translated into functionally distinct effects on nuclear processes. For example, transcriptionally active euchromatic regions are often associated with hyperacetylated histones, while silent heterochromatic regions associate with hypoacetylated forms (Grunstein 1997). Besides acetylation, the histone H3 N terminus is also phosphorylated during different cellular processes. Phosphorylation of serine 10 in histone H3 (H3-S10) is required for proper chromosome condensation and segregation (Wei et al. 1999). The phosphorylation of H3-S10 also correlates with transcriptional activation of immediate-early genes upon mitogen stimulation (Mahadevan et al. 1991). In addition to acetylation and phosphorylation, methylation of histone tails at different residues has been implicated in transcriptional regulation (Jenuwein and Allis 2001; Zhang and Reinberg 2001). It has been shown that the histone methyltransferase (HMTase) responsible for methylation of Arg 2, Arg 17, and Arg 26 in H3 (Ma et al. 2001; Xu et al. 2001) and Arg 3 in H4 (Strahl et al. 2001; Wang et al. 2001b) plays an important role in the transcriptional activation of certain genes. Methylation of Lys 4 in H3 (H3-K4) localizes to heterochromatin boundaries and transcriptionally active loci (Litt et al. 2001; Noma et al. 2001). In contrast, current evidence suggests that H3 Lys 9 (H3-K9) methylation is responsible for the creation of transcriptionally repressive heterochromatin (Rea et al. 2000). Pericentric heterochromatin contains enriched HP1 proteins from specific interaction between methylated H3-K9 and the chromodomain of HP1 (Bannister et al. 2001; Lachner et al. 2001). Clr4 in yeast (Nakayama et al. 2001) and Suv39h in mammals (Aagaard et al. 1999; O'Carroll et al. 2000), which are counterparts of Drosophila melanogaster Su(var)3-9 protein, are major heterochromatic H3-K9 HMTases and play a dominant role in pericentric heterochromatin formation. The pericentric heterochromatin architecture also plays crucial roles in chromosomal segregation as well as in establishing transcriptional repression. According to this notion, the loss of Suv39h HMTases abolished H3-K9 methylation at pericentric heterochromatin, inducing chromosomal instability (Peters et al. 2001).
We previously provided evidence that G9a, a mammalian HMTase, is a candidate for H3-K9 methylation in nonheterochromatic loci (Tachibana et al. 2001). In vitro experiments showed that G9a had a 10- to 20-fold stronger HMTase activity toward H3-K9 compared to Suv39h1, and also that it could methylate Lys 27 of H3 (H3-K27) as well as K9, whereas the targeting site on H3 for Suv39h1 was solely Lys 9. Localization analysis of G9a in nuclei suggested that this HMTase might target sites at transcriptionally active euchromatin rather than repressive pericentric heterochromatin. To investigate the in vivo function(s) of G9a, we generated _G9a_-deficient mice and ES cells. Here, we show that G9a is essential for early embryonic development and plays a dominant role in H3-K9 methylation of euchromatin. Moreover, G9a can exert transcriptional repression in vitro, a function that depends on its HMTase activity. Our data suggest that the euchromatic H3-K9 methylation regulated by G9a is involved in the transcriptional silencing of developmentally regulated genes.
Results
Targeted disruption of G9a gene in mouse germ line
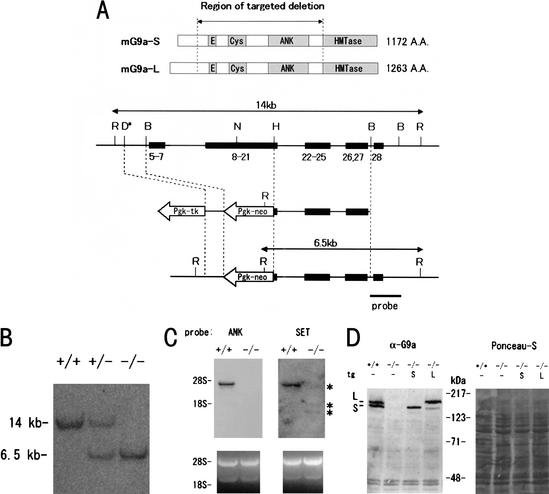
G9a genes were mapped within the major histocompatibility complex class III region in mouse and human (Brown et al. 2001). The murine G9a gene was disrupted by homologous recombination in embryonic stem (ES) cells using a conventional targeting approach that replaces the glutamic acid stretch, adjacent cysteine-rich region, and ankyrin repeats with the Pgk-neomycin gene (Fig. 1A). Successfully targeted ES cell clones were isolated and used to generate chimeric mice that transmitted the mutated allele of G9a through the germ line. ES cells homozygous for the G9a mutation (_G9a_−/−) were also obtained from the heterozygous mutant (G9a+/−) cells by high G418 selection (Fig. 1B). No transcripts were detected with a probe corresponding to the deleted ankyrin repeats, but several truncated and very faint transcripts were detectable with the G9a SET-domain probe in the _G9a_−/− ES cells (asterisks in Fig. 1C). Western blot analysis with specific antibodies against the deleted and undeleted region of C terminus confirmed the absence of G9a protein (Fig. 1D, −/− lane; data not shown). Therefore, it was likely that our G9a targeting results in a null mutation.
Figure 1.
Targeting and genotyping of _G9a_-mutant mice. (A) Schematic representation of the mouse G9a gene locus, the G9a targeting construct, and the targeted allele. Exons are numbered and indicated by black boxes. Two splicing variants of the murine G9a (mG9a-S and mG9a-L) are presented at the top. The G9a targeting construct was designed to delete exons 5 to _Hin_dIII site of 21, which correspond to the glutamic acid stretch to ankyrin (ANK) repeats. R, _Eco_RI; D, _Dra_I (not unique in the 14-kb _Eco_RI fragment); N, _Not_I; H, _Hin_dIII; B, _Bam_HI. (B) Southern blot of _Eco_RI-digested DNA isolated from wild-type, G9a+/−, and _G9a_−/− ES cells. (C) Northern blot analyses of _G9a_−/− ES cells. ANK and SET probes correspond to the region encoding for the ankyrin-repeats and the SET-domain, respectively. (D) Western blot analyses of G9a expression in wild-type and _G9a_−/− ES cells and _G9a_−/− ES cells stably expressing cDNA for the G9a-S or G9a-L isoform.
Western blot analysis of wild-type cells detected two distinct sizes (∼165 and 140 kD) of G9a proteins (Fig. 1D, +/+ lane), neither of which was consistent with the previously reported size of human G9a (112 kD) (Milner and Campbell 1993). Because of this discrepancy, we searched the EST and genomic DNA databases and identified partial DNA sequences that potentially encode larger G9a molecules containing additional N-terminal peptides. Using this DNA sequence information, we then successfully isolated cDNAs corresponding to these two isoforms, designated as G9a-S (GenBank accession no. AB077209) and G9a-L (GenBank accession no. AB077210). When these two cDNAs were introduced into the _G9a_−/− ES cells, α-G9a antibodies clearly detected proteins that corresponded in size to those detected in wild-type cells (Fig. 1D, lanes −/−S and −/−L). It was also revealed that human G9a exists as these two splicing variants (data not shown). A similar conclusion was recently reached by others (Brown et al. 2001).
Lethality and severe growth/developmental defect of G9a-deficient embryos
Mouse lines derived from two independent G9a+/− ES cell lines (#36 and #64) were used for phenotypic analyses. G9a+/− mice were indistinguishable from their wild-type littermates. The G9a+/− mice were intercrossed and the offspring were genotyped by Southern blot hybridization. Among over 200 liveborn offspring analyzed from each line, no homozygous mutant animals were found, strongly suggesting embryonic lethality (data not shown). To determine the time of lethality, embryos of heterozygous mating were isolated at days 8.5 (E8.5) to E12.5 of gestation. We occasionally found _G9a−/−-_genotyped yolk sacs without an embryo proper or completely resorbed embryos at E9.5–E12.5, indicating that the _G9a_−/− embryos likely died within this time frame (Table 1). Body weights of live _G9a_−/− embryos at E8.5 were already mildly reduced, and no further growth was observed at E9.5–E12.5 (Table 2). Since the targeted ES cell line, TT2, is an F1 hybrid of C57BL/6 and CBA, G9a+/− mice were backcrossed sequentially with C57BL/6 to minimize mixed genetic background influences, and offspring of F9 or later generations were used in further studies.
Table 1.
Embryonic lethality of the G9a-mutant mice
| Embryonic day | Genotype | Resorption | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E8.5 | 9 | 25 | 10 | 0 |
| E9.5 | 11 | 19 | 8 (7a + 1b) | 3 |
| E10.5 | 10 | 21 | 6 (4a + 2b) | 6 |
| E11.5 | 11 | 21 | 4 (1a + 3b) | 6 |
| E12.5 | 7 | 15 | 1b | 4 |
Table 2.
Growth arrest of the G9a-mutant embryos
| Embryonic day | Genotype | ||
|---|---|---|---|
| +/+ | +/− | −/− | |
| E8.5 | 0.7 ± 0.4 (9) | 0.7 ± 0.1 (8) | 0.4 ± 0.1 (7) |
| E9.5 | 3.9 ± 0.4 (8) | 3.4 ± 0.7 (11) | 0.4 ± 0.1 (7a) |
| E10.5 | 19.7 ± 2.1 (8) | 17.7 ± 1.7 (6) | 0.5 ± 0.2 (3) |
| E11.5 | 53.0 ± 4.2 (9) | 56.1 ± 3.0 (3) | 0.6 (1) |
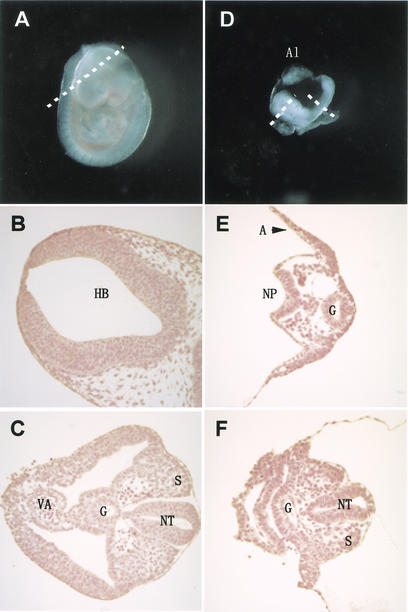
The gross morphology and histological analysis of typical _G9a_−/− embryos at E9.5 are shown in Figure 2A–F. The allantois (Al) extended into the exocoelom, making contact with the chorion. However, the fusion was incomplete and the allantois could be readily detached. _G9a_−/− embryos at E9.5 developed to only the 5–6 somite stage (Fig. 2D) as opposed to 21–25 somites in wild-type siblings (Fig. 2A). The neural groove was unfused at the anterior region and was closed at the tail region. Cardiogenesis was initiated but the primitive blood vessels, which extended from the cephalic region to the most caudal region of these embryos, contained no primitive nucleated red blood cells. Gut (G) diverticulum was well defined in the fore and hind gut regions in embryos. These morphological characteristics observed in E9.5 _G9a_−/− embryos resembled those of E8.0–E8.5 wild-type, and no organ-specific abnormalities were recognized. Several mitotic cells were observed as well as wild-type embryos.
Figure 2.
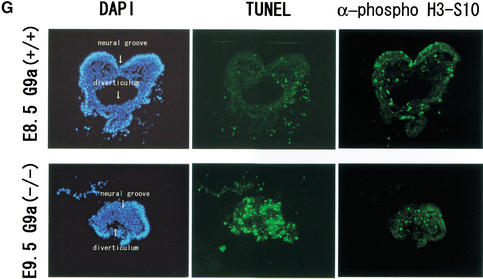
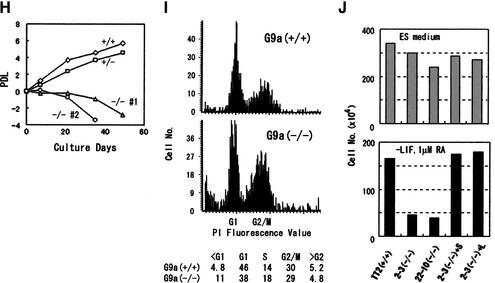
_G9a_−/− embryos were delayed in their development. (A) The wild-type littermate at E9.5. (D) The lateral view of _G9a_−/− embryo at E9.5. The approximate levels of the section presented in B, C, E, and F are indicated as dashed lines. Allantois (Al) develops normally. (B,C) Section through hindbrain (HB in B) to tail region (C). (E,F) Section through forebrain to tail region. S, somites; NT, neural tube; G, gut; VA, vitelline artery; A, amnion; NP, neural plate. (G) TUNEL analysis and α-phospho H3-S10 staining of E9.5 _G9a_−/− (bottom) and the E8.5 wild-type embryos (top) on transversal sections. (H) Growth characteristics of _G9a_−/− PEFs. Here, 104 cells of PEFs derived from individual E9.0 embryos were cultured, and population doublings (PDL) were monitored. (I) Measurement of DNA content of _G9a_−/− PEFs. The wild-type and _G9a_−/− PEFs prepared from E9.5 embryos were cultured for 16 h and the DNA content was subsequently visualized by PI staining. Calculated populations (%) at each cell cycle stage are represented below. (J) Growth defect of the _G9a_-mutant ES cells in differentiation conditions. Here, 105 cells of each ES cell line were cultured in ES medium (upper) or RA (1 μM), no LIF-containing differentiation medium (lower). Vertical bars represent the cell numbers after 4 d of culture. Typical data were picked up from triplicate independent reproducible examinations.
To further examine the cause of _G9a_−/− embryonic growth arrest, we performed immunohistochemical analysis for apoptotic cell death with TdT-mediated dUTP nick end labeling (TUNEL) on sections of E9.5 _G9a_−/− and E8.5 wild-type embryos. Typical transverse sections are shown in Figure 2G. Massive TUNEL-positive cells were readily detected in _G9a_−/− embryos, whereas very few cells were positive in wild-type embryos. Since phosphorylation of H3-S10 is tightly correlated with chromosome condensation during mitosis, we also examined the population of mitotic-stage cells in _G9a_−/− embryos. As seen in wild-type embryos, in which small populations of nuclei were brightly stained, some mitotic cells also existed in _G9a_−/− embryos, and this population was not dissimilar to those in wild-type embryos. The feature of a drastic accumulation of apoptotic cells and the constant existence of mitotic cells in _G9a_−/− embryos was commonly observed in all sections examined.
To determine whether the growth defect of _G9a_−/− embryos was due to extra-embryonic defects, we examined the growth potential of _G9a_−/− primary embryonic fibroblasts (PEFs) in vitro. Similar to the embryonic phenotype, PEFs prepared from the _G9a_−/−embryos at E9.0 showed severe growth defects, whereas the G9a+/− PEFs were indistinguishable from wild-type PEFs (Fig. 2H), suggesting that the _G9a_−/− embryo proper had intrinsic growth defects. Next, we examined the DNA content of the PEFs cultured for 16 h in vitro. As illustrated in Figure 2I, a significant increase in cells with lower DNA contents compared to those of typical G1 phase was observed in _G9a_−/− PEFs, whereas G1 to G2/M ratios were indistinguishable between _G9a_−/− and wild-type. Similar results were obtained from five independently prepared lines of _G9a_−/− PEFs. Therefore, these in vivo and in vitro data suggest that the _G9a_−/− embryonic growth defect is, at least in part, due to apoptotic cell death but not cell cycle arrest.
We used the _G9a_−/− ES cells for further analyses of G9a function. As shown in Figure 2J, no obvious growth defects were observed for _G9a_−/− ES cells during their maintenance in cell culture (Fig. 2J, upper panel). To investigate whether G9a has an important function(s) in more differentiated cells, _G9a_−/− ES cells were induced to differentiate by being cultured with all-trans retinoic acid (RA) in the absence of leukemia inhibitory factor (LIF). With this treatment, _G9a_−/− ES cells exhibited a distinct growth defect (Fig. 2J, bottom panel). These data indicate that, while G9a function may be dispensable for the survival of undifferentiated cells (e.g., epiblasts), it is crucial for more differentiated somatic cells. Exogenous introduction of G9a-S or G9a-L molecules restored normal growth potential to _G9a_−/− ES cells during differentiation. This feature was confirmed by triplicate independent examination. The limited growth defect of differentiating _G9a_−/− ES cells accords well with that of _G9a_−/− somatic cells during embryogenesis.
Loss of H3-K9 methylation leads to accumulation of acetylated H3-K9 and methylated H3-K4
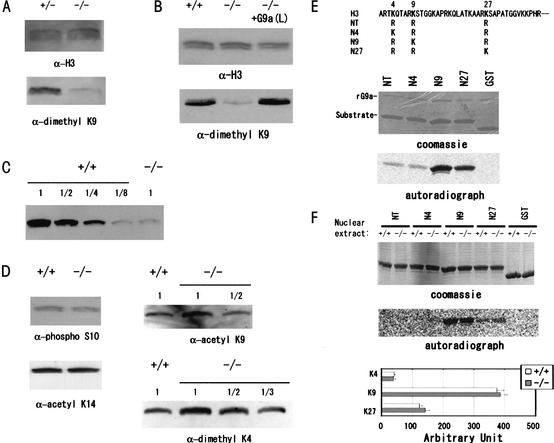
To address the issue of how G9a exerts its HMTase activity in vivo, we examined the covalent modification status of H3 N-terminal tails in _G9a_−/− cells. As shown in Figure 3A, Western blot analysis using α-dimethyl H3-K9 antibodies demonstrated that dimethylated H3-K9 in the _G9a_−/− embryos at E9.5 was drastically decreased. A reduction of dimethyl H3-K9 was also confirmed in two _G9a_−/− ES cell lines (Fig. 3B; data not shown). The defect was completely rescued by the expression of exogenous G9a protein (Fig. 3B, right lane). No significant alteration of dimethyl H3-K9 status was detected between G9a+/+ and G9a+/− in embryos and ES cells (data not shown). Estimation by serial dilution analysis indicated that the amount of dimethyl H3-K9 in _G9a_−/− ES cells was comparable to only one-eighth of that in wild-type ES cells (Fig. 3C), which is much greater than the 50% loss of H3-K9 methylation in Suv39h mutant cells (Maison et al. 2002).
Figure 3.
(A_–_D) Alteration of G9a-dependent covalent H3 modifications detected by Western blot analyses. (A) Purified and precalibrated histones with α-H3 (upper panel) from the G9a+/− and _G9a_−/− embryos were stained with α-dimethyl H3-K9 antibodies. (B) α-dimethyl H3-K9 staining of H3 from wild-type and _G9a_−/− ES cells and _G9a_−/− ES cells expressing exogenous G9a-L. (C) Calibration of the dimethyl H3-K9 content in _G9a_−/− ES cells. The ratio of H3 content between wild-type and mutant were precalibrated and are represented below. (D) Other covalent modification statuses of H3 between wild-type and _G9a_−/− ES cells. All the presented data shown in Fig. 2A–D were reproducible at least twice. (E) HMTase activity of G9a-HMTase domain. Amino acid sequences of GST-fused recombinant H3 N terminus protein as substrates are at the top. Recombinant G9a-HMTase domain could add methyl groups to H3-K9 and H3-K27. (F) H3 HMTase activity in wild-type and _G9a_−/− ES cells. HMTase activity and substrate specificity of nuclear extracts of wild-type and _G9a_−/− ES cells were indistinguishable. HMTase activities of the nuclear extracts toward Lys 4, Lys 9, and Lys 27 were calibrated as Nx–NT.
It has been shown that H3-K9 methylation can influence other modifications of the neighboring residues in H3 tails, such as phosphorylation and acetylation. Methylated H3-K9 can inhibit the phosphorylation of H3-S10 by Ipl1/aurora (Rea et al. 2000). In addition, S10 phosphorylation facilitates the acetylation of Lys 14 in H3 (H3-K14) by the histone acetyltransferase GCN5 (Cheung et al. 2000). Thus, we investigated the acetyl status of H3-K9 and H3-K14, and the phosphorylation status of H3-S10 in _G9a_−/− ES cells (Fig. 3D). H3-S10 phosphorylation and H3-K14 acetylation were indistinguishable in wild-type and _G9a_−/− ES cells. In contrast, H3-K9 acetylation was increased about twofold in _G9a_−/− ES cells, suggesting the existence of competition between methylation and acetylation at H3-K9 in vivo. In addition to these modifications, methylated H3-K4 has been shown to localize to transcriptionally active chromatin (Litt et al. 2001; Noma et al. 2001) and suppress H3-K9 methylation in vitro (Wang et al. 2001a). As shown in Figure 3D, we found a two- to threefold increase in H3-K4 methylation, a result that further indicates the presence of a functional competition between H3-K4 and H3-K9 methylation in vivo.
To date, the catalytically defined set of H3-K9 HMTases in mammals consists of Suv39h (1 and 2) (O'Carroll et al. 2000; Rea et al. 2000), ESET (Schultz et al. 2002; Yang et al. 2002), and G9a. The dominant contribution of G9a to H3-K9 methylation in vivo predicts that total HMTase activity toward H3-K9 might be reduced in _G9a_−/− cells. In addition, the recombinant G9a HMTase domain could transfer methyl groups H3-K27 as well as H3-K9 in vitro (Fig. 3E; Tachibana et al. 2001). Thus, we investigated the HMTase activities toward H3-K9 and H3-K27 in _G9a_−/− ES cells. Surprisingly, nuclear extracts from mutant cells retained HMTase activity toward both H3-K9 and H3-K27. The activity in _G9a_−/− ES cells was comparable to levels observed in wild-type cells (Fig. 3F). These data suggest that, while many proteins exhibit HMTase activities toward H3-K9 and H3-K27 in vitro, only a subset may function in vivo (e.g., Suv39h and G9a). It is likely that other lysine methyltransferases are targeted to proteins other than histones in vivo (Jenuwein 2001).
Global loss of euchromatic H3-K9 methylation in G9a−/− cells
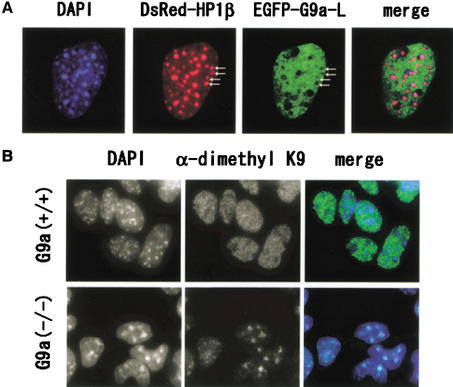
Previously we showed that a truncated human G9a molecule (Milner and Campbell 1993) displayed a nuclear localization profile that was quite different from that of Suv39h1 (Tachibana et al. 2001). We therefore reinvestigated the cellular localization profiles of fluorescent protein (EGFP)-tagged full-length G9a-S and G9a-L in murine fibroblasts with DsRed-tagged human HP1β molecules, which were shown to localize in pericentric heterochromatin and be involved in SUV39H1 complex (Fig. 4A; Aagaard et al. 1999). EGFP-G9a proteins exhibited a nuclear-specific pattern that was broad in interphase nuclei but almost excluded from nucleoli. Importantly, the intensity of EGFP-G9a signals was reduced significantly at pericentric heterochromatin regions, which were DAPI-densely stained and accumulated with HP1β molecules (arrows). The localization profiles were indistinguishable between G9a-S and G9a-L molecules (data not shown). These observations again suggest that the localization profiles of G9a and pericentric heterochromatin-associated Suv39h were quite distinct and mutually exclusive.
Figure 4.
(A) Nuclear localization profiles of transiently expressed EGFP-tagged mouse G9a molecules (G9a-L) and DsRed-tagged human HP-1β molecules in the murine fibroblast cell line C3H10T 1/2 . EGFP-G9a-L molecules exist broadly in interphase nuclei and mostly excluded from HP-1β-enriched pericentric heterochromatin (arrows). (B) Immunohistochemical analyses of _G9a_−/− ES cells with α-dimethyl H3-K9. Nuclei of _G9a_−/− ES cells contained dimethyl H3-K9 only in DAPI-dense regions, whereas these were broadly distributed to both euchromatic and heterochromatic loci in wild-type nuclei.
To further investigate the contributions of G9a to functionally different chromatic regions, we performed immunohistochemical analyses of methylated H3-K9 in wild-type and _G9a_−/− ES cells. Dimethylated H3-K9 was broadly detected in wild-type nuclei, indicating that methylated H3-K9 might exist in many loci, including eu- and heterochromatic regions (Fig. 4B, upper panels). However, the _G9a_−/− cell nucleus contained only large speckles of methylated H3-K9, which were completely overlapping with the DAPI-dense loci (Fig. 4B, bottom panels). This result fits well with the observation that ectopically expressed G9a molecules were excluded from HP1β-enriched pericentric heterochromatin. It is highly likely that methylation of H3-K9 in the pericentric heterochromatin of _G9a_−/− cells is catalyzed mainly by Suv39h HMTases (Peters et al. 2001). Considering that H3 in _G9a_−/− ES cells possessed only one-eighth the amount of dimethylated H3-K9 compared to that of wild-type cells, we conclude that G9a is the major in vivo H3-K9 HMTase that directs methylation of euchromatic regions.
G9a exerts a transcriptionally suppressive function dependent on its HMTase activity
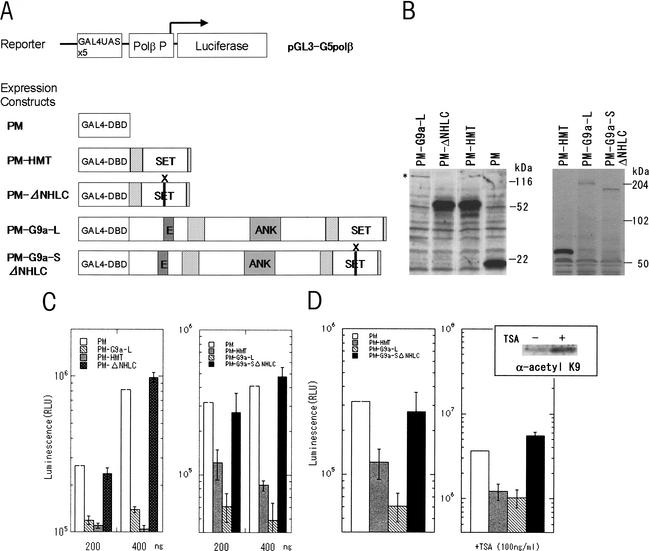
To assess whether G9a-mediated histone methylation functions in transcriptional regulation, we performed reporter gene assays using pGL3-G5polβ, which contains a luciferase gene driven by a DNA pol β promoter proximal to GAL4-binding sites (Sekimata et al. 2001). We transiently introduced pGL3-G5polβ and a construct expressing a GAL4 DNA-binding domain (GAL4-DBD) fused with full-length mG9a-L (PM-G9a-L) or an HMTase domain of G9a (residues 969V-stop) (PM-HMT) into HeLa S2 cells (Fig. 5A,B). We also expressed a GAL4-DBD fused with a dead HMTase full-length mG9a-S (PM-G9a-SΔNHLC, deletion of 1165NHLC1168 of the HMTase domain) or a dead HMTase domain alone (PM-ΔNHLC). Transcription of the reporter gene was significantly inhibited by PM-G9aL and PM-HMT, but not by PM-G9a-SΔNHLC or PM-ΔNHLC (Fig. 5C). A second G9a truncated mutant with crippled HMTase function (1162R to H substitution) also failed to repress reporter gene transcription (data not shown). These results suggest that G9a functions as a negative regulator of transcription through its HMTase activity. To evaluate the involvement of histone deacetylases (HDACs) in the observed transcriptional repression, we performed the experiments in the presence of trichostatin A (TSA), an inhibitor of HDACs (Fig. 5D). Acetylation of total H3-K9 was substantially enhanced by the TSA treatment, but the transcriptional repression was unaffected. Together, these data indicate that the G9a HMTase-mediated repression of gene transcription occurs in a manner independent of the HDAC pathway.
Figure 5.
HMTase-dependent transcriptional repression mediated by G9a. (A) Schematic representation of the used plasmids for in vitro luciferase assays. (B) Protein expression analysis of the GAL4-DBD-fused constructs described in A. Fusion molecules were detected by α-GAL4-DBD. (C) Full-length G9a (PM-G9a-L) and a G9a HMTase domain (PM-HMT) suppress transcription, but dead HMTase full-length G9a (PM-G9a-SΔNHLC ) and a G9a dead HMTase domain (PM-ΔNHLC) cannot. All the transient luciferase assays were performed multiple times and the results presented were reproduced. (D) The G9a-mediated suppression was not relieved in the presence of TSA (100 ng/mL). Upper left panel shows an accumulation of H3-K9 acetylation by the TSA treatment.
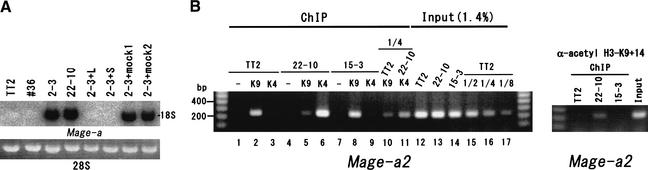
To further investigate the G9a function in transcriptional regulation, we performed oligonucleotide microarray analyses using G9a+/+, _G9a_−/− ES cells, and _G9a_−/− ES cells expressing exogenous G9a (T. Ohta, M. Tachibana and Y. Shinkai, unpubl.). We identified some candidate genes transcription-regulated by G9a, and one of them was a Mage-a gene(s). Human MAGE genes have been isolated as tumor-specific antigen genes (Boon et al. 1994). Eight murine Mage-a genes (Mage-a1 to Mage-a8, highly homologous each other) have been identified, with some of them being expressed not only in tumor cell lines but also in testis and blastocysts (De Plaen et al. 1999). The function of Mage-a genes is currently unknown. As shown in Figure 6A, the expression of Mage-a gene(s) was induced in _G9a_−/− ES cells (2–3 and 22–10) and suppressed by expression of exogenous G9a (2–3+L and +S) but not by a drug-selectable molecule alone (2–3+mock1 and 2). Nucleotide sequencing of the RT-PCR products demonstrated that Mage-a2, Mage-a6, and Mage-a8 were expressed in _G9a_−/− ES cells (data not shown). To further evaluate the Mage-a genes as direct targets of G9a, we performed a chromatin immunoprecipitation (ChIP) analysis. As illustrated in Figure 6B, dimethylated H3-K9 was enriched in chromatin containing the Mage-a2 promoter sequences in G9a+/+ (TT2) and severely decreased in _G9a_−/− ES cells (22–10) (about one-fourth of TT2, Fig. 6B, lanes 5 and 10). In contrast with H3-K9 methylation, dimethylated H3-K4 was enriched in the Mage-a2 promoter region of 22–10 (at least 10-fold enrichment in 22–10 compared to TT2, Fig. 6B, lanes 3 and 11). Exogenous expression of G9a in _G9a_−/− ES cells (15–3) reverted the status of H3-K9 and H3-K4 dimethylation to the G9a+/+ ES cell level. Furthermore, acetylation of H3-K9+K14 at this region was increased significantly in _G9a_−/− ES cells (Fig. 6B, right panel). These data strongly suggest that Mage-a2 gene is silenced by G9a-mediated H3-K9 methylation in ES cells.
Figure 6.
Characterization of Mage-a genes. (A) RNA expression of Mage-a genes. Six micrograms of total RNA prepared from G9a+/+ (TT2), G9a+/− (#36), _G9a_−/− (2–3 and 22–10), 2–3-expressing exogenous G9a-L (2–3-L), G9a-S (2–3-S), or a drug-selectable molecule alone (2–3+mock1 and 2) were separated, blotted to a nylon membrane, and hybridized with a 32P-labeled Mage-a8 cDNA. Expression of Mage-a2, Mage-a6, and Mage-a8 genes was determined in _G9a_−/− ES cells by nucleotide sequencing of the RT-PCR products. The lower panel shows 28S rRNA stained with ethidium bromide in the same gel before blotting. (B) ChIP analyses on the Mage-a2 promoter region. Formaldehyde cross-linked chromatin from TT2, 22–10, and 15–3 (2–3-S) ES cells was immunoprecipitated without (−) or with α-dimethyl H3-K9 (K9), α-dimethyl H3-K4 (K4), or α-acetyl-H3-K9+14 (right panel). An equal amount of Mage-a2 promoter sequence in TT2, 22–10, and 15–3 nucleosomal preparations was determined by PCR from 1.4% of the input ChIP/ PCR reaction (lanes 12_–_14). The linearity of this PCR was confirmed by serial dilution of the TT2 input DNA (lanes 12,15_–_17). The product size of the Mage-a2 PCR is 242 bp. Similar results were obtained with multiple independent experiments.
Discussion
Embryonic lethality in G9a-mutant mice
The early embryonic lethality of _G9a_−/− mice (E9.5–E12.5) is distinct from Suv39h1/2 double mutant mice, which are born at sub-Mendelian ratios (Peters et al. 2001). This finding suggests that Suv39h and G9a HMTase contribute nonoverlapping roles to embryonic growth/development. The developmental arrest of _G9a_−/− embryos at ∼E8.0 coincides with the dramatic reorganization of somatic tissues, which is accompanied by widespread alterations in gene expression profiles and chromatin organization. Thus, it is probable that G9a-mediated methylation of euchromatin is a key component of the mechanism that regulates gene expression during this stage of embryogenesis. Apoptotic cells were increased drastically in growth-arrested _G9a_−/− embryos, and this seemed to be the dominant cause of embryonic growth retardation. Interestingly, they still possessed significant populations of mitotic cells. _G9a_−/− ES cells also displayed growth defects under conditions of cellular differentiation, but not during routine maintenance. These facts suggest that G9a is dispensable for simple proliferating processes but is necessary for some important events during embryonic development or differentiation.
Recent studies have shown that methylation marking on H3-K9 is specifically targeted to the inactivated X chromosomes (Xi) in females (Boggs et al. 2002; Heard et al. 2001). This modification occurs by an Suv39h-HP1-independent pathway, because Suv39h-mutant female cells still possess methylated H3-K9-rich Xi (Peters et al. 2002). We considered the possibility that H3-K9 methylation by G9a is responsible for determining Xi formation. If this model is correct, female _G9a_−/− mice would possess two active X chromosomes, which is known to induce female-specific lethality at a very early developmental stage (Marahrens et al. 1997). However, the developmentally retarded phenotypes of _G9a_−/− male and female embryos at E9.5 were indistinguishable, as shown in Table 2. Thus, this possibility is still open to discussion, but defective Xi formation cannot explain male embryonic lethality in G9a-deficient mice.
Histone methylation of euchromatin
The broad methylation patterns of H3-K9 in the nucleus suggest that its functions are not restricted to pericentric heterochromatin organization in mammals. In _G9a-_deficient cells, the broad methylation of chromatin was abolished. In contrast, Suv39h1/2 double mutant mice retain broader methylation of H3-K9, but they lose methylation at pericentric heterochromatic regions (Peters et al. 2001). Taken together, these findings strongly suggest that G9a and Suv39h HMTases have nonoverlapping functions and target distinct chromosomal loci. Comparisons of Suv39h and G9a protein sequences reveal high similarities in the HMTase enzymatic regions. However, other regions are quite divergent (e.g., chromodomain for Suv39h and ankyrin-repeats for G9a). Thus, it is likely that the unique molecular domains of Suv39h and G9a are responsible for targeting their HMTase functions to pericentric heterochromatin and euchromatin, respectively.
The HP1 protein is also involved in transcriptional silencing and modulation of chromatin architecture (Jones et al. 2000). The chromodomain of HP1 exhibits high affinity for methylated H3-K9 (Bannister et al. 2001; Lachner et al. 2001), which recruits HP1 to pericentric heterochromatin regions following Suv39h-mediated H3-K9 methylation. We speculate that there are two possible functions of G9a-mediated H3-K9 methylation at euchromatin: (1) G9a creates a local heterochromatic architecture in euchromatic regions via interactions with methylated H3-K9 and subpopulations of the HP1 protein. Indeed, recent studies indicate that HP1 might play a crucial role in creating local heterochromatic domains to establish a silent state (Matsuda et al. 2001; Ogawa et al. 2002); (2) Since G9a also methylates H3-K27 in vitro, H3 molecules carrying doubly methylated lysine residues may recruit distinct factors that further influence chromatin structure and gene regulation.
G9a functions as transcriptional repression
Existing data support the notion that H3-K9 methylation may contribute to transcriptional repression (Firestein et al. 2000; Litt et al. 2001; Nakayama et al. 2001; Nielsen et al. 2001; Noma et al. 2001; Vandel et al. 2001). Data of our reporter gene assays imply that G9a exerts a transcriptional repression function in vivo and it is dependent on its HMTase activity. These data strongly suggest that one important function of G9a is gene silencing mediated by H3-K9 methylation in euchromatic regions. The expression profiles of Mage-a2, Mage-a6, and Mage-a8 genes and the dimethylation status of H3-K9 in chromatin containing the Mage-a2 promoter sequences in G9a+/+ and _G9a_−/− ES cells clearly support this notion. To further elucidate the physiological function(s) of HMTase G9a, a crucial next step will be the identification and characterization of entire target genes.
In contrast to H3-K9 methylation, transcriptionally active chromatic regions are associated with methylated H3-K4 (Litt et al. 2001; Noma et al. 2001). Furthermore, preexisting H3-K4 methylation inhibits methylation of H3-K9 in vitro and vice versa (Wang et al. 2001a). A significant increase in methylated H3-K4 in _G9a_−/− cells was observed (Figs. 3D and 6B), suggesting that G9a-mediated H3-K9 methylation negatively regulates H3-K4 methylation in vivo. These findings leave open two possibilities for the mechanisms by which G9a exerts its transcriptionally suppressive function: (1) up-regulation of H3-K9 methylation, or (2) down-regulation of H3-K4 methylation. This type of cross-regulation also exists between H3-K9 methylation and H3-S10 phosphorylation (Rea et al. 2000), and phosphorylation of H3-S10 links to transcriptional activation (Mahadevan et al. 1991). In contrast to Suv39h1/2 double mutant mice (Peters et al. 2001), no accumulation of H3-S10 phosphorylation was observed in _G9a_−/− cells. This finding also indicates that euchromatic H3-K9 methylation is functionally different from that of heterochromatin. Future studies will address whether the histone code of H3-K9 methylation can be translated into different biological outputs in a manner dependent upon their chromosomal locations.
Materials and methods
Generation and genotyping of the G9a-mutant embryos and ES cells
A partial cDNA clone for the mouse G9a gene was isolated from a Uni-ZAP XR mouse testis library (Stratagene) using a radiolabeled human counterpart cDNA (Milner and Campbell 1993) as a probe. The isolated mouse G9a cDNA fragment was used as a probe to screen a C57 Black/6 mouse genomic library (Stratagene). To make a G9a targeting construct, a 4.5-kb genomic fragment, a region from the _Hin_dIII site in exon 21 to the _Bam_HI site downstream of exon 27, was inserted into the modified _Sal_I site of pLNTK, and then a 0.75-kb _Dra_I-_Bam_HI genomic fragment located in the intron between exon 4 and 5 was further subcloned. The G9a targeting construct potentially replaces exons 5-part21 of mouse G9a with the Pgk-neomycin gene.
Next, 1 × 107 ES cells, TT2 line (Yagi et al. 1993) were transfected with 20 μg of _Not_I-linearized _G9a_-targeting construct and selected in ES cell medium containing 0.25 mg/mL G418 and 1.5 μg/mL ganciclovir. Homologous recombinant cells (#36 and #64) were identified by Southern blot analysis of _Eco_RI-digested DNA probed with an exon 28-containing 1.1-kb _Bam_HI genomic DNA fragment and injected into the morula stage of ICR mouse embryos. Established chimeric male mice derived from both clones successfully generated F1 offspring carrying the mutated G9a allele.
PCR genotyping of the _G9a_-mutant mice was carried out using primers external to the short arms of the targeting vector (G3, 5′-GGGCCAGCTCATTCCTCCACTC-3′; mG9a127, 5′-GCAGATGTGATGGCTTGGGGTAG-3′). G9a+/− mice were sequentially backcrossed with the C57BL/6 strain mice, and offspring of F9 or later generation animals were used for further studies. Two independent lines of the G9a homozygous recombinant (_G9a_−/−) ES cells were isolated from line #36 line selected in G418 (3.0 mg/mL)-containing medium.
ES cell culture and transfection
Undifferentiated ES cells were maintained in 10% fetal calf serum and LIF (500 μg/mL)-containing medium. For the growth assays of the retinoic-acid (RA)-induced differentiated cells, cells were cultured in the presence of 1 μM _all-trans-_RA without LIF. Full-length murine G9a cDNA was isolated from a murine thymus cDNA library and subcloned into the expression vector pCAGGS and introduced into ES cells using LIPOFECTAMINE 2000 reagent (GIBCO) according to the manual.
Generation of α-G9a monoclonal antibody
GST-mG9a C-terminal proteins described previously (Tachibana et al. 2001) were used to immunize Armenian hamsters. Monoclonal antibodies were generated by fusion of the immunized spleen cells to the myeloma cell line NS1.
Histological analysis
Embryos were fixed in Bouin fixative, dehydrated through graded ethanol, embedded in paraffin wax and sectioned. Sections were stained with hematoxylin-eosin as described (Nozaki et al. 1999).
For TUNEL and immunohistochemical analysis with sections, embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C for 2 h, and embedded and frozen in OSC compound. TUNEL analysis was performed with a detection kit (Boehringer-Mannheim). For phosphorylated H3-S10 staining of embryos, α- phospho H3-S10 (Cell Signaling) and biotinylated goat antibody against rabbit IgG (Vector) as secondary antibody were used. The antibody complex was visualized by avidin-labeled fluorescein (Vector).
Measurement of DNA content
Primary embryonic fibroblasts (PEFs) were cultured on cover glasses for 16 h. PBS-washed cells were fixed in 70% ethanol for 5 min and stained with propidium iodide (PI) (25 μg/mL) and RNase (200 μg/mL). The relative intensity of PI fluorescence was measured by laser scanning cytometer (LSC101; Olympus).
Protein blot analysis for histone tail modifications
Acid-extracted histones were prepared from E9.5 embryos and ES cells as described (Cheung et al. 2000). Histones were resolved by SDS-PAGE (15%; 30:0.8) gels and transferred to nitrocellulose membranes for Western blotting. The amount of histone H3 was precalibrated with α-H3 antibody (Cell Signaling) and with ponceau-S (Sigma) staining. Covalent modification status of H3 tails was analyzed with α-dimethH3-K9 (Upstate), α-acetylH3-K9 (Cell Signaling), α-phosH3-S10 (described above), α-acetylH3-K14 (Upstate), and α-dimethylH3-K4 (Upstate) antibodies.
HMTase assay of nuclear extract of G9a-mutant ES cells
Nuclear extracts were prepared from wild-type and G9a-mutant ES cells as described (Andrews and Faller 1991) and subsequently used for HMTase assays as described (Tachibana et al. 2001). Briefly, 40 μL of reaction mixture containing 100 μg of the nuclear extracts, 20 μg of recombinant H3-N terminus protein, and 125 nCi _S_-adenosyl-[methyl-14C]-L-methionine in methylase activity buffer (50 mM Tris at pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 250 mM sucrose) was incubated for 60 min at 37°C. The reaction products were separated by 15% SDS-PAGE and visualized by CBB staining. Gels were dried and quantification of methyl-14C was performed using a BAS-5000-Mac imaging analyzer (Fuji Film).
Immunofluorescence analysis
ES cells grown on glass coverslips were fixed with 2% paraformaldehyde, treated with 0.1% Triton X-100, and incubated with the antibodies described above. Primary antibodies were probed by Cy3-conjugated anti-rabbit IgG or FITC-conjugated anti-mouse IgG antibodies (Jackson Immunoresearch Laboratories). Nuclei were counterstained with DAPI, and observed under fluorescence microscopy (Eclipse E600, Nikon). Images were acquired using MetaMorph software (Universal Imaging).
Luciferase assay
Hela S2 cells were plated on 24-well plates (5 × 104/well), cultured overnight, and transfected with 50 ng of pGL3-G5polβ (Sekimata et al. 2001) and 200–400 ng of the indicated expression plasmids. Transfection was carried out using a TransIT-LT1 lipofection reagent (Mirus). After 48-h incubation, cell lysates were prepared and luciferase activity was measured using a Bright-Glu luciferase assay system (Promega). For the HDAC inhibition experiment, the transfected cells were cultured in the presence of 100 ng/mL trichostatin A (TSA) for the last 12 h of incubation. For the expression of a GAL4-DBD fused to G9a-L or a G9a HMTase domain, a full-length or truncated G9a-L cDNA was subcloned into the appropriate site(s) of pM (Clontech) to obtain PM-G9a-L, PM-HMT (GAL4-DBD-G9a969V-end), or PM-ΔNHLC (G9a1165NHLC1168 deletion of PM-HMT). In an in vitro assay, the ΔNHLC molecule showed no HMTase activity. Protein expression of these GAL4-DBD fusion molecules was confirmed by Western blot analysis with α-GAL4-DBD (RK5C1, Santa Cruz).
Northern blot analysis
Six micrograms of total RNAs were separated by 1% agarose-formaldehyde gel electrophoresis, transblotted to a nylon membrane, and probed with 32P-labeled Mage-a8 cDNA.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) analyses were done as described with some modifications (Luo et al. 1998). First, 1 × 107/mL ES cells (in PBS containing 10% FCS) was cross-linked with 1% formaldehyde for 10 min at 37°C. After quenching of the cross-linking reaction with 125 mM glycine, the fixed cells were washed with PBS containing protease inhibitors (1 mM PMSF, 1 μg/mL aprotinin, and 1 μg/mL pepstatin A). The cells were suspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.1) (1 × 107/0.2 mL) and sonicated to average fragment size of 200–1000 bp. Solubilized chromatin was clarified by centrifugation for 10 min at 13,000 rpm at 4°C and diluted 10-fold in ChIP dilution buffer (1% Triton X-100, 1 mM EDTA, 150 mM NaCl, 15 mM Tris-HCl at pH 8.1). The diluted chromatin of 5 × 106 cells was incubated with α-dimethyl H3-K9 and α-dimethyl H3-K4 antibodies for 12–16 h at 4°C. Immune complexes were bound to protein A sepharose beads preblocked with salmon sperm DNA and BSA for 1 h at 4°C. The beads were washed once each with low-salt wash buffer (0.1% SDS, 1% Triton-X100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl at pH 8.1), high-salt wash buffer (500 mM NaCl wash buffer), LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl at pH 8.1), and twice with TE. Immune complexes bound to Protein A beads were treated with 100 μg/mL Proteinase K for 2 h at 56°C, extracted once with phenol/chloroform, and the DNA was precipitated with ethanol plus glycogen as carrier. Precipitated DNA was resuspended in 60 μL of water. DNA was analyzed by PCR using specific primer pairs to Mage-a2 promoter sequences (Mage-a2A, 5′-TTGGTGGACAGGGAAGC TAGGGGA-3′; Mage-a2B, 5′-CGCTCCAGAACAAAATGGC GCAGA-3′). The product size of Mage-a2A/2B PCR is 242 bp.
Acknowledgments
We thank Dr. J. Miyazaki (Osaka University) for pCAGGS plasmid and Dr. M. Sekimata (Fukushima Medical University) for pGL3-G5polβ plasmid. We also thank Drs. N. Yoshida (Kyoto University), S. Mori, and Y. Yokota (Fukui Medical University) for technical assistance with embryo preparation. We thank Drs. T. Matsumoto (Japanese Foundation for Cancer Research), H. Hirata, and R. Kageyama (Kyoto University) for help with immunohistochemical techniques, Dr. T. Shimura (Kyoto University) for help with cell cycle analysis, and Dr. E.M. Oltz (Vanderbilt University) for critically reading this manuscript. This work was supported by a Grant-in Aid from the Ministry of Education, Science, Technology, and Culture of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yshinkai@virus.kyoto-u.ac.jp; FAX 81-75-751-3991.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.989402.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Boon T, Cerottini JC, Van den Eynde B, van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- Brown SE, Campbell RD, Sanderson CM. Novel NG36/G9a gene products encoded within the human and mouse MHC class III regions. Mamm Genome. 2001;12:916–924. doi: 10.1007/s00335-001-3029-3. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- De Plaen E, De Backer O, Arnaud D, Bonjean B, Chomez P, Martelange V, Avner P, Baldacci P, Babinet C, et al. A new family of mouse genes homologous to the human MAGE genes. Genomics. 1999;55:176–184. doi: 10.1006/geno.1998.5638. [DOI] [PubMed] [Google Scholar]
- Firestein R, Cui X, Huie P, Cleary ML. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3–9. Mol Cell Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: Their role in genome organisation and expression. BioEssays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Matsuda E, Agata Y, Sugai M, Katakai T, Gonda H, Shimizu A. Targeting of Kruppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J Biol Chem. 2001;276:14222–14229. doi: 10.1074/jbc.M010663200. [DOI] [PubMed] [Google Scholar]
- Milner CM, Campbell RD. The G9a gene in the human major histocompatibility complex encodes a novel protein containing ankyrin-like repeats. Biochem J. 1993;290:811–818. doi: 10.1042/bj2900811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Takahashi A, Murakami-Sekimata A, Homma Y. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J Biol Chem. 2001;276:42632–42638. doi: 10.1074/jbc.M107048200. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001a;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001b;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]