Active Alu Element “A-Tails”: Size Does Matter (original) (raw)
Abstract
Long and short interspersed elements (LINEs and SINEs) are retroelements that make up almost half of the human genome. L1 and Alu represent the most prolific human LINE and SINE families, respectively. Only a few Alu elements are able to retropose, and the factors determining their retroposition capacity are poorly understood. The data presented in this paper indicate that the length of Alu “A-tails” is one of the principal factors in determining the retropositional capability of an Alu element. The A stretches of the Alu subfamilies analyzed, both old (Alu S and J) and young (Ya5), had a Poisson distribution of A-tail lengths with a mean size of 21 and 26, respectively. In contrast, the A-tails of very recent Alu insertions (disease causing) were all between 40 and 97 bp in length. The L1 elements analyzed displayed a similar tendency, in which the “disease”-associated elements have much longer A-tails (mean of 77) than do the elements even from the young Ta subfamily (mean of 41). Analysis of the draft sequence of the human genome showed that only about 1000 of the over one million Alu elements have tails of 40 or more adenosine residues in length. The presence of these long A stretches shows a strong bias toward the actively amplifying subfamilies, consistent with their playing a major role in the amplification process. Evaluation of the 19 Alu elements retrieved from the draft sequence of the human genome that are identical to the Alu Ya5a2 insert in the NF1 gene showed that only five have tails with 40 or more adenosine residues. Sequence analysis of the loci with the Alu elements containing the longest A-tails (7 of the 19) from the genomes of the NF1 patient and the father revealed that there are at least two loci with A-tails long enough to serve as source elements within our model. Analysis of the A-tail lengths of 12 Ya5a2 elements in diverse human population groups showed substantial variability in both the Alu A-tail length and sequence homogeneity. On the basis of these observations, a model is presented for the role of A-tail length in determining which Alu elements are active.
[The sequence data from this study have been submitted to GenBank under accession nos. AF504933–AF505511.]
The genomes of all higher eukaryotes are littered with copies of mobile elements, including DNA transposons, short and long interspersed elements (SINEs and LINEs), and processed pseudogenes. The human SINE, Alu, is one of the most successful mobile elements, having generated over one million copies in the human genome (Lander et al. 2001). SINEs amplify by using an RNA polymerase III-derived transcript as template, in a process termed retroposition (Rogers and Willison 1983; Weiner et al. 1986). The RNA is reverse transcribed into a DNA molecule that integrates into a new site in the genome, probably using a nicked site in the genomic DNA as a primer (Boeke 1997). Previous studies have indicated that the ORF2 product of L1 elements supplies the endonuclease and reverse transcriptase activities for this integration process (Mathias et al. 1991; Feng et al. 1996). There is a variable-length stretch of homopolymeric adenosine residues or at least an A-rich region at the 3′ end of genomic SINE elements (Deininger et al. 1981). In Alu transcripts, this A stretch is within the transcript because the terminator for the transcription is found in the unique sequences downstream of the Alu element (Shaikh et al. 1997). Thus, each Alu transcript will be very similar in the Alu-containing 5′ end, have a similar 3′ oligo(dA)-rich tail that is encoded by the source gene and then having a unique segment at their 3′ end (Batzer et al. 1990; Shaikh et al. 1997). Thus, although this A-rich region is not a poly(A) tail in the sense used for polyadenylated mRNAs, the term is used in this paper for simplicity. The hypothetical common role of the A-rich regions of LINEs, SINEs and processed pseudogenes in the priming of reverse transcription was one of the first features predicted in these elements (Jagadeeswaran et al. 1981; Weiner et al. 1986). More recently, it has been suggested that the sequence specificity of the L1 endonuclease could nick the genomic integration site to provide a primer that could prime on these A-rich regions for all of these elements (Boeke 1997).
Most Alu amplification occurred >35 million years ago, with the current amplification rate almost 100-fold lower than at the peak of amplification (Shen et al. 1991). This current amplification rate is contributing significantly to human disease (Deininger and Batzer 1999), but must have been more deleterious 40 million years ago (for review, see Batzer and Deininger 2002). Alu elements amplifying at different stages of primate evolution have key diagnostic sequence differences that allow them to be classified into subfamilies (Shen et al. 1991; Deininger et al. 1992). These subfamilies show a generally sequential amplification, with the youngest subfamilies amplifying at a much lower rate. The older subfamilies, which constitute about 85% of the Alu copy number, appear to be incapable of retroposition. One of the most likely explanations for the formation of subfamilies and the changes in amplification rate is that very few Alu elements are capable of amplification at any given time (Deininger et al. 1992). Any element that has made at least one copy is considered a ‘source’ element. The term ‘master’ element is used to refer to elements that may have amplified very efficiently over a relatively long period of time, contributing significantly to the pattern of evolution of the elements (Deininger et al. 1992; Deininger and Batzer 1995; Roy-Engel et al. 2001). Elements capable of very high levels of activity were probably rare during evolution.
The presence of a homopolymeric adenosine run is likely a common structure for priming the SINE and LINE retroposition events (Boeke 1997), but it does not explain why so few Alu elements are capable of amplification. The vast majority of the million plus Alu elements present in the human genome have poly(A) stretches, but only a very few are able to retropose (Deininger and Daniels 1986; Batzer et al. 1990; Deininger et al. 1992). Therefore, the mere presence of an A stretch is not sufficient to confer on an Alu element the ability to retropose efficiently. Multiple factors, such as transcriptional capability and specific SINE interactions (possibly through other RNA binding proteins) with the LINE retrotransposition proteins, have been suggested to affect the amplification process (Schmid and Maraia 1992; Batzer and Deininger 2002). Ordinarily, L1 element proteins show a strong cis preference for the RNA that encodes them (Wei et al. 2001). Thus, Alu elements probably have a specific mechanism to compete effectively with this cis preference. In addition, the presence of an A stretch by itself does not explain why Alu elements amplify so effectively, whereas other polyadenylated RNAs in the cell only form retropseudogenes at much lower rates (Goncalves et al. 2000; Harrison et al. 2002). In this paper, a model is presented in which the length of the Alu A-tail, possibly coupled to interacting with poly(A) binding protein (Sondekoppa Muddashetty et al. 2002; West et al. 2002), may be a critical element that allows Alu elements to compete effectively for the L1 ORF2 product, resulting in their very high amplification rate.
RESULTS
Length Variation of the Alu and L1 “A-Tails”
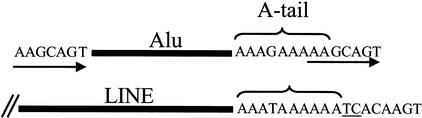
Some of the most recent Alu and L1 inserts have particularly long A-tails and an evaluation was performed to quantitate this observation for both of these families of mobile elements. The length of the A stretch was operationally defined as the number of bases between the last nucleotide of the Alu consensus sequence and the first non-A base in the 3′ flanking direct repeat (for details, see Fig. 1). Determination of the L1 A-tail length was more empirical and less precise because the direct repeats were not defined for most of the elements. In this case, the A-tail length was determined by counting the number of bases between the last nucleotide of the L1 consensus until reaching two consecutive non-A residues, unless another major poly(A) stretch followed. Under these conditions, any errors would more likely involve a few overestimations in the size length. This study was also limited to only the younger Ta subfamily of L1 elements (Boissinot et al. 2000; Sheen et al. 2000; Ovchinnikov et al. 2001) to minimize difficulties identifying the ends of the A-tail.
Figure 1.
Parameters used to determine the length of the A-tail of an Alu element. Alu elements (solid bar) and their A-tails are flanked by direct repeats (arrows) that are created during their insertion into the genome. For this study, the length of the A-tail was considered as the number of bases between the last nucleotide of the Alu consensus sequence and the 3′ flanking direct repeat. If the direct repeat sequence contained adenosine residues in its 5′ end, they were included in the count. The A-tail length for long interspersed elements (LINES) was determined by counting the number of bases between the last nucleotide of the L1 consensus until reaching two consecutive non-adenosine bases, unless another poly(A) stretch followed. In the example, the A-tail length would be considered to be 9 for the Alu and 10 for the LINE. Note that the A-tails may contain other nucleotide residues.
We determined the length of the A-tail from different subfamilies of Alu and L1 elements and compared them to the most recent Alu insertion events. Those elements that resulted in a human disease were considered as the very recent inserts. Several of these are probably de novo events, and most of them should have occurred within a relatively small number of generations because there is probably some selective pressure against the disease-causing alleles that would lead to their rapid loss from the population fairly quickly (Prak and Kazazian 2000). L1 elements that were generated de novo in tissue culture using the L1 retrotransposition assay were also included (Moran et al. 1996). For simplicity, all of these most recent elements will be referred to as the “disease” elements. The Alu elements analyzed were divided into the following subfamilies: Alu J and S were pooled to represent the ‘old’ Alu elements, whereas Alu Yb8, Alu Ya5, and Alu Ya5a2 were studied separately as members of young subfamilies (Roy et al. 2000; Carroll et al. 2001; Roy-Engel et al. 2001). Details of the sources of the Alu and L1 elements analyzed are indicated in Methods.
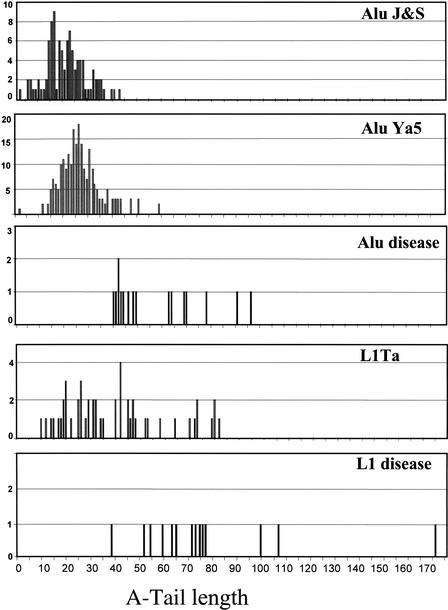
Some of the results of our analysis of the A-lengths of the different Alu and L1 elements are shown in Table 1. The A-tail was significantly longer in the disease inserts, with a difference of almost twofold in the mean length relative to any of the Alu subfamilies. Although, L1 elements have longer A stretches than do Alu elements in general, the same trend in A-length difference was observed. The longer L1 A-tails are consistent with their being generated from a polyadenylated mRNA rather than from an internal A-region as in the SINEs. In addition, the frequency distribution of the A-tail length in the different subfamilies was determined (Fig. 2). Again, there is a striking difference between the disease elements and the other older groups of Alus. The lengths of the A stretches in the disease Alu elements are all over 40 bp, and in the L1 elements only one is <40 bp with the rest being over 53 bp in length. Some of the A-lengths in this group reach as high as 97 bases for Alu and almost 180 bases for L1. For the groups with a large n value (Ya5 and J/S), the data roughly fit a Poisson distribution with a slight skew to the longer A-stretch lengths. This indicates that, once the elements are within the genome, various forces act on the A-tails until they shrink to an equilibrium value. Because the data represent Alu elements that have inserted at a variety of time points, the skewed distribution may indicate that some elements have not yet reached equilibrium. Overall, older Alu elements appear to have shorter A-tails than do their younger counterparts. This correlation was previously observed in LINEs when comparing the A-tail length of a group of older L1-GAG elements to the younger L1H-Ta group (Ovchinnikov et al. 2001).
Table 1.
Mean Length of the 3′ “A-Tail” of Different Alu and L1 Elements
| Repeated element group | Total analyzed | Length A-taila |
|---|---|---|
| Alu J and S | 100 | 21 ± 8 |
| Alu Yb8 | 79 | 28 ± 9 |
| Alu Ya5 | 235 | 26 ± 9 |
| Alu Ya5a2 | 23 | 30 ± 11 |
| Alu “disease”b | 16 | 58 ± 19 |
| L1 Ta | 50 | 41 ± 20 |
| L1 “disease”bc | 14 | 77 ± 33 |
Figure 2.
Histogram of the distribution of the length of A-tail in different subgroups of Alu and L1 elements. Distribution of the A-tail length of Alu elements belonging to old subfamilies (Alu J and S), the young subfamily Alu Ya5, and those reported to cause disease (see text for definition), plus the L1 elements from the young Ta (Ta0 and Ta1) and those linked to disease. Note: The scale for the _Y_-axis (number of elements) is different for each group.
Alu Elements With Long A-Tails Are Uncommon in the Human Genome
To evaluate the abundance and the age of the Alu elements with long A stretches in the human genome, we performed a search of the human genome draft sequence. Only the Alu elements with 40 or more adenosine residues, with no other intervening bases, were retrieved. Fewer than 1000 Alus were identified from the draft sequence (Table 2). The 1000 Alus with long A-tails represents approximately 0.1% (1000/1.1×106) of the total number of Alu elements present in the human genome. The use of a more stringent parameter of 50 or more adenosine residues to perform the search reduces the total number by about 10-fold (Table 2).
Table 2.
Subfamily Distribution of Alu Elements With Long “A-Tails” Retrieved From the Human Genome Draft
| Alu subfamily | A-tail with ≥40 A | A-tail with ≥50 A | ||
|---|---|---|---|---|
| Total members | Percent (%) | Total members | Percent (%) | |
| J | 74 | 7.2 | 7 | 5.6 |
| S | 465 | 45.5 | 51 | 40.5 |
| Y/Yc1 | 313 | 30.6 | 34 | 27.0 |
| Ya5/Ya5a2 | 113 | 11.1 | 26 | 20.6 |
| Yb8/Yb9 | 57 | 5.6 | 8 | 6.3 |
| Total | 1022 | 100 | 126 | 100 |
The distribution of these long A-tailed Alus roughly divides the elements to about half that clearly belong to the currently active Alu subfamilies Y, Ya5, or Yb8 and the other half that belong to the older, inactive Alu subfamilies J, Sx, Sg, or Sc. Evaluation of the proportion of long A-tailed Alu members within each subfamily (using the previously estimated total copy number for each subfamily [Roy-Engel et al. 2001]) showed that the youngest Alu subfamilies have the highest percentage of long A-tail members. The group of most active Alu subfamilies, Ya5/Ya5a2/Ya8, contained the most long A-tail elements with about 4.1% (113/2750) of their members in this category, followed by Alu Yb8/9 with about 3.0% (38/1900), Alu Y with only 0.17% (313/∼190,000), and Alu S and J with 0.06% (539/9× 105). There is an inverse correlation between the age of the Alu subfamily and the proportion of the members with long A-tails in the genome, indicating that loss of A stretches may be a primary, but not the only, inactivating feature in the older subfamilies.
Analysis of the Candidates for the Potential Source Gene of the Alu Ya5a2 in NF1 Patient Loci
If the A-tail length is a critical parameter for Alu retroposition activity, we would predict that the source Alu element responsible for the de novo insertion of an Alu element inactivating the NF1 gene in a neurofibromatosis patient (Wallace et al. 1991) should have an identical sequence to the inserted Alu, as well as possessing a long A-tail. Because the NF1 insertion was in a paternal chromosome, the long A-stretch source Alu would have to be found either in the father's DNA or in the DNA of the patient. A query of the draft of the human genome database in search of the Alu elements with 100% sequence identity to the NF1 Alu retrieved a total of 19 elements. We had previously identified 13 matches to the NF1 insertion event, which is a perfect match to the Ya5a2 consensus, using a less complete version of the human sequence available at the time (Roy et al. 2000).
Of the 19 Ya5a2 elements, only 5 had A-tails of 40 or more bases in the GenBank database. These five and the two with the next longest A-tails were selected to evaluate those specific loci in the genomes of the NF1 patient and the father. The loci were analyzed using polymerase chain reaction (PCR)-generated clones to determine insertion presence or absence of the element and the length of the A stretch. More than one sample from each PCR was sequenced to increase the probability of detecting differences (if any) between the two alleles of each subject or potential size variations generated by the cloning procedure. Only two loci (Table 3) contained Alu elements with the long A-tail in either the patient (35 bases) or the father (53 bases). The 35-base allele may represent the other paternal allele, a maternal allele, or, less likely, the shortening of the allele from one generation to another. In the other case, the locus contained two alleles with very different A-tail lengths in the patient. One of the A-tails was in the 24–26 bp range (the heterogeneity may result from either mosaicism within the patient or, more likely, minor variation created in the PCR and cloning). This allele was similar to the paternal allele(s). However, the patient also had an allele with an A stretch of 45 bases (comparable to the one observed in the draft human genome), which may be an allele inherited from the mother. Because exhaustive measures were not taken to definitively analyze all alleles in these samples, it is impossible to say that there were no other long A-tail alleles. These data show the presence of at least two alleles present in the father or the patient with an A-tail of sufficient length to have served as the source element for the disease-causing insertion. In addition, the A-tail length of a specific Alu element can differ between individuals, indicating a high variability in the population. Evaluation of the degree of insertion polymorphism associated with these elements showed that the three fixed loci with long A-tails had at least one clone comparable to the A-tail length observed in the draft human genomic sequence (Table 3).
Table 3.
“A-Tail” Length of Several Alu Elements With 100% Identity to the Ya5a2 Alu Element Insert in the NF1 Gene
| Locus | Accession number | A-tail length | Allele frequencyc | ||
|---|---|---|---|---|---|
| Drafta | Patient | Parentb | |||
| Ya5a2AD11 | AC090071 | 53 | 35 | 51 | Fixed present |
| Ya5a2AD12 | AC004057 | 46 | 26–28/45 | 24–31 | Fixed present |
| Ya5NBC208 | AL132992 | 37 | 38 | 34 | Fixed present |
| Ya5a2AD7 | AL138681 | 43 | 27–28 | 26–29 | Intermediate |
| Ya5a2AD8 | AL162713 | 53 | 23 | 31 | Intermediate |
| Ya5NBC220c | AC007611 | 39 | 24 | 24–25 | Intermediate |
| Ya5NBC239 | AL133284 | 41 | Emptyd | Emptyd | Low |
Population Diversity of A-Tail Length and Composition
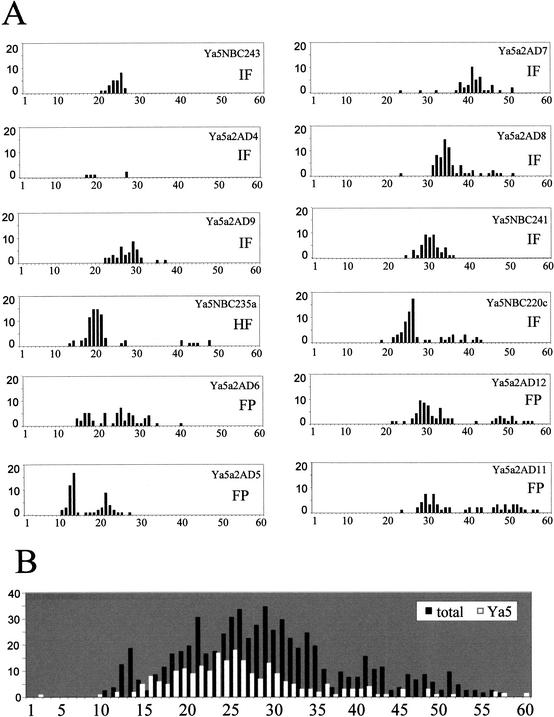
The polymorphism in A-tail lengths from 12 Ya5a2 containing loci was evaluated by sequence analysis from at least 25 and up to 72 PCR-generated clones from multiple individuals from different populations, with the exception of one locus (Ya5a2AD9) in which there are data from only five individuals. These data show extensive population diversity in A-tail length (Fig. 3A). A number of the loci show a similar length distribution to those seen between different elements shown in Figure 2. However, a few of the loci showed a significant number of alleles with A stretches longer than 40. Several appeared to be most consistent with A-tail length distributions centered around two major allele lengths (i.e., Ya5a2AD5, AD11, and AD12). Comparison of the distribution of the Alu Ya5 elements and the combined data of all of the analyzed Ya5a2 loci shows the same distribution, with a more pronounced skew to longer A-tails for the younger Alu Ya5a2 elements (Fig. 3B).
Figure 3.
A-tail length variation within the population. (A) Histograms of the A-tail length distribution of the selected Ya5a2 element from the same locus of different individuals. The histogram reflects the A-tail length (_X_-axis) and the amount of elements for that length (_Y_-axis). The allele frequency for each locus is indicated as fixed present (FP), intermediate (IF), or high frequency (HF); see definitions in Table 3. (B) Histogram of the distribution of the Ya5 elements retrieved form the genome draft sequence (white bars) and the combined data of all the individual Ya5a2 elements (black bars) from different populations analyzed, as listed earlier.
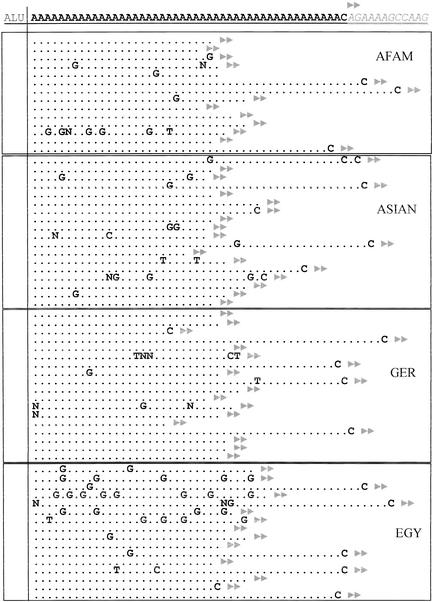
In addition to the changes in length, there are also other mutations appearing in these sequences, which break up the perfect nature of the A stretch. The A-tails for almost half of the Ya5a2 elements analyzed have almost no sequence heterogeneity, with a few rare Alus that have one non-A base. In contrast, the other loci contain Alu elements in which the majority, if not all, contained at least one non-A base in the A stretch. An example of the A-tails of the elements present in one of these loci is shown in Figure 4. Some of the changes appear consistently in individuals from the same population group, indicating a possible founder effect (see Fig. 4). It seems likely that this sequence heterogeneity may also influence the length stability of the elements in which disruption of the A stretch with other bases may prevent length reduction. In some cases, these base changes may serve as nuclei for subsequent microsatellite expansion (Arcot et al. 1995). The A-tail variability may indicate a more dynamic Alu amplification than previously thought, in which different individuals may have different active elements at a given time.
Figure 4.
Sequence alignment of the A-tails of the Alu Ya5a2AD12 from different individuals. The sequence of the A-tail from the Alu element obtained from the human genome draft is shown on the top line. The sequence of the Alu body is not shown but is represented by the word ALU. The 3′ direct repeat is shown in gray italics and is represented by double arrowheads. Nucleotide substitutions at each position are indicated with the appropriate nucleotide or with an N when the nucleotide could not be clearly identified. The four populations analyzed are indicated inside each boxed group: African American (AFAM), Asian (ASIAN), German (GER), and Egyptian (EGY).
DISCUSSION
The Length of the Alu A-Tail and Retroposition Capability
Only a few Alu elements are currently capable of serving as source elements (Batzer and Deininger 1991; Batzer et al. 1992; Liu and Schmid 1993). There are a number of potential features that may limit the retroposition activity of Alu elements and subfamilies. Among these factors are any sequence differences within the Alu element, either of a subfamily or random nature (Liu and Schmid 1993; Chu et al. 1995; Shaikh et al. 1997). The subfamily changes may influence the interaction of the Alu elements with the retroposition machinery, resulting in a selection against the older subfamilies (Matera et al. 1990; Englander et al. 1993; Kim et al. 1994; Labuda and Zietkiewicz 1994; Shaikh and Deininger 1996; Shaikh et al. 1997). The random mutations may result in loss of expression of the older elements, either through promoter mutations or through changes in RNA stability (Alemán et al. 2000; Batzer and Deininger 2002). However, transcription itself only partially explains more than a small part of the limitation in source elements. The majority of Alu RNA transcripts are still transcribed from subfamilies that appear to be incapable of amplification (Shaikh et al. 1997). The 3′ end of the Alu transcripts is the most variable, both in terms of length and homogeneity of the A stretch, and as the unique sequences between the A stretch and the RNA polymerase III terminator. Thus, interactions occurring with this 3′ end would be likely determinants causing selection of a few active master elements.
Both the length and the homogeneity of the A stretch appear to be important for efficient retroposition. Sequence comparison of the BC1 locus, the known master gene for the rodent ID family of SINEs (Kim et al. 1994) from different rodent species relative to the accumulated copy number (Kass et al. 1996) strongly supports this observation (Fig 5). Although the sequence of the BC1 gene and flanking regions in general are well conserved (Deininger et al. 1996), as are their relative expression levels (Sapienza and St Jacques 1986), the 3′ A stretch varies greatly between the different species of rodents. There is a close correlation between A-richness of these BC1 loci and the copy numbers of ID family members in these species (Fig. 5). Of particular note are the presence of only 200 ID copies in guinea pig and the very short A stretches in the BC1 gene, compared with the long A stretches in both Peromyscus species and their high copy numbers. Rat has a copy number closer to 125,000, but previous studies showed that the BC1 RNA gene generates only about 12,000 of the copies (Kim et al. 1994). This apparent requirement for long A-tails in addition to a minimum amount of pure adenosine stretches for efficient retroposition may be determined by the interaction with poly(A) binding protein, as discussed in the following paragraph.
Figure 5.
Sequence alignment of the A-rich regions of the BC1 RNA gene of different rodent species and the copy number. The sequences of the 3′ ends of the BC1 RNA genes, containing the A-rich stretches, are shown with dashes used to improve the alignments. The rodent sequences are as follows: C, Peromyscus californicus (Accession #U33852); D, deer mouse, Peromyscus maniculatus (#U33851); R, rat, Rattus norvegicus (#M16113); M, mouse, Mus musculus (# U01310); G, gerbil, Meriones unguiculatus (#U33852); H, hamster Mesocricetus auratus (#U01309); and GP, guinea pig, Cavia porcellus (#U01304). Variations of the adenosine residues between the species are indicated in bold. Rectangles enclose the longer homopolymeric A stretches within the variant region. BC1 copy number for each species (Kass et al. 1996) is indicated at the right. The asterisk represents the copy number only for the rat BC1 type 1 (Kim et al 1994).
Previous reports hypothesize that poly(A) polymerase may occasionally act on RNA polymerase III transcripts and create the A stretches on SINEs (Chen et al. 1998; Borodulina and Kramerov 2001). However, it seems unlikely that the 3′ end of an Alu transcript would first degrade back to the A stretch followed by a polyadenylation reaction before retroposition. Studies of Alu RNAs have shown no evidence for such polyadenylation being a significant factor (Shaikh et al. 1997), indicating that the Alu A stretches are coded in the genomic elements (Batzer et al. 1990). For all of the new disease-causing insertions to have A stretches of 40 bases or more, their source elements must have had longer A stretches. Therefore, the 1000 Alus with A-tails longer than 40 bp in the initial human genome sequence are possible candidates to serve as source elements. The 126 with 50 A-tail lengths are even better candidates, because this is probably the length for the most efficient, cooperative binding of two molecules of poly(A) binding protein (Smith et al. 1997). However, a long A stretch alone is not sufficient to allow Alu amplification. Elements with mutations mitigating transcription capacity or the interaction with L1 supplied factors, for instance, would not be capable of retroposition even with a long A-tail.
This latter observation would help to explain the apparent amplification inactivity of the old Alu subfamilies despite the presence of some elements with long A-tails. Several of the characterized older Alu elements are highly unlikely to generate pol III transcripts because of 5′ truncation and multiple mutations within their internal A and B boxes. In addition, these older elements have accumulated multiple mutations (both CpG and non-CpG), which may reduce or abolish retropositional capability. To calculate the expected influence of random mutations decreasing the relative retroposition efficiency of the older subfamilies, we used the proportion of Alu transcripts originating from elements of the older Alu S and J subfamilies and the younger Alu Y subfamily (Shaikh et al. 1997). The most actively amplifying subfamily, Alu Ya5, contributed <1% of the transcripts. A “transcript enrichment” factor was calculated by using the percent of each Alu subfamily and the percent of actual transcripts they contribute (Table 4). For example, the Alu Ya5 contributes ∼ 0.3% of the total number of Alus in the genome and contributes 0.8% of the Alu transcripts; thus the enrichment factor would be 2.64 (0.8/0.3), indicating the detection of 2.6-fold more transcripts than that expected just by copy number. We must caution that this transcript enrichment was for expression in cultured cells and not germ cells, and therefore is only an approximation. Multiplication of elements in each subfamily with long A-tails by their relative subfamily transcription rates generates a first correction to the expected amplification capability of these long A-tailed Alus (Table 4). When comparing the actual numbers (observed) of Alu elements in each subfamily from our disease group with our estimates (expected), based on the product of the number of long A-stretch Alus and their relative transcription rate, a reasonable correlation is obtained, with the exception of the expected amplification of some of the older, inactive Alu subfamilies. Thus, the length of the A stretch and transcription rate may be the primary factors, but are not the only factors required for efficient retroposition. Other factors (such as loss of secondary structure, interaction with proteins, etc.) that might negatively impact the amplification capabilities of the older subfamilies in addition to this simple model need to be considered in this model. For example, the ability to interact with proteins, such as the L1 factors, are potential explanations for the discrepancy. In this case, variations digressing away from the consensus sequence more commonly found in older Alu elements may have an important impact in the RNA–protein interaction.
Table 4.
“A-Tail” Length and Transcriptional Activity of the Different Alu Subfamilies
| Alu subfamily | % Alu transcriptsa | % Total Alus | Transcript enrichmentb | Long A-Tail Alus (%)c | Transcription/A-tail Factor | Expected (%)d | Observed (%)e |
|---|---|---|---|---|---|---|---|
| S + J | 66 | 82 | 0.80 | 58 (46) | 36.9 | 4 (23) | 0 (0) |
| Y | 33 | 17 | 1.96 | 34 (27) | 53.0 | 5 (33) | 5 (31) |
| Ya5 | 0.8 | 0.3 | 2.64 | 26 (21) | 54.4 | 5 (34) | 6 (38) |
| Yb8 | 0.5f | 0.2 | 2.50f | 8 (6) | 15.9 | 2 (10)f | 5 (31) |
| Total | 100 | 100 | — | 126 (100) | 160.2 | 16 (100) | 16 (100) |
Alu A-Tail Length Instability
The long A stretches of the newly inserted elements are not evolutionary stable. A clear example of this instability can be observed with the Alu that inserted in the eya1 locus, where the A-tail went from A97 to A31 in one generation (Abdelhak et al. 1997). Our own data show that even the Ya5a2 subfamily, which has an estimated average age of 0.62 million years (0.28–1.08 with 95% confidence) (Roy et al. 2000), is quickly approaching an A-tail distribution similar to that of the older subfamilies. In these latter studies, some cases presented two size distributions of alleles. The longer alleles might be consistent with the original length of the A stretch following insertion. Thus, it is likely that even a period of about one million years has not completely eliminated the initial long A stretches in some loci. Alternatively, the A-tail length may be more dynamic than this simple model of long A stretches shrinking to a smaller size distribution. Other events may allow the A-stretches to amplify to longer lengths (Wang et al. 2001). The general equilibrium may favor the shorter alleles, but occasional growth of A stretches could result in activation of a previously inactive allele, affecting some of the current views on Alu amplification dynamics. These changes in A-tail length could be caused by either strand slippage during replication, unequal recombination, or even possibly gene conversion from an Alu with a long A stretch. Most data on simple-sequence repeats would be consistent with strand slippage being the primary mode of change (Calabrese et al. 2001).
Several potential factors may lead to the overall shortening of A stretches. First, these sequences tend to replicate poorly and therefore either result in selection for shorter alleles or somehow favor shortening by strand slippage. Alternatively, poly(A) stretches have previously been reported to form unusual DNA structures (Cubero et al. 2001; Hizver et al. 2001; McConnell and Beveridge 2001), and this can inhibit progression of the DNA polymerase. Also, Alus in transcribed regions may result in a selection bias if they influenced transcription or transcript stability in some way. One such example was observed in the chloroplasts of Chlamydomonas (Lisitsky et al. 2001). However, when using a reporter construct, the incorporation of Alus with short (20–30 bp) versus long (40–58 bp) A-tails in the transcript showed no significant changes in expression of that reporter gene (data not shown). Whatever the mechanistic cause, however, there is no question that A stretches are quite unstable in evolutionary terms and tend to approach the smaller size distribution relatively rapidly. As these elements get older, the A stretches accumulate mutations, resulting in higher complexity within these sequences. These changes may stabilize the sequence as it reduces the simple sequence nature of the region. Alternatively, microsatellites may form in these regions (Economou et al. 1990; Arcot et al. 1995; Jurka and Pethiyagoda 1995), perhaps resulting in different amplification dynamics. One extreme example is the formation of a triplet repeat in the middle A-rich region of an Alu in the frataxin gene, which can lead to massive, disease-causing amplification of the triplet repeat (Montermini et al. 1997).
Mechanistic Model for the Role of A-Tail Length in Alu Retroposition
Our data support the critical importance of the length of the Alu A stretch for retroposition capability. There is essentially no evidence of activity from the massive number of the older Alu elements where the vast majority contain modest-length A stretches. This indicates that there is a critical threshold length of A stretch for retroposition efficiency, but does not provide evidence for whether that retroposition capability continues to increase rapidly with increasing length beyond that threshold. None of the disease inserts shown in Figure 2 had an A-tail shorter than about 40 bp. If the priming for reverse transcription were to occur randomly at different positions on the A stretch in the RNA, we would expect some of the priming events would create shorter A stretches. The complete absence of short A-tails in the inserts implies that not only do the source RNA(s) have a long A stretch, but also that priming occurs either at, or very near, the 3′ end of the A stretch. They could be preferentially adjacent to the downstream unique sequences, or at least constrained from priming in the first 40 bases of the A stretch in the RNA. Recently, the cytoplasmic poly(A) binding protein (PABP) has been identified as part of the rodent SINE (BC1) ribonucleoprotein (RNP) complex (Sondekoppa Muddashetty et. al 2002; West et. al 2002) and is also complexed with other SINE RNAs. PABP binds poly(A) stretches in a cooperative manner, in which one molecule of PABP binds ∼25 adenosine residues (Smith et al. 1997). Therefore, the longer the A-tail, the more PABP molecules bind, increasing the stability of the protein–RNA interaction. Our A-tail data would be consistent with a requirement for two PABP molecules binding and protecting the first 40–50 bases from the priming event. It is therefore tempting to also consider whether PABP may be an important factor in the efficiency of the SINE retroposition process.
In addition to PABP, SINE RNAs also bind other proteins that bind to the SINE portion of the RNA. Among these are the SRP9/14 proteins that bind the 5′ end of the Alu RNA (Hsu et al. 1995; Chang et al. 1996). SRP9/14 is an important component of the signal recognition particle (SRP), an RNP complex that interacts with the ribosome and targets proteins to the rough endoplasmic reticulum (for reviews, see Walter and Johnson 1994; Bovia and Strub 1996). For Alu elements, these proteins in conjunction with PABP may help target the Alu RNA to the ribosome in close proximity to the L1 RNA, where it may effectively compete for the L1 translation products necessary for the retroposition process. Other SINEs are unlikely to bind SRP9/14 because they are derived from tRNA genes rather than 7SL RNA. However, the proteins that bind to them may serve a similar function.
We propose a model in which the PABP influences SINE retroposition (Fig 6). Many other RNAs in the cell have long A-tails and bind PABP. However, they are almost all mRNA molecules that are capped. The cap may inhibit their retroposition by two possible mechanisms. In one case, the cap targets the mRNA for translation by their own ribosomes, isolating them from the ribosomes that are translating the L1 RNA. The second influence may relate to the other proteins in the cap-binding complex, such as eIF4G and eIF4E. PABP interacts directly with eIF4G, which effectively circularizes the RNA (for reviews, see Gingras et al. 1999). However, the polymerase III-transcribed RNAs are not capped, leaving the A stretch/PABP end free to interact with other molecules, possibly including the cap complex of the L1 RNA (Fig. 6). The proximity of the Alu RNA to the L1 RNA and translation machinery may allow it to efficiently compete for the necessary components required for its retroposition, as proposed previously (Boeke 1997). This might allow the Alu elements to use the cis preference of the L1 translation products (Wei et al. 2001) for their own benefit. In this scenario, almost any RNA polymerase III-transcribed RNA could retropose if it acquired an A stretch long enough to bind PABP effectively. In addition, perhaps pol II-transcribed genes with normal polyadenylation that “escape” the cap complex control could also retropose, for example, 5′ truncated retropseudogenes or endogenous viruses. Although this model is highly speculative, it now seems critical to incorporate A-stretch length into considerations of SINE formation and activity.
Figure 6.
Alu retroposition model. Poly(A) binding protein (PABP) interacts with the A-tails of the Alu and L1 RNA. The SRP9/14 proteins bind the 5′ end of the Alu RNA, which may help target the RNA to the ribosome and translational machinery. The cap complex of L1 RNA interacts with the PABP in the A-tail, circularizing the RNA. In contrast, Alu RNA, being a pol III, has no cap and thus is not circularized, allowing the A-tail/PABP end free to interact with the L1 cap complex. Because the Alu RNA is in close proximity to the L1 RNA when translation of the retroposition factors occurs, it allows the Alu to compete for the required components for its own retroposition. Details of the model are presented in the Discussion.
METHODS
Data Mining and Analysis of the A-Tail Length
The source for members of the Alu Ya5, Ya5a2, and Yb8 subfamilies came from previously generated databases obtained from BLAST searches of the human genome (Roy et al. 2000; Carroll et al. 2001; Roy-Engel et al. 2001). To generate a database of Alu members belonging to older subfamilies, we performed analysis of the elements from three genes, FAA (AC005360), NCF1 (AF184614), and RRM1 (AF107045). They were retrieved from the NCBI GenBank database and analyzed by RepeatMasker2 from the University of Washington Genome Center server (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). The Alu elements from the Sc, Sx, Sg, Sq, Sp, Jo, and Jb subfamilies present in these genes were randomly selected for the analysis of the A-tail length. The list containing the majority of the disease-causing Alu inserts has been previously published (Deininger and Batzer 1999). The Alu elements found in MLVI-2 (Economou-Pachnis and Tsichlis 1985), PROGINS (Rowe et al. 1995), and ACE (Cambien et al. 1992) were excluded, because they are either not clearly causative of disease or were found by linkage analysis rather than by causing a disease. In addition, the full sequence for the Alu elements found in Btk (Lester et al. 1997) or the Factor IX (Wulff et al. 2000) was not available. An additional three examples of disease-causing Alu inserts were found in the following genes: one each in factor IX (Li et al. 2001), factor VIII (Sukarova et al. 2001), and PBGD (Mustajoki et al. 1999). A total of 16 disease Alu elements were analyzed.
A database of 50 Ta L1 elements was collected from the human genome using BLAST on the nr database, selecting for perfect matches to the 26 bases of the Ta consensus sequence (5′-GATGACACATTAGTGGGTGCAGCGCA-3′) with the expected value of −e = 1.0e−40. For recent LINE inserts, the following L1 elements were used: JH-25 (Woods-Samuels et al. 1989), APC (Miki et al. 1992), L1β-thal (Divoky et al. 1996), L1XLCDM (Yoshida et al. 1998), L1RP (Schwahn et al. 1998), two causing disease in mouse L1ORI and L1SPA (Naas et al. 1998), and seven L1 inserts generated from the L1 tissue culture assay (Moran et al. 1996; Wei et al. 2001). All the databases are available on our Web site (http://129.81.225.52).
To search for putative source genes with 100% identity to the Ya5a2 Alu element that inserted into the NF1 gene (Wallace et al. 1991), we searched the GenBank human genome draft database with the BLAST program, as previously described (Roy et al. 2000). To isolate Alus with long A-tails, we performed BLAST searches on the GenBank draft human database with low complexity filter disabled and the maximum number of results returned (-v -b flags) set at 5000 to retrieve more hits and an expected value of e−18; otherwise standard settings were used. The oligos used were TCTC(A × 40 or A × 50). Elements containing an unbroken stretch of 40 or 50 A's were extracted from GenBank accessions and analyzed by RepeatMasker.
Details of the parameters used to determine the length of the A-tails from the Alu and LINE elements are shown in Figure 1.
PCR Amplification
PCR amplification of the different loci containing the candidate Alu elements was performed in 20-μl reactions using an MJ Research PTC 200 thermal cycler with the following conditions: 1X Promega buffer, 1.5 mM MgCl2, 200 μM deoxyribonucleoside triphosphates, 0.25 μM primers, and 1.5 U Taq polymerase (Promega) at 94°C for 2 min; 94°C for 20 sec, X°C for 30 sec (X is the annealing temperature indicated as follows for each primer pair), and 72°C for 1 min, for 30 cycles; 72°C for 20 min. The final PCR products were cloned into pGEMTeasy Vector System I (Promega). The following primers were used to amplify the _NF1_-selected loci [accession number]: Ya5a2AD7 [AL138681]: 5′-TGGACACGCAT GAAAGAAACCCTACC-3′ and 5′-TGGATTAATTCATTCA ACTTCACTACAA-3′at 58°C; Ya5a2AD8 [AL162713]: 5′-TTAGCTGCCATCAACCACCCTTATCA-3′ and 5′-CATGTTTGCTGTCTCCTACTGTCTTCTGC-3′ at 53°C; Ya5AD11 [AC090071]: 5′-TTCTGCTCCAAATAATAAC TACCT-3′ and 5′-AGAATTTAAGACCCTATGGCACCTC-3′ at 50°C; Ya5AD12 [AC004057]: 5′-CAGGCCAGGAGTTTT GAAATTTTATC-3′ and 5′-CATGCCTCTTCTCACCTGTT TCAACCA-3′ at 58°C; Ya5a2AD4 [AC023932]: 5′-TGACG GGAGAAGTTAACAGT-3′ and 5′-TGCACCTGACAGAAA ATCT-3′ at 55°C; Ya5a2AD5 [AC026839]: 5′-GCATTAAGAA TGTGGACCAT-3′ and 5′- TGTAGTTGGAAGCCCTTAAT-3′ at 60°C; Ya5a2AD6 [AL355580]: 5′-ATTATTCCTAGTGAGGGGA ATGAA-3′ and 5′-ACTTCCTATGATTCCATCTCCAA-3′ at 60°C; and Ya5a2AD9 [AC079456]: 5′-TTTATTGCCAGAA GCTTTCGT-3′ and 5′-AAGCGCTTCCTTCATAAATCA-3′ at 60°C. For Ya5NBC243, Ya5NBC235a, Ya5NBC239, Ya5NBC241, Ya5NBC220c, and Ya5NBC208, primers and conditions have been previously published (Roy et al. 2000). DNA sequences (∼600 individual sequences) from the Alu elements listed earlier were determined in a diverse panel of human genomes. These DNA sequences were derived from Alu-containing PCR products cloned using TOPO TA cloning kits (Invitrogen) and sequenced using an ABI 3100 automated DNA sequencer. These sequences were assigned GenBank accession numbers AF504933–AF505511.
WEB SITE REFERENCES
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.
http://129.81.225.52; Deininger Lab Web page.
Acknowledgments
A.M.R. was supported by a Brown Foundation fellowship from the Tulane Cancer Center. Parts of these data were used as the foundation for the research project presented by O.O.O. in the International Science Fair in Kentucky, May 2002. This research was supported by the Louisiana Board of Regents Millennium Trust Health Excellence Fund HEF (2000–05)-05 (M.A.B. and P.L.D.), (2000–05)-01 (M.A.B. and P.L.D.), and (2001–06)-02 (M.A.B.), National Institutes of Health RO1 GM45668 (P.L.D.) and GM59290 (M.A.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pdeinin@tulane.edu; FAX (504) 588-5516.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.384802\. Article published online before print in August 2002.
REFERENCES
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Levi-Acobas F, Cruaud C, Le Merrer M, Mathieu M, et al. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet. 1997;6:2247–2255. doi: 10.1093/hmg/6.13.2247. [DOI] [PubMed] [Google Scholar]
- Alemán C, Roy-Engel AM, Shaikh TH, Deininger PL. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 2000;28:4755–4761. doi: 10.1093/nar/28.23.4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. A human-specific subfamily of Alu sequences. Genomics. 1991;9:481–487. doi: 10.1016/0888-7543(91)90414-a. [DOI] [PubMed] [Google Scholar]
- ————— Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Kilroy GE, Richard PE, Shaikh TH, Desselle TD, Hoppens CL, Deininger PL. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 1990;18:6793–6798. doi: 10.1093/nar/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Bazan HA, Kim J, Morrow SL, Shaikh TH, Arcot SS, Deininger PL. Large-scale subcloning of bacteriophage λ ZAP clones. Biotechniques. 1992;12:370–371. [PubMed] [Google Scholar]
- Boeke JD. LINEs and Alus—The polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Borodulina OR, Kramerov DA. Short interspersed elements (SINEs) from insectivores. Two classes of mammalian SINEs distinguished by A-rich tail structure. Mamm Genome. 2001;12:779–786. doi: 10.1007/s003350020029. [DOI] [PubMed] [Google Scholar]
- Bovia F, Strub K. The signal recognition particle and related small cytoplasmic ribonucleoprotein particles. J Cell Sci (Pt 11) 1996;109:2601–2608. doi: 10.1242/jcs.109.11.2601. [DOI] [PubMed] [Google Scholar]
- Calabrese PP, Durrett RT, Aquadro CF. Dynamics of microsatellite divergence under stepwise mutation and proportional slippage/point mutation models. Genetics. 2001;159:839–852. doi: 10.1093/genetics/159.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambien F, Poirier O, Lecerf L, Evans A, Cambou JP, Arveiler D, Luc G, Bard JM, Bara L, Ricard S, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature. 1992;359:641–644. doi: 10.1038/359641a0. [DOI] [PubMed] [Google Scholar]
- Carroll ML, Roy-Engel AM, Nguyen SV, Salem AH, Vogel E, Vincent B, Myers J, Ahmad Z, Nguyen L, Sammarco M, et al. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J Mol Biol. 2001;311:17–40. doi: 10.1006/jmbi.2001.4847. [DOI] [PubMed] [Google Scholar]
- Chang DY, Hsu K, Maraia RJ. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sinha K, Perumal K, Gu J, Reddy R. Accurate 3' end processing and adenylation of human signal recognition particle RNA and alu RNA in vitro. J Biol Chem. 1998;273:35023–35031. doi: 10.1074/jbc.273.52.35023. [DOI] [PubMed] [Google Scholar]
- Chu WM, Liu WM, Schmid CW. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 1995;23:1750–1757. doi: 10.1093/nar/23.10.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero E, Luque FJ, Orozco M. Theoretical studies of d(A:T)-based parallel-stranded DNA duplexes. J Am Chem Soc. 2001;123:12018–12025. doi: 10.1021/ja011200t. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. SINE master genes and population biology. In: Maraia R, editor. The impact of short, interspersed elements (SINEs) on the host genome. Georgetown, TX: R.G. Landes; 1995. pp. 43–60. [Google Scholar]
- ————— Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Daniels G. The recent evolution of mammalian repetitive DNA elements. Trends Genet. 1986;2:76–80. [Google Scholar]
- Deininger PL, Jolly D, Rubin C, Friedmann T, Schmid CW. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981;151:17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Tiedge H, Kim J, Brosius J. Evolution, expression, and possible function of a master gene for amplification of an interspersed repeated DNA family in rodents. In: Cohn WE, Moldave K, editors. Progress in nucleic acid research and molecular biology. San Diego: Academic Press; 1996. pp. 67–88. [DOI] [PubMed] [Google Scholar]
- Divoky V, Indrak K, Mrug M, Brabec V, Huisman THJ, Prchal JT. A novel mechanism of β-thalassemia. The insertion of L1 retrotransposable element in β globin IVSII. Blood. 1996;88:148a. [Google Scholar]
- Economou EP, Bergen AW, Warren AC, Antonarakis SE. The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome. Proc Natl Acad Sci. 1990;87:2951–2954. doi: 10.1073/pnas.87.8.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou-Pachnis A, Tsichlis PN. Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res. 1985;13:8379–8387. doi: 10.1093/nar/13.23.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander EW, Wolffe AP, Howard BH. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J Biol Chem. 1993;268:19565–19573. [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Goncalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Hegyi H, Balasubramanian S, Luscombe NM, Bertone P, Echols N, Johnson T, Gerstein M. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 2002;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine–thymine tract and its role in gene regulation. Proc Natl Acad Sci. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K, Chang DY, Maraia RJ. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J Biol Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran P, Forget BG, Weissman SM. Short, interspersed repetitive DNA elements in eukaryotes: Transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981;26:141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: Compilation and analysis. J Mol Evol. 1995;40:120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Kass DH, Kim J, Deininger PL. Sporadic amplification of ID elements in rodents. J Mol Evol. 1996;42:7–14. doi: 10.1007/BF00163205. [DOI] [PubMed] [Google Scholar]
- Kim J, Martignetti JA, Shen MR, Brosius J, Deininger PL. The rodent BC1 RNA gene is a master gene for ID element amplification. Proc Natl Acad Sci. 1994;91:3607–3611. doi: 10.1073/pnas.91.9.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda D, Zietkiewicz E. Evolution of secondary structure in the family of 7SL-like RNAs. J Mol Evol. 1994;39:506–518. doi: 10.1007/BF00173420. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lester T, McMahon C, VanRegemorter N, Jones A, Genet S. X-linked immunodeficiency caused by insertion of Alu repeat sequences. J Med Gen Suppl. 1997;34:S81. [Google Scholar]
- Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, Careri D, Pinto MT, Kasper CK, Sommer SS. Frequency of recent retrotransposition events in the human factor IX gene. Hum Mutat. 2001;17:511–519. doi: 10.1002/humu.1134. [DOI] [PubMed] [Google Scholar]
- Lisitsky I, Rott R, Schuster G. Insertion of polydeoxyadenosine-rich sequences into an intergenic region increases transcription in Chlamydomonas reinhardtii chloroplasts. Planta. 2001;212:851–857. doi: 10.1007/s004250000449. [DOI] [PubMed] [Google Scholar]
- Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Hellmann U, Schmid CW. A transpositionally and transcriptionally competent Alu subfamily. Mol Cell Biol. 1990;10:5424–5432. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- McConnell KJ, Beveridge DL. Molecular dynamics simulations of B '-DNA: Sequence effects on A-tract-induced bending and flexibility. J Mol Biol. 2001;314:23–40. doi: 10.1006/jmbi.2001.4926. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, et al. The Friedreich ataxia GAA triplet repeat: Premutation and normal alleles. Hum Mol Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Muddashetty, R.S., Khanam, T., Kondrashov, A., Bundman, M., Iacoangeli, A., Kremershothen, J., Cuning, K., Barnekow, A., Huttenhoffer, A., Tiedge, H., et al. 2002. Poly(A) binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. (in press). [DOI] [PubMed]
- Mustajoki S, Ahola H, Mustajoki P, Kauppinen R. Insertion of Alu element responsible for acute intermittent porphyria. Hum Mutat. 1999;13:431–438. doi: 10.1002/(SICI)1098-1004(1999)13:6<431::AID-HUMU2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Troxel AB, Swergold GD. Genomic characterization of recent human LINE-1 insertions: Evidence supporting random insertion. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak ET, Kazazian HH., Jr Mobile elements and the human genome. Nat Rev Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- Rogers JH, Willison KR. A major rearrangement in the H-2 complex of mouse t haplotypes. Nature. 1983;304:549–552. doi: 10.1038/304549a0. [DOI] [PubMed] [Google Scholar]
- Rowe SM, Coughlan SJ, McKenna NJ, Garrett E, Kieback DG, Carney DN, Headon DR. Ovarian carcinoma-associated TaqI restriction fragment length polymorphism in intron G of the progesterone receptor gene is due to an Alu sequence insertion. Cancer Res. 1995;55:2743–2745. [PubMed] [Google Scholar]
- Roy AM, Carroll ML, Nguyen SV, Salem AH, Oldridge M, Wilkie AO, Batzer MA, Deininger PL. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 2000;10:1485–1495. doi: 10.1101/gr.152300. [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL. Alu insertion polymorphisms for the study of human genomic diversity. Genetics. 2001;159:279–290. doi: 10.1093/genetics/159.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C, St Jacques B. ‘Brain-specific’ transcription and evolution of the identifier sequence. Nature. 1986;319:418–420. doi: 10.1038/319418a0. [DOI] [PubMed] [Google Scholar]
- Schmid CW, Maraia R. Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Genet Dev. 1992;2:874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Deininger PL. The role and amplification of the HS Alu subfamily founder gene. J Mol Evol. 1996;42:15–21. doi: 10.1007/BF00163206. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD. Reading between the LINEs: Human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Smith BL, Gallie DR, Le H, Hansma PK. Visualization of poly(A)-binding protein complex formation with poly(A) RNA using atomic force microscopy. J Struct Biol. 1997;119:109–117. doi: 10.1006/jsbi.1997.3864. [DOI] [PubMed] [Google Scholar]
- Sukarova E, Dimovski AJ, Tchacarova P, Petkov GH, Efremov GD. An Alu insert as the cause of a severe form of hemophilia A. Acta Haematol. 2001;106:126–129. doi: 10.1159/000046602. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wang T, Lerer I, Gueta Z, Sagi M, Kadouri L, Peretz T, Abeliovich D. A deletion/insertion mutation in the BRCA2 gene in a breast cancer family: A possible role of the Alu-polyA tail in the evolution of the deletion. Genes Chromosomes Cancer. 2001;31:91–95. doi: 10.1002/gcc.1110. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: Cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Deininger P, Efstradiatis A. The reverse flow of genetic information: Pseudogenes and transposable elements derived from nonviral cellular RNA. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- West, N.C., Roy-Engel, A.M., Imataka, H., Sonenberg, N., and Deininger, P.L. 2002. Shared components of SINE RNPs. J. Mol. Biol. (in press). [DOI] [PubMed]
- Woods-Samuels P, Wong C, Mathias SL, Scott AF, Kazazian HH, Jr, Antonarakis SE. Characterization of a nondeleterious L1 insertion in an intron of the human factor VIII gene and further evidence of open reading frames in functional L1 elements. Genomics. 1989;4:290–296. doi: 10.1016/0888-7543(89)90332-7. [DOI] [PubMed] [Google Scholar]
- Wulff K, Gazda H, Schroder W, Robicka-Milewska R, Herrmann FH. Identification of a novel large F9 gene mutation—An insertion of an Alu repeated DNA element in exon e of the factor 9 gene. Hum Mutat. 2000;15:299. doi: 10.1002/(SICI)1098-1004(200003)15:3<299::AID-HUMU23>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Nakamura A, Yazaki M, Ikeda S, Takeda S. Insertional mutation by transposable element, L1, in the DMD gene results in X-linked dilated cardiomyopathy. Hum Mol Genet. 1998;7:1129–1132. doi: 10.1093/hmg/7.7.1129. [DOI] [PubMed] [Google Scholar]