Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis (original) (raw)
Abstract
Published and new samples of Aboriginal Australians and Melanesians were analyzed for mtDNA (n = 172) and Y variation (n = 522), and the resulting profiles were compared with the branches known so far within the global mtDNA and the Y chromosome tree. (i) All Australian lineages are confirmed to fall within the mitochondrial founder branches M and N and the Y chromosomal founders C and F, which are associated with the exodus of modern humans from Africa ≈50–70,000 years ago. The analysis reveals no evidence for any archaic maternal or paternal lineages in Australians, despite some suggestively robust features in the Australian fossil record, thus weakening the argument for continuity with any earlier Homo erectus populations in Southeast Asia. (ii) The tree of complete mtDNA sequences shows that Aboriginal Australians are most closely related to the autochthonous populations of New Guinea/Melanesia, indicating that prehistoric Australia and New Guinea were occupied initially by one and the same Palaeolithic colonization event ≈50,000 years ago, in agreement with current archaeological evidence. (iii) The deep mtDNA and Y chromosomal branching patterns between Australia and most other populations around the Indian Ocean point to a considerable isolation after the initial arrival. (iv) We detect only minor secondary gene flow into Australia, and this could have taken place before the land bridge between Australia and New Guinea was submerged ≈8,000 years ago, thus calling into question that certain significant developments in later Australian prehistory (the emergence of a backed-blade lithic industry, and the linguistic dichotomy) were externally motivated.
Keywords: human evolution, population genetics
Australia was probably occupied by humans at least 50,000 years ago (1), at a time when lowered sea levels created a land bridge between Australia and neighboring New Guinea (NG) and when the region was separated from the Eurasian land mass by only narrow straits such as Wallace's Line (Fig. 1). Australia's archaeological record remains mysterious. To begin with, Australia harbors among the oldest modern human fossils outside Africa dating to ≈46,000 years (2, 3), despite the large geographic distance from the African homeland of mankind (4). Moreover, the earliest known Australian skeletons, at Lake Mungo, are gracile, whereas some younger skeletal finds (e.g., at Kow Swamp) have robust morphology (5). Some modern Australian aboriginals retain elements of this robustness, for example, in the form of pronounced brow ridges (supraorbital tori) (6, 7). Various explanations can be put forward for the inconsistent morphological record, for example that local Homo erectus of Southeast Asia admixed into the modern human gene pool to a lesser or greater extent (5, 8), or that there have been multiple migrations to Australia that gave rise to the differing morphologies at different times, such as hypothetical new migrants from India (9), or that Australia has been genetically isolated for a sufficiently long time to produce marked continent-/Australian-specific features (10–14).
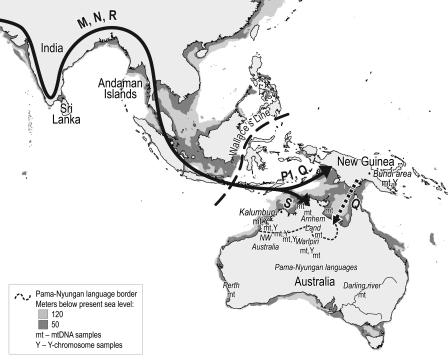
Fig. 1.
Coastlines of Australia and NG ≈50,000 years ago. After the initial spread of H. sapiens out of Africa to Sahul (the formerly connected land mass of Australia and NG), the principal processes are differentiation of the mitochondrial DNA clades Q and S. Subsequent to that process, there is little migration within Sahul other than Q from NG to Australia. The genetic isolation of Australia is in the main very clearly evident already before the Sahul land bridge disappears ≈8,000 years ago. See Results and Discussion for further details.
Archaeological data indicate the intensification of density and complexity of different stone tools in Australia during the Holocene period and the emergence of backed-blade stone-tool technology (15). The first Dingoes (Canis lupus dingo) also appear at about the same time (3,500–4,000 years ago) and were proposed to have been introduced by new human arrivals, from India (16), along with new stone tool types (17). This debate is ongoing (15, 18, 19).
Recent molecular studies on humans have likewise yielded a diversity of interpretations, ranging from a deep but undated split distinguishing Australians even from their immediate neighbors to the north in NG (20) to a very recent immigration event within the Holocene in the past 10,000 years (21, 22).
Using new Australian and NG samples screened for mtDNA and Y chromosome variation and benefiting from the increasing genetic sample coverage available for Australia [Fig. 1 (4, 20–32)], we can now attempt to clarify some of the salient features of the record of Australian population history and confirm its considerable isolation.
Results and Discussion
African Ancestry of Australian and NG Y and mtDNA Types.
We carried out a phylogenetic analysis of our Australian and NG complete mtDNA sequences and compared the resulting branches with the Asian mtDNA tree, as known so far (Fig. 2). The result confirms that both Australian and NG maternal lineages consist exclusively of the known out-of-Africa founder types M and N, dated to ≈50–70,000 years ago, and their derivatives (24–27, 29, 30, 33–37). This mitochondrial finding is mirrored in our Y chromosome data (Fig. 3), where we observe the paternal lineages in Australians and New Guineans to fall into either branches C or F, proposed to be the earliest out-of-Africa founder types (31). These results indicate that Australians and New Guineans are ultimately descended from the same African emigrant group 50–70,000 years ago, as all other Eurasians. In other words, these data provide further evidence that local H. erectus or archaic Homo sapiens populations did not contribute to the modern aboriginal Australian gene pool, nor did Australians and New Guineans derive from a hypothetical second migration out of Africa (38), nor is there any suggestion of a specific relationship with India (9, 21, 22).
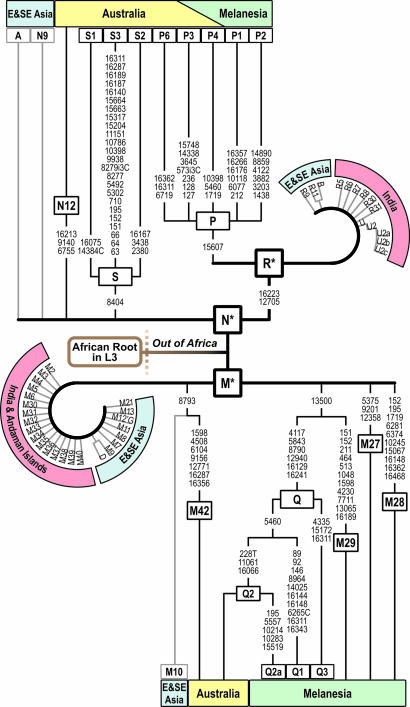
Fig. 2.
Simplified tree of autochthonous Near Oceanian mtDNA branches. East and Southeast Asian, and Indian specific clusters are added for comparison. Mutations relevant to Australia, Melanesia, and NG are shown along the branches. Only branches identified by at least two complete mtDNA sequences are included. For data and a detailed tree, see SI Fig. 4.
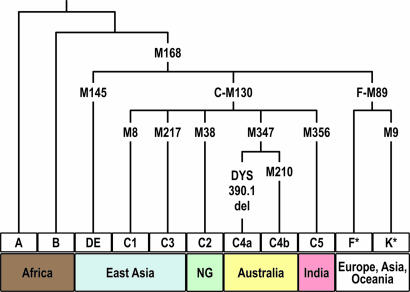
Fig. 3.
Simplified Y chromosomal phylogeny including the recently discovered Australia-specific marker M347. For data and a detailed tree, see SI Fig. 5.
Comparing the Australian complete mtDNA sequences within the context of the Asian phylogeny (25, 26, 38–45), we find that the Australians do not share any derived branches with Asians more recent than the founding types M, N, and R (Fig. 2). Similarly, our increased resolution of regionally differentiated Y chromosomal types, C5 in India, C4 in Australia, and C2 in NG provide evidence of significant long-term isolation (Fig. 3). Although the confirmed existence of F* chromosomes in India (13, 46) suggests they may also exist in Australia and NG, incomplete molecular analysis for types G–J in some previous studies (28, 47) leaves the issue of the presence of basal F* chromosomes in Australia and NG unresolved. The implication is that the migration rate of the founders from Africa along the Indian Ocean has been rapid relative to the mutation rate of the complete mtDNA genome [one mutation in ≈5,000 years; see Mishmar _et al._ (37)]. These findings support the relatively rapid migration of the Eurasian founder types to Southeast Asia (45) and, as we can now confirm, all the way to Australia. It should be noted that migration in this context refers not simply to travel but also to successful colonization. Applying the given mutation rate to the M, N, and R founders, the migration from southwestern Asia to Australia would have taken <5,200 years at 95% confidence, assuming a Poisson mutation process. This migration speed is in the same order of magnitude as estimated for other prehistoric continental settlements (48).
Australian and NG Founder Lineages.
An important result in our high-resolution mtDNA data is the discovery that Australians and New Guineans not only share the same M and N founders dating from the African exodus but furthermore within M share a characteristic variant nucleotide position 13500, which is widespread in Australia, NG, and neighboring Melanesia but not found elsewhere in the world. Taken together with the fact that the ancestral node, but not the derived lineages, is shared between Australia and NG/Melanesia (Fig. 2), we argue for a single founder group settling the whole region of Australia and NG ≈50,000 years ago. Strongly supporting evidence for this view comes from the N portion of the mtDNA phylogeny (Fig. 2), where a major deep subclade P is found in both Australia and NG/Melanesia but not elsewhere, with the time-depth estimates for P again ranging around the 50,000-year mark (Table 1).
Table 1.
Age estimates for mtDNA branches found in Australians, New Guineans, and Melanesians
| Region | Hg | N | ρ | SE | Age, yr |
|---|---|---|---|---|---|
| Aus/Mel | M | 50 | 7,9 | 1,1 | 53,400 ± 7,500 |
| Aus/Mel | Q'M29 | 27 | 6,6 | 1,4 | 44,300 ± 9,800 |
| Aus/Mel | Q | 22 | 4,7 | 1,0 | 32,000 ± 6,500 |
| Mel | Q1 | 11 | 3,2 | 0,9 | 21,500 ± 6,100 |
| Aus/Mel | Q2 | 4 | 4,5 | 1,4 | 30,400 ± 9,300 |
| Mel | Q3 | 7 | 3,1 | 0,8 | 21,300 ± 5,500 |
| Mel | M29 | 5 | 2,8 | 1,2 | 18,900 ± 8,300 |
| Mel | M27 | 7 | 5,9 | 1,4 | 39,600 ± 9,800 |
| Mel | M28 | 8 | 3,0 | 1,0 | 20,300 ± 6,500 |
| Mel | M28a | 6 | 1,7 | 0,7 | 11,300 ± 4,500 |
| Aus | M42 | 6 | 6,0 | 1,3 | 40,600 ± 9,000 |
| Aus/Mel | N | 51 | 7,9 | 1,1 | 53,200 ± 7,300 |
| Aus | N12 | 4 | 2,5 | 1,1 | 16,900 ± 7,200 |
| Aus | S | 12 | 3,8 | 0,8 | 25,400 ± 5,200 |
| Aus | S1 | 4 | 3,3 | 1,1 | 22,000 ± 7,700 |
| Aus | S2 | 4 | 2,3 | 0,8 | 15,200 ± 5,100 |
| Aus/Mel | R | 33 | 8,6 | 1,2 | 58,400 ± 8,400 |
| Aus/Mel | P | 31 | 7,6 | 0,9 | 51,700 ± 5,800 |
| Mel | P1 | 6 | 4,5 | 1,0 | 30,400 ± 6,500 |
| Mel | P2 | 7 | 1,9 | 0,6 | 12,600 ± 4,000 |
| Aus/Mel | P3 | 5 | 5,8 | 1,2 | 39,200 ± 8,200 |
| Aus/Mel | P4 | 8 | 9,8 | 1,9 | 65,900 ± 13,200 |
| Aus | P4b | 3 | 7,0 | 1,7 | 47,300 ± 11,700 |
| Mel | P4a | 5 | 3,8 | 1,1 | 25,700 ± 7,500 |
Within Australia, the ancient mtDNA branch S (27) stands out, because it is found in 34% of our Australians [supporting information (SI) Table 2], and it is well represented in other regions of Australia, as detailed in Fig. 1 (4, 21, 23–27, 29) and has so far not been detected elsewhere in the world, based on the available global mtDNA database of >40,000 sequences (49). This branch is distinguished from the root of macrohaplogroup N by a transition at nucleotide position 8404. Nearly all Australians who do not have mtDNA type S nevertheless harbor deep mtDNA branches specific to Australia, several of which are described in this study (SI Fig. 4). These deep and continent-specific branches indicate substantial isolation since the first colonization of Australia. Although NG and Australia were not separated until 8,000 years ago, we can estimate the time depth for the arrival in Australia both qualitatively and quantitatively. Qualitatively, as argued above, the relatively nested phylogenetic structure, with no mutation events separating the M, N, and R founders around the Indian Ocean even at the highly resolved level of the complete mtDNA sequence, indicates an arrival in Australia soon after the African exodus, the latter dated to 50–70,000 years ago (14, 36, 48). Quantitatively, the absolute date estimates for the founder clusters in Near Oceania yield dates of up to 58,000 ± 8,000 years ago (Table 1).
Occurrence of a “New Guinean” Lineage in Northern Australia.
There is an important exception to the general pattern of Australian-specific lineages in Australian aboriginals, and this concerns mtDNA branch Q. Thus far, Q has been considered as having a geographic distribution restricted to NG and Melanesia (25, 26, 30, 33–35, 50). Surprisingly, in our northern Australian Kalumburu sample, we now find an Aboriginal Australian mtDNA lineage bearing all of the basic mutations characteristic of haplogroup Q. This Australian Q lineage does not appear to be a recent arrival from NG (nor indeed a case of sample confusion), because the lineage does not belong to any of the common and widespread Q subclusters known so far from NG and Melanesia. The Australian Q instead branches deeply within Q to a depth of five mtDNA mutations. The mutational time of separation of this Q lineage from existing NG Q branches is estimated at 30,400 ± 9,300 years (Table 1). The geographically restricted appearance of Q in northern Australia may suggest a secondary arrival of settlers from NG well before the land bridge between Australia and NG was submerged ≈8,000 years ago.
Apart from this potential signal of secondary migration into Australia, there seem to be no further lineages either on the Australian Y or mtDNA tree that would provide clear evidence for extensive genetic contact since the first settlement, except possibly for a P3 sublineage shared between Australia and NG (Fig. 2). Thus, Australia appears to have been largely isolated since initial settlement, in agreement with one interpretation of the fossil record (10, 11). In particular, there are no lineages exclusively shared between Australia and India that might have indicated common ancestry as originally proposed by Huxley (9). Indeed, we have identified a new Y marker M347 (Fig. 3), which distinguishes all Australian C types from Indian or other Asian C types and adds weight to the rejection of the Huxley hypothesis. NG, in contrast, does carry a clear imprint of new arrivals at least along its coasts, where the “Austronesian” B mtDNA type has been established (51, 52).
This conclusion may have a negative bearing on the much-discussed emergence of a new stone tool industry in Australia, the “small tool” tradition, characterized by backed blades (15, 53). There is currently no evidence in Australia to associate this change in the material culture record with the arrival of new maternal and paternal lineages.
A major question that has not been addressed here and awaits resolution is the intriguing linguistic landscape of Australia, where seven-eighths of the continent is dominated by a single language family (Pama-Nyungan), whereas all other language families are concentrated in the northwestern region of Australia [Fig. 1 (54)]. Our samples from Kalumburu are from the linguistically diverse northern zone, where we have identified potential secondary gene flow into Australia as evidenced by a mitochondrial Q lineage distantly related to current NG Q lineages. The secondary migration ≈30,000 years ago associated with the arrival of the Q lineage would be considered too early, in the view of most linguists, to account for this dichotomy. Future more exhaustive genetic surveys of the Australian continent may one day resolve whether the Australian linguistic landscape can be better understood with the identification of such potential contact events. At present, it may seem preferable to seek an explanation for the dichotomy in terms of events and processes internal to Australia.
Conclusions
The mitochondrial and Y chromosomal results presented here point toward one early founder group settling both Australia and NG soon after the exodus from Africa ≈50–70,000 years ago, at a time when the lowered sea levels joined the two islands into one land mass, necessitating sea travel only across narrow straits such as Wallace's Line. The deep and specific phylogenetic lineages today within this former landmass indicate a small founding population size and subsequent isolation of Australia and, to a lesser extent, of NG, from the rest of the world. These founder events and the lack of contact could underlie the divergent morphological development seen in the Australian human fossil record and could also help explain the remarkably restricted range of Pleistocene Australian lithic industries and bone artifacts compared with contemporaneous cultures elsewhere in the world (55).
Materials and Methods
Samples.
In total, 172 Australian and Melanesian mtDNAs and 522 Y chromosome profiles were used in this study. Samples were obtained with informed consent. The following mtDNA sequences were generated: 32 sampled Aboriginal Australians from Kalumburu in northwestern Australia and 48 NG highlanders from the Bundi area (Fig. 1). Four of the Australian individuals had been characterized by Y chromosome short tandem repeat analysis (20). In addition, mtDNA sequences were generated from the following DNA samples described in Kivisild et al. (27): two Aboriginal Australian samples (Oc06 and Oc10), two NG samples (Oc01 and Oc16), and two Melanesian samples (Oc03 and Oc04). Extended Y chromosomal profiles were generated for the males within these samples (6/32 Aboriginal Australians and 19/48 New Guineans).
Previously published mtDNA profiles were included as follows: 33 complete or nearly complete mtDNA sequences from Australia (25–27, 29, 30); 32 complete or nearly complete mtDNA sequences from NG (25–27, 30, 33, 34); and 27 complete or nearly complete mtDNA sequences from Melanesia (25, 27, 30, 34, 35).
Published Eurasian and Near Oceanian Y chromosomal haplotypes in the present study include: 102 Aboriginal Australians (28, 31, 32); 395 New Guineans (28, 31, 32, 47); 1,021 individuals from Southeast Asian populations (28, 31, 32, 47, 56); 1,141 individuals from the Indian subcontinent and Pakistan (13, 31, 32, 46); 358 individuals from East Asian populations (13, 28, 31, 32); and 1,065 individuals from Northeast and Central Asian populations (31, 32, 56) (see SI Fig. 5 for further details).
mtDNA Typing.
The first hypervariable segment (HVS-1) of mtDNA (nps 16024–16383) and the stretch 57–302 of HVS-2 were sequenced directly from both strands in all samples. Additionally, two macrohaplogroup M and N defining mutations, namely 10398 A>G and 10400 C>T, were genotyped in all samples by RFLP (DdeI 10394 and AluI 10397 respectively). M types were further analyzed, by direct sequencing, for the Q and M29 marker 13500C (33, 50) and N types were checked for the S marker 8404C (27) and the P marker 15607G (33, 50). Additional coding region markers were analyzed in S and P mtDNA types (SI Table 3). The haplotypes defined by control region sequences and coding region SNPs were further grouped by their mutational motifs under following subhaplogroups: B4a1a1, M7b1, P1, P2, P3, P4b, Q1, and Q2 (see SI Table 3 and SI Fig. 4 for further details) (30, 33, 43, 51).
Most NG (40/48) and approximately one-half of Aboriginal Australian (14/32) mtDNA haplotypes could be sufficiently well characterized using existing mtDNA haplogroup nomenclature (SI Tables 2 and 3). Of the 26 mtDNA control region sequences that did not show clear affiliation to previously described haplogroups, nine Australian and NG individuals were selected for complete mtDNA sequencing. All recently characterized mutations that were found during the complete mitochondrial genome sequencing were typed in individuals with similar or identical mtDNA control region sequences (SI Table 3).
Multiplex SNP Assay.
A mtDNA multiplex PCR was designed and performed in a reaction volume of 25 μl containing 1× PCR buffer, 6.5 mM MgCl2, 600 mM each dNTP, 0.01–0.2 mM of each primer (SI Table 4), and 2 units of AmpliTaq Gold DNA polymerase (Applied Biosystems, Tartu, Estonia). The thermal cycling program was: denaturation at 95°C for 10 min followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 65°C for 30 s, followed by 6 min at 65°C.
Excess primers and dNTPs were removed by addition of 1 μl (1 unit/μl) of shrimp alkaline phosphatase and 0.02 μl (10 units/μl) of Exonuclease I (Amersham Pharmacia Biotech, Piscataway, NJ) to 2.5 μl of PCR product and incubating the mixture at 37°C for 30 min followed by 80°C for 15 min.
Single-base extension (SBE) reactions were performed in 5 μl with 1 μl of purified PCR product, 3 μl of SNaPshot (Applied Biosystems), or SNuPe (Amersham Biosciences, Piscataway, NJ) reaction mix, 0.5 μl of SBE primer mix (0.01–0.3 mM each primer; see SI Table 5), and 0.5 μl of water. The SBE primer mix was diluted in 160 mM ammonium sulfate (Sigma–Aldrich, Helsinki, Finland) to minimize primer-dimer artifacts. Excess nucleotides were removed by addition of 1 μl (1 unit/μl) shrimp alkaline phosphatase to the SBE mix and incubation at 37°C for 20 min followed by incubation at 80°C for 15 min. Two microliters of SBE product were mixed with 18 μl of Hi-Di formamide (Applied Biosystems) and 0.1 μl of GeneScan-120 Liz internal size standard (Applied Biosystems), and analyzed by capillary electrophoresis using ABI Prism 3730XL Genetic Analysers with 50 cm capillary arrays and POP-6 polymer (Applied Biosystems) or a MegaBACE Analysis System (Amersham Biosciences). Full methodological and theoretical details are available elsewhere (57, 58).
Y Chromosome Typing.
Eighteen Y chromosomal markers (M4, M9, M11, M38, M45, M70, M89, M130, M147, M175, M177, M208, M210, M214, M230, M231, M347, and M356) (SI Fig. 5) were typed in 25 of the Kalumburu and Bundi samples. One previously unpublished biallelic M347 marker is reported here. M347 was amplified by using primers (F, 5′-AAGTGGAGGGTATGTTTCAGCC-3′; R, 5′-GGCAACAATAGGCAGATGGCTC-3′) specific for a single 558-bp amplicon. The thermal cycling program was: denaturation at 95°C for 3 min followed by 36 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 40 s, followed by 5 min at 72°C. Nucleotide position 374 A>G (ancestral>derived) variant was sequenced directly by using one of the same external primers. The following were additionally genotyped: haplogroup C* lineages (altogether 13 individuals) from the study by Kivisild et al. (46) were typed for the presence of the M356 marker reported by Sengupta et al. (13); NG haplogroup K* lineages from the study by Underhill et al. (31, 32) were typed for the presence of M230 marker; and the M347 marker was typed in Australian C lineages from the study by Underhill et al. (31, 32).
Coalescence Age Estimation.
Phylogenetic trees were constructed manually and confirmed by using the Network software (www.fluxus-engineering.com) (59, 60). Coalescence ages of mtDNA haplogroups were calculated by the rho (ρ) statistic as described by refs. 61 and 62 by using the coding region mutation rate of one synonymous transition per 6,764 years (27).
Supplementary Material
Supporting Information
Abbreviations
NG
New Guinea
SBE
single-base extension.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EF495214–EF495222 (complete mtDNA sequences), EF524341–EF524420 (mtDNA HVS-1 sequences), and EF524421–EF524500 (partial HVS-2 sequences)].
References
- 1.Stringer C. Philos Trans R Soc London B. 2002;357:563–579. doi: 10.1098/rstb.2001.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowler J, Johnston H, Olley J, Prescott J, Roberts R, Shawcross W, Spooner N. Nature. 2003;421:837–840. doi: 10.1038/nature01383. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell JF, Allen J. J Arch Sci. 2004;31:835–853. [Google Scholar]
- 4.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 5.Bräuer G. Mellars P, Stringer C. The Human Revolution: Behavioural and Biological Perspectives on the Origins of Modern Humans. Princeton, NJ: Princeton Univ Press; 1989. pp. 123–154. [Google Scholar]
- 6.Larnach SL, Macintosh NWG. The Craniology of the Aborigines of Queensland. Sydney, Australia: University of Sydney; 1970. [Google Scholar]
- 7.Larnach SL. Australian Aboriginal Craniology. Sydney, Australia: University of Sydney; 1978. [Google Scholar]
- 8.Thorne AG, Wolpoff MH. Sci Am. 1992;266:76–79. 82–3. doi: 10.1038/scientificamerican0492-76. [DOI] [PubMed] [Google Scholar]
- 9.Huxley TH. J Ethnol Soc London. 1870;2:404–412. [Google Scholar]
- 10.Pardoe C. Arch Oceania. 1991;26:79–85. [Google Scholar]
- 11.Brown P. Philos Trans R Soc London B. 1992;337:235–242. doi: 10.1098/rstb.1992.0101. [DOI] [PubMed] [Google Scholar]
- 12.Lahr M. The Evolution of Modern Human Diversity. Cambridge, UK: Cambridge Univ Press; 1996. [Google Scholar]
- 13.Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A, et al. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards MB, Bandelt H-J, Kivisild T, Oppenheimer S. In: Human Mitochondrial DNA, the Evolution of Homo sapiens. Bandelt H-J, Macaulay V, Richards M, editors. Berlin: Springer; 2006. pp. 225–265. [Google Scholar]
- 15.Mulvaney DJ, Kamminga J. Prehistory of Australia. Washington, DC: Smithsonian Institution Press; 1999. [Google Scholar]
- 16.Gollan K. In: Recent Advances in Indo-Pacific Prehistory. Misra VN, Bellwoood P, editors. New Delhi, India: Oxford & IBH; 1985. pp. 439–443. [Google Scholar]
- 17.Glover IC, Presland G. In: Recent Advances in Indo-Pacific Prehistory. Misra VN, Bellwoood P, editors. New Delhi, India: Oxford & IBH; 1985. pp. 185–195. [Google Scholar]
- 18.Corbett L. The Dingo in Australia and Asia. Ithaca, NY: Comstock/Cornell; 1995. [Google Scholar]
- 19.Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E, Lundeberg J. Proc Natl Acad Sci USA. 2004;101:12387–12390. doi: 10.1073/pnas.0401814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forster P, Kayser M, Meyer E, Roewer L, Pfeiffer H, Benkmann H, Brinkmann B. Mol Biol Evol. 1998;15:1108–1114. doi: 10.1093/oxfordjournals.molbev.a026018. [DOI] [PubMed] [Google Scholar]
- 21.Redd AJ, Stoneking M. Am J Hum Genet. 1999;65:808–828. doi: 10.1086/302533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redd A, Roberts-Thomson J, Karafet T, Bamshad M, Jorde L, Naidu J, Walsh B, Hammer M. Curr Biol. 2002;12:673–677. doi: 10.1016/s0960-9822(02)00789-3. [DOI] [PubMed] [Google Scholar]
- 23.van Holst Pellekaan S, Frommer M, Sved J, Boettcher B. Am J Hum Genet. 1998;62:435–449. doi: 10.1086/301710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huoponen K, Schurr TG, Chen Y, Wallace DC. Hum Immunol. 2001;62:954–969. doi: 10.1016/s0198-8859(01)00294-4. [DOI] [PubMed] [Google Scholar]
- 25.Ingman M, Gyllensten U. Genome Res. 2003;13:1600–1606. doi: 10.1101/gr.686603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 27.Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, et al. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kayser M, Brauer S, Cordaux R, Casto A, Lao O, Zhivotovsky LA, Moyse-Faurie C, Rutledge RB, Schiefenhoevel W, Gil D, et al. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- 29.van Holst Pellekaan SM, Ingman M, Roberts-Thomson J, Harding RM. Am J Phys Anthropol. 2006;131:282–294. doi: 10.1002/ajpa.20426. [DOI] [PubMed] [Google Scholar]
- 30.Friedlaender JS, Friedlaender FR, Hodgson JA, Stoltz M, Koki G, Horvat G, Zhadanov S, Schurr TG, Merriwether DA. PLoS ONE. 2007;2:e248. doi: 10.1371/journal.pone.0000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley R, Oefner PJ, Cavalli-Sforza LL. Annu Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 32.Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, et al. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 33.Friedlaender J, Schurr T, Gentz F, Koki G, Friedlaender F, Horvat G, Babb P, Cerchio S, Kaestle F, Schanfield M, et al. Mol Biol Evol. 2005;22:1506–1517. doi: 10.1093/molbev/msi142. [DOI] [PubMed] [Google Scholar]
- 34.Pierson MJ, Martinez-Arias R, Holland BR, Gemmell NJ, Hurles ME, Penny D. Mol Biol Evol. 2006;23:1966–1975. doi: 10.1093/molbev/msl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merriwether DA, Hodgson JA, Friedlaender FR, Allaby R, Cerchio S, Koki G, Friedlaender JS. Proc Natl Acad Sci USA. 2005;102:13034–13039. doi: 10.1073/pnas.0506195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson E, Forster P, Richards M, Bandelt HJ. Am J Hum Genet. 1997;61:691–704. doi: 10.1086/515503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L. Science. 2005;308:996. doi: 10.1126/science.1109987. [DOI] [PubMed] [Google Scholar]
- 39.Kong Q-P, Yao Y-G, Sun C, Bandelt H-J, Zhu C-L, Zhang Y-P. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palanichamy M, Sun C, Agrawal S, Bandelt H-J, Kong Q-P, Khan F, Wang C-Y, Chaudhuri T, Palla V, Zhang Y-P. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, et al. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 42.Sun C, Kong QP, Palanichamy MG, Agrawal S, Bandelt HJ, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP. Mol Biol Evol. 2006;23:683–690. doi: 10.1093/molbev/msj078. [DOI] [PubMed] [Google Scholar]
- 43.Kivisild T, Tolk H-V, Parik J, Wang Y, Papiha SS, Bandelt H-J, Villems R. Mol Biol Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. (erratum 20:162) [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M, Cabrera VM, Gonzalez AM, Larruga JM, Takeyasu T, Fuku N, Guo L-J, Hirose R, Fujita Y, Kurata M, et al. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, et al. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 46.Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk H-V, Stepanov V, et al. Am J Hum Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurles ME, Sykes BC, Jobling MA, Forster P. Am J Hum Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forster P. Philos Trans R Soc London B. 2004;359:255–264. doi: 10.1098/rstb.2003.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Röhl A, Brinkmann B, Forster L, Forster P. Int J Legal Med. 2001;115:29–39. doi: 10.1007/s004140100217. [DOI] [PubMed] [Google Scholar]
- 50.Forster P, Torroni A, Renfrew C, Röhl A. Mol Biol Evol. 2001;18:1864–1881. doi: 10.1093/oxfordjournals.molbev.a003728. [DOI] [PubMed] [Google Scholar]
- 51.Trejaut JA, Kivisild T, Loo JH, Lee CL, He CL, Hsu CJ, Lee ZY, Lin M. PLoS Biol. 2005;3:e247. doi: 10.1371/journal.pbio.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoneking M, Jorde LB, Bhatia K, Wilson AC. Genetics. 1990;124:717–733. doi: 10.1093/genetics/124.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans N, Jones R. In: Archaeology and Linguistics: Aboriginal Australia in Global Perspective. McConvell P, Evans N, editors. Melbourne, Australia: Oxford Univ Press; pp. 385–418. [Google Scholar]
- 54.Crystal D. The Cambridge Encyclopedia of Language. Cambridge, UK: Cambridge Univ Press; 1987. [Google Scholar]
- 55.Mellars P. Science. 2006;313:796–800. doi: 10.1126/science.1128402. [DOI] [PubMed] [Google Scholar]
- 56.Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF. Am J Hum Genet. 2001;69:615–628. doi: 10.1086/323299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez JJ, Endicott P. Nat Protocols. 2006;1:1370–1378. doi: 10.1038/nprot.2006.247. [DOI] [PubMed] [Google Scholar]
- 58.Endicott P, Metspalu M, Stringer C, Macaulay V, Cooper A, Sanchez JJ. PLoS ONE. 2006;1:e81. doi: 10.1371/journal.pone.0000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bandelt H-J, Forster P, Sykes BC, Richards MB. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bandelt H-J, Forster P, Röhl A. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 61.Forster P, Harding R, Torroni A, Bandelt H-J. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 62.Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information