Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes (original) (raw)
Abstract
The evolutionary origin of telomerases, enzymes that maintain the ends of linear chromosomes in most eukaryotes, is a subject of debate. _Penelope_-like elements (PLEs) are a recently described class of eukaryotic retroelements characterized by a GIY-YIG endonuclease domain and by a reverse transcriptase domain with similarity to telomerases and group II introns. Here we report that a subset of PLEs found in bdelloid rotifers, basidiomycete fungi, stramenopiles, and plants, representing four different eukaryotic kingdoms, lack the endonuclease domain and are located at telomeres. The 5′ truncated ends of these elements are telomere-oriented and typically capped by species-specific telomeric repeats. Most of them also carry several shorter stretches of telomeric repeats at or near their 3′ ends, which could facilitate utilization of the telomeric G-rich 3′ overhangs to prime reverse transcription. Many of these telomere-associated PLEs occupy a basal phylogenetic position close to the point of divergence from the telomerase-PLE common ancestor and may descend from the missing link between early eukaryotic retroelements and present-day telomerases.
Keywords: reverse transcriptase, telomerase, transposable elements
Genomic DNA in many eukaryotes is composed, to a large extent, of transposable elements (TEs), especially retrotransposons, which multiply via an RNA intermediate copied into DNA by reverse transcriptase (RT) and inserted into new sites by an endonuclease (EN)/integrase. Although RT creates new copies, DNA cleavage is essential for TE proliferation, i.e., insertion into previously unoccupied sites. Integrases of retrovirus-like (LTR) retrotransposons insert dsDNA into chromosomes, whereas EN of non-LTR retrotransposons generate the 3′OH-end that primes cDNA synthesis directly onto the chromosome (target-primed reverse transcription). The only known eukaryotic RT-containing genes lacking EN domains are telomerase RTs (TERTs), which are not TEs but specialized ribonucleoprotein enzymes maintaining telomeres by repeated copying of a short segment of an unlinked template RNA, primed by the 3′OH end of a linear chromosome (see refs. 1–5 for review).
_Penelope_-like elements (PLEs) are a widespread but not very extensively studied class of eukaryotic TEs characterized by a single ORF coding for RT and an unusual GIY-YIG EN domain also found in bacterial group I introns, and by the presence of spliceosomal introns in several members (4, 6). They occupy a special place in retroelement phylogeny by sharing a common ancestor with TERTs (4). PLEs insert relatively randomly throughout the genome, preferring AT-rich targets (6). Indeed, the element-encoded EN, in which the conserved residues are essential for transposition, exhibits some sequence preferences but no pronounced sequence specificity (7).
Rotifers of the class Bdelloidea, a large taxon of multicellular freshwater invertebrates considered to be anciently asexual (8, 9), contain a distinct group of PLEs, called Athena (4), carrying spliceosomal introns within highly conserved RT motifs. Two Athena copies initially obtained from a genomic library of the bdelloid Philodina roseola were missing the entire EN domain and contained short stretches of reverse-complement telomeric repeats, (TCACCC)3–5, near their 3′ termini. This finding prompted us to investigate the universality of the EN domain absence and possible telomeric associations in this special group of PLEs in two bdelloid species, Adineta vaga and P. roseola, representing two families that separated tens of millions of years ago (9).
Results
To find out whether Athena elements are indeed located at telomeres, we developed a method for constructing telomere-enriched plasmid minilibraries containing inserts of chromosomal DNA, originally located either at telomeres or at sites of chromosome breakage, which does not rely on any prior knowledge of sequences at the chromosome ends (Fig. 1; Materials and Methods). Three independent minilibraries were prepared for A. vaga, which has 12 chromosomes and an ≈500-Mbp genome (10, 11). Random sequencing of minilibrary clones identified (TGTGGG)n as A. vaga telomeric repeats. We obtained 44 different telomere sequences ending with (TGTGGG)n [supporting information (SI) Table 1], indicating chromosome end polymorphism. Notably, two telomeres (designated M and N) contained 5′ truncated but otherwise intact ORFs of two Athena variants, designated Athena-AvM and -AvN (Fig. 2a). Telomeric clones were also obtained by probing the A. vaga genomic fosmid library with (TGTGGG)4. Fosmid sequencing revealed several Athena variants, forming head-to-tail interspersed tandem arrays at the chromosome termini (Fig. 3a). The variant Athena-AvO was first identified on fosmids, and its 3′ UTR was then shown to match telomeres O1-O3 (Figs. 2a and 3a; SI Table 1). Subtelomeric Athena copies are 5′ truncated by addition of reverse-complement telomeric repeats, and their coding sequences are typically followed by one to three shorter stretches of reverse-complement telomeric repeats (three to five repeat units). Similarly oriented but decayed Athena copies were found in the adjacent proximal region of a fosmid, forming arrays up to seven deep (not shown). All complete Athena elements code for both an RT and an upstream ORF1 with several nuclear localization signals and a coiled-coil motif (Fig. 2a). In no case, however, could we detect an associated EN domain: the C-terminal region is ≈100–150 amino acids shorter than in EN-containing PLEs (SI Fig. 6). Although it is formally possible that the RT domain per se may exhibit a cryptic EN activity, this possibility appears unlikely.
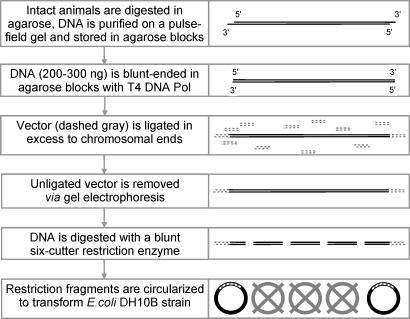
Fig. 1.
Flow chart of the chromosome end enrichment procedure (see Materials and Methods for details).
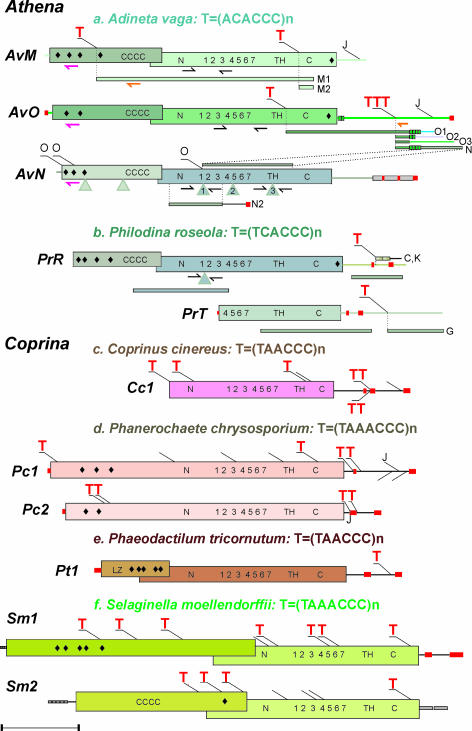
Fig. 2.
Structural organization of telomere-associated retroelements. Each red letter T indicates point of PLE 5′ truncation and addition of reverse-complement telomeric repeats at a chromosome end; 5′ truncation points within individual copies are shown by thin diagonal lines; reverse-complement telomeric repeat units are specified for each species. Noncoding sequences are shown by a thin line; PLE ORFs are shown by an open rectangle with the N- and C-terminal domains (N, C) and the central region, which includes the seven core RT motifs (RT1–RT7) and the thumb domain (TH). J, 5′ truncation point in an upstream copy when joined to a full-length downstream copy, forming a “pseudoLTR” (see also SI Fig. 7); O, point of addition of Athena-AvO to -AvN at the O1, O3, and N1 telomeres containing both elements in the same orientation. Small red boxes mark the position of short internal telomeric repeat stretches; larger boxes mark longer tandem repeats (shown in SI Fig. 7); introns are denoted by triangles. Telomeric minilibrary clones from telomeres M1–M2, O1–O3, and N1–N2 in A. vaga and C, K in P. roseola (also listed in SI Table 1) are aligned with the corresponding Athena sequences. Also shown is the position of _Athena_-specific primers used for RT-PCR (black, paired), STELA (orange), and 5′ RACE (purple) (see Fig. 3 b–d for experiments). Only Athena variants found at telomeres are shown; additional diverged variants were identified on sequenced cosmids/fosmids but have not yet been found at telomeres and are not presented here. ◆, nuclear localization signals; cccc, coiled-coil domains; LZ, leucine zipper motif. (Scale bar, 1 kb.).
Fig. 3.
Characterization of bdelloid Athena elements. (a) Structure of telomeres M1, O3, and O4 in _Athena_-containing fosmids obtained from the A. vaga genomic library. Color codes and ORFs are as in Fig. 2; telomeres are in red; truncated Athena copies are delimited with ∼ (vertical or horizontal). There are 10 and 8 48-bp repeats between AvO and AvN in the O3 and O4 telomeres, respectively. Juno1.4 is a slightly 3′ truncated copy of an LTR retrotransposon in an inverse orientation (41). (Scale bar, 1 kb.) (b) STELA. The rationale (12) is shown on the top. A telorette oligo is annealed to the G-rich overhang and, after ligation, a specific telomere is amplified with the teltail primer and the primer in the subtelomeric region. The EtBr-stained gel shows amplification of telomeres M and O with the corresponding Athena primers (Fig. 2a; Materials and Methods); below is the same gel probed with (TGAGGG)4 for visualization of telomeric repeat-containing amplicons. As a control, lanes marked (Telorette−) contained no telorette oligos in the ligation mix. Amplification of telomeres M1, O1, and O3 was confirmed by cloning and sequencing of total PCR products. (c) 5′ RACE for AvN and AvO. Arrows indicate the position of RNA start sites relative to ORF1, obtained by sequencing of the corresponding amplicons. The level of AvM transcription (d) was insufficient to generate a RACE product. (d) RT-PCR of A. vaga poly(A)+ RNA with AvM, AvO, and AvN primers (see Fig. 2a). All upper bands correspond in sequence to the unspliced message; lower bands are spliced at the predicted intron boundaries (AvN) or result from cryptic splicing (AvO).
We next sought to confirm by an alternative technique that the Athena elements are located at chromosome ends. We chose a PCR-based technique called single telomere length analysis (STELA), which was developed to measure single telomere length variation (12). Primers were designed to amplify telomeres containing Athena-AvM and -AvO (Figs. 2 a and b and 3b). Sequencing of cloned amplicons confirmed their exact correspondence to the telomere M1 for AvM primers and to telomeres O1-O3 for AvO primers. The length of the amplified telomeric repeat tracts (up to 65 repeat units) can be as short as three to four units, and occasional incorporations of a variant repeat were observed in the proximal region, indicating that telomeric tracts are subject to cycles of expansion and contraction, during which considerable telomere shortening may occur.
It was also of interest to find out whether the Athena variants that code for a full-length ORF can be transcribed, and whether the transcription start site is located at or near the 5′ end of the element to give rise to a full-length protein. RT-PCR experiments yielded bands of the expected size and sequence for the three Athena variants depicted in Fig. 2a, including spliced forms of the intron-containing AvN, for which an unspliced product was also detected (Fig. 3d). Transcription start sites were determined for AvO and AvN by 5′ RACE (Materials and Methods), and sequencing of individual amplicons confirmed that the RNA start sites in each case are positioned upstream of the first ATG codon of ORF1, with a single predominant start site for AvN and several start sites for AvO (Fig. 3c; SI Fig. 7).
The telomere cloning procedure was also applied to P. roseola, a species with 13 chromosomes, two of which are dot chromosomes (10). Its estimated genome size of ≈2,000 Mbp (11) exceeds that of A. vaga, and exhaustive cloning of chromosome ends is more challenging because of the lower ratio of chromosome ends to random breaks. From three independent P. roseola minilibraries, we obtained 20 (TGAGGG)n-containing telomeres (SI Table 1), one of which matched the Athena-PrT variant found on two of three sequenced cosmids (Fig. 2b). Two other telomeric clones had weaker matches to the transcribed and spliced (4) Athena-PrR variant, also present on two cosmids (PrR*, Fig. 1b; SI Fig. 7). In addition, we recovered five Athena clones not capped with telomeric repeats (Fig. 2b; SI Table 1), which may have originated either from sites of chromosome breakage or from exposed chromosome termini not yet capped by telomeric repeats.
The sequenced P. roseola cosmids, similar to A. vaga telomeric fosmids, exhibited a high density of Athena elements, all characteristically lacking an EN domain. Two cosmids carried four variants each, together with various DNA transposons (13), and one consisted almost entirely of six Athena variants, intact followed by decayed. As in A. vaga, many Athena copies were truncated at the 5′ end with reverse-complement telomeric repeats and carried short stretches of such repeats downstream of the RT ORF. The _Athena_-containing cosmid inserts, which in this case do not carry terminal telomeric repeats because of the library construction method, were used as probes for fluorescent in situ hybridization to P. roseola embryo nuclei (SI Fig. 8). Each cosmid yielded, on average, four strong and two weak telomeric hybridization signals, the latter at the two ends of a dot chromosome. No hybridization to internal sites was detected, although the sensitivity of the technique allows one to visualize only fragments as large as 30–40 kb (14). Labeling of several ends agrees with the telomere cloning data, whereas other more diverged Athena variants that may be present at other ends may have insufficient homology to the probe to generate observable signal. Six additional cloned A. vaga and four P. roseola telomeres (SI Table 1) were also suspected to be formed by terminal addition of as-yet-unknown diverged variants; they contain identical subterminal segments 0.3–2 kb in length.
To find out how many copies of each Athena variant are present in the A. vaga genome, we performed an exhaustive screen of the genomic library with Athena probes and compared the number of positive fosmids with the number of fosmids containing the A. vaga hsp82 gene, of which there are four copies (15). This method, in contrast to in situ and telomeric minilibrary screening, is biased against chromosome termini, which are strongly underrepresented in genomic libraries, but would detect all internal copies, even short ones. We find that, for each tested Athena variant, the number of hybridizing fosmids per genome is even less than that for hsp82 (SI Table 2). Most of these fosmids, however, also hybridize to the telomeric repeat probe, indicating they likely originate from subtelomeric locations and contain remnants of former telomeres.
To find out whether telomere-associated EN-deficient retroelements are a unique feature of bdelloid genomes or represent a more general phenomenon, we searched publicly available databases for PLEs with similar properties. Among numerous PLE ORFs assembled from diverse eukaryotes, we identified EN-deficient ORFs in genomes of representatives of three other kingdoms: fungi (inky cap mushroom Coprinus cinereus and the white rot fungus Phanerochaete chrysosporium); plants (spike moss Selaginella moellendorffii); and stramenopiles, or heterokonts (pennate diatom Phaeodactylum tricornutum) (Fig. 2 c–f). Strikingly, all of them exhibit the same connections with species-specific telomeric repeats: most of the copies contain short stretches of such repeats at or near the 3′ termini, and are 5′ truncated by a longer stretch of telomeric repeats comprising the chromosome end (Fig. 2 c–f; SI Table 3). The fungal Coprina elements are somewhat distinct in having a single long ORF and a slightly extended C terminus (Fig. 2 c and d; SI Fig. 6), whereas the protist and plant elements, like Athena, possess an upstream ORF1, which exhibits poor conservation (as opposed to RT), low amino acid sequence complexity, and no discernable sequence motifs other than nuclear localization signals and coiled-coil domains (Fig. 2 c–f). In all of these elements, the 5′ end is apparently present at a single genomic location, so that the full-length elements may essentially be regarded as single-copy genes (SI Table 4).
Remarkably, comparison of available PLE sequences shows that sequence similarities between PLEs and TERTs can be extended beyond the seven core RT1-RT7 motifs into the N- and C-terminal domains, with the N termini alignable for at least 200 amino acids and the C termini of TERTs and EN(−) PLEs ending at approximately the same position, which serves as the EN addition point in EN(+) PLEs (SI Fig. 6). The extended alignment provides an opportunity to refine PLE-TERT phylogenetic relationships, previously investigated at the level of core RT only (4, 16). An initial snapshot of the phylogenetic data structure within the combined PLE-TERT data set was obtained by NeighborNet analysis (SI Fig. 9_a_), and the suggested topology was then evaluated by other phylogenetic methods such as likelihood distance-based analysis with bootstrap networks (Fig. 4) and maximum-likelihood analysis under the best-fitting model (SI Fig. 9_b_). Of the two major PLE groups with the GIY-YIG domain found in animals, Penelope/Poseidon and Neptune (6, 17), the Penelope group forms a well supported late-branching clade, whereas the position of the Neptune group is less certain. All telomere-associated EN(−) PLEs can be roughly assigned to two major groups, Coprina and Athena, with Coprina elements appearing as the earliest-branching clades since the divergence of PLEs and TERTs from the common ancestor, possibly predating EN acquisition. In our previous analysis of the core RT domain (4), Athena elements formed a sister clade to Neptune, but this placement by Bayesian analysis may have been overconfident, because it is not observed in neighbor-joining or maximum-likelihood analyses, and statistical tests demonstrate that the branching order of Athena and Neptune elements cannot be determined with confidence (SI Table 5). These tests also reject late-branching position for Coprina elements, thereby placing their origin early in eukaryotic evolution. Two alternatives for Athena origin may be considered: initial lack of EN, or its secondary loss. The latter appears somewhat less likely, because several independent EN losses by precise truncation would have had to occur in each of the Athena variants.
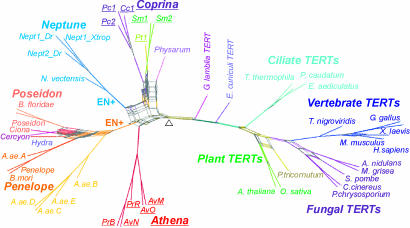
Fig. 4.
Bootstrap network of 46 PLE and TERT sequences based on maximum-likelihood (ML) distances estimated with a WAG substitution matrix plus an eight-category gamma rate heterogeneity correction. The data set included 700 characters from the core RT and its N-terminal and C-terminal extensions (SI Fig. 6). A 370-aa RT fragment of an early branching PLE was found in the slime mold, Physarum polycephalum (Amoebozoa), but no evidence is yet available for its association with telomeres because of insufficient genome coverage. This PLE contains an insertion between motifs RT3 and RT4 called IFD (insertion into the fingers domain), which is found only in TERTs and is important for TERT function, apparently stabilizing very short DNA-RNA hybrids (42). EN(−) retroelements shown in Fig. 2 (AvM, AvO, AvN, PrR, Cc1, Pc1, Pc2, Pt1, Sm1, and Sm2) are underlined; EN(+) indicates the presence of EN domain in Neptune and Poseidon/Penelope groups (full element and species names are given in SI Fig. 6). The Coprina group may or may not be monophyletic. Triangle indicates the midpoint. For clade support values, see SI Fig. 9_b_.
Discussion
Several telomere-associated non-LTR retrotransposons have been described: HeT-A, TAHRE, and TART in Drosophila (18, 19), SART and TRAS in Bombyx mori (20), and GilM and GilT in Giardia lamblia (21). Most of them have an intact EN domain, raising the possibility of EN-mediated specific insertion into a subterminal target, shown directly for SART and TRAS (20). In our case, however, the lack of an associated EN domain, characteristic patterns of telomeric repeat distribution at the 5′ and 3′ termini, orientation preference, and similarity to TERTs strongly argue in favor of terminal addition to exposed chromosome ends. The lack of EN activity leaves these elements with little choice other than using the available 3′-OH at the chromosome ends to prime reverse transcription. The shortness of the telomeric repeat stretch between PLEs and the adjacent genomic DNA (SI Fig. 7) indicates that, before PLE addition, telomere length is considerably reduced, which is likely associated with loss of the normal capping structure. Utilization of free chromosome ends would not completely rule out occasional insertion at internal sites, e.g., in the course of dsDNA break repair, as observed for mammalian L1 non-LTR retrotransposons with a disabled EN domain (22), at replication forks (23), or upon action of ENs coded elsewhere. All of these processes, however, would be insufficient for effective spread of EN-deficient PLEs, and the overwhelming majority of insertions do occur at telomeres.
Our model for EN-independent terminal retrotransposition, which accommodates most of the observed structural features, is presented in Fig. 5. Notably, terminal retrotransposition exhibits the same polarity as in telomeric repeat addition by TERTs. cDNA synthesis is accompanied by telomerase-mediated addition of telomeric repeats to the variably truncated 5′ end at sites with three to four nucleotide microhomologies to the telomeric repeat unit. At the target-priming stage, reverse-complement telomeric repeats in the 3′ UTR could facilitate annealing between the template and the telomeric G-rich 3′ overhang. Primer-template annealing is required for integration of non-LTR retrotransposons in B. mori (20) and may also facilitate L1 integration in mammals (24). The occurrence of several short telomeric repeat stretches within each 3′ UTR may have resulted from occasional acquisitions of additional downstream sequences after terminal transposition and readthrough transcription, similar to 3′ transduction in L1 elements (25). Elements that apparently do not require 3′ telomeric repeats for attachment, such as AvM, might be capable of extending severely eroded telomeres, which have already lost their telomeric repeats. The ORF1 product may be hypothesized to play a role in targeting, as shown for Drosophila HeT-A and B. mori SART elements (20, 26) and/or in primer-template annealing, as shown for mammalian L1 ORF1, which also contains a coiled-coil domain and a basic region (27).
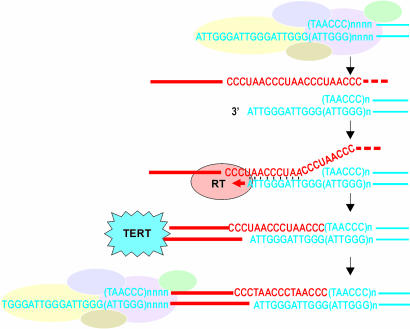
Fig. 5.
Model for EN-independent terminal retrotransposition. Red, retroelement sequences; blue, chromosomal DNA; pale ovals, proteins that normally form caps at the telomeres. Priming at the G-rich 3′ overhang is facilitated by annealing with reverse-complement telomeric repeats in the 3′ UTR of the RNA template. In the absence of telomeric repeats, annealing at microhomologies could be assisted by ORF1. Telomeric repeats are added by telomerase, after which the normal capping structure is restored. Note that the second-strand synthesis would not require special mechanisms other than routine DNA replication as occurs during C-rich strand synthesis (not to scale).
Although EN(−) retroelements may simply be transposing to telomeres to minimize damage to host genes, their low replicative capacity, resulting from inability to generate insertion sites on their own, is not very likely to ensure their survival as “selfish DNA” (28), which should replicate more efficiently than host DNA. Rather, it may be hypothesized that these low copy number elements, essentially confined to the chromosome termini, were occasionally preserved in evolution as a supplement to the telomerase-based system, providing extra protection against terminal DNA loss. In the early days of eukaryotic evolution, when primordial RNA-dependent DNA polymerases have not yet become associated with ENs to give rise to “selfish” retrotransposons that later conquered most eukaryotic genomes, movement of reverse transcripts could have been limited to the free DNA ends. Over time, an ancestral retroelement could have evolved into a telomerase catalytic subunit upon disruption of linkage between RT and its template RNA, which would then become a subunit of the telomerase holoenzyme. In the evolutionary history of eukaryotes, telomere-associated PLEs may therefore be regarded as descendants of the missing link between ancient EN(−) retroelements and the present-day telomerases, shedding light on the fundamental problem of evolution of telomerase-based maintenance of linear chromosome ends.
Materials and Methods
Construction of Telomere-Enriched Plasmid Minilibraries.
High-molecular-weight (HMW) chromosomal DNA was prepared by embedding rotifers into 0.7% low melting point (LMP) agarose blocks, digesting with Proteinase K (Invitrogen, Carlsbad, CA) at 55°C for 30 h in 1× digestion buffer (50 mM NaCl/50 mM Tris·HCl, pH 8.0/100 mM EDTA/1% Sarcosyl/2 mM spermine/2 mM spermidine), and removing broken DNA by pulsed-field gel electrophoresis with the following parameters: 0.7% LMP agarose (SeaPlaque, FMC Bioproducts, Rockland, ME), 5 V/cm, switch time 50–250 sec, switch angle 120°, run time 18 h, 0.5× TAE buffer at 12°C [BioRad (Hercules, CA) CHEF-DR III System]. HMW DNA (>1.9 Mbp) was excised from the gel compression zone and stored in agarose blocks in 50 mM Tris·HCl, pH 8.0/50 mM EDTA. For cloning, blocks were dialyzed against 50 mM Tris·HCl, pH 7.5/10 mM MgCl2 for 5 h at 4°C on a shaker, transferred to 0.75-ml tubes, and supplemented with a soaking solution of DTT, dNTPs, BSA, MgCl2, Tris·HCl, pH 7.5, and T4 DNA polymerase (NEB, Ipswich, MA) to bring their concentrations in agarose blocks to 5 mM, 0.25 mM each, 100 μg/ml, 10 mM, 50 mM, and 3 units/100 μl, respectively. After soaking for 4 h on ice, tubes were transferred to 14°C for 1 h to activate T4 DNA polymerase and then back on ice. Blocks were carefully removed and dialyzed against 25 mM Tris·HCl, pH 8.0/50 mM EDTA for 10 h to remove salt, dNTPs, and T4 DNA polymerase. Blocks were transferred to fresh 0.75-ml tubes, agarose was melted for 5 min at 65°C, supplemented with 2 μg/100 μl of pBluescript II SK− (Stratagene, La Jolla, CA) linearized with HincII, and dephosphorylated with shrimp alkaline phosphatase (Promega, St. Louis, MO). Extreme care was taken to add the vector as slowly and gently as possible to minimize HMW DNA breakage. Vector was allowed to diffuse in melted agarose for 3 h at 37°C, and agarose was supplemented with a mixture of DTT, ATP, BSA, MgCl2, Tris·HCl, pH 7.5, and T4 DNA ligase (High-concentration; Invitrogen) to their final concentrations of 10 mM, 1 mM, 50 μg/ml, and 15 Weiss units/100 μl, respectively. Again, extreme care was taken not to cause breakage of HMW DNA. The ingredients were allowed to diffuse in melted agarose for 30 min at 37°C, and the tubes were transferred to 14°C for 24 h to allow ligation. After ligation, extreme care was no longer necessary. Blocks were melted for 5 min at 65°C, agarose was mixed by pipetting, transferred on ice, permitted to solidify, and equilibrated with 0.5× TAE buffer for 3 h. Unligated vector was removed from genomic DNA by four rounds of electrophoresis (two forward and two reverse) in 0.5% LMP agarose, 0.5× TAE at 4°C. Genomic DNA in agarose was digested with β-agarase I (NEB) in 0.5× TAE supplemented with 1× NEBuffer III and 100 μg/ml BSA. DNA was digested to completion with HincII (10 units/100 μl), extracted with phenol-chloroform and chloroform, EtOH-precipitated, and dissolved in 72 μl of H2O. The solution was supplemented with 10 mM DTT, 1 mM ATP, 10 mM MgCl2, 50 mM Tris·HCl, pH 7.5, and 15 Weiss units of T4 DNA ligase in the final volume of 100 μl. After ligation for 16 h at 14°C, DNA was extracted with phenol-chloroform and chloroform, EtOH-precipitated, and dissolved in 4 μl of water to transform 20 μl of DH10B electrocompetent Escherichia coli (Invitrogen) in BioRad Gene Pulser (2 kV, 25 μF, 200 Ohm, and 2-mm-wide cuvette). Inserts were sequenced with M13 forward and reverse primers to determine the telomeric end and, if no internal tandem repeats were present, sequenced to completion by primer walking from the nontelomeric end. The procedure was initially tested on Drosophila melanogaster genomic DNA and resulted in cloning of a telomere-associated retrotransposon HeT-A (not shown).
Cloning, Sequencing, and Hybridization.
Telomere sequences were also obtained by screening the A. vaga genomic fosmid library prepared from sheared embryo DNA (15) with (TGTGGG)4 telomeric repeat probe. End sequencing of hybridizing fosmids was used to determine whether the insert contains telomeric repeats at one end. Genomic P. roseola cosmid library, prepared by partial _Sau_3AI digestion (14), was used to select _Athena_-containing clones by hybridization to a PCR-generated mixed Athena probe described in ref. 4. _Athena_-containing fosmids/cosmids were sheared by sonication, subcloned into pBluescript II SK−, and sequenced on ABI3730XL. Cosmids used as FISH probes were purified by using NucleoBond Maxi Kit (Clontech, Mountain View, CA), labeled by nick-translation to incorporate the red fluorophore Alexa 568-dUTP (Molecular Probes, Invitrogen), under conditions adjusted to yield 100- to 300-nt fragments, and FISH was performed as in ref. 14. Cultures of A. vaga and P. roseola maintained in the laboratory descend from a single egg isolated 10 and 15 years ago, respectively.
STELA.
Rotifer genomic DNA (0.5 μg) was used for STELA (12) with the following modifications: the total volume of ligation mix was 15 μl; 25 pmol of each telorette oligo (GTGACGCTATCATAACGCTCCCCACACCC, GTGACGCTATCATAACGCTCCCACACCCA) were used together; after ligation, genomic DNA was separated from unligated oligonucleotides on Sephacryl S500, extracted with phenol/chloroform and chloroform, precipitated with EtOH, and resuspended in 30 μl of H2O. One microliter of resuspended DNA was used for PCR with Expand Long Template PCR system (Roche, Indianapolis, IN) with primers teltail (GTGACGCTATCATAACGCTC) and AvM (TGGTAGGCTTTCAAGGCTG) or AvO (ACGTTTCGTCCGTTCTACC). PCR products were separated in agarose gels and either analyzed by Southern blotting or cloned and sequenced.
RNA Manipulations.
Total RNA was extracted from ≈104 rotifers with 1 ml of TRIzol reagent (Invitrogen). Poly(A) fraction was prepared with Oligotex RNA Midi kit (Qiagen, Chatsworth, CA). Poly(A)+ RNA (1 μg) was treated with DNaseI (Invitrogen), extracted with phenol/chloroform, precipitated with EtOH, and reverse-transcribed with SuperScript III (Invitrogen) in the total volume of 10 μl, with or without RT added. After heat inactivation, reactions were diluted 5-fold, and 1 μl was used for PCR with Platinum Taq High Fidelity Polymerase (Invitrogen) by using the same cycling conditions: 2 min at 94°C (20 min at 94°C, 1 min at 53°C, 30 min at 68°C) × 38; 5 min at 68°C. The following pairs of primers were used: AvM, CGAAGCAACGAAAACAATCA and GATAATTTCTTTCTTAATGCCG; AvO, ACGATATCTTCATCGCAGCA and CACAGTTCCGAAATCCAACA; AvN intron 1, TCGACAAAATGATGCCAAAG and CTGATTGTTTATTTGCTAACTC; AvN intron 3, TACGAGTCGTCCGCTTGTGT and GTGGTTGACCGGAGTTTGAC. PCR products were resolved on 1.2% LMP agarose gels, excised, digested with β-agarase I (NEB), extracted with phenol/chloroform, precipitated with EtOH and sequenced. For 5′ RACE, poly(A)+ RNA (100 ng) was used for first-strand synthesis with _Athena_-specific primers R1-AvO (CAGGAGGAGCACCAGGAAT) or R1-AvN (GATCATAATAACTTTGGTAGAGA). Upon extension, reactions were treated with RNase H and RNase T1 for 30 min at 37°C. Extension products were extracted with phenol/chloroform, EtOH-precipitated, and resuspended in H2O. cDNAs were tailed with TdT (NEB) supplemented with 0.2 mM dCTP. After heat inactivation, reactions were diluted 5-fold, and 1 μl was used for nested PCR with Platinum Taq as above, using primers RACE_AUAP (AGTGACCGTATCATTTGGCTG) and R2-AvO (GTCCTTGGCTTCAAGGTCTG) or R2-AvN (CTTTTTTCTTCTTGATTGGATGAT). PCR products were separated on agarose gels and sequenced.
Bioinformatics.
The whole-genome shotgun (WGS) sequence (AACS00000000) of Coprinus cinereus (also known as Coprinopsis cinerea) strain Okayama-7 no. 130 was produced by the Broad Institute of the Massachusetts Institute of Technology and Harvard University (http://fungal.genome.duke.edu and www.broad.mit.edu/annotation/fungi/coprinus_cinereus). The WGS assembly (AADS00000000) of a homokaryotic P. chrysosporium strain RP-78 (29) and the WGS reads of Phaeodactylum tricornutum and Selaginella moellendorffii were produced by the U.S. Department of Energy Joint Genome Institute (www.jgi.doe.gov). PLEs were identified by TBLASTN searches of WGS assemblies and subsequent BLASTN searches of trace archives. Reads containing five or more telomeric repeat units were retrieved and sorted into telomeric clusters. Mate pairs from every cluster were used as queries in BLASTN searches of WGS assemblies to verify that each cluster forms a scaffold in only one direction. A similar approach was used in Li et al. (30). The longest Coprina fragments are contained in GenBank entries AACS01000397.1 (Cc1), AADS01000564 and AY916132 (Pc1), and AADS01000820 (Pc2). Consensus sequences for Cc1, Pc1, Pc2, Pt1, Sm1, and Sm2 were deposited in Repbase Update (31).
Phylogenetic Analysis.
For phylogenetic inference, we used the region 540-1280 of the alignment shown in SI Fig. 6 and provided as SI Dataset. The best-fitting model of protein sequence evolution was selected by using ProtTest 1.3 (32) among a set of 80 candidate models constituted by all combinations of the empirical amino acid substitution matrices (JTT, mtREV, mtMam, mtArt, Dayhoff, WAG, rtREV, cpREV, Blosum62, and VT) with a gamma distribution with eight rate categories (+G8), a proportion of invariable sites (+I), and observed amino acid frequencies (+F). All statistical criteria selected rtREV+I+G8(+F) (33) as the best-fitting model, with WAG+I+G8(+F) (34) coming a close second; other models performed significantly worse. ProtTest also calculated the observed amino acid frequencies and the rate heterogeneity parameter α. Evaluation of the phylogenetic data structure using phylogenetic networks was done with NeighborNet (35), implemented in SplitsTree 4.6 (36). Likelihood distance-based phylogenetic trees were inferred by applying the BioNJ algorithm (37) in SplitsTree 4.6 on ProteinML distances computed by using the WAG model and the α and θ parameter values previously estimated by ProtTest. NeighborNet networks (35) were constructed from the same distance estimates. Bootstrap proportions were also obtained from 1,000 replicates by using the same distance correction. Bootstrap networks were then constructed from all splits that occurred in any of the 1,000 bootstrap replicates. Phylogenetic network construction allows one to visualize conflicting signals and areas of uncertainty in the data set. The topology obtained by NeighborNet was also obtained in neighbor-joining analyses by MEGA 3.1 (38) (JTT substitution model; pairwise deletion; gamma distributed rates; and 100 bootstrap replications). For maximum-likelihood analysis under the best-fitting model, we used Treefinder (39) under rtREV+G8+F, substituting the amino acid frequencies of rtREV with observed frequencies calculated by ProtTest. Likelihood-based statistical tests of alternative topologies were conducted with TreePuzzle 5.2 (40) under WAG+I+G8+F model.
Supplementary Material
Supporting Information
Acknowledgments
We thank M. Meselson (Harvard University) for encouragement and support throughout the course of this work and critical reading of the manuscript, A. Pokrovski (Tufts University, Medford, MA) for help with SI Table 2, and J. Hur (Harvard University) and J. Mark Welch, Marine Biological Laboratory for providing genomic libraries. We thank the National Institutes of Health for financial support and the National Science Foundation for continued financial support (Grant MCB-0614142).
Abbreviations
TE
transposable element
RT
reverse transcriptase
EN
endonuclease
TERT
telomerase RT
PLE
_Penelope_-like element
HMW
high molecular weight
WGS
whole-genome shotgun
STELA
single telomere length analysis.
Note Added in Proof.
While this manuscript was under review, the paper by Morrish et al. (43) reported that upon transfection of a human LINE-1 retrotransposon with a disabled endonuclease domain into V3 CHO cells, which exhibit a deprotected telomere phenotype, as many as 30% of L1 retrotranspositions may be directed to such dysfunctional telomeres. Although in CHO cells, these transpositions are typically followed by chromosome rearrangements, utilization of chromosome ends as substrates for EN-independent retrotransposition underscores the ancestral link between EN-free RTs and the ends of eukaryotic linear chromosomes.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF484951–EF485020).
See Commentary on page 9107.
References
- 1.Blackburn EH. Nat Struct Biol. 2000;7:847–850. doi: 10.1038/79594. [DOI] [PubMed] [Google Scholar]
- 2.Eickbush TH. Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura TM, Cech TR. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 4.Arkhipova IR, Pyatkov KI, Meselson M, Evgen'ev MB. Nat Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 5.Kazazian HH., Jr Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 6.Evgen'ev MB, Arkhipova IR. Cytogenet Genome Res. 2005;110:510–521. doi: 10.1159/000084984. [DOI] [PubMed] [Google Scholar]
- 7.Pyatkov KI, Arkhipova IR, Malkova NV, Finnegan DJ, Evgen'ev MB. Proc Natl Acad Sci USA. 2004;101:14719–14724. doi: 10.1073/pnas.0406281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Normark BB, Judson O, Moran N. Biol J Linn Soc. 2003;79:69–84. [Google Scholar]
- 9.Mark Welch D, Meselson M. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 10.Mark Welch JL, Meselson M. Hydrobiologia. 1998;387/388:403–407. [Google Scholar]
- 11.Mark Welch D, Meselson M. Biol J Linn Soc. 2003;79:85–91. [Google Scholar]
- 12.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 13.Arkhipova IR, Meselson M. Proc Natl Acad Sci USA. 2005;102:11781–11786. doi: 10.1073/pnas.0505333102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mark Welch J, Mark Welch D, Meselson M. Proc Natl Acad Sci USA. 2004;101:1618–1621. doi: 10.1073/pnas.0307677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hur JH. PhD dissertation. Cambridge, MA: Harvard University; 2006. [Google Scholar]
- 16.Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 17.Arkhipova IR. Syst Biol. 2006;55:875–885. doi: 10.1080/10635150601077683. [DOI] [PubMed] [Google Scholar]
- 18.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 19.George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genome Res. 2007;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara H, Osanai M, Matsumoto T, Kojima KK. Chromosome Res. 2005;13:455–467. doi: 10.1007/s10577-005-0990-9. [DOI] [PubMed] [Google Scholar]
- 21.Arkhipova IR, Morrison HG. Proc Natl Acad Sci USA. 2001;95:11495–11502. [Google Scholar]
- 22.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 23.Zhong J, Lambowitz AM. EMBO J. 2003;22:4555–4565. doi: 10.1093/emboj/cdg433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulpa DA, Moran JV. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 25.Moran JV, DeBerardinis RJ, Kazazian HH. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 26.Rashkova S, Athanasiadis A, Pardue ML. J Virol. 2003;77:6376–6384. doi: 10.1128/JVI.77.11.6376-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. J Mol Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Doolittle WF, Sapienza C. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 29.Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein K, Ramaiya P, Detter J, Larimer F, et al. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Rehmeyer CJ, Staben C, Farman ML. Bioinformatics. 2005;15:1695–1698. doi: 10.1093/bioinformatics/bti181. [DOI] [PubMed] [Google Scholar]
- 31.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 32.Abascal F, Zardoya R, Posada D. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 33.Dimmic MW, Rest JS, Mindell DP, Goldstein RA. J Mol Evol. 2002;55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 34.Whelan S, Goldman N. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 35.Bryant D, Moulton V. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 36.Huson DH, Bryant D. Mol Biol Evol. 2006;23:254–257. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 37.Gascuel O. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. Brief Bioinformat. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 39.Jobb G, von Haeseler A, Strimmer K. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 41.Gladyshev EG, Meselson M, Arkhipova IR. Gene. 2006;390:136–145. doi: 10.1016/j.gene.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lue NF, Lin YC, Mian IS. Mol Cell Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information