Amplification of a 280-Kilobase Core Region at the ERBB2 Locus Leads to Activation of Two Hypothetical Proteins in Breast Cancer (original) (raw)
Abstract
Amplification of the ERBB2 oncogene at 17q12 is clinically the most relevant genetic aberration in breast cancer and several studies have linked ERBB2 activation to poor clinical outcome. The development of targeted antibody-based therapy for **ERBB2**-overexpressing tumors and the possible role of ERBB2 as a predictor of chemotherapy treatment response have further emphasized the essential role of ERBB2 in breast cancer. Here, we performed a detailed characterization of the molecular events occurring at the ERBB2 amplicon in primary breast tumors. Analysis of the amplicon structure in 330 breast tumors by fluorescence in situ hybridization to a tissue microarray revealed a 280-kb common region of amplification that contains 10 transcribed sequences, including eight known genes. The expression levels of these 10 transcripts were determined in 36 frozen samples of grade-matched **ERBB2**-amplified and -nonamplified (as determined by fluorescence in situ hybridization) primary breast tumors by using quantitativereal-time reverse transcriptase-polymerase chain reaction. A highly significant association between amplification and expression levels was observed for six of these genes, including ERBB2 and two uncharacterized hypothetical proteins, MGC9753 and MGC14832. These results support the recent findings on the influence of copy number on gene expression levels and highlight novel genes that might contribute to the clinical behavior of **ERBB2**-amplified breast tumors.
Several recurrent regions of amplification containing a defined target gene have been identified in breast cancer, clinically most important being the amplification and overexpression of the ERRB2 oncogene at 17q12, which occurs in 10 to 34% of breast tumors. 1 Many studies have demonstrated a relationship between ERBB2 gene/protein abnormalities and poor clinical outcome of the patients, 1 thus implicating an essential role for ERBB2 in breast cancer. Recently ERRB2 oncogene has received additional attention as a target for novel and specific antibody-based tumor therapy. 1-4 This targeted therapy has been successfully used for treatment of patients with advanced breast cancer, however, for currently unknown reasons, all patients with ERBB2 overexpression do not respond favorably to this treatment. 1-4 The ERBB2 oncogene has also been recently implicated as a possible predictor of response to adjuvant chemotherapy. The possibility to modify treatment strategies for breast cancer patients based on the ERBB2 status of the tumor has been especially appealing. 1,5-7
These recent discoveries have directed further interest toward the ERBB2 oncogene, especially to the identification of additional genetic factors that might influence the treatment responses of _ERBB2_-positive tumors. 6,7 Several genes, located adjacent to ERBB2, have been shown to be frequently amplified and overexpressed together with ERBB2 indicating that the 17q12 amplification leads to simultaneous activation of multiple genes. 8-15 These co-amplified and overexpressed genes might be likely candidates for factors that have an impact on the treatment responses observed in the _ERBB2_-amplified tumors.
Previously, we performed a detailed characterization of the structure of the ERBB2 amplicon in breast cancer cell lines to obtain a comprehensive view on the molecular events occurring at this locus. 16 We were able to define a minimal common region of amplification at 17q12, restricted to a less than 0.5-Mb region around the ERBB2 locus. We next explored the molecular consequences of amplification and showed that expression levels of most of the genes located in this minimal common region of amplification were elevated. 16 Although studies, such as those performed using comparative genomic hybridization, have illustrated that genetic aberrations observed in cell-line model systems are highly representative of those occurring in primary tumors, 17,18 it is essential to confirm results obtained from such model systems in primary tumors. Such confirmation is especially relevant in the case of amplicons because previous studies have suggested that the size of amplicons decreases as a function of time. The initial amplicons are thought to be large but during subsequent cell cycles, progressively smaller regions are selected for leading to a reduction in the amplicon size. 19,20 In addition, it is also critically important to explore whether the consequences of amplification on gene expression levels in primary tumors are similar to those observed in cell lines or whether the transcriptional regulation in primary tumors is more stringent and less dependent on gene copy number.
In the present study, we performed a detailed evaluation of the ERBB2 amplicon structure in a large series of primary breast tumors using the tissue microarray technology and explored the molecular consequences of amplification on gene expression levels.
Materials and Methods
Tissue Microarray
The tissue microarray used in this study has been described previously and included 612 formalin-fixed, paraffin-embedded primary breast cancers from the years 1985 to 1995 obtained from patients with clinicopathological information. 21 The use of these specimens was approved by the Ethics Committee of the University of Basel.
Fluorescence in Situ Hybridization (FISH)
Bacterial artificial chromosome (BAC) clones were identified by performing sequence similarity searches against the nr and htgs databases using the blastn program. 16 The identity of the clones was confirmed by polymerase chain reaction (PCR). BAC DNAs were labeled with SpectrumOrange-dUTP (Vysis, Inc., Downers Grove, IL) by random priming and a SpectrumGreen-labeled chromosome 17 centromere probe (Vysis) was used as a reference. FISH to normal metaphase chromosomes was done to verify that the probes recognized a single copy target at 17q12-q21. FISH to tissue microarray slides was done as described. 22 Briefly, the tissue microarray sections were treated according to the Paraffin Pretreatment Reagent kit protocol (Vysis), denaturated at 94°C for 5 minutes in Tth-buffer [10 mmol/L Tris-HCl, pH 8.9, 0.1 mol/L KCl, 1.5 mmol/L MgCl, 50 μg/ml bovine serum albumin, 0.05% Tween 20 (v/v)], treated with Proteinase K (10 μg/ml in phosphate-buffered saline) at 37°C for 10 minutes, dehydrated, and air-dried. After an overnight hybridization, the slides were washed in 0.4× standard saline citrate/0.3% Nonidet P-40 at 72°C for 3 minutes, and then counterstained with 4′,6-diamidino-2-phenylindole in anti-fade solution. Hybridization signals were evaluated using a Zeiss fluorescence microscope (Carl Zeiss, Inc., Thornwood, NY). The number of cells counted varied from one tumor sample to another, with a minimum of 50 cells analyzed for each case. Strict criteria were used to define amplification to ensure the accuracy of scoring. Specimens containing a threefold or higher increase in the number of test probe signals, as compared with the chromosome 17 centromere signals, in at least 10% of the tumor cells were considered to be amplified.
Frozen Tumor Samples
Freshly frozen specimens from 36 primary breast cancers were obtained from Tampere University Hospital (Tampere, Finland). The use of these samples was approved by the Ethics Committee of the Pirkanmaa Hospital District. Tumors were selected using information on the ERBB2 protein expression status (as determined by routine immunohistochemistry) and the 17q12 amplification status was verified by FISH. The amplified (n = 15) and nonamplified (n = 21) tumor groups were matched according to standard clinicopathological features (Table 1) ▶ .
Table 1.
Clinicopathological Characteristics of 36 Primary Breast Tumors
| Variable | Amplified* | Nonamplified* | P value† |
|---|---|---|---|
| All tumors | 15 | 21 | |
| ER | |||
| Positive | 10 | 16 | |
| Negative | 5 | 5 | 0.228 |
| PR | |||
| Positive | 7 | 13 | |
| Negative | 8 | 8 | 0.626 |
| Histological grade | |||
| II | 1 | 7 | |
| III | 14 | 14 | 0.186 |
| Cell proliferation | |||
| Ki67 < 45% | 7 | 12 | |
| Ki67 ≥ 45% | 8 | 9 | 0.705 |
| Lymph node status | |||
| Positive | 8 | 7 | |
| Negative | 7 | 14 | 0.590 |
| Tumor size | |||
| <2 cm | 6 | 12 | |
| ≥2 cm | 9 | 7 | 0.844 |
Quantitative Reverse Transcriptase (RT)-PCR
Hematoxylin and eosin-stained tissue sections were prepared from individual frozen tumor samples and were used to select a representative area from each tumor where a 2-mm core biopsy was obtained for RNA isolation. Total RNA was isolated from core biopsies using Qiagen RNeasy MiniKit (Qiagen Inc., Valencia, CA). Samples were treated with RNase Free DNase I (Epicenter, Madison, WI) at 37°C for 30 minutes, followed by inactivation of the enzyme at 65°C for 15 minutes. First-strand cDNA synthesis was performed using Superscript II reverse transcriptase and random hexamer primers (Invitrogen, Carlsbad, CA). Molecular beacon probe sets for eight known genes (ERBB2, GRB7, MLN64, PNMT, NEUROD2, ZNFN1A3, TCAP, PPP1R1B), two hypothetical proteins (MGC14832, MGC9753), and the housekeeping gene TPB as well as specific double-stranded DNA standards for each gene were obtained from Gorilla Genomics, Inc. (Alameda, CA). Each probe set contains PCR primers for specific amplification of the target gene as well as a fluorogenic molecular beacon probe for measurement of the accumulation of the specific PCR product. The molecular beacon probe hybridizes to the target sequence and emits fluorescent light, the amount of which reflects the quantity of the target gene. Quantitative real-time PCR analyses were performed using the LightCycler equipment (Roche, Mannheim, Germany). 23 The reactions contained 2 μl of 10× PCR Buffer, 0.4 μl 50× Probe Mix (Gorilla Genomics, Inc.), 0.4 μl Titanium _Taq_DNA polymerase (Clontech Laboratories, Inc., Alameda, CA), and 1 μl of cDNA sample or standard, adjusted to 20 μl with sterile H2O. The PCR program consisted of initial denaturation at 95°C followed by 45 cycles of denaturation at 95°C for 10 seconds, annealing at 55°C for 10 seconds, and elongation at 72°C for 6 seconds. Quantitative analysis was performed using the LightCycler software according to the manufacturer’s instructions. Briefly, the fit-point method was used to determine the crossing point values, representing the cycle number where fluorescence level for each sample reaches a specific cut-point. A dilution series of the DNA standard was used as a template in each PCR run to prepare a standard curve by plotting the crossing point of each standard against the logarithmic value of its concentration. The concentrations of the unknown samples were then determined by setting their crossing points to the standard curve. The expression levels of studied genes were normalized by the housekeeping gene TBP.
Statistical Analyses
Comparisons of the clinicopathological parameters and median gene expression levels between amplified and nonamplified tumor groups were done using the paired Student’s _t_-test and the nonparametric Mann-Whitney test, respectively. All P values are two-tailed.
Results
Defining the Minimal Common Region of Amplification in Primary Breast Tumors
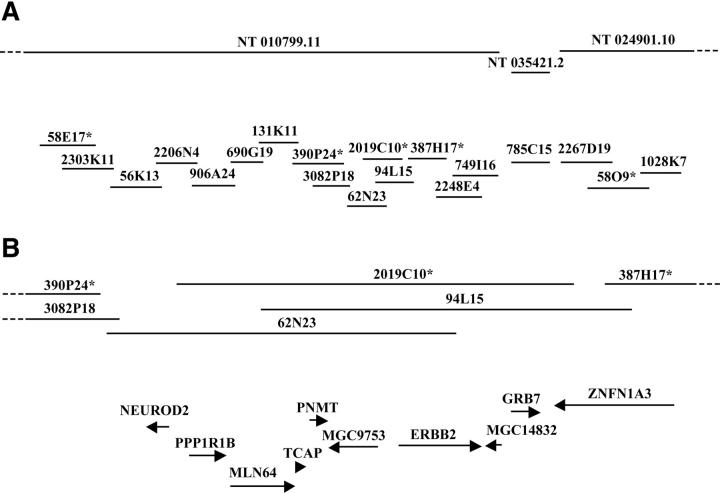
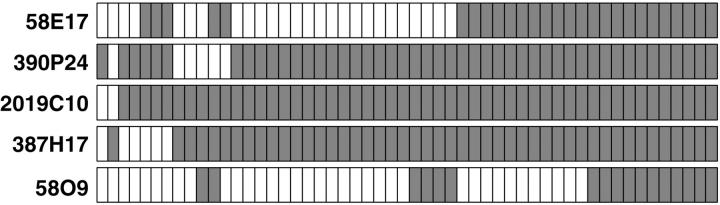
Based on our previous results in breast cancer cell lines 16 and the information available in the human genome databases (www.ncbi.nlm.nih.gov/genome/guide/human/ and www.genome.ucsc.edu), five BAC clones located within a 2-Mb region at 17q12-q21 were selected for copy number analysis in which 612 primary breast tumors were assessed using FISH to tissue microarray (Figure 1A) ▶ . Of 612 tumors, 330 (54%) were evaluable with all five BAC probes. Uninformative cases consisted of missing or unrepresentative tumor samples in the array section or tumors in which analysis failed with one or more probes. The frequencies of copy number increases ranged from 5.2 to 15.5%, with the lowest frequency observed with clone 58O9 and highest with 2019C10. Fifty-three tumors (16.1%) showed amplification with at least one of the probes (Figure 2) ▶ . The majority of amplified tumors (46 of 53, 86.8%) showed co-amplification of at least three neighboring probes (Figure 2) ▶ , indicating that the amplicon in this region is usually large and contiguous. BAC 2019C10 was most frequently amplified and, consistent with our results from breast cancer cell lines, 16 defined the minimal common region of the amplification.
Figure 1.
Physical map of the 17q12-q21 region. A: Genomic contigs (obtained from www.ncbi.nlm.nih.gov/genome/guide/human/) and BAC clones are symbolized with horizontal lines. The clones marked with an asterisk were used for copy number analyses. B: Schematic representation of the 280-kb minimal region of amplification located between clones 390P24 and 387H17. Transcripts mapping to the minimal region are represented with arrows and their orientation is indicated. The maps were not drawn in scale.
Figure 2.
Amplicon mapping in primary breast tumors by FISH to tissue microarray. Amplification patterns of five BAC clones is shown for 53 primary breast tumors that had amplification with at least one of the clones tested. Columns correspond to individual tumor samples and rows to each clone. Open boxes represent no copy number increase and shaded boxes indicate amplification.
Expression Levels of Minimal Region Genes in Amplified and Nonamplified Tumors
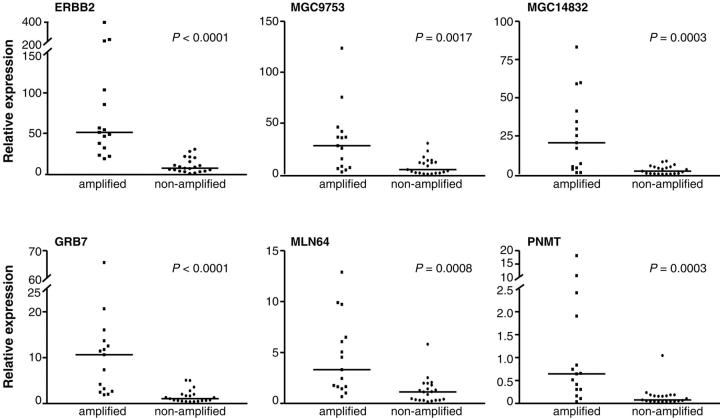
According to human genome databases (www.ncbi.nlm.nih.gov/genome/guide/human/ and www.genome.ucsc.edu), the size of the minimal region of amplification is ∼280 kb and it contains nine transcripts (Figure 1B) ▶ , seven of which represent known genes (NEUROD2, MLN64, TCAP, PNMT, ERBB2, GRB7, PPP1R1B) and two that code for hypothetical proteins (MGC9753, MGC14832). In addition, this region includes part of the ZNFN1A3 gene (Figure 1B) ▶ . Next, we used quantitative real-time RT-PCR to measure the expression levels of these transcripts in 36 primary breast tumors, 15 of which were amplified by FISH. The quantitative RT-PCR showed that the expression levels of TCAP and NEUROD2 were very low or absent in all tumor samples (data not shown). The median value of expression was calculated for the rest of the genes in the amplified and nonamplified tumor groups. Six genes (ERBB2, GRB7, PNMT, MLN64, MGC9753, and MGC14832) showed a highly significant correlation between the amplification status and the median expression level (Figure 3) ▶ . The most significant difference was observed for ERBB2 and GRB7 (P < 0.0001) that showed >7 and >14 times higher median expression levels in the amplified than in the nonamplified tumor group. A fraction of amplified tumors showed expression levels similar to those seen in the nonamplified tumors. However, for ERBB2 and GRB7, the expression levels in all tumors with amplification were above the median expression level of the nonamplified tumors whereas for MLN64, MGC9753, and MGC14832, 13 of the 15 amplified tumors had expression levels above the median of the nonamplified group. The expression levels for PNMT, although statistically associated with the amplification status, were clearly elevated in only a small portion of the amplified tumors (Figure 3) ▶ . The ZNFN1A3 and PPP1R1B genes did not show a statistically significant difference in the expression pattern between amplified and nonamplified tumors (data not shown).
Figure 3.
Expression analyses of ERBB2, MGC9753, MGC14832, GRB7, MLN64, and PNMT by quantitative RT-PCR. Expression levels were normalized by the housekeeping gene TBP and are shown for the amplified and nonamplified tumor groups. Median value of expression is indicated by a horizontal line.
Discussion
Numerous studies have illustrated the amplification and overexpression of the ERBB2 oncogene in human breast cancer and its association to poor clinical outcome, thus emphasizing an essential role for ERBB2 in breast cancer pathogenesis. 1 Because of the development of targeted therapy against ERBB2 and the possible role of ERBB2 as a predictor of chemotherapy treatment response, identification of factors that might modulate the clinical behavior of _ERBB2-_amplified tumors has become increasingly important. 6,7 Co-amplification and increased expression of several other genes adjacent to ERBB2 have been reported. 8-15 These co-amplified and overexpressed genes might be likely candidates for factors that have an impact on the treatment responses observed in the _ERBB2-_amplified tumors. Recently, we showed that in breast cancer cell lines the minimal common region of 17q12 amplification is restricted to a single BAC clone. 16 Here we explored the extent of the 17q12 amplicon in a large set of primary breast tumors and identified a core region of amplification spanning only ∼280 kb. This result confirms our previous data from cell lines 16 and demonstrates that the structure of the ERBB2 amplicon is surprisingly constant in breast cancer. Similar small amplicons have also been reported for example at 20q13.2 in breast cancer and at 14q13 in esophageal carcinomas. 24,25 Although the size of an amplicon might be dependent on its chromosomal location and/or the tumor type, the recent identification of small amplicons is also likely to reflect improved methodologies and the availability of highly accurate and detailed mapping information.
The minimal common region of amplification at 17q12 contains a total of 10 transcripts, including the ERBB2 oncogene. Quantitative real-time RT-PCR analysis revealed that only six of these transcripts showed a strong statistically significant correlation between amplification and expression levels indicating that increased gene copy number does not inevitably lead to elevated expression. In addition to ERBB2, expression levels of five other genes, GRB7, MLN64, PNMT, MGC9753, and_MGC14832_, were consistently elevated in tumors with amplification. This overall statistical correlation was not absolute because for each gene a fraction of amplified tumors showed expression levels similar to those seen in the nonamplified tumors. However, it is well-known that overexpression of cancer-related genes, such as ERBB2, often occurs through other mechanisms than amplification and therefore the nonamplified group is also likely to contain tumors with genuine elevated expression levels, making direct comparisons between individual samples difficult.
Our current observations on the correlation between amplification and expression levels are in good agreement with recent microarray-based studies that have implicated gene copy number alterations as significant determinants of global gene expression patterns. 26,27 The data from the microarray studies indicate that up to 62% of amplified genes show elevated expression levels. The present results verify that this global notion is also valid in the case of a single amplified region. In addition, our data also confirm that even within a specific amplicon all genes do not show increased expression levels, emphasizing the issue that other factors than copy number are crucial for the regulation of transcription levels. The fact that amplification leads to elevated expression of several but not all genes within an amplicon argues against the traditional concept in which there is a single target gene for each amplicon. However, it cannot be directly assumed that all of these genes with elevated expression levels would have important and independent roles in tumorigenesis. It is indeed possible that increased expression of a particular gene represents a simple by-product of co-amplification and does not confer any advantage for the cancer cell. However, it is also possible that the coordinated effect of dysregulated expression of several genes is essential for the selective growth advantage of the tumor cells.
The GRB7 and MLN64 genes have been previously shown to be co-amplified and overexpressed with ERBB2 10-12,15,16 and, based on their function, increased expression of these genes might be important for cancer pathogenesis. GRB7 codes for a SH2 domain-containing growth factor receptor tyrosine kinase that has been shown to function in cell migration, thus suggesting a possible role for GRB7 in metastasis. 28 MLN64 shares significant homology with the steroidogenic acute regulatory protein and it has been proposed to facilitate steroid hormone production in cancer cells. 29 Although statistically associated with gene amplification, the expression levels of PNMT were rather low and not consistently elevated in the amplified tumor group, therefore decreasing its relevance as a putative cancer gene. On the contrary, the hypothetical proteins MGC9753 and MGC14832 showed highly significant association between gene amplification and expression levels. Unfortunately, their sequence shows no similarity to any currently known genes or proteins, therefore leaving their function and possible role in cancer unresolved.
In conclusion, we have defined a 280-kb minimal region of amplification at the ERBB2 locus in breast cancer. The amplification was significantly associated with increased expression of 6 of the 10 genes located within this region. In addition to the ERBB2 oncogene, GRB7 and MLN64 have been previously implicated to have a likely role in cancer. The potential contribution of two hypothetical proteins, MGC9753 and MGC14832, to the development and progression of breast cancer or to the clinical behavior of _ERBB2-_amplified tumors needs to be evaluated. Our results also strongly support the recent findings that gene amplification is a major determinant of gene expression levels.
Acknowledgments
We thank Ms. Kati Rouhento for skillful technical assistance.
Footnotes
Address reprint requests to Anne Kallioniemi, Laboratory of Cancer Genetics, Institute of Medical Technology, University of Tampere, P.O. Box 67, Tampere, FIN-33014 Finland. E-mail: anne.kallioniemi@uta.fi.
Supported in part by grants from the Academy of Finland, the Foundation for Finnish Cancer Institute, the Medical Research Fund of the Tampere University Hospital, the Pirkanmaa Cancer Society, the Pirkanmaa Cultural Foundation, the Finnish Breast Cancer Group, and the Research and Science Foundation of Farmos.
References
- 1.Ross JS, Fletcher JA: The HER-2/neu oncogene: prognostic factor, predictive factor and target for therapy. Semin Cancer Biol 1999, 9:125-138 [DOI] [PubMed] [Google Scholar]
- 2.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ: Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 1999, 17:2639-2648 [DOI] [PubMed] [Google Scholar]
- 3.Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz CC, Dantis L, Sklarin NT, Seidman AD, Hudis CA, Moore J, Rosen PP, Twaddell T, Henderson IC, Norton L: Phase II study of weekly intravenous trastuzumab (Herceptin) in patients with HER2/neu-overexpressing metastatic breast cancer. Semin Oncol 1999, 26:78-83 [PubMed] [Google Scholar]
- 4.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, Shak S, Stewart SJ, Press M: Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002, 20:719-726 [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Park C: HER-2 and choice of adjuvant chemotherapy in breast cancer. Semin Oncol 2001, 28:332-335 [DOI] [PubMed] [Google Scholar]
- 6.Clark GM: Should selection of adjuvant chemotherapy for patients with breast cancer be based on erbB-2 status? J Natl Cancer Inst 1998, 90:1320-1321 [DOI] [PubMed] [Google Scholar]
- 7.Orr MS, O’Connor PM, Kohn KW: Effects of c-erbB2 overexpression on the drug sensitivities of normal human mammary epithelial cells. J Natl Cancer Inst 2000, 92:987-994 [DOI] [PubMed] [Google Scholar]
- 8.van de Vijver M, van de Bersselaar R, Devilee P, Cornelisse C, Peterse J, Nusse R: Amplification of the neu (c-erbB-2) oncogene in human mammary tumors is relatively frequent and is often accompanied by amplification of the linked c-erbA oncogene. Mol Cell Biol 1987, 7:2019-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith WN, Douglas F, Wishart GC, McCallum HM, George WD, Kaye SB, Brown R: Co-amplification of erbB2, topoisomerase II alpha and retinoic acid receptor alpha genes in breast cancer and allelic loss at topoisomerase I on chromosome 20. Eur J Cancer 1993, 29A:1469-1475 [DOI] [PubMed] [Google Scholar]
- 10.Stein D, Wu J, Fuqua SA, Roonprapunt C, Yajnik V, D’Eustachio P, Moskow JJ, Buchberg AM, Osborne CK, Margolis B: The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J 1994, 13:1331-1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC: Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11–q21.3 region of chromosome 17. Genomics 1995, 28:367-376 [DOI] [PubMed] [Google Scholar]
- 12.Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R: Two distinct amplified regions at 17q11–q21 involved in human primary breast cancer. Cancer Res 1996, 56:3886-3890 [PubMed] [Google Scholar]
- 13.Zhu Y, Qi C, Jain S, Le Beau MM, Espinosa R, Atkins GB, Lazar MA, Yeldandi AV, Rao MS, Reddy JK: Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc Natl Acad Sci USA 1999, 96:10848-10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergameschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO: Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 1999, 23:41-46 [DOI] [PubMed] [Google Scholar]
- 15.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams CF, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D: Molecular portraits of human breast tumours. Nature (Lond) 2000, 406:747-752 [DOI] [PubMed] [Google Scholar]
- 16.Kauraniemi P, Bärlund M, Monni O, Kallioniemi A: New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res 2001, 61:8235-8240 [PubMed] [Google Scholar]
- 17.Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y, Gooden GC, Ethier SP, Kallioniemi A, Kallioniemi OP: Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res 2000, 60:4519-4525 [PubMed] [Google Scholar]
- 18.Larramendy ML, Lushnikova T, Bjorkqvist AM, Wistuba II, Virmani AK, Shivapurkar N, Gazdar AF, Knuutila S: Comparative genomic hybridization reveals complex genetic changes in primary breast cancer tumors and their cell lines. Cancer Genet Cytogenet 2000, 119:132-138 [DOI] [PubMed] [Google Scholar]
- 19.Stark GR, Debatisse M, Giulotto E, Wahl GM: Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 1989, 57:901-908 [DOI] [PubMed] [Google Scholar]
- 20.Windle BE, Wahl GM: Molecular dissection of mammalian gene amplification: new mechanistic insights revealed by analyses of very early events. Mutat Res 1992, 276:199-224 [DOI] [PubMed] [Google Scholar]
- 21.Bärlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi OP, Kallioniemi A: Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res 2000, 60:5340-5344 [PubMed] [Google Scholar]
- 22.Andersen CL, Hostetter G, Grigoryan A, Sauter G, Kallioniemi A: Improved procedure for fluorescence in situ hybridization on tissue microarrays. Cytometry 2001, 45:83-86 [DOI] [PubMed] [Google Scholar]
- 23.Wittwer CT, Ririe KM, Andrew RV, David DA, Gundry RA, Balis UJ: The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques 1997, 22:176-181 [DOI] [PubMed] [Google Scholar]
- 24.Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, Jay KE, Froula J, Cloutier T, Kuo WL, Yaswen P, Dairkee S, Giovanola J, Hutchinson GB, Isola J, Kallioniemi OP, Palazzolo M, Martin C, Ericsson C, Pinkel D, Albertson D, Wu-Bo L, Gray JW: Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci USA 1998, 95:8703-8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, Glover TW, Beer DG: The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res 2002, 62:5273-5279 [PubMed] [Google Scholar]
- 26.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO: Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA 2002, 99:12963-12968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A: Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 2002, 62:6240-6245 [PubMed] [Google Scholar]
- 28.Shen TL, Han DC, Guan JL: Association of Grb7 with phosphoinositides and its role in the regulation of cell migration. J Biol Chem 2002, 277:29069-29077 [DOI] [PubMed] [Google Scholar]
- 29.Akiyama N, Sasaki H, Ishizuka T, Kishi T, Sakamoto H, Onda M, Hirai H, Yazaki Y, Sugimura T, Terada M: Isolation of a candidate gene, CAB1, for cholesterol transport to mitochondria from the c-ERBB-2 amplicon by a modified cDNA selection method. Cancer Res 1997, 57:3548-3553 [PubMed] [Google Scholar]