Peroxisome Proliferator-Activated Receptor α Regulates a MicroRNA-Mediated Signaling Cascade Responsible for Hepatocellular Proliferation (original) (raw)
Abstract
Activation of peroxisome proliferator-activated receptor α (PPARα) leads to hepatocellular proliferation and liver carcinomas. The early events mediating these effects are unknown. A novel mechanism by which PPARα regulates gene expression and hepatocellular proliferation was uncovered. MicroRNA (miRNA) expression profiling demonstrated that activated PPARα was a major regulator of hepatic miRNA expression. Of particular interest, let-7C, an miRNA important in cell growth, was inhibited following 4-h treatment and 2-week and 11-month sustained treatment with the potent PPARα agonist Wy-14,643 in wild-type mice. let-7C was shown to target c-myc via direct interaction with the 3′ untranslated region of c-myc. The PPARα-mediated induction of c-myc via let-7C subsequently increased expression of the oncogenic mir-17-92 cluster; these events did not occur in _Ppar_α-null mice. Overexpression of let-7C decreased c-myc and mir-17 and suppressed the growth of Hepa-1 cells. Furthermore, using the human PPARα-expressing mouse model, which is responsive to Wy-14,643 effects on β-oxidation and serum triglycerides but resistant to hepatocellular proliferation and tumorigenesis, we demonstrated a critical role for let-7C in liver oncogenesis. Wy-14,643 treatment did not inhibit let-7C or induce c-myc and mir-17 expression. These observations reveal a let-7C signaling cascade critical for PPARα agonist-induced liver proliferation and tumorigenesis.
The peroxisome proliferator-activated receptors (PPARs) belong to the steroid hormone nuclear receptor (NR) superfamily, consisting of 49 members in the mouse genome. Three isoforms of PPAR have been identified, PPARα, PPARβ/δ, and PPARγ, all encoded by separate genes. PPARs are liganded transcription factors that upon activation heterodimerize with retinoid X receptor and bind to direct repeat 1 peroxisome proliferator response elements. Subsequent recruitment of coactivators initiates PPAR-dependent gene transcription. PPARα is expressed mainly in the liver, heart, and muscle and is a major regulator of fatty acid transport and catabolism and energy homeostasis (15). Several synthetic lipid-lowering agents, including the widely prescribed hypolipidemic drugs fenofibrate (TriCor), gemfibrozil (Gemcor, Lopid), and clofibrate (Atromid-S), activate PPARα and lower plasma triglyceride levels (27, 43). In contrast to the beneficial effects of PPARα, PPARα agonists cause liver tumors in rodents via PPARα-dependent mechanisms (21, 44, 47). Targeted deletion of the mouse PPARα established that the receptor was required for the lipid-lowering activity and hepatocarcinoma in response to PPARα ligands. _Ppar_α-null mice treated with PPARα-selective agonists failed to exhibit classical responses, including peroxisome proliferation, the induction of lipid-metabolizing enzymes, and a decrease of serum triglycerides (29). In addition, no hepatocellular proliferation or liver tumors were observed in any _Ppar_α-null mice fed the potent PPARα agonist Wy-14,643 for 11 months, while all Wy-14,643-treated wild-type (WT) mice developed liver tumors (43). The mechanism of PPARα ligand-induced tumorigenesis is not known. PPARα agonists were shown to increase oxidative stress through overproduction of reactive oxygen species and increase hepatic cell proliferation, both of which are critical for the production of hepatocellular carcinomas (45, 59). PPARα ligands act via a nongenotoxic mechanism, and like other nongenotoxic carcinogens, they are particularly troublesome because long-term rodent bioassays are required for their detection (6). The PPARα agonists gemfibrozil and clofibrate have been used to lower serum triglyceride levels in human patients for over 15 and 30 years, respectively, yet epidemiological studies of patients who received these drugs did not demonstrate a significant increase in cancer incidence (2). However, these studies should be viewed with caution, as the onset of cancer following exposure to carcinogens in humans is expected to take over 20 years. In addition, fibrates are relatively weak PPARα ligands; therefore, it remains a major concern of regulatory agencies whether PPARα agonists or newer, high-affinity developmental drugs will be harmful to humans upon chronic exposure. Thus, it is critical to identify the PPARα-dependent gene regulatory pathways that promote cell proliferation and tumorigenesis.
Increasing evidence has implicated several microRNAs (miRNAs) in tumorigenesis (8, 9, 18, 24, 26, 36, 50). miRNAs represent a large class of noncoding RNAs that are transcribed in the nucleus as single primary transcripts or large polycistronic transcripts encoding several miRNAs. Intergenic miRNAs are thought to be transcribed by an independent promoter, whereas miRNAs found in introns of protein-encoding genes may use the same promoter as the proximal coding gene (7, 29). Following transcription of the primary miRNA, the large transcript is processed into a 65-nucleotide hairpin structure, referred to as the precursor miRNA. The precursor miRNA is exported into the nucleus, where it is processed into a 22-nucleotide RNA structure by the endonuclease known as dicer (22, 30). The mature miRNA interacts with the RNA-induced protein complex and bind to 3′ untranslated regions (UTR) of target genes via complementary base pairing (17, 33, 35). Following binding to target genes, miRNAs negatively regulate gene expression by modulating the stability or translation of mRNA. miRNAs have diverse functions, including the regulation of metabolism, embryogenesis, and oncogenesis (1, 4). Expression of miRNAs is tightly controlled in a tissue- and development-specific manner; however, there is relatively little information on transcription factors that regulate their expression. To date, only c-Myc, cyclic AMP response element-binding protein, and myogenic transcription factors have been shown to regulate specific miRNAs in mammalian cells (41, 46, 54, 63).
The present study uncovered a novel paradigm for PPARα gene regulation and clearly demonstrates PPARα as a bona fide regulator of hepatic miRNA expression. In addition, this study identifies a Wy-14,643-induced miRNA-signaling cascade important in liver proliferation.
MATERIALS AND METHODS
Animals.
Seven- to eight-week-old WT and _Ppar_α-null mice were housed five animals per cage in a temperature- and light-controlled environment. For 2-week and 11-month treatments, pelleted mouse chow containing either 0% (control) or 0.1% Wy-14,643 (Bioserv, Frenchtown, NJ) was prepared and provided to mice ad libitum. For the 4-h treatments Wy-14,643 was dissolved in carboxymethyl cellulose (Sigma, St. Louis, MO) and water, and the mice were treated by gavage at 50 mg/kg. All animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the National Cancer Institute Animal Care and Use Committee.
Plasmid construction.
The c-myc 3′ UTR containing the let-7C binding site (pGL3-c-Myc UTR) was amplified from genomic DNA using primers containing XbaI sites: 5′-TTTCCCTCTAGATCTGTGGAGAAGAGGCAAACCC-3′ (forward) and 5′-TTTCCCTCTAGATGAACCAGGGAATGGCTTCG-3′ (reverse). The amplicon was cloned into the XbaI site of the pGL3 promoter vector (Promega, Madison, WI), and the construct was confirmed by sequencing.
Luciferase assay.
Hepa-1 cells were plated in 24-well plates (5 × 104 cells/well) and cultured in Dulbecco's modified essential medium (DMEM) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). The cells were cotransfected with the pGL3-c-Myc UTR and either scrambled RNA oligonucleotide (QIAGEN, Valencia, CA) or 5 nM, 10 nM, or 25 nM let-7C RNA oligonucleotide (Ambion, Austin TX) using the HiPerfect transfection reagent (QIAGEN). At 72 h posttransfection, a standard dual luciferase assay was used and normalized to a cotransfected control reporter (Promega).
RNA analysis.
RNA was extracted from liver or Hepa-1 cells using the TRIzol reagent (Invitrogen, Carlsbad, CA) and was separated on a 15% denaturing polyacrylamide gel. miRNAs were detected by 32P end labeling of antisense probes to miRNA sequences. Quantitative real-time reverse transcriptase PCR (qPCR) was performed with cDNA generated from 1 μg total RNA with a SuperScript III reverse transcriptase kit (Invitrogen) using either random hexamers or gene-specific primers. Primers were designed for qPCR using Primer Express software (Applied Biosystems, Foster City, CA), and sequences are available upon request. qPCRs were carried out using SYBR green PCR master mix (Applied Biosystems) in an ABI Prism 7900HT sequence detection system (Applied Biosystems). Values were quantified using the comparative threshold cycle method, and samples were normalized to β-actin.
Western blot analysis.
Livers samples or Hepa-1 cells were lysed in 0.1% Triton X-100 buffer. The liver or cell lysate was separated and transferred using a standard protocol. The membranes were incubated with an antibody against c-Myc (Santa Cruz Biotechnology Inc, Santa Cruz, CA), and the signals obtained were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Chemicon International, Temecula, CA).
Chromatin immunoprecipitation (ChIP).
Freshly isolated livers were ground to a fine powder under liquid nitrogen and cross-linked in 1% formaldehyde in 1× phosphate-buffered saline at 37°C for 20 min, or formaldehyde was added to a final concentration of 1% directly to the media in the case of Hepa-1 cells. Cross-linking was terminated with 0.125 M glycine, and the cell pellet was washed twice with 1× phosphate-buffered saline. Nuclei were isolated and lysed in a sodium dodecyl sulfate (SDS) lysis solution (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS, and protease inhibitors). Chromatin was sheared by sonication, and the nucleus lysate was cleared by centrifugation at 50,000 × g for 30 min. The soluble chromatin was diluted 10-fold (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], and 167 mM NaCl) and immunoprecipitated with primary antibody to c-Myc (Santa Cruz). The antibody/protein/DNA complex was isolated using magnetic beads conjugated with protein A (New England Biolabs, Ipswich, MA). Following several washes, the protein/DNA complex was eluted (50 mM NaHCO3, 1% SDS) from the magnetic beads, and cross-linking was reversed by incubation at 65°C for 6 h. The samples were incubated with proteinase K for 1 h at 45°C. Following protein digestion, the DNA was purified using phenol-chloroform/isoamyl alcohol extraction, and 2 to 5 μl of sample was used for PCR. Oligonucleotide primers were as follows: for c-myc binding site 1, 5′-TTGGAATGGGGCTCGGAAAGTG-3′ (forward) and 5′-TATCCTTGGGCAATGCTTCGGG (reverse); for c-myc binding site 2, 5′-GGGAAGCCTACTGTAAAAGCCAAC-3′ (forward) and 5′-GAGGAGCCAATAGCCAGAAGTTC-3′ (reverse).
MTT assay.
Hepa-1 cells were seeded in 24-well plates (20,000 cells/well). Cells were transfected in medium containing 0.1% FBS in DMEM with scrambled oligonucleotide or 5 nM, 10 nM, or 25 nM let-7 oligonucleotide using the HiPerfect transfection reagent. At 72 h posttransfection the cells were incubated in the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reagent (5 mg/ml). The medium with the MTT reagent was removed after 15 to 30 min, and the cells were solubilized in dimethyl sulfoxide (Fisher Biotech, Fairlawn, NJ). The plates were read at a wavelength of 570 nm.
BrdU labeling assay.
Hepa-1 cells were seeded in six-well plates (1 × 105 cells/well). Cells were transfected in medium containing 0.1% FBS in DMEM with scrambled oligonucleotide or 5 nM, 10 nM, or 25 nM let-7 oligonucleotide or cotransfected with pBABE-c-myc or empty vector and scrambled oligonucleotide or let-7 oligonucleotide (10 nM) using the HiPerfect transfection reagent. At 72 h posttransfection, the cells were labeled with 10 μM of bromodeoxyuridine (BrdU) (Sigma). BrdU-labeled cells were trypsinized, fixed, treated with DNase I, and stained with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody using a BrdU flow kit (BD Pharmingen San Diego, CA). Stained cells were then quantified by flow cytometry.
Apoptosis assay.
Hepa-1 cells were seeded in six-well plates (1 × 105 cells/well). Cells were transfected in medium containing 0.1% FBS in DMEM with scrambled oligonucleotide or 25 nM let-7 oligonucleotide using the HiPerfect transfection reagent. At 72 h posttransfection, adherent cells were harvested by mild trypsinization and pooled with detached cells. The cells were stained with FITC-conjugated anti-annexin V antibody using an annexin V-FITC apoptosis detection kit (BD Pharmingen). Stained cells were then quantified by flow cytometry.
PI staining.
Hepa-1 cells were seeded in six-well plates (1 × 105 cells/well). Cells were transfected in medium containing 0.1% FBS in DMEM with scrambled oligonucleotide or 5 nM, 10 nM or 25 nM let-7 oligonucleotide using the HiPerfect transfection reagent. At 72 h posttransfection, adherent cells harvested by mild trypsinization were pooled with detached cells. Cells were incubated with propidium iodide (PI) for 5 min on ice. PI-stained cells were subsequently quantified by flow cytometry.
miRNA microarray.
Total RNA isolation was performed as explained above. RNA quality control, labeling, hybridization, and scanning were performed by LC Sciences (Houston, TX) using the latest probe content in the Sanger miRBase. The microarray experiment design was a treatment-control comparison with five mice per group. Three microarray experiments were performed; each independent experiment used a total of 10 mice (five control diet-fed mice and five Wy-14,643-treated mice). Preliminary statistical analysis was performed by LC sciences on raw data normalized by the locally weighted regression method on the background-subtracted data. Further statistical comparisons were performed using analysis of variance. miRNAs that were modulated >1.5-fold with a P value of <0.01 were considered significant and are shown in Table 1.
TABLE 1.
miRNA microarray
| miRNA | Chromosome(s)a | Fold difference (Wy/Vehb) | Overlapping transcript(s) |
|---|---|---|---|
| mmu-let-7c | 15 and 16 | 0.03 | Intergenic |
| mmu-mir-376b | 12 | 0.03 | Intergenic |
| mmu-mir-182 | 6 | 18.25 | Intergenic |
| mmu-mir-34a | 4 | 8.11 | Intergenic |
| mmu-mir-200c | 6 | 6.23 | Intergenic |
| mmu-mir-203 | 12 | 0.17 | Intergenic |
| mmu-mir-689 | 1 and 16 | 5.94 | Intergenic and zinc finger CCCHc type containing 7A; intron 19 |
| mmu-mir-30e | 4 | 0.18 | Nuclear transcription factor Y gamma; intron 3 |
| mmu-mir-378 | 18 | 4.20 | PPARγ, coactivator 1 beta; intron 1 |
| mmu-mir-100 | 9 | 0.26 | Intergenic |
| mmu-mir-705 | 6 | 3.78 | RAB11 family interacting protein 5; intron 5 |
| mmu-mir-148a | 6 | 0.27 | Intergenic |
| mmu-mir-146 | 11 | 0.31 | Intergenic |
| mmu-mir-143 | 18 | 0.31 | Intergenic |
| mmu-mir-130a | 2 | 0.32 | Intergenic |
| mmu-mir-99a | 16 | 0.32 | Intergenic |
| mmu-mir-125b | 9 and 16 | 0.33 | RIKEN cDNA 2610203C20 gene; intron 2 and intergenic |
| mmu-mir-106a | X | 2.66 | RP23-354J21.1 |
| mmu-mir-17-5p | 14 | 2.36 | Intergenic |
| mmu-mir-20b | X | 2.33 | Intergenic |
| mmu-mir-106b | 5 | 2.25 | Minichromosome maintenance deficient 7; intron 13 |
| mmu-mir-107 | 19 | 2.25 | Pantothenate kinase 1; intron 5 |
| mmu-mir-125a | 17 | 2.20 | Intergenic |
| mmu-mir-101a | 4 | 0.47 | RP23-32L21.4; exon 2 |
| mmu-mir-93 | 5 | 2.11 | Minichromosome maintenance deficient 7; intron 13 |
| mmu-mir-103 | 11 | 2.00 | Pantothenate kinase 3; intron 5 |
| mmu-mir-20a | 14 | 2.00 | Intergenic |
Gene expression profiling.
An Agilent 22 K mouse 60-mer oligonucleotide microarray (Agilent Technologies, Santa Clara, CA) containing more than 20,800 unique well-characterized gene/expressed sequence tag features were used. All procedures for hybridization and for slide and image processing were carried out according to the manufacturer's instructions. The slides were washed, dried, and scanned using an Agilent G2565AA microarray scanner (Agilent Technologies). The procedures were repeated for a replicate experiment with independent hybridization and processing. The data were processed and analyzed as previously described (60).
Data analysis.
Results are expressed as means ± standard deviations (SD). P values were calculated by independent t test. A P value of <0.05 was considered significant.
RESULTS
Wy-14,643 regulates hepatic miRNA expression via a PPARα-dependent pathway.
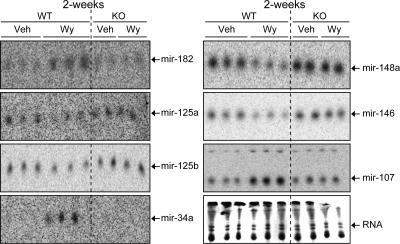
WT mice were fed either a control diet or a diet containing 0.1% Wy-14,643, a specific PPARα agonist, for 2 weeks. Following treatment, a genome-wide miRNA expression profile was assessed in the liver. The microarray contained the latest probe content from the Sanger miRBase. miRNAs that gave average signals above those of negative control probes were considered detectable. Twenty-seven miRNAs were significantly regulated following Wy-14,643 treatment (Table 1). To confirm the microarray data, Northern blot analysis was performed on several miRNAs that were regulated following Wy-14,643 treatment (Table 1). The results from the Northern blot analysis correlated well with the microarray data; in addition, the Northern blot analysis demonstrated PPARα-dependent regulation of hepatic miRNA expression (Fig. 1). It has been proposed that a single miRNA may regulate 100 to 200 gene transcripts (32); therefore, the present study may have discovered a major pathway by which PPARα controls gene expression. Taken together, these data demonstrate for the first time that a ligand-activated NR regulates the expression of miRNAs.
FIG. 1.
Wy-14,643 regulates miRNA expression in the liver via a PPARα-dependent mechanism. Northern blot analysis of mir-182, mir-125a, mir-125b, mir-34a, mir-148a, mir-146, and mir-107 following 2-week Wy-14,643 treatment (Wy) in WT and _Ppar_α-null mice (KO) compared to vehicle only (Veh).
Wy-14,643 inhibits let-7C expression.
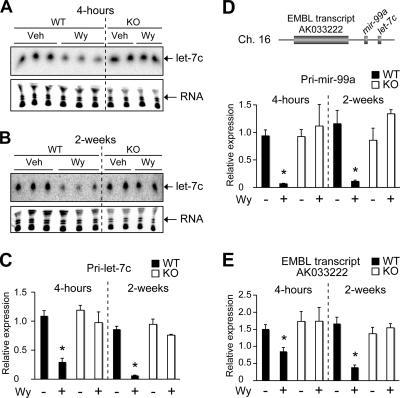
let-7C was decreased in the liver following Wy-14,643 treatment (Table 1). The let-7 family of miRNAs were shown to be potential tumor suppressors (31, 62), and let-7 was recently shown to inhibit expression of the oncogene ras (26). To directly investigate the role of PPARα in regulation of let-7C, WT and _Ppar_α-null mice were treated with Wy-14,643 for 4 h and 2 weeks. Northern blot analysis demonstrated a suppression of let-7C miRNA following 4-h Wy-14,643 treatment and 2-week sustained treatment, whereas no decrease was observed in the _Ppar_α-null mice (Fig. 2A and B). In addition, when the longer primary let-7C transcript (pri-let-7C) was assessed by qPCR, its expression was also decreased following 4-h and 2-week Wy-14,643 treatments (Fig. 2C). The chromosomal positional relationship of let-7C was assessed and found to be downstream of mir-99a and EMBL transcript AK033222 (Fig. 2D, top). AK033222 (Fig. 2E) and pri-mir-99a (Fig. 2D, bottom) were coordinately regulated with let-7C, suggesting a mechanism in which a large primary transcript is first produced and then further processed to form mature miRNAs. In addition, pri-let-7C, AK033222, and pri-mir-99a were regulated in a PPARα-dependent manner, as revealed by the finding that Wy-14,643 had no effect on pri-let-7C, AK033222, or pri-mir-99a in either 4-h- or 2-week-treated _Ppar_α-null mice (Fig. 2C to E). Interestingly, when the neuron-specific mature mir-99a was assessed by Northern blot analysis, no expression in the liver was observed, suggesting a posttranscriptional tissue-specific processing of miRNAs (data not shown) (28).
FIG. 2.
Wy-14,643 inhibits let-7C expression in the liver via a PPARα-dependent mechanism. (A and B) Northern blot analysis of let-7C following 4-h (A) and 2-week (B) Wy-14,643 treatment (Wy) in WT and _Ppar_α-null mice (KO) compared to vehicle only (Veh). Lower panels show ethidium bromide staining of small RNAs. (C to E) qPCR analysis of pri-let-7C (C), pri-mir-99a (D), and EMBL transcript AK033222 (E) expression. Each bar represents the mean plus SD from three independent experiments. *, P < 0.05 compared to vehicle-treated animals.
let-7C regulates c-myc expression.
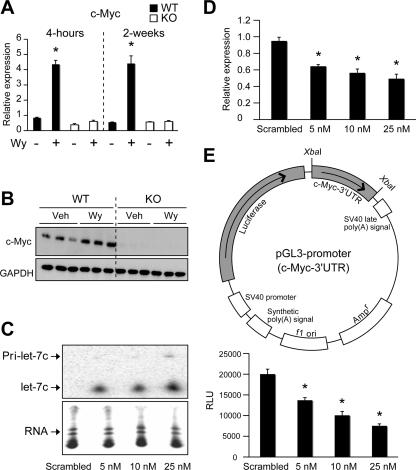
To identify novel putative targets of let-7C important in growth regulation, several computational miRNA target prediction databases were assessed (49). Interestingly, c-myc, a gene known to be induced following Wy-14,643 treatment via an undefined mechanism, was shown to be a putative target of let-7C (21). To confirm previous results, 4-h and 2-week Wy-14,643-treated WT and _Ppar_α-null livers were assessed for c-myc gene induction. A clear induction of c-myc mRNA was observed as early as 4 h after ligand treatment in a PPARα-dependent manner (Fig. 3A). In addition, c-myc protein expression was induced following Wy-14,643 treatment as assessed by Western blot analysis (Fig. 3B). It is now recognized that miRNAs can cause mRNA degradation, and let-7C has been demonstrated to degrade mRNA via partial base paring to its targets (3, 53, 57). Indeed, the let-7C mRNA degradation target is present in the 3′ UTR of c-myc mRNA. Therefore, c-myc mRNA levels were assessed following overexpression of let-7C in the mouse hepatoma cell line Hepa-1. Increasing let-7C expression in Hepa-1 cells (Fig. 3C) decreased c-myc gene expression in a dose-dependent manner (Fig. 3D). To determine if this was a direct mechanism, the 3′ UTR of c-myc was cloned into the luciferase-expressing pGL3-promoter vector using a unique XbaI site just downstream of the luciferase stop codon (Fig. 2E, top). Luciferase activity was suppressed by overexpressing let-7C in Hepa-1 cells (Fig. 3E, bottom). These results suggest that c-myc mRNA is a novel direct target of let-7C-mediated gene silencing in liver-derived cell lines.
FIG. 3.
let-7C inhibits c-myc gene expression. (A) qPCR analysis of c-myc following 4-h and 2-week Wy-14,643 treatment in WT and _Ppar_α-null mice (KO). Each bar represents the mean plus SD. *, P < 0.05 compared to vehicle-treated animals. (B) Western blot analysis of c-myc following 8-h Wy-14,643 treatment in WT and _Ppar_α-null mice (KO). (C and D) Northern blot analysis of let-7C (C) (lower panels show ethidium bromide staining of small RNAs) and qPCR analysis of c-myc following overexpression of let-7C (D). Each bar represents the mean plus SD from three independent experiments. *, P < 0.05 compared to scrambled-oligonucleotide-transfected samples. (E) Schematic diagram of the pGL3-promoter c-myc 3′ UTR construct (top). Standard luciferase assay in Hepa-1 cell cotransfected with the c-myc 3′ UTR pGL3 construct and either scrambled oligonucleotide or 5 nM, 10 nM, or 25 nM let-7C oligonucleotide. The data was normalized to an internal control, and each bar represents the mean ± SD from three independent experiments. *, P < 0.05 compared to scrambled-oligonucleotide-transfected samples.
PPARα regulates the c-_myc_-activated miRNA cistron.
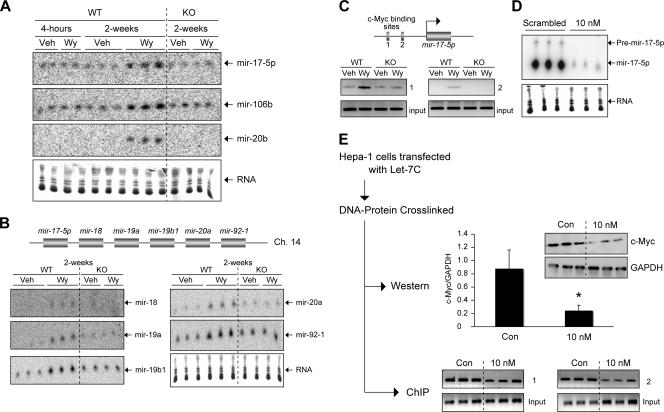
The miRNA expression profile demonstrated an increase in miRNAs known to be regulated by c-Myc (mir-106a, mir-106b, mir-17-5p, mir-20a, and mir-20b) (Table 1) (41). When several of these miRNAs were assessed by Northern blot analysis, Wy-14,643 was found to induce their expression through a PPARα-dependent mechanism only in mice treated for 2 weeks, suggesting a secondary mechanism downstream of PPARα activation (Fig. 4A). Interestingly, mir-17-5p, an miRNA belonging to the mir-17-92 polycistronic cluster (Fig. 4B, top), was shown to be important in cell proliferation (16, 24, 25). The mir-17-92 cistron has also been implicated in enhanced cell cycle progression, blockade of tumor cell apoptosis, and increased neovascularization (16, 24, 25, 41). In addition, the c-Myc-regulated miRNA cluster was shown to be overexpressed in human B-cell lymphomas (25) and lung cancers (24). Northern blot analysis found, in addition to mir-17-5p, induction of all the members of the mir-17-92 polycistron following 2-week Wy-14,643 treatment. Conserved c-Myc binding sites were identified adjacent to the mir-17 cluster. ChIP experiments were performed with livers of WT and PPARα-null mice following Wy-14,643 treatment to determine whether c-myc bound directly to the mouse mir-17 genomic locus. Wy-14,643 induced c-myc binding to these sites in WT mice, as measured by an in vivo ChIP assay, whereas no increase in binding following Wy-14,643 treatment was observed in the _Ppar_α-null mice (Fig. 4C, bottom). The data clearly demonstrated in vivo association of c-Myc to the mir-17 genomic cluster and provided strong evidence that mir-17-92 polycistrons are directly regulated by c-Myc via PPARα. To assess whether let-7C could regulate mir-17-5p expression, let-7C was overexpressed in mouse Hepa-1 cells. Northern blot analysis demonstrated that overexpression of let-7C decreased the expression of mir-17-5p (Fig. 4D). Next, to determine whether the decrease in mir-17-5p expression was due to a decrease in c-myc association with the mir-17 genomic locus, ChIP analysis for c-Myc was performed in Hepa-1 cells following overexpression of let-7C. Hepa-1 cells were transfected with let-7C precursor molecules, and the DNA-protein complex was cross-linked. A small aliquot was used for Western blot analysis, and the rest was processed for ChIP analysis using antibody to c-Myc. Consistent with the gene expression data (Fig. 3D), let-7C decreased c-myc protein expression (Fig. 4E, top), which correlated to a decrease in c-myc association with both c-myc binding sites on the mir-17 genomic locus (Fig. 4E, bottom). The above data suggest the existence of an oncogenic PPARα-driven miRNA signaling cascade.
FIG. 4.
Wy-14,643 increases expression of c-_myc_-activated miRNAs via a PPARα-dependent mechanism. (A) Northern blot analysis of mir-17-5p, mir-106b, and mir-20b following 4-h and 2-week Wy-14,643 (WY) treatment compared to vehicle only (Veh). Lower panels show ethidium bromide staining of small RNAs. (B) Schematic diagram of the mir-17-92 polycistron (top). Northern blot analysis of mir-18, mir-19a, mir-19b1, mir-20a, and mir-92-1 following 2-week Wy-14,643 (Wy) treatment compared to vehicle only (Veh). Lower panels show ethidium bromide staining of small RNAs. (C) ChIP analysis of c-myc binding sites upstream of the mir-17-5p locus. (D) Northern blot analysis of mir-17-5p following overexpression of let-7C (10 nM) in Hepa-1 cells. Lower panels show ethidium bromide staining of small RNAs. (E) Western blot analysis and ChIP assay performed on parallel samples following overexpression of let-7c (10 nM) in Hepa-1 cells. Western analysis of c-myc and GAPDH and enhanced chemiluminescence quantitation normalized to GAPDH; each bar represents the mean plus SD. *, P < 0.05 compared to scrambled-oligonucleotide-transfected samples (top). ChIP assay of c-myc binding sites upstream of the mir-17-5p locus (bottom).
let-7C regulates cell proliferation in liver-derived Hepa-1 cells.
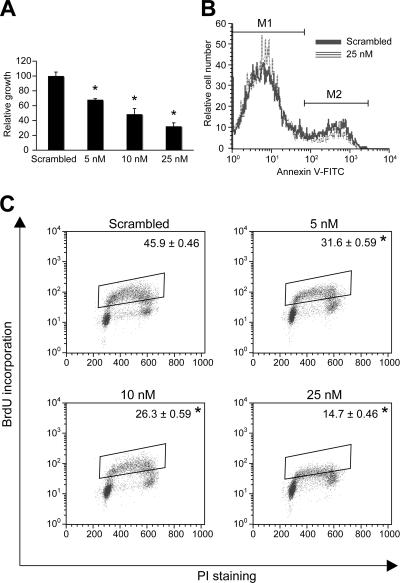
To gain insight into the functional consequence of let-7C regulation by PPARα, let-7C was overexpressed in Hepa-1 cells, and cell growth was assessed. Hepa-1 cells were transfected with let-7C (5 nM, 10 nM, or 25 nM) or scrambled RNA oligonucleotide, and at 72 h posttransfection, cell growth was monitored by MTT assay (Fig. 5A). Cell growth was inhibited in a dose-dependent manner following let-7C transfection. To assess the contribution of apoptosis and antiproliferation to let-7C-mediated growth inhibition, BrdU incorporation and annexin V staining were assessed. let-7C decreased BrdU incorporation in a dose-dependent manner (Fig. 5C). Cell cycle analysis indicated growth arrest; an increase in the number of cells in G0/G1 and a decrease in the numbers of cells in S phase and G2/M were observed following overexpression of let-7C (see Table S1 in the supplemental material). The same doses of let-7C had no effect on cell apoptosis, as assessed by annexin V staining, or on cell toxicity, as assessed by trypan blue or PI staining (Fig. 5B and C and data not shown). In addition, cotransfection of let-7C and c-myc increased cell proliferation in Hepa-1 cells compared to cells transfected with let-7C alone (see Table S2 in the supplemental material), suggesting that c-myc is a critical downstream effector of let-7C. The combined results provide strong evidence that let-7C and c-myc control hepatocellular proliferation.
FIG. 5.
let-7C overexpression inhibits cellular proliferation. (A) Cell growth analysis by MTT assay in Hepa-1 cells transfected with scrambled oligonucleotide or 5 nM, 10 nM, or 25 nM let-7C oligonucleotide; each bar represents the mean plus SD. *, P < 0.05 compared to scrambled-oligonucleotide-transfected samples. (B and C) Annexin V (B) and BrdU (C) quantitation by flow cytometry following let-7C overexpression. The data are means plus SD from three independent experiments. *, P < 0.05 compared to scrambled-oligonucleotide-transfected samples.
let-7C is not suppressed in mice resistant to Wy-14,643-induced liver tumorigenesis.
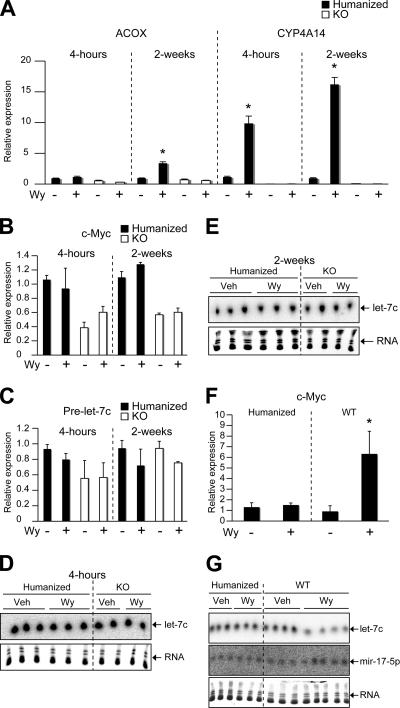
Chronic Wy-14,643 treatment yields a 100% liver tumor incidence in WT mice at 11 months, whereas no tumors are observed in _Ppar_α-null mice (21, 44, 47). In addition, deregulation of c-myc has frequently been observed in several cancers (34). Long-term overexpression of c-myc leads to hepatocellular carcinomas in mouse models (39), suggesting a pivotal role of c-myc in liver oncogenesis. To this end, a mouse line in which the mouse PPARα was replaced by the human PPARα (hPPARα mice) was used to assess the importance of let-7C in Wy-14,643-induced liver tumors. These mice express human PPARα specifically in the liver under the control of the doxycycline-regulatable system in a _Ppar_α-null background. The hPPARα and WT mice respond to treatment with Wy-14,643, as revealed by induction of genes critical in β-oxidation and a decrease in serum triglyceride levels (13). However, during the first 2 weeks of Wy-14,643 treatment, only the WT mice and not the hPPARα mice exhibited significant hepatocellular proliferation (13). Furthermore, hPPARα mice were shown to be resistant to Wy-14,643-induced liver tumor formation. Wy-14,643 treatment, which yields a 100% liver tumor incidence in WT mice at 11 months of age, induced a very low incidence of hepatocellular tumors (5%) in hPPARα mice (38). Microarray gene expression profiling was used to identify highly discriminant gene sets in the liver between 4-h Wy-14,643-treated WT and hPPARα mice (see Tables S3 and S4 in the supplemental material). The patterns of gene expression following Wy-14,643 in WT and hPPARα mice were generally similar, and differential changes shown to be statistically significant are listed in Tables S3 and S4. Interestingly, c-myc was shown to be a major differentially expressed gene in the liver in WT and hPPARα mice following 4-h Wy-14,643 treatment (see Table S3). Microarray data demonstrated that c-myc was specifically upregulated in the Wy-14,643-induced-tumor-susceptible WT mice, whereas no induction was found in the tumor-resistant hPPARα mice. To verify the microarray data, qPCR was performed. hPPARα mice demonstrated induction of mRNAs encoding acyl coenzyme A oxidase and cytochrome P450 Cyp4a14, two well-characterized PPARα target genes, and this was comparable to changes seen in WT mice as previously demonstrated (13). However, expression of the c-myc mRNA was not increased following either 4-h or 2-week Wy-14,643 treatment (Fig. 6A and B). let-7C expression was assessed in the hPPARα mice, and no change in pri-let-7C or mature let-7C was observed in the hPPARα mice following 4-h and 2-week Wy-14,643 treatment (Fig. 6C to E). Consistent with these data, 11-month chronically treated WT and hPPARα mice demonstrated that let-7C was suppressed only in the WT mice, whereas hPPARα mice demonstrated no significant change with respect to let-7C expression (Fig. 6G). In addition, c-_myc_-regulated mir-17-5p was induced only in the WT mice following Wy-14,643 treatment, correlating with c-myc expression (Fig. 6F and G). Together, these data suggest the importance of let-7C regulation in Wy-14,643-induced liver tumorigenesis.
FIG. 6.
let-7C is not suppressed following Wy-14,643 treatment in hPPARα mice. (A) qPCR analysis of acyl coenzyme A oxidase (ACOX) and cytochrome P450 4A14 (CYP4A14). (B and C) c-myc (B) and pre-Let7C (C) following 4-h and 2-week Wy-14,643 treatment in hPPARα and _Ppar_α-null (KO) mice. (D and E) Northern blot analysis of let-7C following 4-h (D) and 2-week (E) Wy-14,643 treatment in hPPARα and _Ppar_α-null mice (KO). (F and G) qPCR analysis of c-myc (F) and Northern blot analysis of let-7C and mir-17-5p (G) following 11-month treatment in hPPARα and WT mice. Each bar represents the mean plus SD from three independent samples. *, P < 0.05 compared to vehicle or untreated animals.
DISCUSSION
Twenty-seven miRNAs were significantly regulated in the liver following 2-week Wy-14,643 treatment. The present study confirmed several changes identified by microarray using Northern blot analysis. In particular, the current study focused on let-7C, a family of miRNAs that were shown to be critical in the developmental timing of Caenorhabditis elegans. In addition, let-7 was markedly reduced in lung cancers (50) and shown to be localized to chromosomal sites implicated in several other cancers (9). Overexpression of let-7 also inhibited growth of a lung cancer cell line (50). The PPARα agonist Wy-14,643 suppressed let-7C expression in WT mice but not in _Ppar_α-null mice, indicating that the repression was PPARα dependent. A previous report demonstrated that a large portion of the mammalian intergenic miRNAs are first transcribed as long pre-miRNAs and then processed (23). The present study provides further evidence for this mechanism. Pri-let-7 and upstream transcripts pri-mir-99a and AK033222 were inhibited following Wy-14,643 treatment, suggesting that let-7C may be transcribed as part of a longer primary transcript in a PPARα-dependent manner. Currently, the mechanism involved in PPARα-dependent repression of let-7C is unclear, and additional experiments are required to identify upstream regulatory elements that control let-7C expression. However, due to the rapid inhibition of let-7C expression following Wy-14,643 treatment, it is believed to be a direct mechanism. In addition, from the lack of any significant difference in basal let-7C expression levels between WT and _Ppar_α-null mice, it appears to be an active transrepression mechanism, where the receptor is recruited to the genomic regulatory region following ligand treatment. Gene repressive mechanisms of liganded PPARα have remained unclear; however, several NRs involved in transcriptional repression of proinflammatory genes have been shown to inhibit transcription through protein-protein interaction or by direct recruitment of cofactors to the promoter (37). Recently, two NRs, liver X receptor and PPARγ, were shown to repress gene expression following ligand treatment in a context-specific manner (19, 42). The mechanism involves sumoylation of the NR, thus placing the receptor in a conformation allowing corepressor interaction. Future studies will be required to determine whether these mechanisms are relevant in PPARα inhibition of let-7C.
When the antisense molecules designed to inhibit the activity of let-7C were transfected into Hepa-1 cells, no derepression of c-myc levels was observed (data not shown). This may be attributed to the low endogenous levels of let-7C in transformed cell lines compared to the liver. In addition, c-myc is highly expressed in Hepa-1 cells, and thus, it may be difficult to observe increases above these high levels. However, in the reciprocal experiments in which let-7C was overexpressed, it negatively regulated c-myc gene expression, inhibited cell growth, and caused cell cycle arrest. The c-myc gene has been demonstrated to be essential in the growth and regeneration of the liver. Overexpression of c-myc results in chronic hepatic proliferation and increased incidence of liver cancers (39). Coexpression of let-7C and c-myc partially alleviated the decrease in cell growth observed in cells transfected with let-7C alone, suggesting that other genes in addition to c-myc may play a role in let-7C cell cycle arrest. let-7 family members have been demonstrated to inhibit Ras expression (26). The ras proto-oncogene was shown to be essential in cell proliferation and cell survival (11, 14). Increased expression of ras is frequently observed during tumorigenesis (5, 48, 55, 56). Interestingly, several PPARα agonists have been shown to induce ras (12, 20). In addition, transgenic mice overexpressing ras or carrying a mutant ras demonstrate a decrease in tumor latency following chronic PPARα agonist treatment (40, 51, 52). Currently, the relative contributions of ras and c-myc in Wy-14,643-induced cellular proliferation are not known; however, it is likely that both play an integral role in hepatocyte proliferation.
The critical downstream signals in c-_myc_-induced oncogenesis are undefined; however, several genes important for c-_myc_-induced cell proliferation and transformation are targets for the mir-17 cluster (32, 61). Also, the acute effects of c-myc on tumor angiogenesis are attributed solely to the mir-17-92 cluster (16). Therefore, the Wy-14,643-induced cell proliferation in hepatocytes following reciprocal regulation of let-7C and c-myc may be partially mediated via an increase in mir-17-92 cluster. Interestingly, let-7 and mir-17-92 polycistronic cluster were inversely expressed in lung cancers; the let-7 miRNA family was downregulated (50), whereas the mir-17-92 cluster was overexpressed in (24). The present study provides a mechanistic link explaining the expression patterns of let-7 and mir-17 in lung cancers and proposes a novel miRNA signaling cascade initiated by PPARα in Wy-14,643-induced hepatocellular proliferation.
As discussed in the introduction, the consequences of chronic exposure to peroxisome proliferators in humans are unknown. Therefore, the use of potent PPARα activators in humans is avoided, particularly if long-term exposures are expected. To determine whether PPARα agonists are carcinogenic in humans, the mechanisms of cellular proliferation and tumorigenesis following PPARα agonist treatment need to be determined. The sustained inhibition of let-7C and the coordinate increase of c-myc and mir-17 expression in mice treated with Wy-14,643 for 11 months strongly implicate let-7C as a critical tumor suppressor in Wy-14,643-induced hepatocarcinogenesis. In addition, the hPPARα model, which was demonstrated to be a critical tool for predicting cancer risk in humans exposed to PPARα agonists (13, 38), did not demonstrate a decrease in let-7C at 4 h, 2 weeks, or 11 months of Wy-14,643 treatment, as seen in the tumor-susceptible WT mice. c-myc was not induced following 4-h, 2-week, or 11-month Wy-14,643 treatment, demonstrating a causal relationship between let-7C and c-myc expression and Wy-14,643-induced tumor formation. Thus, the present study strongly implicates let-7C as a critical tumor suppressor in Wy-14,643-induced hepatocarcinogenesis. In addition, let-7C and c-myc may be valuable as early surrogate markers to predict human cancer risks posed by current fibrate drugs or aid in assessing the carcinogenic properties of novel lipid-lowering fibrate drugs that target PPARα. Currently, the molecular mechanism leading to the resistance of Wy-14,643-induced tumors in hPPARα mice is unknown. However, c-myc appears to be a major discriminant gene between WT and hPPARα mice following a 4-h Wy-14,643 treatment, which the present study has clearly shown to be regulated by let-7C via a PPARα-dependent pathway.
Recently, transgenic mice containing a constitutive active PPARα in hepatocytes (LAP-VP16PPARα) were generated. While these mice demonstrated increased hepatocellular proliferation, no liver tumors were observed (58). Interestingly, no difference in basal let-7c expression was observed between WT mice and the LAP-VP16PPARα mice. However Wy-14,643 treatment decreased let-7c expression in the transgenic mice (data not shown). These data suggest that ligand treatment is needed for inhibition of let-7c and indicate that PPARα agonists may regulate genes in addition to that for the VP16PPARα fusion protein or that PPARα activation in nonparenchymal cells is critical for tumorigenesis and let-7c expression. These possibilities are being assessed.
miRNAs regulate a large numbers of genes; therefore, identifying the mechanisms which control their expression during tumorigenesis is critical. To our knowledge, the present study is the first to identify PPARα or any liganded NRs as a transcription factor important for miRNA regulation. The NR superfamily members are important for a variety of physiological processes, and it is possible that the present study may have uncovered an integral role of these transcription factors in miRNA regulation that in turn control physiological homeostasis altered by NR activation. Interestingly, genome-wide ChIP tiling arrays to assess estrogen receptor binding sites identified binding of this NR to unexplored regions of the genome distant from any known promoters (10); it will be of great interest to assess if this is a universal mechanism shared by other NRs.
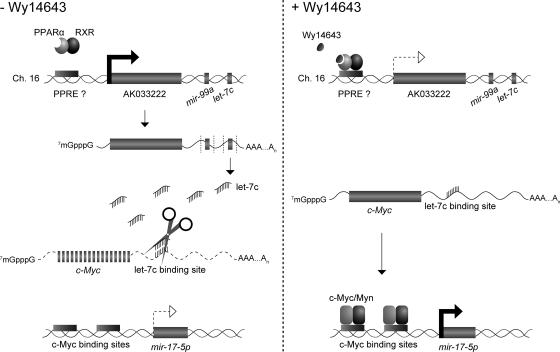
In conclusion, a novel signaling cascade regulated by PPARα is proposed. Basal-level expression of let-7C inhibits c-myc. Upon Wy-14,643 treatment, let-7C expression is inhibited, and a subsequent increase in c-myc is observed. Following an increase in c-myc, the level of the mir-17-92 polycistronic cluster is increased. Collectively, this pathway may lead to increased hepatocellular proliferation and tumorigenesis (Fig. 7).
FIG. 7.
Model of PPARα-regulated miRNA signaling cascade in the liver. In the absence of PPARα agonists, increased basal expression of let-7C causes degradation of c-myc. Activation of PPARα inhibits let-7C expression, leading to increase in c-myc expression. Subsequent binding of c-myc upstream of the oncogenic mir-17 miRNA cluster induces expression of the polycistronic miRNA.
Supplementary Material
[Supplemental material]
Acknowledgments
This study was funded by the National Cancer Institute Intramural Research Program. Y.M.S. was supported by a postdoctoral fellowship from the American Cancer Society (PF-06-014-01-CNE).
We thank Shioko Kimura for critical review of the manuscript.
We have no competing financial interest.
Footnotes
▿
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Ambros, V. 2001. microRNAs: tiny regulators with great potential. Cell 107**:**823-826. [DOI] [PubMed] [Google Scholar]
- 2.Ashby, J., A. Brady, C. R. Elcombe, B. M. Elliott, J. Ishmael, J. Odum, J. D. Tugwood, S. Kettle, and I. F. Purchase. 1994. Mechanistically-based human hazard assessment of peroxisome proliferator-induced hepatocarcinogenesis. Hum. Exp. Toxicol. 13(Suppl. 2)**:**S1-S117. [DOI] [PubMed] [Google Scholar]
- 3.Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122**:**553-563. [DOI] [PubMed] [Google Scholar]
- 4.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116**:**281-297. [DOI] [PubMed] [Google Scholar]
- 5.Bos, J. L., E. R. Fearon, S. R. Hamilton, M. Verlaan-de Vries, J. H. van Boom, A. J. van der Eb, and B. Vogelstein. 1987. Prevalence of ras gene mutations in human colorectal cancers. Nature 327**:**293-297. [DOI] [PubMed] [Google Scholar]
- 6.Butterworth, B. E., and S. R. Eldridge. 1995. A decision tree approach for carcinogen risk assessment. Prog. Clin. Biol Res. 391**:**49-70. [PubMed] [Google Scholar]
- 7.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10**:**1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin, G. A., C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, and C. M. Croce. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99**:**15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin, G. A., C. Sevignani, C. D. Dumitru, T. Hyslop, E. Noch, S. Yendamuri, M. Shimizu, S. Rattan, F. Bullrich, M. Negrini, and C. M. Croce. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 101**:**2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38**:**1289-1297. [DOI] [PubMed] [Google Scholar]
- 11.Chang, F., L. S. Steelman, J. G. Shelton, J. T. Lee, P. M. Navolanic, W. L. Blalock, R. Franklin, and J. A. McCubrey. 2003. Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway. Int. J. Oncol. 22**:**469-480. [PubMed] [Google Scholar]
- 12.Cherkaoui Malki, M., Y. C. Lone, M. Corral-Debrinski, and N. Latruffe. 1990. Differential proto-oncogene mRNA induction from rats treated with peroxisome proliferators. Biochem. Biophys. Res. Commun. 173**:**855-861. [DOI] [PubMed] [Google Scholar]
- 13.Cheung, C., T. E. Akiyama, J. M. Ward, C. J. Nicol, L. Feigenbaum, C. Vinson, and F. J. Gonzalez. 2004. Diminished hepatocellular proliferation in mice humanized for the nuclear receptor peroxisome proliferator-activated receptor alpha. Cancer Res. 64**:**3849-3854. [DOI] [PubMed] [Google Scholar]
- 14.Cox, A. D., and C. J. Der. 2003. The dark side of Ras: regulation of apoptosis. Oncogene 22**:**8999-9006. [DOI] [PubMed] [Google Scholar]
- 15.Desvergne, B., L. Michalik, and W. Wahli. 2006. Transcriptional regulation of metabolism. Physiol. Rev. 86**:**465-514. [DOI] [PubMed] [Google Scholar]
- 16.Dews, M., A. Homayouni, D. Yu, D. Murphy, C. Sevignani, E. Wentzel, E. E. Furth, W. M. Lee, G. H. Enders, J. T. Mendell, and A. Thomas-Tikhonenko. 2006. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 38**:**1060-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18**:**504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eis, P. S., W. Tam, L. Sun, A. Chadburn, Z. Li, M. F. Gomez, E. Lund, and J. E. Dahlberg. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 102**:**3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghisletti, S., W. Huang, S. Ogawa, G. Pascual, M. E. Lin, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2007. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARγ. Mol. Cell 25**:**57-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldsworthy, T. L., S. M. Goldsworthy, C. S. Sprankle, and B. E. Butterworth. 1994. Expression of myc, fos and Ha-ras associated with chemically induced cell proliferation in the rat liver. Cell Prolif. 27**:**269-278. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez, F. J., J. M. Peters, and R. C. Cattley. 1998. Mechanism of action of the nongenotoxic peroxisome proliferators: role of the peroxisome proliferator-activator receptor alpha. J. Natl. Cancer Inst. 90**:**1702-1709. [DOI] [PubMed] [Google Scholar]
- 22.Gregory, R. I., K. P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432**:**235-240. [DOI] [PubMed] [Google Scholar]
- 23.Gu, J., T. He, Y. Pei, F. Li, X. Wang, J. Zhang, X. Zhang, and Y. Li. 2006. Primary transcripts and expressions of mammal intergenic microRNAs detected by mapping ESTs to their flanking sequences. Mamm. Genome 17**:**1033-1041. [DOI] [PubMed] [Google Scholar]
- 24.Hayashita, Y., H. Osada, Y. Tatematsu, H. Yamada, K. Yanagisawa, S. Tomida, Y. Yatabe, K. Kawahara, Y. Sekido, and T. Takahashi. 2005. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65**:**9628-9632. [DOI] [PubMed] [Google Scholar]
- 25.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435**:**828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120**:**635-647. [DOI] [PubMed] [Google Scholar]
- 27.Jonkers, I. J., A. H. Smelt, and A. van der Laarse. 2001. Hypertriglyceridemia: associated risks and effect of drug treatment. Am. J. Cardiovasc. Drugs 1**:**455-466. [DOI] [PubMed] [Google Scholar]
- 28.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12**:**735-739. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. S., T. Pineau, J. Drago, E. J. Lee, J. W. Owens, D. L. Kroetz, P. M. Fernandez-Salguero, H. Westphal, and F. J. Gonzalez. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15**:**3012-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y., K. Jeon, J. T. Lee, S. Kim, and V. N. Kim. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21**:**4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, Y. S., and A. Dutta. 2006. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr. Opin. Investig. Drugs 7**:**560-564. [PubMed] [Google Scholar]
- 32.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115**:**787-798. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J., M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J. J. Song, S. M. Hammond, L. Joshua-Tor, and G. J. Hannon. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305**:**1437-1441. [DOI] [PubMed] [Google Scholar]
- 34.Matsumura, I., H. Tanaka, and Y. Kanakura. 2003. E2F1 and c-Myc in cell growth and death. Cell Cycle 2**:**333-338. [PubMed] [Google Scholar]
- 35.Meister, G., M. Landthaler, A. Patkaniowska, Y. Dorsett, G. Teng, and T. Tuschl. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15**:**185-197. [DOI] [PubMed] [Google Scholar]
- 36.Michael, M. Z., S. M. O'Connor, N. G. van Holst Pellekaan, G. P. Young, and R. J. James. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1**:**882-891. [PubMed] [Google Scholar]
- 37.Moehren, U., M. Eckey, and A. Baniahmad. 2004. Gene repression by nuclear hormone receptors. Essays Biochem. 40**:**89-104. [DOI] [PubMed] [Google Scholar]
- 38.Morimura, K., C. Cheung, J. M. Ward, J. K. Reddy, and F. J. Gonzalez. 2006. Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor alpha to Wy-14,643-induced liver tumorigenesis. Carcinogenesis 27**:**1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami, H., N. D. Sanderson, P. Nagy, P. A. Marino, G. Merlino, and S. S. Thorgeirsson. 1993. Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis. Cancer Res. 53**:**1719-1723. [PubMed] [Google Scholar]
- 40.Nesfield, S. R., C. J. Clarke, D. J. Hoivik, R. T. Miller, J. S. Allen, K. Selinger, and M. J. Santostefano. 2005. Evaluation of the carcinogenic potential of clofibrate in the rasH2 mouse. Int. J. Toxicol. 24**:**301-311. [DOI] [PubMed] [Google Scholar]
- 41.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435**:**839-843. [DOI] [PubMed] [Google Scholar]
- 42.Pascual, G., A. L. Fong, S. Ogawa, A. Gamliel, A. C. Li, V. Perissi, D. W. Rose, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437**:**759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters, J. M., R. C. Cattley, and F. J. Gonzalez. 1997. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis 18**:**2029-2033. [DOI] [PubMed] [Google Scholar]
- 44.Peters, J. M., C. Cheung, and F. J. Gonzalez. 2005. Peroxisome proliferator-activated receptor-alpha and liver cancer: where do we stand? J Mol. Med. 83**:**774-785. [DOI] [PubMed] [Google Scholar]
- 45.Rao, M. S., and J. K. Reddy. 1996. Hepatocarcinogenesis of peroxisome proliferators. Ann. N. Y. Acad. Sci. 804**:**573-587. [DOI] [PubMed] [Google Scholar]
- 46.Rao, P. K., R. M. Kumar, M. Farkhondeh, S. Baskerville, and H. F. Lodish. 2006. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA 103**:**8721-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy, J. K., D. L. Azarnoff, and C. E. Hignite. 1980. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature 283**:**397-398. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Viciana, P., O. Tetsu, K. Oda, J. Okada, K. Rauen, and F. McCormick. 2005. Cancer targets in the Ras pathway. Cold Spring Harbor Symp. Quant. Biol. 70**:**461-467. [DOI] [PubMed] [Google Scholar]
- 49.Sethupathy, P., M. Megraw, and A. G. Hatzigeorgiou. 2006. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods 3**:**881-886. [DOI] [PubMed] [Google Scholar]
- 50.Takamizawa, J., H. Konishi, K. Yanagisawa, S. Tomida, H. Osada, H. Endoh, T. Harano, Y. Yatabe, M. Nagino, Y. Nimura, T. Mitsudomi, and T. Takahashi. 2004. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64**:**3753-3756. [DOI] [PubMed] [Google Scholar]
- 51.Torrey, C. E., H. G. Wall, J. A. Campbell, P. Kwanyuen, D. J. Hoivik, R. T. Miller, J. S. Allen, M. J. Jayo, K. Selinger, and M. J. Santostefano. 2005. Evaluation of the carcinogenic potential of clofibrate in the FVB/Tg.AC mouse after dermal application—part II. Int. J. Toxicol. 24**:**327-339. [DOI] [PubMed] [Google Scholar]
- 52.Torrey, C. E., H. G. Wall, J. A. Campbell, P. Kwanyuen, D. J. Hoivik, R. T. Miller, J. S. Allen, M. J. Jayo, K. Selinger, P. M. Savina, and M. J. Santostefano. 2005. Evaluation of the carcinogenic potential of clofibrate in the FVB/Tg.AC mouse after oral administration—part I. Int. J. Toxicol. 24**:**313-325. [DOI] [PubMed] [Google Scholar]
- 53.Valencia-Sanchez, M. A., J. Liu, G. J. Hannon, and R. Parker. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20**:**515-524. [DOI] [PubMed] [Google Scholar]
- 54.Vo, N., M. E. Klein, O. Varlamova, D. M. Keller, T. Yamamoto, R. H. Goodman, and S. Impey. 2005. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 102**:**16426-16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogelstein, B., C. I. Civin, A. C. Preisinger, J. P. Krischer, P. Steuber, Y. Ravindranath, H. Weinstein, P. Elfferich, and J. Bos. 1990. RAS gene mutations in childhood acute myeloid leukemia: a Pediatric Oncology Group study. Genes Chromosomes Cancer 2**:**159-162. [DOI] [PubMed] [Google Scholar]
- 56.Vogelstein, B., E. R. Fearon, S. R. Hamilton, S. E. Kern, A. C. Preisinger, M. Leppert, Y. Nakamura, R. White, A. M. Smits, and J. L. Bos. 1988. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319**:**525-532. [DOI] [PubMed] [Google Scholar]
- 57.Wu, L., J. Fan, and J. G. Belasco. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 103**:**4034-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, Q., S. Ito, and F. J. Gonzalez. 28 February 2007. Hepatocyte-restricted constitutive activation of PPARα induces hepatoproliferation but not hepatocarcinogenesis. Carcinogenesis. doi: 10.1093/carcin/bgm046. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 59.Yeldandi, A. V., M. S. Rao, and J. K. Reddy. 2000. Hydrogen peroxide generation in peroxisome proliferator-induced oncogenesis. Mutat. Res. 448**:**159-177. [DOI] [PubMed] [Google Scholar]
- 60.Yim, S. H., J. M. Ward, Y. Dragan, A. Yamada, P. C. Scacheri, S. Kimura, and F. J. Gonzalez. 2003. Microarray analysis using amplified mRNA from laser capture microdissection of microscopic hepatocellular precancerous lesions and frozen hepatocellular carcinomas reveals unique and consistent gene expression profiles. Toxicol. Pathol. 31**:**295-303. [DOI] [PubMed] [Google Scholar]
- 61.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4**:**R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, B., X. Pan, G. P. Cobb, and T. A. Anderson. 2007. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2**:**-12. [DOI] [PubMed]
- 63.Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436**:**214-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]