SATB1-Binding Sequences and Alu-Like Motifs Define a Unique Chromatin Context in the Vicinity of Human Immunodeficiency Virus Type 1 Integration Sites (original) (raw)
Abstract
Retroviral integration has recently been shown to be nonrandom, favoring transcriptionally active regions of chromatin. However, the mechanism for integration site selection by retroviruses is not clear. We show here the occurrence of _Alu_-like motifs in the sequences flanking the reported viral integration sites that are significantly different from those obtained from the randomly picked sequences from the human genome, suggesting that unique primary sequence features exist in the genomic regions targeted by human immunodeficiency virus type 1 (HIV-1). Additionally, these sequences were preferentially bound by SATB1, the T lineage-restricted chromatin organizer, in vitro and in vivo. Alu repeats make up nearly 10% of the human genome and have been implicated in the regulation of transcription. To specifically isolate sequences flanking the viral integration sites and also harboring both _Alu_-like repeats and SATB1-binding sites, we combined chromatin immunoprecipitation with sequential PCRs. The cloned sequences flanking HIV-1 integration sites were specifically immunoprecipitated and amplified from the pool of anti-SATB1-immunoprecipitated genomic DNA fragments isolated from HIV-1 NL4.3-infected Jurkat T-cell chromatin. Moreover, many of these sequences were preferentially partitioned in the DNA associated tightly with the nuclear matrix and not in the chromatin loops. Strikingly, many of these regions were disfavored for integration when SATB1 was silenced, providing unequivocal evidence for its role in HIV-1 integration site selection. We propose that definitive sequence features such as the _Alu_-like motifs and SATB1-binding sites provide a unique chromatin context in vivo which is preferentially targeted by the HIV-1 integration machinery.
SATB1 (special AT-rich sequence-binding protein 1) orchestrates the maintenance of chromatin architecture in a cell type-specific manner by organizing it into domains via periodic anchoring of base-unpairing regions (BURs) to the nuclear matrix (12). In thymocyte nuclei, SATB1 forms a cage-like “network” pattern circumscribing heterochromatin and selectively tethers BURs to its network, resulting in coordinated regulation of distant genes (12). In SATB1-deficient thymocytes, multiple genes, including cytokine receptor genes, are derepressed at inappropriate stages of T-cell development in a spatiotemporal manner (2). SATB1 regulates large chromatin domains by acting as a “docking site” for several chromatin-remodeling enzymes in T cells (31, 54). SATB1 can act as either an activator or a repressor of a large number of genes, depending upon its posttranslational modifications (33). Gene-profiling studies demonstrated that SATB1 dysregulates more than 10% of genes and therefore acts as a global regulator of gene expression (33). This prompted us to explore the regulatory potential of SATB1 by isolating and characterizing all of its genomic targets. Interestingly, sequence analysis of some of the isolated targets revealed that they were similar, if not identical, to the reported human immunodeficiency virus type 1 (HIV-1) integration sequences, suggesting a role for SATB1, the T-cell-specific chromatin organizer, in target site selection.
An early obligatory event in HIV-1 pathogenesis is the integration of cDNA into the human genome which is catalyzed by preintegration complexes (PICs) (22). These complexes contain viral DNA; several viral proteins, including integrase and matrix; and a few cellular proteins (22). The integration reaction requires specific repeated sequences at the ends of the viral cDNA (22). Although the mechanisms of retroviral DNA integration have been well established, the mechanism of target site selection and the sequence requirements for integration, if any, in the host genome are not well defined. The base composition of the regions surrounding integration sites has been shown to affect retroviral target site selection (19, 56). Recently, Holman and Coffin reanalyzed the sequence databases generated by genome-wide studies of genomic sequences at and around integrations of HIV, murine leukemia virus (MLV), and avian leukosis and sarcoma virus (ALSV). Their statistical analysis showed certain base preferences at and near the integration sites (23). The host genome is assembled into a compact but heterogeneous higher-order chromatin structure (52). Studies of in vitro integrations using naked template DNA have indicated a preference for certain sequences (7, 10, 27, 45); however, the body of evidence also suggests that the primary sequence per se may not be the only requirement (39, 41, 49, 51). Because of the heterogeneity of the chromatin, the site of integration of HIV into the genome could have dramatic effects on its transcriptional activation (26). Centromeric alphoid repeats are disfavored for HIV integration (13). Various features of host DNA have been targeted by the retroviral integration machinery, many of which are the characteristics of nuclear-matrix attachment regions (MARs), and indeed they have been proposed to be targeted by retroviruses for integration (39). Recent investigations of HIV-1 integrations into the human genome have indicated that it favors active genes and local hot spots (35, 47). Introns were preferred over exons for integration, and all targeted genes were predicted to be transcribed by RNA polymerase II (47). Comparative analysis of sets of DNA sequences from the integration sites of different retroviruses also suggested a role for host chromatin proteins (40).
To better understand how HIV-1 selects integration sites within the T-cell genome, we analyzed motifs and patterns in the genomic sequences directly surrounding the cloned integration sites. Our analysis of the genomic sequences flanking known integration sites revealed _Alu_-like motifs that may promote chromatin organization favorable to the integration machinery. Chromatin immunoprecipitation (ChIP)-PCR analysis of HIV-1-infected T cells demonstrated association of SATB1 with the genomic regions flanking integration sites. Additionally, the cloned sequences flanking HIV-1 integration sites from the studies of Schröder et al. (47) were found in SATB1-immunoprecipitated chromatin. Our studies suggest that SATB1-mediated assembly of chromatin in T cells may play a role in integration site selection by HIV-1.
MATERIALS AND METHODS
HIV-1 infection.
CEM-GFP (green fluorescent protein), a CD4+ reporter T-cell line (21), was infected at a multiplicity of infection of 1 by incubation with an NL4.3 virus isolate (1) for 4 h at 37°C in the presence of 1 μg/ml Polybrene. The cells were then washed with phosphate-buffered saline (PBS), transferred to fresh RPMI 1640 with 10% fetal calf serum, and incubated at 37°C in a CO2 incubator. The progress of infection was visualized by GFP expression and monitored by analysis of p24 antigen in the culture supernatant with a p24 antigen enzyme-linked immunosorbent assay kit (Perkin-Elmer Life Science). The cells were harvested at 48 h postinfection for isolation of genomic DNA and for ChIP.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously, under a condition of protein excess (28). Binding reactions were performed with a 10-μl total volume containing 10 mM HEPES (pH 7.9), 1 mM dithiothreitol, 100 mM KCl, 2.5 mM MgCl2, 10% glycerol, 0.5 μg of double-stranded poly(dI-dC), 10 μg of bovine serum albumin, and 10 to 100 ng of recombinant SATB1. Samples were pre incubated at room temperature for 5 min prior to the addition of a 32P-labeled probe. Gel-purified, 32P-labeled, PCR-amplified products of in vivo and in vitro integration sequences were used as probes and correspond to approximately 5 ng of DNA. In competition assays, we also added a 10-fold or 100-fold amount of homologous and heterologous gel-purified PCR products, as well as a well-characterized MAR sequence containing seven copies of the 25-bp core of the immunoglobulin H (IgH) MAR (29). After 15 min of incubation at room temperature, the products of these binding reactions were resolved by 6 to 8% native polyacrylamide gel electrophoresis. The gels were dried under vacuum and exposed to X-ray film. The differences in the intensities of the probe bands reflect the differences in labeling efficiency due to the base composition of the sequences. Binding affinities were estimated in the form of dissociation constants (_Kd_s) by performing EMSA analysis under a condition of protein excess as previously described (15, 17).
Genomic DNA isolation and PCR.
Human peripheral blood mononuclear cells (PBMCs) were isolated from blood of normal seronegative donors by layering on a Ficoll gradient. Cells were harvested by centrifugation and washed in 1× PBS, and DNA was isolated with a genomic DNA isolation kit (QIAGEN). Diluted DNA was PCR amplified in 100-μl reaction mixtures containing 50 mM KCl, 10 mM Tris-HCl, 1.5 mM MgCl2, 0.1% Triton X-100, 1.0 U of Taq DNA polymerase (Promega), and 1 μM each primer pair with 1 cycle of 95°C for 5 min and 30 cycles of 95°C for 1 min, 48°C for 1 min, and 72°C for 1 min. PCR products were resolved by native polyacrylamide gel electrophoresis, stained with SYBR gold (Molecular Probes), and visualized under UV illumination. For preparation of labeled probes, amplification reaction mixtures were supplemented with 1 μl of [32P]dCTP and labeled PCR products were purified by gel elution by standard procedures. One nanogram of labeled DNA probe was used in each binding reaction mixture. Reverse transcription (RT)-PCR analysis was performed by using the kit according to the manufacturer's instructions. Total RNA was extracted from cultured cells with TRI Reagent (Sigma). Quantitative PCRs were performed with SYBR green IQ Supermix (Bio-Rad) and an ICycler IQ real-time thermal cycler (Bio-Rad). The _n_-fold changes in the level of SATB1 expression were calculated from the threshold cycle (CT) values as follows: _n_-fold change = 2−(ΔCT), where ΔCT = CT,SATB1 − CT,GAPDH.
Bioinformatic analyses.
Multiple alignments were performed with a locally installed Clustal program, ClustalX version 1.86 (42). For identification of consensus motifs, the integration sequences were analyzed by MEME, a bioinformatic tool that calculates consensus sequences from a given set of data (3). Details of the parameters used for determination of the consensus and generation of random sequence database are available on request.
ChIP.
Jurkat cells or control (uninfected) and CEM-GFP cells infected with the HIV-1 NL4.3 isolate were cross-linked for 15 min at 37°C by adding formaldehyde (to a final concentration of 1%) directly to the culture medium, and ChIPs were carried out as previously described (32). Briefly, cells were cross-linked with 1% formaldehyde for 10 min, followed by subsequent washes with wash buffer 1 (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 7.5], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM sodium butyrate, 1 μg/ml each aprotinin, pepstatin, and leupeptin) and wash buffer 2 (0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 7.5], 1 mM PMSF, 10 mM sodium butyrate, 1 μg/ml each aprotinin, pepstatin, and leupeptin). The cell pellet was resuspended in lysis buffer (150 mM NaCl, 25 mM Tris-HCl [pH 7.5], 5 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1 mM PMSF, 10 mM sodium butyrate, 1 μg/ml each aprotinin, pepstatin, and leupeptin) and lysed by sonication. The sonicated sample was clarified by centrifugation at 20,000 × g in a microcentrifuge at 4°C for 10 min. The clear supernatant containing soluble cross-linked chromatin was used for immunoprecipitation with anti-SATB1 (16), anti-PARP, anti-p53, and anti-HMG-I(Y) (all from Santa Cruz Biotechnology). Control immunoprecipitations were performed with normal rabbit IgG and mouse monoclonal IgG1 (Upstate Biotechnology). After immunoprecipitation, chromatin-antibody complexes were eluted by adding 2% sodium dodecyl sulfate, 0.1 M NaHCO3, and 10 mM dithiothreitol and incubating the mixture for 10 min at room temperature. Reversal of cross-linking was performed by addition of 0.05 volume of 4 M NaCl and incubation for 4 h at 65°C, followed by phenol-chloroform extraction and ethanol precipitation. One-fiftieth of the DNA from each pool was PCR amplified in 50-μl reaction mixtures containing 50 mM KCl, 10 mM Tris-HCl, 1.5 mM MgCl2, 0.1% Triton X-100, 1.0 U of Taq DNA polymerase (Promega), and 1 μM each primer pair with 1 cycle of 95°C for 5 min and 30 cycles of 95°C for 1 min, 48 to 62°C for 1 min, and 72°C for 1 min. PCR products were resolved by native polyacrylamide gel electrophoresis, stained with SYBR gold (Molecular Probes), and visualized under UV light.
Isolation of chromatin loop and nuclear-matrix-associated DNAs.
DNA from chromatin fractionated into loops and nuclear matrix was isolated as previously described (48). Briefly, Jurkat cells were washed with PBS, followed by gentle lysis of cells with CSK buffer 1 {0.5% Triton X-100, 10 mM PIPES [piperazine-N,_N_′-bis(2-ethanesulfonic acid), pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM PMSF, 1× protease inhibitor cocktail (Sigma Chemical Co.)}. Nuclei were then resuspended in CSK buffer II (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 2 M NaCl, 1 mM dithiothreitol, 1× protease inhibitor cocktail), followed by DNase I digestion for 4 h at 37°C. The reaction mixture was then centrifuged at 12,000 rpm for 15 min. Supernatant containing chromatin loops and pellet containing undigested nuclear-matrix-associated chromatin were deproteinized by proteinase K treatment for 2 h at 56°C. The DNA was recovered by ethanol precipitation and is referred to as loop and matrix fractions, respectively.
Localization of SATB1-binding sites by FISH.
Amplified fluorescence in situ hybridization (FISH) for detection of single-copy loci was performed by the tyramide signal amplification method as previously described (11). By this method, we monitored single-copy loci with short 300- to 600-bp DNA sequences that bind in vivo to SATB1 within T-cell nuclear matrices and histone-depleted nuclei, generating “halos” due to distended chromatin loops. Briefly, nuclear halos were prepared by high-salt treatment of isolated nuclei and nuclear matrices were further prepared by digesting the bulk of the extended chromatin loops with restriction enzymes as previously described (11). Specific probes against the integration sites or SATB1-binding sequences were generated by labeling respective PCR-amplified DNAs with biotin-14-dCTP (Invitrogen). Labeled probes were used for hybridization and detection with the TSA Biotin System in accordance with the manufacturer's (Perkin-Elmer) instructions.
Short hairpin RNA-mediated knockdown of SATB1.
CEM-GFP cells (1 × 107) were transfected separately with 10 μg pSUPER vector or pSUPER-shSATB1 construct DNA by using SiMPORTER transfection reagent (Upstate Biotechnology). Cells were maintained for 48 h in RPMI 1640 with 10% fetal calf serum and incubated at 37°C in a CO2 incubator. An aliquot of 1 × 106 cells was used to prepare RNA with the TRI Reagent (Sigma), followed by cDNA preparation with 1 μg of total RNA. The relative expression of SATB1 was quantitated by real-time PCR as described above, except that the _n_-fold changes in the expression level of SATB1 were calculated from the threshold cycle (CT) values as follows: _n_-fold change = 2−Δ(ΔCT), where ΔCT = CT,SATB1 − CT,GAPDH, and Δ(ΔCT) = CT,siSATB1 − CT,control. The rest of the transfected CEM-GFP cells were then used for infection with HIV-1 NL4.3 as described above. Cells were harvested at 48 h postinfection for isolation of genomic DNA.
RESULTS AND DISCUSSION
Isolation of SATB1-binding sites.
Since SATB1 organizes T-cell chromatin in a unique manner (12) that may reflect upon its regulatory potential, we wished to isolate genomic binding sequences for SATB1. The cage-like manner in which SATB1 occupies the nuclear volume actually suggests that SATB1 may bind to a significant portion of the chromatin, and this organization may dictate the regulation of chromatin domains in T cells. SATB1 preferentially binds to genomic sequences with an ATC context (15); however, no consensus has been defined yet. We therefore initiated a genome-wide analysis of SATB1-binding sequences in a human lymphoblastoid Jurkat T-cell line and PBMCs by the ChIP strategy and cloned the isolated DNAs. The clones were then sequenced, and the sequences were used for BLAST analysis of the human genome to map them. Surprisingly, one of the sequences mapped to the 11q13 locus, which is reported as the integration hot-spot region for HIV-1 (47). In fact, the sequence of the ChIP clone we obtained was virtually identical to a portion of the 2.5-kb hot-spot region deposited as BH 609658 (47) (data not shown). The first genome-wide mapping and analysis of integration target sites suggested that HIV-1 prefers to integrate within intronic regions of transcriptionally active genes (47). Interestingly, this study also revealed the presence of regions with clustering of integration sites termed integration hot spots (47). The integration site choice of MLV turned out to be similar with respect to the activity status of genes; however, the transcription start sites of active genes were preferred (35). In contrast, ASLV integration sites were distributed more randomly throughout the genome, with a very weak bias toward transcriptionally active genes and no bias for transcription start sites (40). If there are regional hot spots and a particular conformation of chromatin is favored, then we reasoned that there must be a common signature embedded within the DNA sequence itself that generates a chromatin conformation preferred by the PIC. The analysis of chromosomal regions preferred for integration also suggested a role for chromatin proteins (40).
SATB1 binds preferentially to the sequences flanking in vivo integration sites.
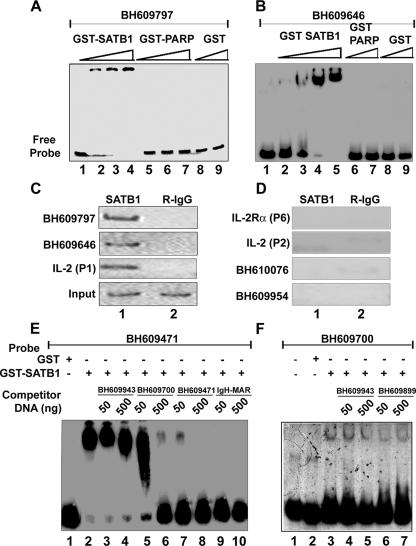
Our initial observation that SATB1, a T lineage-restricted MAR-binding protein, bound to a sequence from one of the proposed HIV-1 integration hot-spot regions in vivo prompted us to analyze the binding potential of other sequences. We essentially used the sequence information from the BH series (47) after separating the sequences from virus-host DNA junction clones generated by integration reactions with naked cell-free DNA as the template (in vitro) or a chromatin template in live cells (in vivo). From the sequences deposited by Bushman and colleagues, we randomly selected a few in vivo and a few in vitro sequences and designed primers for their PCR amplification. PCR amplification of each of these sequences was performed with genomic DNA isolated from Jurkat cells or PBMCs and specific primer pairs. EMSA with labeled DNAs from two representative in vivo integration clones, BH609797 and BH609646, indicated that SATB1 bound them tightly in vitro (Fig. 1A and B, respectively, lanes 2 to 4). As controls, we used glutathione _S_-transferase (GST)-PARP, another DNA-binding protein (Fig. 1A, lanes 5 to 7, and 1B, lanes 6 and 7), and GST alone (Fig. 1A and B, lanes 8 and 9), both of which did not bind at all. Additional EMSA analysis with labeled DNAs corresponding to different integration clones from the BH series indicated that SATB1 bound 80% (12 out of 15) in vivo sequences, as opposed to 20% (2 out of 10) in vitro sequences (data not shown). For accurate comparison of binding affinities, we next estimated the dissociation constants (_Kd_s) with SATB1 for all in vivo integration clones (data not shown). The Kd values were in the range of 2.5 to 60 nM, compared to the 1 nM of the IgH MAR heptamer (Table 1). Thus, SATB1 seems to bind preferentially to the in vivo integration sequences and to at least some of them with an affinity comparable to that of the IgH MAR, which contains the well-characterized BUR motif (29). It is reported that SATB1 does not bind to all of its genomic targets with the same affinity (12, 15).
FIG. 1.
SATB1 specifically binds to HIV-1 integration sequences in vitro and in vivo. In vitro binding of SATB1 to HIV-1 integration clones BH609797 (A) and BH609646 (B) was demonstrated by EMSA as described in Materials and Methods. Briefly, 10 to 100 ng of recombinant purified GST-SATB1, GST-PARP, or GST was incubated as indicated with radiolabeled, PCR-amplified DNA probes in the presence of 1 μg of competitor DNA. Protein-DNA complexes were resolved on native polyacrylamide gels. In vivo binding of SATB1 was demonstrated by PCR amplification of DNA isolated by ChIP (C and D) with anti-SATB1 (lane 1) or rabbit IgG (R-IgG; lane 2). Distal promoter region P1 of _IL_-2 was used as a positive control for SATB1 binding, whereas proximal promoter region P2 of _IL_-2 and region P6 of _IL_-_2R_α were used as negative controls. ChIP analysis of two representative in vivo integration clones, BH609797 and BH609646 (C), and two representative in vitro integration clones, BH610076 and BH609954 (D), is depicted. The specificity of binding of SATB1 to HIV-1 in vivo integration clones BH609471 (E) and BH609700 (F) was demonstrated by competition EMSA. Briefly, 2 μg of recombinant purified GST-SATB1 (lanes 2 to 10 in panel E and lanes 3 to 7 in panel F) or GST alone (lane 1 in panel E and lane 2 in panel F) was incubated with radiolabeled, PCR-amplified DNA probes in the presence of 1 μg of poly(dI-dC) competitor DNA. Protein-DNA complexes were resolved on 6% native polyacrylamide gels. Binding reactions were competed for by adding a 10-fold (50 ng) or 100-fold (500 ng) excess of unlabeled, PCR-amplified homologous or heterologous DNA to the binding reaction mixture as indicated.
TABLE 1.
SATB1-binding affinities of various HIV-1 integration sequencesa
| Serial no. | DNA clone name | K d (nM) |
|---|---|---|
| 1 | BH609824 | 10.0 |
| 2 | BH609708 | >50.0 |
| 3 | BH609874 | >40.0 |
| 4 | BH609617 | 5.0 |
| 5 | BH609700 | >40.0 |
| 6 | BH609907 | 5.0 |
| 7 | BH609792 | >50.0 |
| 8 | BH609769 | 4.0 |
| 9 | BH609551 | 8.0 |
| 10 | BH609864 | 15.0 |
| 11 | BH609471 | 2.5 |
| 12 | BH609797 | 15.0 |
| 13 | BH609614 | 60.0 |
| 14 | BH609642 | 10.0 |
| 15 | BH609451 | 15.0 |
| 16 | BH609820 | 25.0 |
| 17 | BH609475 | >40.0 |
| 18 | IgH MAR-WT(25)7 | 1.0 |
We next performed ChIP assays to monitor the binding of SATB1 to these sequences in vivo. In vitro binding with naked DNA substrates may not always reflect the in vivo occupancy of SATB1 at the same site. Since we hypothesized a role for chromatin architecture in integration target choice by the PIC, it was essential to monitor binding of SATB1 to these sites in vivo. As in the case of the 11q13 hot spot for integration, we found that SATB1 bound both the BH609797 and BH609646 in vivo integration sequences from the Bushman study in vivo (Fig. 1C, lane 1 in the top two parts). As controls for the ChIP assay, we used sequences from the upstream portions of _IL_-2 and _IL_-_2R_α that were characterized with respect to their in vivo occupancy by SATB1 (32). As expected under these conditions, the distal P1 region of the _IL_-2 promoter was bound by SATB1 in vivo but not the proximal P2 region and also not the P6 region of the _IL_-_2R_α promoter. We also performed ChIP analysis for DNAs from two representative in vitro integration clones, BH610076 and BH609954, and found that these sites are not bound by SATB1 in vivo (Fig. 1D, lane 1 in the bottom two parts). To further verify the specificity of binding, we performed a competition assay wherein binding was competed for by homologous and heterologous unlabeled DNAs. As probes, we used in vivo integration sequence BH609471, which binds with high affinity, and BH609700, which binds with low affinity (data not shown). The binding of SATB1 to BH609471 was not affected by addition of unlabeled DNA corresponding to heterologous in vitro integration clone BH609943 (Fig. 1E, lanes 3 and 4). A homologous in vivo integration clone, BH609700, competed for binding in a dose-dependent manner (Fig. 1E, lanes 5 and 6). However, when unlabeled BH609471 itself was used at a 10- or 100-fold excess, it competed for binding by SATB1 effectively (Fig. 1E, lanes 7 and 8, respectively). The 25-bp core of the IgH MAR sequence exhibits very high base-unpairing potential (29) and therefore is bound specifically by SATB1. When we used a heptameric combination of this sequence in the competition assay, the binding of SATB1 to labeled BH609471 was completely abolished (Fig. 1E, lanes 9 and 10). In the case of low-affinity-binding in vivo integration clone BH609700, competition with a 10- or 100-fold excess of two heterologous in vitro integration sequences, BH609943 and BH609899, that are not bound by SATB1 (data not shown) did not abolish the complexes formed between SATB1 and BH609700 (Fig. 1F, lanes 3 to 7), suggesting that this binding is highly specific and dependent on the sequence context. These results further prove that SATB1 preferentially binds to the in vivo integration sequences.
No binding of any host factor(s) to the retroviral integration sequences has previously been demonstrated; no specific primary sequence patterns or motifs have been identified in sequences flanking the integration sites. Three independent genome-wide studies on integration site preferences of HIV, MLV, and ASLV suggested a role for the transcriptional activity status of chromatin (35, 40, 53). Recent investigation of the influence of the transcriptional status of the metallothionein gene on integration site choice by ASLV in quail cells demonstrated that ASLV disfavors transcriptionally active genes. Specifically, integration of the viral genome was favored in an uninduced gene and was significantly inhibited when the same gene was induced (36). Thus, despite an apparent preference for integration of retroviral DNA into transcribed regions of the host genome, increased transcription can be inhibitory to the integration process (36). HIV-1 and HIV-based vectors showed a strong bias toward integration into active genes and gene-rich regions of chromosomes (40, 46, 47). MLV does not favor integration into transcription units but favors integration in the vicinity of transcription start sites (40, 53). ASLV differs strikingly from these two; it does not favor integration near transcription start sites, nor does it favor active genes (40). Collectively, these studies suggest that PICs of retroviruses may interact with chromatin-associated factors and/or transcriptional cofactors to facilitate integration (20). Corroborating this notion, Ciuffi et al. recently demonstrated that the HIV integrase-interacting protein LEDGF/p75 has an impact on retroviral integration site selection. This was achieved by comparing the genome-wide distributions of 4,118 unique integration sites in three cell lines depleted of LEDGF/p75 and in matched controls (14). The frequency of integration in transcription units was reduced in all three cell lines in which LEDGF/p75 was silenced, compared to the paired controls (14). Although the reduction in integration frequency within transcription units was modest, this observation underlines the impact of chromatin-associated proteins on retroviral integration. Studies on the closely related Ty retrotransposons of yeast revealed that interactions with bound chromosomal proteins can tether the Ty integration machinery to chromosomes and thereby direct integration to nearby sites (6, 44, 55). In fact, Bushman proposed a similar “docking” model to explain integration by retroviruses (9). The chromatin-associated protein SATB1 therefore could serve as a docking site for the HIV-1 PIC. Therefore, study of SATB1-binding sites would be of importance for understanding the chromatin context targeted by retroviral PICs. Genomic regions flanking HIV-1 integration sites seem to be enriched in SATB1-binding sequences and therefore may contribute toward a T-cell-specific higher-order chromatin organization. We therefore searched for hidden motifs and patterns in reported sequences flanking HIV-1 integration sites.
_Alu_-like motifs are enriched in sequences flanking the reported HIV-1 integration sequences.
We initially performed a gapped alignment of sets of cloned integration sequences in the NCBI database with ClustalX (42). Multiple alignments of in vivo integration sequences revealed a striking pattern. We found that these sequences share extended homologous regions which are spread across the lengths of the sequences. The sequence similarity appeared to be present in “chunks” of similar sequences in all of the sequences taken for alignment (data not shown). Sequences of these blocks of homology seem to differ from the ATC context that is typically observed with known SATB1-binding sequences (data not shown) (12, 15). Such chunks of homologies were characteristically absent from the alignments of sequences from in vitro integrations (data not shown). Additionally, an unrooted phylogenetic tree plotted with these sequences showed one major branch of related sequences, which comprised more than 60% of the sequences. As controls we used sets of random DNA sequence data generated with a Markov chain simulator (43; data not shown). Strikingly, the unrooted phylogenetic trees for in silico-generated random sequences or sequences picked randomly from the human genome displayed virtually no relatedness among individual sequences (data not shown).
Since we observed significant homology in regions of the integration sites, we then investigated whether there exist any consensus motifs among them by using the online tool MEME (3). We identified three consensus sequences of 31 to 50 bp among these homologous regions within the integration sites, with an average occurrence per sequence of close to 1 (Table 2, rows 1 to 3). Furthermore, realignment of the motifs with the respective sequences by the motif alignment search tool (MAST) revealed that the integration regions are composed of multiple consensus sequences that are either arranged tandemly or interspersed (data not shown). These consensus elements were compared with known repeats in the human genome and were found to be _Alu_-like elements. We performed a similar analysis for 452 2-kb sequences picked randomly from the human genome. The motifs obtained in the sequences flanking integration sites are significantly different from those obtained from the randomly picked sequences (Table 2, rows 4 to 6), suggesting that unique primary sequence features exist in the genomic regions targeted by HIV-1. The width and average number of occurrences of each of the motifs are comparable within the two data sets, and both also have very high E values. It can thus be concluded that such occurrences of motifs are not chance events and are highly statistically significant. To perform a completely “unfiltered” scan for motifs within the genomic sequences, repeat masking was not performed prior to a motif search. However, the derived motifs were then searched for within the repeat-masked sequences with MAST and were undetectable.
TABLE 2.
Consolidated data obtained for the consensus motifs from the databases of sequences flanking the integration sites and randomly picked human sequences of the same lengtha
| Motif | Sequence | Width (bases) | No. of sites | Avg no. of occurrences |
|---|---|---|---|---|
| 1 | GGCGCGCGCCTGTAATCCCAGCACCTCGGGAGGCCGAGGCGGGGGGATCA | 50 | 500 | 1.17 |
| 2 | CCCCGGGTGGCGGGGATTGCAGGGATCTGCGATCACGCCAAGC | 43 | 500 | 1.17 |
| 3 | CCAGCCTGGGCAACAGAGTGAGACCCCGTCT | 31 | 461 | 1.07 |
| 4 | TGCCTCAGCCTCCCAAATAGCTGGGATTACAGGCGTGAGCCACCACGCCC | 50 | 450 | 0.99 |
| 5 | AGACCAGCCTGGGCAACATAGTGAAACCCCGTCTCTACAAAAAAAAAAAA | 50 | 450 | 0.99 |
| 6 | GCAGTGGCGCGATCTCGGCTCACTGCAACCTCCGCCTCCCGGGTTCAAGC | 50 | 348 | 0.77 |
The Alu repeats constitute about 5 to 10% of the human genome (4). Alu elements affect the genome in several ways, causing insertion mutations, recombination between elements, gene conversion, and alterations in gene expression (4). The Alu repeats have been implicated in transcription and transcription control (30, 34). In support of this, Alu repeats have been shown to be enriched in histone H3 lysine 9 methylation (30). Alu elements are each a dimer of similar, but not identical, fragments with a total size of about 300 bp and originate from the 7SL RNA gene. Each element contains a bipartite promoter for RNA polymerase III, a poly(A) tract located between the monomers, a 3′-terminal poly(A) tract, and numerous CpG islands and is flanked by short direct repeats. The chromatin context of the Alu repeats is important for their function (38), and the Alu elements themselves can play a role in chromosomal rearrangement (37). Interestingly, analysis of HIV-1 proviral integrations in isolates derived both from integrations in infected individuals and from cultured cells revealed a significant propensity of HIV-1 to integrate at or near the Alu repeats (50). Additionally, genome-wide analysis of HIV integration sites by the Bushman group found 15.9% of the in vivo integration sites to be in Alu repeats (47). Therefore, it was not very surprising that our analysis picked up _Alu_-like motifs in the sequences flanking HIV-1 integration sites. Additionally, mapping of genomic positions of integration sites revealed that HIV-1 preferentially integrates within the transcribed and GC-rich regions of the human genome (19). The high GC content of Alu repeats may therefore constitute another feature facilitating their preferential targeting by the PIC.
SATB1-associated chromatin contains cloned sequences flanking HIV-1 integration sites.
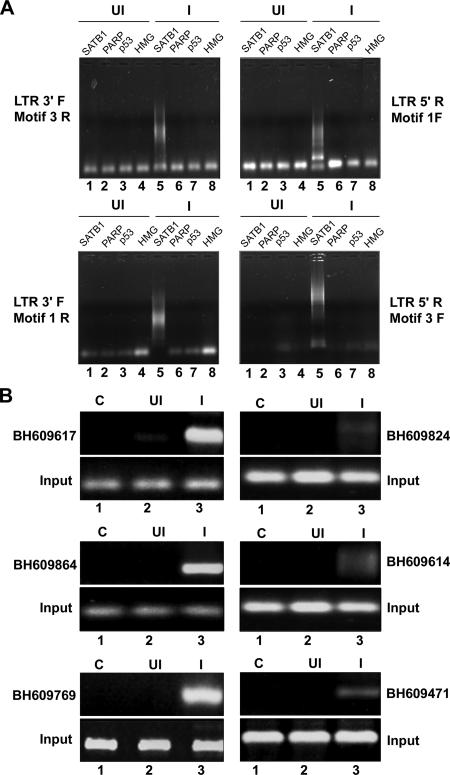
To test for the presence of _Alu_-like motifs in HIV integration sites enriched in SATB1 binding, we performed ChIP with SATB1-specific antibodies and subsequently amplified the Alu repeats and HIV long terminal repeat (LTR) sequences in recovered DNA by PCR with specifically designed primers corresponding to the _Alu_-like motifs (motifs 1 to 3, Table 2) and the LTRs of the NL4.3 isolate of HIV-1. As controls we used antibodies specific to p53, PARP, and HMG-I(Y). With combinations of _Alu_-like motif-specific and LTR-specific primers, we observed that PCR-amplified products were obtained specifically in the anti-SATB1-immunoprecipitated chromatin (Fig. 2A, lane 5 in all parts), suggesting that out of the four chromatin proteins tested, namely, SATB1, PARP, p53, and HMG-I(Y), only SATB1 is specifically associated with the regions flanking HIV-1 integrations sites. We subsequently confirmed that many of the cloned integration sequences from the BH series were specifically amplified in PCRs with specific primers and purified DNA template from anti-SATB1-immunoprecipitated chromatin (Fig. 2B, lane 3 in all parts). Control reaction mixtures with anti-SATB1-immunoprecipitated chromatin from uninfected cells did not yield any amplification product, confirming the specificity of the ChIP-PCRs. Collectively, these results demonstrated that the SATB1-associated chromatin harbors many regions of the genome that constitute HIV-1 integration sites.
FIG. 2.
ChIP-PCR analysis of DNA flanking integration sites from the pool of SATB1-bound DNA. (A) ChIP analysis of regions flanking HIV-1 integration sites. In vivo association of SATB1 with the regions flanking integration sites was monitored by ChIP-PCR as described in Materials and Methods. DNA fragments isolated from infected (I) and control (uninfected [UI]) cell chromatin after immunoprecipitation with antibodies against four chromatin proteins [SATB1, PARP, p53, and HMG-I(Y)] were used as templates for PCR amplification with primer sets containing an LTR-specific primer and a primer specific for an _Alu_-like motif. The combinations of primers used are indicated at the sides. (B) PCR amplification of reported integration sequences (BH series) in the SATB1-immunoprecipitated and PCR-amplified DNA pool with the LTR 3′F and Motif 3R primers. We selected the indicated clones that bind SATB1 in vitro and confirmed in vivo association with SATB1 by PCR with primer sets corresponding to each of them and ChIP-PCR-amplified DNA as the template.
Regions flanking HIV-1 integration sites are associated with the nuclear matrix in vivo.
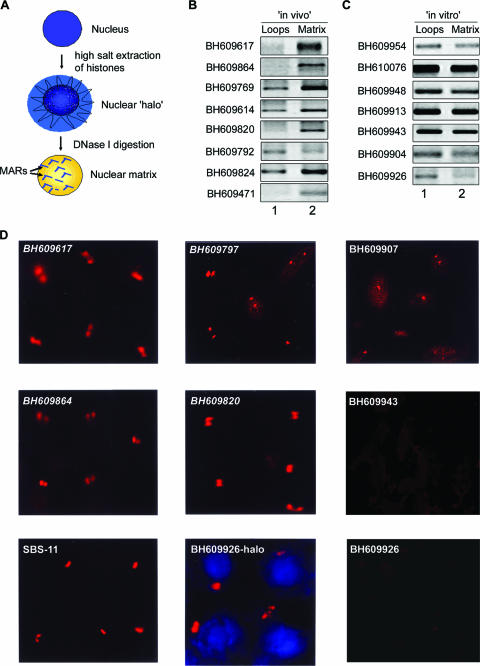
Chromatin is anchored to the nuclear matrix by matrix/scaffold attachment regions (M/SARs), thereby organizing genomic DNA into topologically distinct loop domains that are important in replication and transcription (45). M/SARs are often closely associated with transcriptional promoters and enhancers of several genes and have been shown to generate long-range chromatin accessibility (24). Juxtaposition with M/SARs correlates with transcriptional augmentation (5). SATB1 is a cell type-specific MAR-binding protein (16). We therefore reasoned that if genomic regions flanking HIV-1 integration sites are enriched in SATB1-associated chromatin, then they should also be anchored to the nuclear matrix in vivo. To test this, we performed a partitioning assay designed to separately isolate (i) genomic DNA associated with the nuclear matrix and (ii) that of chromatin loops. The DNA isolated from the loop and matrix fractions was then used as the template for PCR amplification with primer sets corresponding to various cloned integration sites from the BH series. This analysis revealed selective enrichment of most of the cloned in vivo integration sites in the nuclear matrix fraction compared to that of the loops (Fig. 3B). Indeed, clone BH609471, which exhibited the highest binding affinity for SATB1 (Table 1), partitioned completely in the matrix fraction and was not detected in the loop fraction (Fig. 3B, lowermost part). This selective partitioning was not observed with all of the non-SATB1-binding in vitro integration clones tested, and they were amplified to comparable levels from both the loop and matrix fractions (Fig. 3C), suggesting that their association with the nuclear matrix is presumably governed by the ability of SATB1 to bind to these sequences in vivo. To unequivocally demonstrate the association of SATB1-binding sequences with the nuclear matrix, biotin-labeled PCR products representing few in vivo and in vitro integration clones were then used as probes for high-resolution FISH of in situ-prepared nuclear matrices after the bulk of the chromatin loops had been digested and removed (scheme depicted in Fig. 3A). Genomic DNA that was tightly anchored to the matrix, corresponding to the base of the chromatin loops, hybridized with the SATB1-binding sequences in Jurkat cells (Fig. 3D). This hybridization at the base of chromatin loops was totally undetectable with the BH609943 and BH609926 (Fig. 3D) in vitro integration sequences, which are not bound by SATB1 in vitro, indicating that SATB1 may play a role in anchoring these genomic regions to the nuclear matrix in vivo. Additionally, in vitro integration clone BH609926 hybridized with the nuclear halo (faint blue 4′,6′-diamidino-2-phenylindole [DAPI]-stained area outside of the DAPI-intense nucleus, Fig. 3D, middle part at the bottom) containing the extended chromatin loops. This result corroborated that of the matrix-loop partitioning assay, wherein the sequence BH609926 partitioned predominantly in the loop fraction. Thus, the SATB1-binding in vivo and in vitro integration clones seem to preferentially associate with the nuclear matrix in vivo. Since these sequences have the same primary sequence motifs as other integration sites, we predict that SATB1 may actively tether most, if not all, integration sites to the nuclear matrix in vivo.
FIG. 3.
Regions flanking HIV-1 integration sites partition preferentially in the chromatin fraction associated with the nuclear matrix. (A) Schematic representation of the protocol used for the preparation of nuclear halos and matrices. For details, see Materials and Methods. (B and C) The matrix-loop partitioning assay was performed as described in Materials and Methods. PCR amplification of the indicated in vivo (B) or in vitro (C) integration sequences with template DNA from the loop (lane 1) or matrix (lane 2) fraction is depicted. The PCR products were resolved on 1% agarose and visualized by staining with ethidium bromide. (D) Amplified single-locus FISH revealed that cloned sequences flanking in vivo HIV-1 integration sites are associated specifically with the nuclear matrix in situ. Briefly, nuclear matrices were prepared from Jurkat T cells in situ and PCR-amplified and biotin-labeled DNA probes corresponding to the integration clones (BH series) were used for hybridization and detected by amplified FISH as previously described (11). The in vivo integration clones are in italics. As a positive control for matrix hybridization, we used the reported SATB1-binding sequence SBS-11 (15) as depicted at the bottom left. BH609926-halo represents hybridization of the biotin-labeled probe corresponding to this in vitro integration clone to the nuclear halo comprising distended chromatin loops. The same probe did not hybridize with the nuclear matrix preparation, as depicted at the bottom right.
SATB1 is differentially expressed in various cell lines used for HIV infection.
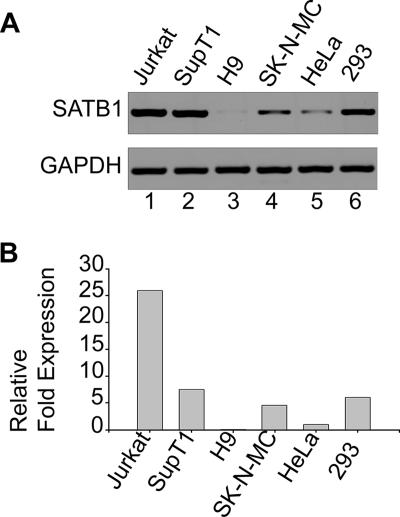
SATB1 is known to be a T lineage-restricted chromatin organizer (2, 12, 54). SATB1 is also known to be expressed in cells that are naturally infected with HIV-1, such as the PBMCs (32). Since we hypothesized that SATB1 and proteins with similar functions could play a role in the organization of host cell chromatin that facilitates the integration process, we wished to determine if SATB1 is expressed in different cell types and to what levels. We therefore monitored the level of expression of SATB1 in various T-cell and non-T-cell lines, especially in those that were used for investigations reporting genomic integration sites for retroviruses. We isolated total RNA from Jurkat, SupT1, H9, SK-N-MC, HeLa, and HEK 293 cells and performed RT-PCR analysis of the level of SATB1 expression with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control (Fig. 4A). The expression levels in different cell lines were compared by quantitating the relative _n_-fold expression levels by real-time RT-PCR analysis (Fig. 4B). We observed that SATB1 was expressed at the highest levels in Jurkat cells, while it was expressed at almost undetectable levels in H9 cells. Among all of the other cell lines, intermediate levels of expression were observed. The relative expression level of SATB1 or functionally similar proteins may govern the integration site preference of retroviruses by modulating the chromatin loop architecture. There exists a homolog of SATB1 called SATB2 that is expressed in certain cell types that do not express SATB1 (8, 18). Different cell types seem to express a protein(s) similar to SATB1 in function that governs cell type-specific chromatin organization. Thus, it may be argued that the interaction(s) of specific components of the retroviral PIC with specific host chromatin proteins may mediate target site selection.
FIG. 4.
SATB1 is differentially expressed in various cell lines. (A) Total RNA was isolated and RT-PCR analysis was performed as described in Materials and Methods. RT-PCR products for SATB1 (upper part) from Jurkat (lane 1), SupT1 (lane 2), H9 (lane 3), SK-N-MC (lane 4), HeLa (lane 5), and HEK 293 (lane 6) cells were electrophoresed on a 1% agarose gel and visualized by staining with ethidium bromide. GAPDH was used as an internal control for normalization (lower part). (B) Quantitation of relative levels of SATB1 expression in different cell lines was performed by real-time RT-PCR analysis as described in Materials and Methods. SATB1 expression levels in different cell lines were expressed as _n_-fold changes in comparison with that of HeLa cells, which was set at 1 arbitrary U.
Knockdown of SATB1 alters integration site choice by HIV-1.
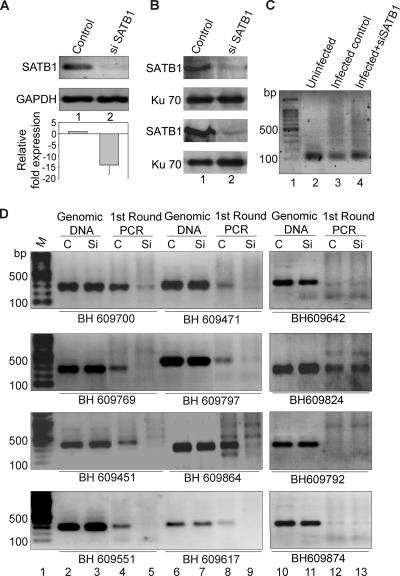
To directly address the role of SATB1 in HIV integration site selection, we monitored the presence of reported integration sites from the BH series (47) that are SATB1 targets (this work) in regions flanking HIV integration sites in CEM T cells depleted of SATB1. The level of expression of SATB1 in CEM cells is comparable to that in Jurkat cells (data not shown). SATB1 expression was knocked down in CEM cells by transfecting pSUPER-shSATB1 (Fig. 5A). The knockdown of SATB1 was quantitated by real-time RT-PCR and calculated to be 14-fold (Fig. 5A). We also performed immunoblot analysis of extracts from the siSATB1-transfected cells versus that of empty-vector-transfected cells and found a reduction of SATB1 of about 10-fold, even at the protein level (Fig. 5B). Control (pSUPER vector-transfected) and siSATB1 (pSUPER-shSATB1-transfected) cells were then infected with HIV-NL4.3. Genomic DNA from these cells was used for isolation of regions flanking HIV integration sites by PCR amplification with primer sets containing an LTR-specific primer (LTR3′F) and a primer specific for an _Alu_-like motif (motif 3R). A control PCR with genomic DNA from uninfected cells did not yield the typical smear of amplified PCR products obtained with DNA from infected cells (Fig. 5C, compare lane 2 with lanes 3 and 4). These PCR-amplified products from the first round were then used as templates to PCR amplify specific BH series sequences, with genomic DNA serving as a control for the second round of PCRs (Fig. 5D, lanes 2, 3, 6, 7, 10, and 11). In vivo integration clones BH609700, BH609471, BH609769, BH609797, BH609451, BH609864, BH609551, and BH609617 were specifically amplified in the reaction mixtures with DNA from the first-round PCR of the control cells (Fig. 5D, lanes 4 and 8). There was a marked reduction in the PCR amplification of the BH series sequences from the first-round PCR products in the siSATB1 cells (Fig. 5D, lanes 5 and 9), suggesting that the integration site choice is altered when SATB1 is knocked down. We also observed that many of the reported integration sites were actually not targeted by the virus during infection, as deduced from the lack of PCR amplification product in the second-round PCR, even in the control cells (lane 12). In vivo integration clones BH609642, BH609792, and BH609874 failed to be amplified in the reaction mixtures with DNA from first-round PCRs. Since our data suggest that SATB1 binds these regions in vitro, collectively these observations suggest that HIV integration may not always occur at the same site(s) within the genome. However, in only one instance (BH609824) did we find that the second-round PCR product in control infected cells was not affected by SATB1 knockdown (Fig. 5D, second part from the top, lane 13), suggesting that a few integration events may occur in genomic regions that are not bound by SATB1 in vivo. It is unlikely that SATB1, or any other protein that seems to be involved in the same process, is the sole determinant of integration site selection by HIV. Our data nevertheless provide compelling evidence of a role for SATB1 in HIV-1 integration site selection.
FIG. 5.
Integration of HIV-1 at SATB1-binding regions is disfavored upon siRNA-mediated knockdown of SATB1. PCR analysis of regions flanking HIV-1 integration sites is presented. (A) RT-PCR validation of SATB1 knockdown. Total RNA was isolated and RT-PCR analysis was performed as described in Materials and Methods. RT-PCR products for SATB1 (upper part) from control (lane 1) and shSATB1-transfected (lane 2) HIV-1-infected CEM cells were electrophoresed on 1% agarose gel and visualized by staining with ethidium bromide. GAPDH was used as an internal control for normalization (lower part). The knockdown of SATB1 expression was quantitated by real-time RT-PCR analysis (histogram below). (B) Immunoblot analysis of SATB1 expression in control (lane 1) and siSATB1 (lane 2) cells is presented in two replicates to monitor consistent knockdown of the expression of SATB1. Expression of the Ku 70 subunit of the Ku autoantigen was used as a control (lower part). (C) First round of PCR amplification to isolate the genomic regions flanking the HIV-1 integration sites. Genomic DNA was isolated from HIV-1-infected CEM cells that were transfected with the pSUPER vector (control, lane 3) or pSUPER-shSATB1 (siSATB1, lane 4) and PCR amplified with primer sets containing an LTR-specific primer and a primer specific for one of the _Alu_-like motifs. Genomic DNA from uninfected CEM cells was used as a control for the PCRs (lane 2). (D) PCR amplification of reported integration sequences (BH series). The second-round PCRs were performed with primers specific for indicated BH series clones and purified DNA templates in the form of genomic DNA or first-round PCR products from control (C, pSUPER vector-transfected) cells and siSATB1 (Si, pSUPER-shSATB1-transfected) cells. Lane 1, 100-bp DNA ladder. The templates used were genomic DNA from control (lanes 2, 6, and 10) or siSATB1 (lanes 3, 7, and 11) cells and first-round PCR products from control (lanes 4, 8, and 12) or siSATB1 (lanes 5, 9, and 13) cells. All PCR products were electrophoresed on 1% agarose gels and visualized by staining with ethidium bromide.
Collectively, these results demonstrate that the SATB1-associated chromatin harbors multiple regions of the genome that constitute HIV-1 integration sites. In the absence of SATB1, chromatin organization may be altered in such a manner that does not promote integration near SATB1-binding sites. This could be due to a lack of interaction between a component(s) of the PIC and SATB1, to dynamic changes in chromatin loop domains, or both. We have, indeed, demonstrated that SATB1 collaborates with the nuclear-matrix-associated promyelocytic leukemia protein to organize the major histocompatibility complex class I locus into a distinct higher-order chromatin loop structure (31). Furthermore, gamma interferon treatment and silencing of either SATB1 or promyelocytic leukemia protein dynamically alters the chromatin architecture, leading to an altered expression profile of a subset of major histocompatibility complex class I genes (31). Thus, the organization of the higher-order chromatin “loopscape” by SATB1 and its interaction partners may be an important determinant of the retroviral integration site selection process. Here we show that silencing of SATB1 disfavors certain regions of genome for HIV-1 integration. Elucidation of the molecular mechanism of this phenomenon requires further investigation of the interactions of the HIV-1 proteins with SATB1 and genome-wide comparative analysis of integration sites in the presence or absence of SATB1.
Our analyses of the HIV-1 integration sites revealed unique signatures embedded within these sequences. First, they consist of multiple repeats of _Alu_-like consensus sequences and are specifically bound by SATB1. SATB1-binding sites consist of an AT-rich consensus element often flanked by GC-rich sequences (P. K. Purbey and S. Galande, unpublished data). Since SATB1 organizes T-cell chromatin into a unique cage-like architecture that excludes heterochromatin, it is possible that the integration machinery of HIV-1 may specifically target such regions for promoting its own replication and transcription. If HIV integration occurs in a chromatin context of alphoid repeats, it produces latent infection (25). Thus, it is evident that the chromatin context of the HIV integration site is important for its own life cycle. Additionally, HIV integrations favor the entire length of the transcriptional regions whereas MLV integrations are distributed evenly upstream and downstream of the transcriptional start site (53). Interestingly, in contrast to the findings of Schröder et al. (47), integration sites deposited by Wu et al. (53) did not show any kind of clustering (hot spots). This bias could be attributed to the differences in the expression levels of SATB1 in the cell lines used in these two studies; the SupT1 cells used by the Bushman group express higher levels of SATB1 compared to the HeLa or H9 cells used by the Burgess group (Fig. 4). The size of chromosomal regions favorable for integration (∼100 kb) (40) also closely matches the average size of a chromosomal loop, which further argues for a role for higher-order chromatin organization in retroviral integration. Different retroviruses seem to have distinct patterns of integration site selection within the human genome, suggesting that there may be local recognition of chromosomal features and implying a role for chromosomal proteins (40).
No consensus sequences have been determined in the primary flanking sequences of target site DNA in any of the retroviral integration site studies performed so far. The base preferences reported by Holman and Coffin (23) actually correspond to the final step in proviral integration, when the integrase recognizes and cleaves host DNA. However, it can be gleaned that the PIC may have to first tether itself at specific sites within the chromatin and then integrase may actually be able to find preferred bases in the vicinity in a manner akin to the Ty retrotransposons (9). Our results demonstrate that HIV-1 prefers to integrate in T-cell chromatin specifically at sites that are enriched in specific consensus sequences and repeating patterns. A primary sequence arrangement of this kind may itself promote chromatin organization in a unique architectural pattern in vivo that remains to be investigated.
Acknowledgments
We are grateful to G. C. Mishra for support and encouragement and T. Kohwi-Shigematsu for the gift of the SATB1 antibody, pGEX-PARP-DBD, and the SBS-11 clone. We thank the CDAC supercomputing facility at Pune and Bangalore, where part of the computational analysis was performed.
The molecular clone NL 4.3 and the CEM-GFP reporter cell line were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. P.K., S.M., P.K.P., and D.N. are supported by fellowships from the Council of Scientific and Industrial Research, India. D. S. Ravi is supported by a fellowship from the University Grants Commission, New Delhi, India. Work in the laboratory of S.G. is partly supported by a grant from the Department of Biotechnology, Government of India. S. Galande is an International Senior Research Fellow of the Wellcome Trust, United Kingdom.
Footnotes
▿
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koeing, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59**:**284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, J. D., D. H. Yasui, H. Niida, T. Joh, D. Y. Loh, and T. Kohwi-Shigematsu. 2000. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14**:**521-535. [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymer, p. 28-36. In R. Altman, D. Brutlag, P. Karp, R. Lathrop, and D. Searls (ed.), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Press, Menlo Park, CA. [PubMed]
- 4.Batzer, M. A., and P. L. Deininger. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3**:**370-379. [DOI] [PubMed] [Google Scholar]
- 5.Bode, J., C. Benham, A. Knopp, and C. Mielke. 2000. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr. 10**:**73-90. [PubMed] [Google Scholar]
- 6.Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93**:**1087-1089. [DOI] [PubMed] [Google Scholar]
- 7.Bor, Y. C., M. D. Miller, F. D. Bushman, and L. E. Orgel. 1996. Target-sequence preferences of HIV-1 integration complexes in vitro. Virology 222**:**283-288. [DOI] [PubMed] [Google Scholar]
- 8.Britanova, O., S. Akopov, S. Lukyanov, P. Gruss, and V. Tarabykin. 2005. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 21**:**658-668. [DOI] [PubMed] [Google Scholar]
- 9.Bushman, F. D. 2003. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115**:**135-138. [DOI] [PubMed] [Google Scholar]
- 10.Bushman, F. D., T. Fujiwara, and R. Craigie. 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249**:**1555-1558. [DOI] [PubMed] [Google Scholar]
- 11.Cai, S., and T. Kohwi-Shigematsu. 1999. Intranuclear relocalization of matrix binding sites during T cell activation detected by amplified fluorescence in situ hybridization. Methods 19**:**394-402. [DOI] [PubMed] [Google Scholar]
- 12.Cai, S., H. J. Han, and T. Kohwi-Shigematsu. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34**:**42-51. [DOI] [PubMed] [Google Scholar]
- 13.Carteau, S., C. Hoffman, and F. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 72**:**4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11**:**1287-1289. [DOI] [PubMed] [Google Scholar]
- 15.de Belle, I., S. Cai, and T. Kohwi-Shigematsu. 1998. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 141**:**335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson, L. A., T. Joh, Y. Kohwi, and T. Kohwi-Shigematsu. 1992. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70**:**631-645. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson, L. A., C. D. Dickinson, and T. Kohwi-Shigematsu. 1997. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J. Biol. Chem. 272**:**11463-11470. [DOI] [PubMed] [Google Scholar]
- 18.Dobreva, G., J. Dambacher, and R. Grosschedl. 2003. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 17**:**3048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elleder, D., A. Pavlicek, J. Paces, and J. Hejnar. 2002. Preferential integration of human immunodeficiency virus type 1 into genes, cytogenetic R bands and GC-rich DNA regions: insight from the human genome sequence. FEBS Lett. 51**:**285-286. [DOI] [PubMed] [Google Scholar]
- 20.Engelman, A. 2005. The ups and downs of gene expression and retroviral DNA integration. Proc. Natl. Acad. Sci. USA 102**:**1275-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gervaix, A., D. L. West, M. Leoni, D. D. Richman, F. Wong-Staal, and J. Corbeil. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. USA 94**:**4653-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindmarsh, P., and J. Leis. 1999. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 63**:**836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holman, A. G., and J. M. Coffin. 2005. Symmetrical base preferences surrounding HIV-1 and avian sarcoma/leukosis. Proc. Natl. Acad. Sci. USA 102**:**6103-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenuwein, T., W. C. Forrester, L. A. Fernandez-Herrero, G. Laible, M. Dull, and R. Grosschedl. 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385**:**269-272. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22**:**1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20**:**1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura, Y., Y. M. H. Lee, and J. M. Coffin. 1992. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc. Natl. Acad. Sci. USA 89**:**5532-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohwi-Shigematsu, T., I. de Belle, L. A. Dickinson, S. Galande, and Y. Kohwi. 1998. Identification of base-unpairing region (BUR)-binding proteins and characterization of their in vivo binding sequences. Methods Cell Biol. 53**:**323-354. [DOI] [PubMed] [Google Scholar]
- 29.Kohwi-Shigematsu, T., and Y. Kohwi. 1990. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry 29**:**9551-9560. [DOI] [PubMed] [Google Scholar]
- 30.Kondo, Y., and J. P. Issa. 2003. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 278**:**27658-27662. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, P. P., O. Bischof, P. K. Purbey, D. Notani, H. Urlaub, A. Dejean, and S. Galande. 2007. Functional interaction between PML and SATB1 regulates chromatin loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 9**:**45-56. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, P. P., P. K. Purbey, D. S. Ravi, D. Mitra, and S. Galande. 2005. Displacement of SATB1-bound HDAC1 corepressor by HIV-1 transactivator induces expression of interleukin-2 and its receptor in T cells. Mol. Cell. Biol. 25**:**1620-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar, P. P., P. K. Purbey, C. K. Sinha, D. Notani, A. Limaye, R. S. Jayani, and S. Galande. 2006. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell 22**:**231-243. [DOI] [PubMed] [Google Scholar]
- 34.Leem, S. H., N. Kouprina, J. Grimwood, J. H. Kim, M. Mullokandov, Y. H. Yoon, J. Y. Chae, J. Morgan, S. Lucas, P. Richardson, C. Detter, T. Glavina, E. Rubin, J. C. Barrett, and V. Larionov. 2004. Closing the gaps on human chromosome 19 revealed genes with a high density of repetitive tandemly arrayed elements. Genome Res. 14**:**239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack, K. D., X. Jin, S. Yu, R. Wei, L. Kapp, C. Green, B. Herndier, N. W. Abbey, A. Elbaggari, Y. Liu, and M. S. McGrath. 2003. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J. Acquir. Immune Defic. Syndr. 33**:**308-320. [DOI] [PubMed] [Google Scholar]
- 36.Maxfield, L. F., C. D. Fraize, and J. M. Coffin. 2005. Relationship between retroviral DNA-integration-site selection and host cell transcription. Proc. Natl. Acad. Sci. USA 102**:**1436-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeil, N. 2004. AluElements: repetitive DNA as facilitators of chromosomal rearrangement. J. Assoc. Genet. Technol. 30**:**41-47. [PubMed] [Google Scholar]
- 38.Medstrand, P., L. N. van de Lagemaat, and D. L. Mager. 2002. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 12**:**1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mielke, C., K. Maass, M. Tummler, and J. Bode. 1996. Anatomy of highly expressing chromosomal sites targeted by retroviral vectors. Biochemistry 35**:**2239-2252. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2**:**E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mñller, H. P., and H. E. Varmus. 1994. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 13**:**4704-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85**:**2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pesole, G., S. Luini, and M. D'Souza. 2000. PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics 16**:**450. [DOI] [PubMed] [Google Scholar]
- 44.Pryciak, P. M., and H. E. Varmus. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69**:**769-780. [DOI] [PubMed] [Google Scholar]
- 45.Roberge, M., and S. M. Gasser. 1992. DNA loops: structural and functional properties of scaffold-attached regions. Mol. Microbiol. 6**:**419-423. [DOI] [PubMed] [Google Scholar]
- 46.Sandmeyer, S. 2003. Integration by design. Proc. Natl. Acad. Sci. USA 100**:**5586-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hot spots. Cell 110**:**521-529. [DOI] [PubMed] [Google Scholar]
- 48.Seo, J., M. M. Lozano, and J. P. Dudley. 2005. Nuclear matrix binding regulates SATB1-mediated transcriptional repression. J. Biol. Chem. 280**:**24600-24609. [DOI] [PubMed] [Google Scholar]
- 49.Shih, C. C., J. P. Stoye, and J. M. Coffin. 1988. Highly preferred targets for retrovirus integration. Cell 53**:**531-537. [DOI] [PubMed] [Google Scholar]
- 50.Stevens, S. W., and J. D. Griffith. 1996. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J. Virol. 70**:**6459-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Withers-Ward, E. S., Y. Kitamura, J. P. Barnes, and J. M. Coffin. 1994. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 8**:**1473-1787. [DOI] [PubMed] [Google Scholar]
- 52.Wolffe, A. P., and D. Guschin. 2000. Chromatin structural features and targets that regulate transcription. J. Struct. Biol. 129**:**102-122. [DOI] [PubMed] [Google Scholar]
- 53.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300**:**1749-1751. [DOI] [PubMed] [Google Scholar]
- 54.Yasui, D., M. Miyano, S. Cai, P. Varga-Weisz, and T. Kohwi-Shigematsu. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419**:**641-645. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, Y., J. Dai, P. G. Fuerst, and D. F. Voytas. 2003. Controlling integration specificity of a yeast retrotransposon. Proc. Natl. Acad. Sci. USA 100**:**5891-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoubak, S. 1994. Regional specificity of HTLV-1 proviral integration in human genome. Gene 143**:**155-163. [DOI] [PubMed] [Google Scholar]